Abstract
Anti-angiogenic therapies for cancer such as VEGF neutralizing antibody bevacizumab have limited durability. While mechanisms of resistance remain undefined, it is likely that acquired resistance to anti-angiogenic therapy will involve alterations of the tumor microenvironment. We confirmed increased tumor-associated macrophages in bevacizumab-resistant glioblastoma patient specimens and two novel glioblastoma xenograft models of bevacizumab resistance. Microarray analysis suggested downregulated macrophage migration inhibitory factor (MIF) to be the most pertinent mediator of increased macrophages. Bevacizumab-resistant patient glioblastomas and both novel xenograft models of resistance had less MIF than bevacizumab-naive tumors, and harbored more M2/protumoral macrophages that specifically localized to the tumor edge. Xenografts expressing MIF-shRNA grew more rapidly with greater angiogenesis and had macrophages localizing to the tumor edge which were more prevalent and proliferative, and displayed M2 polarization, whereas bevacizumab-resistant xenografts transduced to upregulate MIF exhibited the opposite changes. Bone marrow-derived macrophage were polarized to an M2 phenotype in the presence of condition-media derived from bevacizumab-resistant xenograft-derived cells, while recombinant MIF drove M1 polarization. Media from macrophages exposed to bevacizumab-resistant tumor cell conditioned media increased glioma cell proliferation compared with media from macrophages exposed to bevacizumab-responsive tumor cell media, suggesting that macrophage polarization in bevacizumab-resistant xenografts is the source of their aggressive biology and results from a secreted factor. Two mechanisms of bevacizumab-induced MIF reduction were identified: (1) bevacizumab bound MIF and blocked MIF-induced M1 polarization of macrophages; and (2) VEGF increased glioma MIF production in a VEGFR2-dependent manner, suggesting that bevacizumab-induced VEGF depletion would downregulate MIF. Site-directed biopsies revealed enriched MIF and VEGF at the enhancing edge in bevacizumab-naive patients. This MIF enrichment was lost in bevacizumab-resistant glioblastomas, driving a tumor edge M1-to-M2 transition. Thus, bevacizumab resistance is driven by reduced MIF at the tumor edge causing proliferative expansion of M2 macrophages, which in turn promotes tumor growth.
Introduction
Anti-angiogenic therapy holds promise for treating malignancies such as glioblastoma, a devastating brain cancer whose median survival of under two years from diagnosis1 creates an urgent need for effective treatments. Unfortunately, while initial responses to anti-angiogenic therapy are often significant, these agents have limited durations of response.2 Half of glioblastomas treated with bevacizumab, a neutralizing antibody targeting vascular endothelial growth factor (VEGF), develop acquired resistance after initial response.3 Acquired resistance to anti-angiogenic therapy confers a poor prognosis,1, 4 making it a significant problem preventing anti-angiogenic therapies from fulfilling their therapeutic promise.
Increased tumor-associated macrophages (TAMs) have been described in preclinical5 and clinical6 specimens of glioblastomas whose tumors progress during bevacizumab treatment and correlate with a poor prognosis in these patients7 but the links between VEGF blockade, TAM recruitment and resistance have yet to be established. We sought to identify a comprehensive mechanism defining the role of macrophages in bevacizumab resistance that accounts for tumor-secreted cytokines regulating TAMs, spatial heterogeneity in TAM localization and the evolution of our understanding that TAMs fall into two subtypes: M1 classically-activated macrophages with pro-inflammatory antitumoral function and M2 alternatively-activated macrophages, which promote tumor growth and invasion.8, 9, 10
Results
Increased myeloid macrophages in bevacizumab-resistant glioblastomas
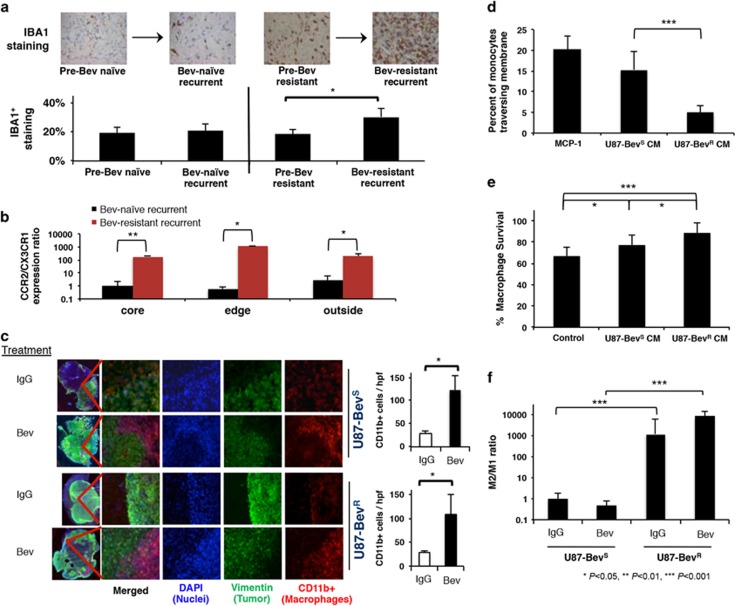
Immunostaining revealed that bevacizumab-resistant glioblastomas had more IBA1+ cells, a marker of myeloid-derived macrophages than microglia,11 than their paired pre-treatment bevacizumab-naive glioblastomas (P=0.02), while no such increase occurred when glioblastomas recurred without bevacizumab treatment (P=0.8) (n=8/group) (Figure 1a). To confirm that these IBA1+ cells expanding in resistant glioblastomas were marrow-derived macrophages recruited through the disrupted blood–brain barrier of glioblastoma rather than native microglia, we performed absolute qPCR for microglia marker CX3CR1 and myeloid macrophage marker CCR212, 13 on CD11b+ cells isolated from site-directed biopsies of the central core, enhancing edge and infiltrated white matter of patient glioblastomas as illustrated in Supplementary Figure S1. The ratio of CCR2 to CX3CR1 expression was greater in all sites of bevacizumab-resistant (n=3; one after monotherapy and two after combination therapy) versus bevacizumab-naive (n=5) glioblastomas (P=0.006–0.02) (Figure 1b), consistent with these increased IBA1+ cells in bevacizumab-resistant glioblastoma being marrow-derived.
Figure 1.
Increased M2 macrophages at the tumor edge associated with bevacizumab-resistant glioblastoma. (a) Human glioblastomas (n=8/group) exhibited increased IBA1+ macrophages upon becoming bevacizumab-resistant compared to paired pre-treatment bevacizumab-naive specimens (P=0.02), while no such increase occurred in bevacizumab-naive glioblastomas after recurrence compared with before (P=0.8) Y-axis represents percent of the high-powered field that was immunopositive as derived by ImageJ software. (b) Absolute qPCR for microglia marker CX3CR1 and myeloid macrophage marker CCR2 on CD11b+ cells isolated from site-directed biopsies of the central core, enhancing edge, and infiltrated white matter of patient glioblastomas revealed that the ratio of CCR2:CX3CR1 expression increased robustly in all sites of bevacizumab-resistant (n=3) versus bevacizumab-naive (n=5) glioblastomas (P=0.006 central core; P=0.01 enhancing edge; P=0.02 infiltrated white matter) (c) U87-BevS and U87-BevR intracranial xenografts exhibited increased CD11b+ macrophages (red) upon treatment with bevacizumab compared with treatment with IgG (P=0.01). These macrophages were recruited to the tumor (green) edge (n=5/group). (d) THP-1 human monocytes exhibited decreased chemotaxis in response to U87-BevR conditioned media (CM) compared with U87-BevS CM (P<0.0001) (n=4/group), whereas (e) murine bone marrow derived macrophages had greater cell numbers when cultured in U87-BevR conditioned media for 48 h compared with U87-BevS conditioned media (P=0.04), with both U87-BevR (P<0.001) and U87-BevS (P=0.04) conditioned media increasing macrophage numbers (n=8/group). (f) qPCR of FACS-sorted CD11b+ TAMs isolated from intracranial U87-BevR and U87-BevS xenograft (n=5/group) revealed increased M2 macrophages in U87-BevR intracranial xenografts relative to U87-BevS intracranial xenografts, independent of treatment (P<0.0001), as revealed by M2/M1 polarization ratios determined by multiplying the qPCR fold increases in three different M2 primers divided by the qPCR fold increases in three different M1 primers normalized to results for U87-BevS xenografts treated with IgG.
We then corroborated these findings in two different intracranial xenograft models of bevacizumab resistance established by our group. First, we analyzed CD11b+ cells in patient-derived intracranial bevacizumab-sensitive SF7300 and bevacizumab-resistant SF7796 xenografts, which were derived from samples taken at the enhancing edge of patient tumors and whose bevacizumab responsiveness mirrors that of the patient glioblastoma they derive from.14 We found that bevacizumab increased CD11b+ cells significantly in both bevacizumab sensitive SF7300 (P=0.01) and bevacizumab-resistant SF7796 (P=0.002) xenografts (Supplementary Figure S2), but the increase in CD11b+ cells was significantly larger in bevacizumab-resistant SF7796 than bevacizumab-sensitive SF7300 xenografts (111 vs 44%, P=0.01). Second, we analyzed TAMs in intracranial xenografts derived from U87-BevR and U87-BevS, isogeneic cell line-derived bevacizumab-resistant and sensitive xenograft models, respectively, whose establishment we described.14, 15 Immunostaining revealed increased CD11b+ cells in bevacizumab-treated sensitive and resistant xenografts relative to those treated with IgG control antibody (Figure 1c; P=0.01). Interestingly, the CD11b+ cells associated with bevacizumab treatment localized to the tumor edge (Figure 1c). Furthermore, the increased CD11b+ cells seen in bevacizumab-treated sensitive and resistant xenografts proved to be a mixture of marrow-derived macrophages recruited from the circulation and native microglia based on absolute qPCR of isolated tumoral CD11b+ cells for microglia marker CX3CR1 and myeloid macrophage marker CCR2 (Supplementary Figure S3a).12, 13 Bevacizumab increased expression of marrow-derived TAM marker CCR2 relative to microglial marker CX3CR1 in U87-BevS xenografts (P=0.01); however, IgG-treated U87-BevR xenografts had a greater CCR2:CX3CR1 ratio and bevacizumab did not increase this ratio (P=0.6) (Supplementary Figure S3a). Furthermore, bevacizumab lowered expression of microglia marker TMEM119 16 in the CD11b+ cells in U87-BevS (P=0.001) and U87-BevR (P=0.002) xenografts (Supplementary Figure S3b).
Mechanisms of TAM recruitment after bevacizumab treatment
We then investigated potential mechanisms of these findings of increased TAMs after bevacizumab treatment of sensitive and resistant xenografts from both models. We first performed monocyte chemotaxis assays using conditioned media (CM) from U87-BevR and U87-BevS cells to determine whether tumor cells from resistant versus sensitive xenografts exhibited differential chemotaxis of recruited monocytes. The chemotactic response of human THP-1 monocytes to monocyte chemotactic protein-1 (MCP-1/CCL2), an important monocyte-attracting chemokine, was reduced after pre-treatment with U87-BevR CM versus U87-BevS CM (P=0.03) (Supplementary Figure S4). Furthermore, the chemotactic response of THP-1 monocytes was weaker to U87-BevR CM than to U87-BevS CM (P<0.0001) (Figure 1d). In contrast, CM from U87-BevR cells increased the number of bone marrow-derived murine macrophages relative to CM from U87-BevS cells (P=0.04) (Figure 1e). Thus, while the total number of TAMs increased comparably in sensitive U87-BevS versus resistant U87-BevR intracranial xenografts after bevacizumab treatment, tumor cells from the sensitive xenografts achieved this increase through more monocyte chemotaxis while tumor cells from resistant xenografts achieved this increase by promoting proliferation of differentiated macrophages.
Macrophage polarization after bevacizumab resistance
We then determined whether the increased TAMs seen in both sensitive and resistant xenografts after bevacizumab treatment were differentially polarized towards the M1 anti-tumoral or M2 protumoral subtype in sensitive versus resistant xenografts. To do so, we isolated CD11b+ macrophages from intracranial U87-BevR versus U87-BevS xenografts using fluorescence activated cell sorting (FACS) followed by quantitative PCR (qPCR) analysis of expression of three M1 (Nos2=inducible nitric oxide synthase, CXCL10 and IL-1β) and three M2 markers (Arginase 1, TGF-β, and MMP9), from which we derived an M2/M1 ratio of the expression of the three M2 markers divided by expression of the three M1 markers. Macrophages from U87-BevR xenografts exhibited higher M2/M1 ratios than macrophages infiltrating U87-BevS xenografts independent of whether they were treated with IgG or bevacizumab (P<0.0001), suggesting durable M2 polarization of the macrophages once the resistant phenotype had been established (Figure 1f).
Screening for mediators of macrophage changes in bevacizumab resistance
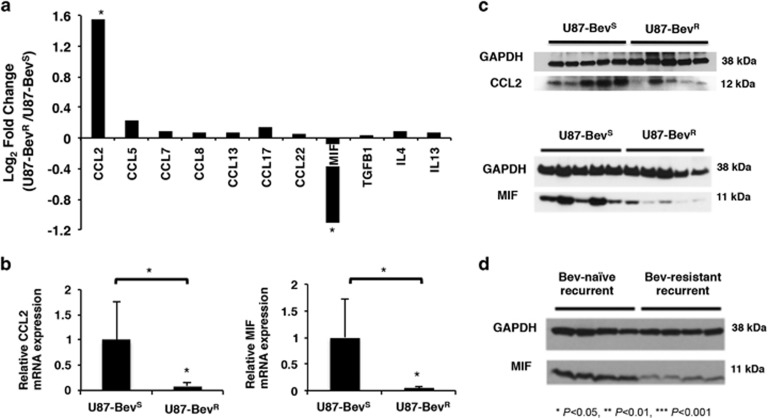
To investigate mediators of the increased number of macrophages and M2 polarization in bevacizumab-resistant xenografts, we performed a microarray analysis comparing U87-BevR to U87-BevS xenografts and specifically analyzed tumor-secreted cytokines with known biologic effects on monocytes or macrophages (Supplementary Table S1; Figure 2a). Of these cytokines, only MCP-1/CCL2 and macrophage migration inhibitory factor (MIF) expression changed significantly during the evolution of bevacizumab resistance in U87-BevR (raw P<0.0001), the former increasing and the latter decreasing. While CCL2 contributes to macrophage recruitment,17 qPCR (Figure 2b) and western blot (Figure 2c) of U87-BevR and U87-BevS xenografts revealed CCL2 mRNA (P<0.05) and protein levels to actually decrease in U87-BevR xenografts, explaining the reduced chemotaxis of monocytes to U87-BevR CM than to U87-BevS CM (Figure 1c). Thus, CCL2 was not the source of the increased macrophages we noted in bevacizumab-resistant xenografts.
Figure 2.
Decreased MIF expression associated with bevacizumab-resistant glioblastoma. (a) Microarray analysis of U87-BevR relative to U87-BevS (n=3/group) was performed to assess tumor-secreted factors known to influence macrophages, with only two changes being statistically significant (*raw P<0.001): upregulated CCL2 and downgregulated MIF. (b) qPCR revealed reduced CCL2 and MIF transcript (P<0.05 each), while (c) Immunoblotting revealed reduced CCL2 and MIF protein in U87-BevR xenografts (n=5) compared to U87-BevS intracranial xenografts (n=5). (d) Immuoblotting revealed reduced MIF in bevacizumab-resistant patient glioblastomas (n=4) compared to bevacizumab-naive patient glioblastomas (n=4).
We then further investigated the MIF downregulation identified in the microarray of our bevacizumab-resistant xenografts because MIF has potential roles in macrophage recruitment and polarization.18 We confirmed the microarray results, finding significant reduction in MIF levels in U87-BevR compared to U87-BevS xenografts at the transcriptional (P<0.05; Figure 2b) and protein (Figure 2c) levels. Similarly, tumoral MIF in patient-derived SF7300 bevacizumab-responsive xenografts was higher than in patient-derived SF7796 bevacizumab-resistant xenografts (Supplementary Figure S5). We then corroborated these findings from both of our xenograft models of bevacizumab resistance in patient specimens. MIF expression in bevacizumab-resistant glioblastoma patient specimens (n=4; two treated with monotherapy and two with bevacizumab plus another agent) was lower than in bevacizumab-naive patient glioblastomas (n=4) (Figure 2d).
MIF depletion drives features of bevacizumab-resistant glioblastoma
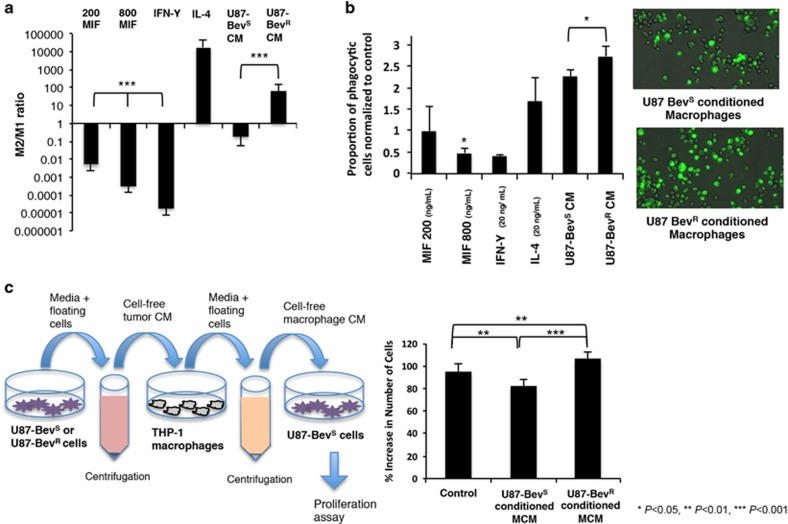
To determine whether the M2 polarization noted in bevacizumab-resistant xenografts reflected altered factor(s) secreted by tumor cells, we performed bone marrow-derived macrophage polarization assays. First, murine macrophages were incubated with recombinant cytokines or tumor cell CM. We then assessed M1 vs M2 polarization by using qPCR to determine expression of the three M1 and three M2 markers listed above to calculate M2/M1 ratios. CM from cultured U87-BevR cells yielded more M2-differentiated macrophages compared to CM from U87-BevS cells (P<0.0001; Figure 3a). Recombinant MIF at 0.2 and 0.8 μg/ml converted macrophages to an M1 phenotype in a dose-dependent manner (P<0.0001; Figure 3a). Thus, the loss of MIF expression in U87-BevR xenografts proved to be a possible source of the differential effects of the CM from U87-BevR versus U87-BevS cells on murine macrophages.
Figure 3.
In cultured cells, MIF promotes M1 polarization, causing macrophages to be less phagocytic and exert a less pro-tumoral effect. (a) Murine bone marrow-derived macrophage precursors were matured for 5 days and rested for 3 days prior to an 18 hour polarization in media containing 200 ng/ml and 800 ng/ml MIF, 20 ng/ml IFN-γ (for M1 polarization), 20 ng/ml IL-4 (for M2 polarization), or conditioned media from U87-BevR or U87-BevS cells (n=3/group). M1/M2 polarization ratio was assessed by multiplication of the qPCR fold increases in three different M1 primers divided by the qPCR fold increases in three different M2 primers normalized to results when incubating with media alone. Recombinant MIF drove M1 polarization in a dose-dependent manner (P<0.0001), while, unlike media from U87-BevS cells, media from U87-BevR cells drove an M2 macrophage polarization (P<0.0001). (b) The phagocytic activity of THP-1 monocyte-derived macrophages without stimulation, cultured with cytokines (recombinant MIF, IFN-γ, and IL-4), or cultured with conditioned media (CM) was assessed via uptake of fluorescent heat-killed E. coli with subsequent measurement of the proportion of fluorescent cells (n=6/group). M1 polarized macrophages were less phagocytic whereas M2 polarized macrophages were more phagocytic relative to unstimulated control macrophages. Macrophages treated with 800 ng/ml MIF were less phagocytic relative to control (P=0.005). Macrophages treated with CM from U87-BevR cells were more phagocytic than macrophages treated with CM from U87-BevS (P=0.01) (example images shown to the right). (c) Sequential conditioned media (SCM) experiments were performed as illustrated to the left. Briefly, media from U87-BevR or U87-BevS cells was applied to THP-1-derived macrophages, and the media was then taken from those macrophages and applied to U87 cells, with numbers of U87 cells counted 48 h later (n=24/group). U87-BevR-derived macrophage conditioned media (U87-BevR MCM) stimulated a significantly greater expansion of U87 cells than U87-BevS-derived macrophage conditioned media (U87-BevS MCM) (P<0.0001), with U87-BevR MCM and U87-BevS MCM stimulating more (P=0.005) and less (P=0.003) U87 expansion than control media, respectively.
To confirm that the M1 and M2 polarization induced by MIF and U87-BevR CM, respectively, corresponded to the expected functional differences between these two macrophage subtypes, we performed macrophage phagocytosis assays on THP-1 human monocytes differentiated into macrophages and treated with recombinant MIF or CM from U87-BevR or U87-BevS cells. Recombinant MIF reduced phagocytosis of these macrophages (P=0.005), while U87-BevR CM increased their phagocytosis relative to U87-BevS CM (P=0.01; Figure 3b; Supplementary Figure S6). This finding was consistent with the description of M2 macrophages as more phagocytic than M1 macrophages.19, 20 Next, we performed a sequential conditioned media experiment to investigate the reciprocal impact of these macrophage changes induced by U87-BevR versus U87-BevS CM on cultured glioma cells (Figure 3c). CM from U87-BevR or U87-BevS cells was transferred to macrophages derived from THP-1 monocytes, followed by collecting the macrophage CM and applying it to U87-MG cells, the parental cell line from which U87-BevR and U87-BevS were derived. The media from macrophages exposed to U87-BevR CM caused more proliferation (P<0.0001; Figure 3c) but no change in cell motility (P=0.6; Supplementary Figure S7) of U87-MG cells than the media from macrophages exposed to U87-BevS CM (P<0.0001) (Figure 3c). Thus, the M2 macrophage polarization induced by U87-BevR cells promoted the tumor cell proliferation that is part of the aggressive bevacizumab-resistant phenotype.4
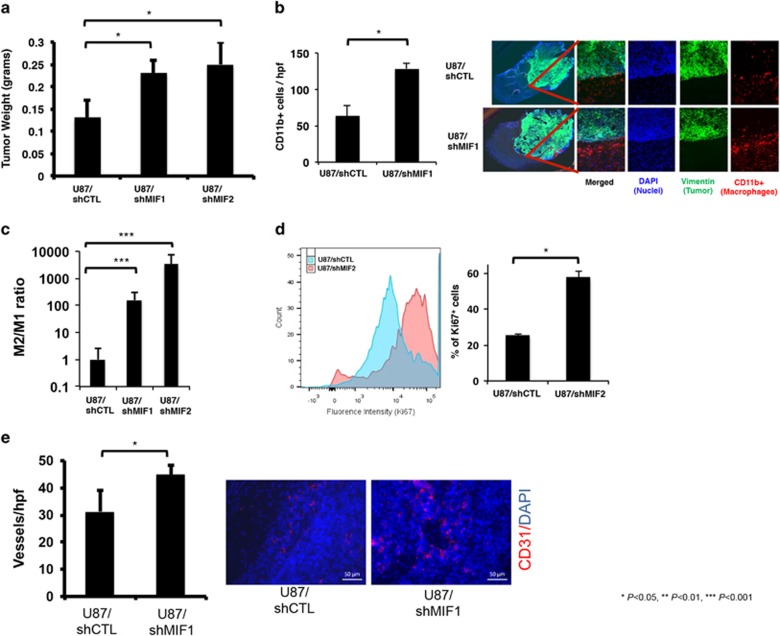
We then determined whether the loss of tumor-secreted MIF induced by anti-angiogenic therapy played a role in the increased number of macrophages and M2 polarization characterizing the resistant phenotype in vivo. We therefore engineered U87-MG cells to express control sequence-targeting shRNA (U87/shCTL) or each of two different sequences of MIF-targeted shRNAs (Supplementary Figure S8). U87/shMIF intracranial xenografts exhibited increased tumor weight (P=0.04 U87/shMIF1, P=0.03 U87/shMIF2; Figure 4a; Supplementary Figure S9) and more CD11b+ cells (P=0.01; Figure 4b) than U87/shCTL xenografts, with the CD11b+ cells in U87/shMIF xenografts accumulating at the tumor edge (Figure 4b). Compared to macrophages in U87/shCTL xenografts, macrophages in U87/shMIF xenografts had higher M2/M1 ratios derived from qPCR of the same three M1 and three M2 genes analyzed in Figure 1f (P<0.0001; Figure 4c). A larger proportion of the C11b+ TAMs in U87/shMIF xenografts expressed proliferation marker Ki67 by FACS compared to macrophages in U87/shCTL xenografts (P<0.05) (Figure 4d). These findings suggest that MIF knockdown increased the number of pro-tumoral macrophages by promoting their M2 polarization and proliferation post-differentiation, as occurred in bevacizumab-resistant xenografts. And, contrary to prior reports in other cell types,21, 22, 23, 24, 25, 26 intracranial U87/shMIF xenografts exhibited a greater vessel density (P=0.03; Figure 4e; Supplementary Figure S9) than intracranial U87/shCTL xenografts, while transducing U87 or LN229 glioma cells with MIF shRNA either increased (U87/shMIF1 P=0.02 and U87/shMIF2 P=0.01) or did not change (LN229/shMIF1 P=0.8 and LN229/shMIF2 P=0.4) their invasiveness (Supplementary Figure S10). These findings suggested that MIF depletion could promote compensatory angiogenesis during bevacizumab resistance and could contribute to or at least not offset the increased invasiveness seen during bevacizumab resistance.14, 15
Figure 4.
MIF reduction increases tumor-associated macrophages and drives M2 polarization in vivo. Intracranial xenografts established from U87 glioma cells transduced with two different MIF-targeted shRNA sequences versus control-targeted shRNA sequences exhibited (n=3/group) (a) larger tumor weights (P=0.04 U87/shMIF1 and P=0.03 U87/shMIF2); (b) more TAMs (red) (P=0.01), with the TAMs preferentially localizing to the tumor (green) periphery; (c) increased M2 TAM polarization based on elevated M2/M1 ratios (P<0.0001) in CD11b+ FACS-sorted TAMs analyzed by qPCR; (d) more proliferative TAMs based on FACS revealing a larger percentage of CD11b+ TAMs to be positive for the proliferation marker Ki67 (P<0.05); and (e) more vessel density based on CD31 vessel staining (P=0.03). Scale bars, 50 μm.
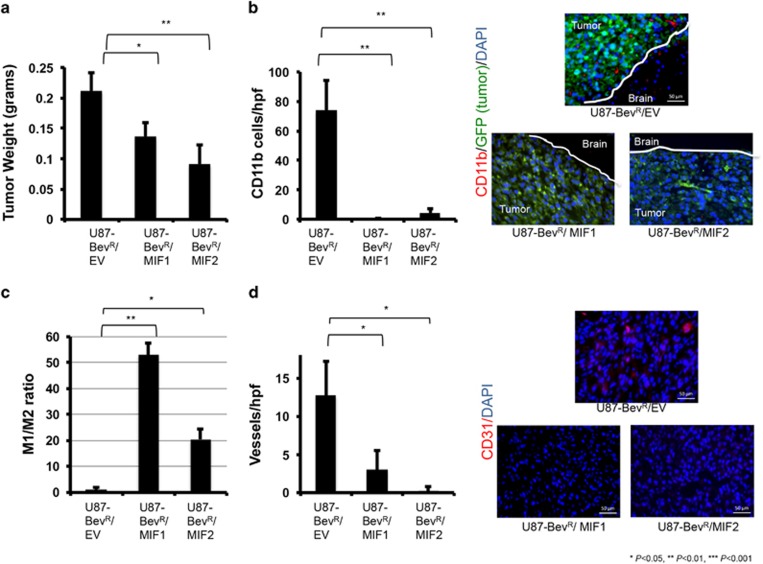
We then investigated the reciprocal question: the impact of overexpressing MIF in resistant U87-BevR cells (Supplementary Figure S11) on tumor growth and TAM polarization. Xenografts derived from two U87-BevR-derived clones transduced to overexpress MIF exhibited decreased tumor weight (P=0.03 U87/BevR-MIF1, P=0.009 U87/BevR-MIF2) (Figure 5a), decreased TAMs (P=0.006; Figure 5b), increased M1/M2 ratio (P=0.002 U87/BevR-MIF1, P=0.01 U87/BevR-MIF2; Figure 5c), and decreased angiogenesis (P=0.046 U87/BevR-MIF1 and P=0.04 U87/BevR-MIF2; Figure 5d). Thus, MIF restoration reversed some of the features of tumor growth and the tumor microenvironment seen with bevacizumab resistance.
Figure 5.
MIF overexpression in bevacizumab-resistant cells decreases tumor-associated macrophages and drives M1 polarization in vivo. Intracranial xenografts established from U87-BevR glioma cells transduced to overexpress MIF in two different clones relative to U87-BevR cells transduced with empty vector (EV) exhibited (n=3/group) (a) decreased tumor weight (P=0.03 U87-BevR/MIF1 and P=0.009 U87-BevR/MIF2); (b) increased CD11b+ TAMs (P=0.006 for both U87-BevR/MIF1 and U87-BevR/MIF2 compared to U87-BevR/EV) (CD11b+ cells red, GFP+ tumor cells green, DAPI rendering nuclei blue); (c) increased M1/M2 ratio (P=0.002 U87/BevR-MIF1, P=0.01 U87/BevR-MIF2); and (d) decreased vascularity (P=0.046 U87-BevR/MIF1 and P=0.04 U87-BevR/MIF2). Scale bars, 50 μm.
Mechanisms of MIF depletion during bevacizumab resistance
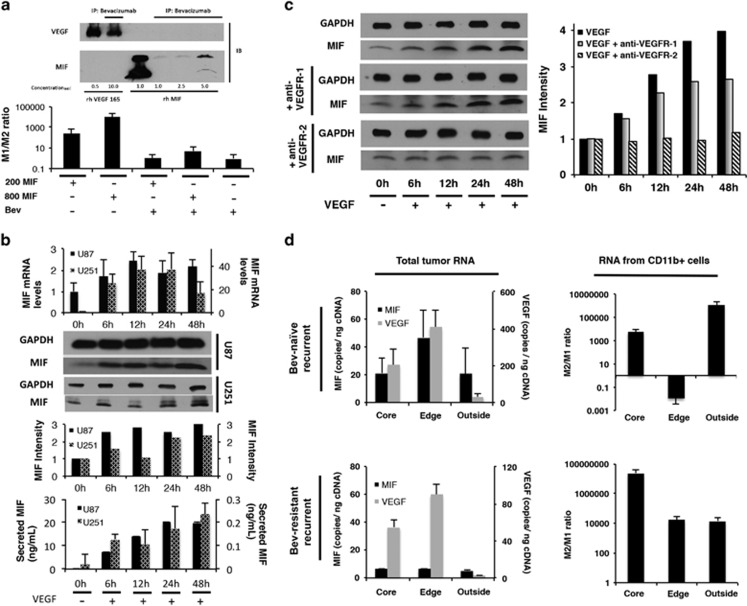
Having established the importance of MIF downregulation in driving M2 macrophage polarization and proliferation in bevacizumab-resistant glioblastomas, we then characterized the mechanisms of MIF downregulation in bevacizumab-resistant glioblastomas. To do so, we investigated the direct interaction between MIF and bevacizumab, as well as the relationship between MIF and VEGF, the target of bevacizumab. We used immunoprecipitation to perform an in vitro protein-protein binding assay with bevacizumab and recombinant MIF, which revealed that pure bevacizumab binds recombinant MIF in a dose-dependent manner (Figure 6a). This direct binding, unlikely to involve accessory proteins because studies were performed using pure recombinant proteins free of other molecules, offers one potential mechanism by which bevacizumab could reduce MIF levels. To confirm that the bevacizumab-MIF binding identified by immunoprecipitation was a functional interaction in which bevacizumab sequestered MIF and reduced its functional effects, we repeated the macrophage polarization assay from Figure 3a in the presence of recombinant MIF and/or bevacizumab. Bevacizumab blocked the ability of recombinant MIF to drive M1 polarization as assessed using qPCR to measure the M1/M2 ratio of the fold change in the three M1 and three M2 markers described above (Figure 6a).
Figure 6.
Dual mechanisms of bevacizumab-induced MIF depletion. (a) Immunoprecipitation revealed that bevacizumab bound MIF, with bevacizumab binding of VEGF serving as a positive control (upper portion of figure). Bevacizumab blocked the ability of recombinant MIF to drive M1 polarization in cultured bone marrow cells (lower portion of figure). (b) Treatment of cultured U87 and U251 cells with 100 ng/ml recombinant VEGF for varying time points up to 48 hours revealed that VEGF increased MIF transcription by qPCR, intracellular protein by western blot of whole cell lysate, and protein secretion by ELISA (P<0.01), offering a potential mechanism by which bevacizumab could reduce tumoral MIF. (c) Cultured U87 cells were treated with VEGF in the presence of blocking antibodies targeting VEGF receptors-1 and 2 (VEGR-1 and VEGFR-2). Blocking VEGFR-2 eliminated VEGF-induced MIF expression, while blocking VEGFR-1 had no effect on VEGF-induced MIF expression. (d) Site-directed biopsies taken from four bevacizumab-naive recurrent glioblastomas revealed increased MIF and VEGF RNA copies at the enhancing edge relative to the central core and FLAIR bright non-enhancing periphery, as assessed by absolute quantification qPCR (P=0.03). In contrast, site-directed biopsies taken from a bevacizumab-resistant recurrent glioblastoma revealed the same regional pattern in VEGF RNA levels, but loss of the spike in MIF RNA that typically occurs at the enhancing edge (P=0.02). Absolute quantification qPCR of column purified CD11b+ cells from bevacizumab-naive versus recurrent glioblastoma revealed elevated M2/M1 ratio at the central core and infiltrated white matter in both tumor types, but at the enhancing edge the bevacizumab-naive tumor had predominantly M1 macrophages, while the resistant tumor had predominantly M2 macrophages (P=0.008-0.01).
MIF secretion occurs through a unique mechanism in which MIF is initially stored in vesicles, and then released through an ATP-binding cassette transporter ABCA1. MIF secretion can therefore be regulated at the level of transcription as well as secretion. Using qPCR, immunoblotting, and ELISA, we showed that VEGF drives MIF expression through both short-term post-translational effects and longer-term transcriptional effects in U87-MG and U251 glioblastoma cells (Figure 6b), offering a mechanism through which VEGF depletion caused by prolonged bevacizumab treatment can reduce MIF expression. To determine which VEGF receptor mediated VEGF-driven MIF expression, U87-MG cells were treated with VEGF in the presence of blocking antibodies targeting VEGF receptors-1 and 2 (VEGR-1 and VEGFR-2). Blocking VEGFR-2 eliminated VEGF-induced MIF expression, while blocking VEGFR-1 did not alter VEGF-induced MIF expression (Figure 6c; Supplementary Figs. S12-S13).
Because macrophages localized to the tumor edge in our intracranial bevacizumab-resistant xenografts, we investigated the spatial variability in macrophage infiltration and associated factors in patient specimens. We analyzed VEGF and MIF expression by absolute quantification qPCR in site-directed biopsies from the central core, enhancing edge, and FLAIR bright infiltrated white matter outside of enhancement from patient bevacizumab-naive glioblastomas, obtained as illustrated in Supplementary Figure S1. VEGF and MIF expression peaked together at the enhancing edge relative to the central core or infiltrated white matter (n=4 cases; P=0.01–0.02; Figure 6d), consistent with the VEGF regulation of MIF expression that we noted in cultured U87-MG cells. Site-directed biopsies from the same locations in bevacizumab-resistant glioblastomas (n=3; same cases as Figure 1b) revealed that VEGF mRNA once again elevated at the enhancing edge (P=0.03). This finding is expected in bevacizumab-treated tumors because bevacizumab targets VEGF protein; therefore, VEGF regional transcriptional patterns should be preserved in bevacizumab-resistant glioblastomas. However, given the binding efficiency of bevacizumab to VEGF (Figure 6a), nearly all of the VEGF protein should be neutralized by bevacizumab in a treated tumor.27 In contrast to the preserved regional variation in VEGF expression, MIF mRNA was suppressed in all regions of bevacizumab-resistant glioblastomas, including at the enhancing edge where it normally peaked (P=0.02 vs bevacizumab-naive enhancing edge; Figure 6d). This suggests that bevacizumab neutralization of VEGF eliminating VEGF-induced MIF transcription, one of two mechanisms of bevacizumab-induced MIF depletion we identified, was a significant mechanism in bevacizumab-resistant patient tumors. We then isolated CD11b+ macrophages through column purification of site directed biopsies from bevacizumab-naive and bevacizumab-resistant glioblastomas. We performed absolute quantification qPCR to calculate regional M2/M1 ratios from the three M1 and three M2 markers described above. We found that bevacizumab-naive and resistant glioblastomas both had M2 macrophages predominating relative to M1 in the central core and infiltrated white matter. Within the enhancing edge, however, where MIF expression was lost during bevacizumab resistance, bevacizumab-resistant glioblastomas were enriched for M2 macrophages, while bevacizumab-naive glioblastomas were enriched for M1 macrophages (P<0.0001 bevacizumab-naive versus bevacizumab-resistant M2/M1 at enhancing edge; Figure 6d).
Discussion
We found MIF depletion during anti-angiogenic therapy to cause both the increased TAMs and increased M2 polarization of these macrophages that bevacizumab-resistant glioblastomas exhibit. Our findings suggest a sequence in which bevacizumab reduces tumor cell MIF expression, which typically occurs at the edge of bevacizumab-naive glioblastoma, early during treatment. This reduced MIF leads to increased proliferation of TAMs at the tumor edge while the treatment is still effective, which we found in our two xenograft models of bevacizumab sensitivity versus resistance and also found to be unique to bevacizumab treatment as compared to other treatments in our patient specimens. Eventually, the increased macrophages at the tumor edge become M2 polarized, which is also due to the loss of MIF at this location. The M2 macrophages at the tumor edge then promote tumor cell proliferation and the invasive growth that characterizes bevacizumab resistance.
We identified two mechanisms that could explain how VEGF depletion could trigger MIF depletion, loss of VEGF-induced MIF transcription and bevacizumab neutralization of MIF. Loss of VEGF-induced MIF transcription is supported by our site-directed biopsies from bevacizumab-resistant glioblastomas. The ability of bevacizumab to bind VEGF, shown by our protein-protein binding assay, is also supported by protein BLAST analysis, which although only demonstrating 31% protein homology between MIF and VEGF, confirmed homology between amino acids 48–76 of MIF and amino acids 29–51 of VEGF, the latter of which contains residues implicated in bevacizumab binding.28 While these effects on MIF would occur commonly and in a relatively short-term upon initiating bevacizumab treatment, MIF reduction is likely only severe enough to trigger TAM polarization changes and ensuing effects on tumor growth in a subset of tumors treated for longer time. One of the two mechanisms we identified for bevacizumab-induced MIF depletion could arise with VEGFR2-targeted therapies, as these therapies would not directly deplete MIF the way that we found bevacizumab could but the effect of these therapies on tumor cell VEGFR2 could prevent VEGF-induced MIF secretion based on our data. Future studies are needed to confirm whether MIF depletion occurs with VEGFR2-targeted therapy or is unique to VEGF-targeted anti-angiogenic therapy.
We found that the increased TAMs occurring during MIF reduction was due to increased macrophage proliferation rather than increased recruitment of circulating monocytes. This proliferative expansion of macrophages is a signature of type-2 inflammation associated with CD4+ T helper 2 (TH2) cells and M2 macrophages.29 Further work is needed to clarify if bevacizumab resistance increases intratumoral TH2 cells.
Additionally, we found that the increased TAMs associated with anti-angiogenic therapy resistance occurred primarily at the enhancing tumor edge. Recent studies have found that microenvironmental variations affect the spatial distribution and density of macrophage subtypes in inflammatory conditions.30, 31 Similarly, spatial variability in TAM polarization correlates with hypoxia in non-brain tumors in mice.32 Our findings of regional variation in MIF levels, with MIF peaking at the enhancing edge of patient bevacizumab-naive glioblastomas, would suggest MIF as an important source of regional variation of macrophage polarization. Increased MIF expression at the enhancing edge of glioblastoma could create a regional M1 polarization zone that somewhat confines tumor cells to the enhancing region, a confinement that is lost when bevacizumab depletes MIF, thereby softening the tumor border through M2 polarization and ultimately leading to invasive resistance.
The ability of tumor-secreted MIF to cause reduced angiogenesis, reduced tumor growth, and neutral to reductive effects on tumor cell invasiveness that we demonstrated, all anti-tumoral functions lost in bevacizumab-resistant glioblastomas, must be reconciled with studies showing pro-tumoral MIF effects on tumor cells such as promoting invasion and proliferation23, 24, 25, 26 or on the microenvironment such as promoting angiogenesis21, 22 or escape from NK cell-mediated cytotoxicity.33 Even our finding that MIF drives M1 TAM polarization, while consistent with reports in adipose cells in MIF knockout mice34 and a study correlating glioblastoma TAM expression of MIF receptor CD74 with M1 polarization,35 must be resolved with reports suggesting that MIF can drive pro-tumoral M2 macrophage polarization.18, 36
There are several potential explanations for these conflicting findings. First, tumor cell-secreted23, 24, 25, 26 versus stromal cell-secreted18, 37, 38 MIF could exert different effects on tumor cells or their microenvironment. Second, the cytokine milieu, which is affected by tumor type and therapeutic response, might alter MIF function. Third, there may be a dose-response relationship in which the collective effect of MIF on tumors is pro-tumoral at both extremely low and high levels. Just as VEGF depletion itself has direct adverse effects due to VEGF’s pro-angiogenic effects being balanced by its anti-invasive role,39 suggesting that intermediate tumoral VEGF levels may be optimal, achieving a balance between MIF’s pro-tumoral versus anti-tumoral functions may require intermediate tumoral MIF levels. In the case of VEGF blockade, this would support intermediate dosing strategies.40 Fourth, post-translational modifications of MIF like glycosylation or N-cysteinylation, driven partly by local tumoral levels of reactive oxygen species (ROS),41 could shift the pro-tumoral versus anti-tumoral balance in MIF functions. This would mean that therapies targeting MIF, which have been investigated preclinically, should be used with caution and it would be preferable to target a MIF form that preferentially exerts its pro-tumoral effects, such as the oxidized MIF currently being targeted in a clinical trial (ClinicalTrials.Gov; NCT01765790).
Further work is needed to clarify where in the spectrum of other described mechanisms of resistance to anti-angiogenic therapy our mechanism falls. These other mechanisms include vessel co-option42 driven by actin-related protein 2/3 complex (Arp2/3);43 tumor cell invasion driven by c-Met14, 44 and β1 integrin;15 hypoxia-driven expression of hyaluronic acid and sulfated glycosaminoglycans (sGAGs);45 increased angiopoietin-2 expression by endothelial cells;46 upregulation of tumor cell DLL4-Notch signaling;47 increased tumor cell expression of VEGFR-2;48 increased tumor cell expression of branched-chain amino acid transaminase1;49 Tie2-expressing monocytes;5, 6, 44 evolution of a mesenchymal phenotype;5, 14, 50 and increased hypoxia-inducible protein 2 (HIG2).51 It remains unclear if these multiple mechanisms occur in parallel or if resistance to anti-angiogenic therapy is heterogeneous. Clinically, answering this question is challenging because bevacizumab is typically used in combination with DNA damaging chemotherapy.
Continued validation of our mechanism would open up novel therapeutic opportunities. While some have suggested combining anti-angiogenic therapy with immunomodulatory approaches such as anticancer vaccines (ClinicalTrials.gov, NCT01814813) or T-cell targeted approaches,52 our work suggests a role for combining anti-angiogenic therapy with strategies promoting M1 macrophage polarization such as antibodies targeting colony-stimulating factor 1 receptor (CSF1R), which are undergoing clinical trials.53 Such an approach could prevent the entrenchment of bevacizumab resistance and improve the clinical efficacy of VEGF-targeted anti-angiogenic therapy, allowing this modality to fulfill its tremendous therapeutic promise.
Materials and methods
Cell culture
U87-MG (ATCC HTB-14), U251 (Sigma-Aldrich, Milwaukee, WI, USA), and LN229 (ATCC CRL2611) human glioblastoma cells and THP1 human monocytes (ATCC TIB-202) were passaged fewer than six months and verified by providing companies using short tandem repeat (STR) profiling and confirmed to be Mycoplasma free. Cells for culture were obtained from U87-BevR, U87-BevS, SF7300, and SF7796 xenografts generated as described.14, 15 Glioblastoma cells were cultured in DMEM High Glucose/F-12 50/50% mixture media with 10% FBS and 1% penicillin/streptomycin at 37 °C. THP1 cells were cultured in RPMI-1640 medium with 10% FBS, L-glutamine (2 mmol/l), 2-mercaptoethanol (55 μM; Gibco) and 1% penicillin/streptomycin at 37 °C.
Conditioned media
CM from subconfluent U87-MG or THP-1 cells growing for 48 h was centrifuged at 400 g for 5 min, and the supernatant was collected and stored at −20 °C. Sequential conditioned media (SCM) was prepared by applying U87-BevR/U87-BevS CM to differentiated macrophages; this media was then collected and applied to U87-BevS cells (n=24 technical replicates/group).
MIF shRNA and cDNA
Lentiviral clones containing two shRNA sequences against human MIF and a control sequence (GE Dharmacon), lentivirus packaging plasmid psPAX2, envelope plasmid VSV-G, and transfection reagent FuGENE6 (Promega) were used to produce viral particles from HEK293T cells. Lentivirus was added to U87-MG and LN229 cells in DMEM media supplemented with 10% FBS and polybrene (Santa Cruz Biotechnology) per manufacturer’s protocol. To overexpress MIF, human MIF cDNA was generated as a PCR product (primers in Supplementary Table S2) from a plasmid containing MIF cDNA (Origene) in the pENTR/D-TOPO Cloning Kit (Invitrogen), sequenced, and cloned into the pLenti6.3/V5-DEST Gateway Vector (Life Technologies, Carlsbad, CA, USA). Cultured U87-BevR cells were transduced with empty or MIF-containing vector, from which U87-BevR/EV (empty vector) along with U87-BevR/MIF1 and U87-BevR/MIF2, two clones overexpressing MIF relative to U87-BevR, were selected, with resulting cells GFP+ due to the vector GFP gene.
Microarray analysis
Tumor chunks previously harvested from generational xenografts had been flash frozen at explantation and stored in liquid nitrogen. Samples (n=3 technical replicates/group) were retrieved and dissociated using passage through a 21-gauge sterile syringe and a QiaShredder (Qiagen, Germantown, ML, USA). Dissociated tissue was processed for RNA using the RNeasy kit (Qiagen), following manufacturer’s protocol. RNA was tested for quality (RIN scores>8) using the RNA 6000 chip with the Bioanalyzer (Agilent). RNA was converted to labeled cRNA using the TargetAmp-Nano Labeling Kit for Illumina Expression BeadChip (EpiCentre), following manufacturer’s protocol. Labeled cRNA was kept at -20 °C, with chip hybridization performed by the UCSF Genome Core Facility (GCF).
Bioinformatics
Data (.idat files) from GCF underwent standard QC and processing through the UCSF Bioinformatics Core, and were deposited in GEO (Accession number=GSE81465). BLAST protein comparisons were rendered through http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins.
qPCR
RNA isolated with an RNeasy kit (Qiagen) was reverse-transcribed into cDNA with Superscript III (Invitrogen). Power Syber Green Master Mix (Applied Biosystems) was used with primers described in Supplementary Table S2. Quantitative real-time PCR was performed on an Applied Biosystems 7900 HT cycler (Supplementary Methods). The product of expression of three M1 markers (Nos2=inducible nitric oxide synthase, CXCL10, and IL-1β) was divided by the product of expression of three M2 markers (Arginase 1, TGF-β, and MMP9) for M1/M2 ratios and inverted for M2/M1 ratios.
Western blot
Human tissue and cellular preparations were harvested in radio immunoprecipitation buffer (RIPA) containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM beta-glycorophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin (RIPA Buffer, 10x, CST#9806, Cell Signaling, Danvers, MA, USA) and one tablet each of PhosStop and Complete Mini (Roche, Indianapolis, IN, USA). Insoluble materials were removed by centrifugation at 14 000 r.p.m. for 20 min at 4 °C. Protein concentration was determined using the bicinchronic acid (BCA) assay (Pierce Biotechnology, Rockford, IL, USA). Samples were prepared with 10–30 μg of protein in RIPA buffer with 4 × LDS loading buffer (Cat#LP0001, Life Technologies). Samples were electrophoresed on SDS–PAGE gels, transferred to PVDF membranes, probed with primary antibodies overnight, then secondary antibodies for 1 h at room temperature (Supplementary Table S3). Membranes were developed using Clarity Western ECL substrate (Cat#10026385 Rev A, Bio-Rad) and radiographic film.
Immunoprecipitation
Immunoprecipitation is described in Supplementary Methods.
Animal studies
Animal experiments were approved by the UCSF IACUC (approval #AN105170-02). 200 000 cells were implanted intracranially into the right frontal lobes of athymic mice (3-4 weeks, female) stereotactically. Mice were treated intraperitoneally with 10 mg/kg IgG control antibody (cat# I4506, Sigma) or bevacizumab (Genentech, San Francisco, CA, USA) twice weekly. For U87-BevR vs U87-BevS, we used five technical replicates/group to give over 90% power to detect 50% more CD11b+ cells with bevacizumab treatment. Randomization was not used because tumor size could not be ascertained non-invasively. Treating personnel were blinded to treatment group by having different personnel prepare solutions and code treatment groups. For U87/shCTL vs U87/shMIF and U87-BevR/EV vs U87-BevR/MIF, n=3 technical replicates/group were used to give over 90% power to detect 75% change in CD11b+ cells. Resulting tumors were (1) dissociated into single cell suspensions in 10% FBS/1% PFA for flow cytometry; (2) perfused with PBS followed by 1% PFA, after which brains were isolated and kept in 4% PFA overnight, sunk in 30% sucrose, embedded in OCT, and cut on a cryostat for immunostaining; or (3) homogenized, then lysed into RIPA buffer for western blotting.
Immunofluorescence
Immunofluorescence staining of de-paraffinized tissues used antibodies described in Supplementary Table S3, followed by DAPI coverslipping medium (010020, Southern Biotech., Birmingham, AL, USA). Slides were imaged using the Zeiss AxioObserver described above. Images were photographed and CD11b+ cells manually counted by two observers blinded to treatment group.
Bone marrow experiments
Murine bone marrow cells were harvested from the femurs and tibia of BALB/c mice and cultured for 16 hours in 25 ng/ml M-CSF. Non-adherent cells, representing macrophage precursors, were matured for 5 days in RPMI-1640 supplemented with 10% FBS and 25 ng/ml M-CSF and rested for 3 days in RPMI-1640 supplemented with 10% FBS. Macrophage polarization was performed by culturing macrophages for 18 more hours in RPMI-1640 with 5% FBS and 20 ng/ml IFN-γ (for M1 polarization) (315-05, Peprotech), 20 ng/ml IL-4 (for M2 polarization) (214-14, Peprotech), CM from U87-BevR or U87-BevS cells, recombinant murine MIF (300-69, Peprotech) and/or 40 μg/ml bevacizumab (Genentech).
Phagocytosis, monocyte chemotaxis, and matrigel invasion assays
Are described in Supplementary Methods.
Flow cytometry
Live cells were spun, counted, and resuspended in FACS buffer. Cells were then incubated with a cell viability assay (Cat# L34959 Thermo Fisher Scientific, Waltham, MA, USA) and blocked with either mouse Fc block (anti-mouse CD16/32) (Cat# 553141, BD Biosciences, Bedford, MA, USA) or human Fc block (Cat# 564219, BD Biosciences, Bedford, MA, USA) per manufacturers’ protocols. Cells were incubated with fluorescent antibodies (Supplementary Table S3) along with isotype controls, for 30 min at 4 °C in the dark. Cells were washed and resuspended in FACS buffer. FACS was performed on BD FACS Aria3. Data were analyzed with BD FlowJo software (Tree Star Inc., Ashland, OR, USA).
Isolating CD11b+ cells from site-directed biopsies
With UCSF IRB approval (#11-06160), biopsies were performed after obtaining informed consent utilizing a three-dimensional intraoperative navigation system for stereotactic sampling of the FLAIR bright infiltrated margin outside the enhancement, the enhancing edge of the tumor, and the central tumor core inside the enhancement before tumor resection commenced (Supplementary Figure 1). Regional biopsy tissues were dissociated using the gentleMACS dissociator and the human tumor dissociation kit (Miltenyi Biotec, Auburn, CA, USA). CD11b+ cells were isolated from the dissociated samples using microbeads (Miltenyi Biotec) per manufacturer’s protocol. Other human tissue was obtained through the UCSF Brain Tumor tissue bank, which banked tissue after informed patient consent.
Statistics
For continuous variables, a Shapiro-Wilk test was used to confirm a normal distribution and homogeneity of variances was verified, which occurred for each analyzed variable. A student’s t-test (paired or unpaired depending on data) was then used to compare the variables. For multiple comparisons of normally distributed variables with homogeneous variances, two way ANOVA was used to analyze for main effects and interactions. Interobserver variability for manual counting of immunofluorescence was assessed using SPSS VARCOMP analysis. P values are two-tailed and P<0.05 was considered significant. For microarray statistics, see Supplementary Methods.
Acknowledgments
MKA was supported by the American Brain Tumor Association (ABTA), the James S. McDonnell Foundation, American Cancer Society (ACS), University of California Cancer Research Coordinating Committee and the NIH (5K02NS64167 and 1R01NS079697). BC was supported by the Howard Hughes Medical Institute (HHMI) fellowship, the American Association of Neurological Surgeons (AANS) and ABTA. DH was supported by the Howard Hughes Medical Institute (HHMI) fellowship. WC was supported by the UCSF School of Medicine (SOM) Pathways Explore Summer Grant.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Clark AJ, Lamborn KR, Butowski NA, Chang SM, Prados MD, Clarke JL et al. Neurosurgical management and prognosis of patients with glioblastoma that progresses during bevacizumab treatment. Neurosurgery 2012; 70: 361–370. [DOI] [PubMed] [Google Scholar]
- Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer 2008; 8: 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisl TN, Kim L, Moore K, Duic P, Royce C, Stroud I et al. Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. J Clin Oncol 2009; 27: 740–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLay M, Jahangiri A, Carbonell WS, Hu YL, Tsao S, Tom MW et al. Microarray analysis verifies two distinct phenotypes of glioblastomas resistant to antiangiogenic therapy. Clin Cancer Res 2012; 18: 2930–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao Y, Liang J, Holmes L, Zurita AJ, Henry V, Heymach JV et al. Glioblastoma resistance to anti-VEGF therapy is associated with myeloid cell infiltration, stem cell accumulation, and a mesenchymal phenotype. Neuro Oncol 2012; 14: 1379–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrusiewicz K, Liu D, Cortes-Santiago N, Hossain MB, Conrad CA, Aldape KD et al. Anti-vascular endothelial growth factor therapy-induced glioma invasion is associated with accumulation of Tie2-expressing monocytes. Oncotarget 2014; 5: 2208–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu-Emerson C, Snuderl M, Kirkpatrick ND, Goveia J, Davidson C, Huang Y et al. Increase in tumor-associated macrophages after antiangiogenic therapy is associated with poor survival among patients with recurrent glioblastoma. Neuro Oncol 2013; 15: 1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 2004; 25: 677–686. [DOI] [PubMed] [Google Scholar]
- Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000prime Reports 2014; 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Gabrusiewicz K, Heimberger A. The controversial role of microglia in malignant gliomas. Clin Dev Immunol 2013; 2013: 285246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Imai Y, Ohsawa K, Nakajima K, Fukuuchi Y, Kohsaka S. Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res Mol Brain Res 1998; 57: 1–9. [DOI] [PubMed] [Google Scholar]
- Mizutani M, Pino PA, Saederup N, Charo IF, Ransohoff RM, Cardona AE. The fractalkine receptor but not CCR2 is present on microglia from embryonic development throughout adulthood. J Immunol 2012; 188: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saederup N, Cardona AE, Croft K, Mizutani M, Cotleur AC, Tsou CL et al. Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PloS one 2010; 5: e13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahangiri A, De Lay M, Miller LM, Carbonell WS, Hu YL, Lu K et al. Gene expression profile identifies tyrosine kinase c-Met as a targetable mediator of antiangiogenic therapy resistance. Clinical Cancer Research 2013; 19: 1773–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell WS, DeLay M, Jahangiri A, Park CC, Aghi MK. beta1 integrin targeting potentiates antiangiogenic therapy and inhibits the growth of bevacizumab-resistant glioblastoma. Cancer Res 2013; 73: 3145–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA 2016; 113: E1738–E1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Toi M, Saji H, Muta M, Bando H, Kuroi K et al. Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clin Cancer Res 2000; 6: 3282–3289. [PubMed] [Google Scholar]
- Yaddanapudi K, Putty K, Rendon BE, Lamont GJ, Faughn JD, Satoskar A et al. Control of tumor-associated macrophage alternative activation by macrophage migration inhibitory factor. J Immunol 2013; 190: 2984–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppstadter J, Seif M, Dembek A, Cavelius C, Huwer H, Kraegeloh A et al. M2 polarization enhances silica nanoparticle uptake by macrophages. Front Pharmacol 2015; 6: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidi M, Gotti E, Bologna L, Miranda E, Rimoldi M, Sica A et al. M2 macrophages phagocytose rituximab-opsonized leukemic targets more efficiently than m1 cells in vitro. J Immunol 2009; 182: 4415–4422. [DOI] [PubMed] [Google Scholar]
- Choudhary S, Hegde P, Pruitt JR, Sielecki TM, Choudhary D, Scarpato K et al. Macrophage migratory inhibitory factor promotes bladder cancer progression via increasing proliferation and angiogenesis. Carcinogenesis 2013; 34: 2891–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta Y, Pasupuleti V, Du W, Welford SM. Macrophage migration inhibitory factor secretion is induced by ionizing radiation and oxidative stress in cancer cells. PloS One 2016; 11: e0146482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ren Y, Wu QC, Lin SX, Liang YJ, Liang HZ. Macrophage migration inhibitory factor enhances neoplastic cell invasion by inducing the expression of matrix metalloproteinase 9 and interleukin-8 in nasopharyngeal carcinoma cell lines. Chin Med J 2004; 117: 107–114. [PubMed] [Google Scholar]
- Meyer-Siegler KL, Iczkowski KA, Leng L, Bucala R, Vera PL. Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J Immunol 2006; 177: 8730–8739. [DOI] [PubMed] [Google Scholar]
- Piette C, Deprez M, Roger T, Noël A, Foidart J-M, Munaut C. The dexamethasone-induced inhibition of proliferation, migration, and invasion in glioma cell lines is antagonized by macrophage migration inhibitory factor (MIF) and can be enhanced by specific MIF inhibitors. J Biol Chem 2009; 284: 32483–32492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Nishihira J, Yoshiki T, Kondo M, Sato Y, Sasaki F et al. Macrophage migration inhibitory factor promotes tumor invasion and metastasis via the Rho-dependent pathway. Clin Cancer Res 2005; 11: 1050–1058. [PubMed] [Google Scholar]
- Hu YL, Jahangiri A, De Lay M, Aghi MK. Hypoxia-induced tumor cell autophagy mediates resistance to anti-angiogenic therapy. Autophagy 2012; 8: 979–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller YA, Chen Y, Christinger HW, Li B, Cunningham BC, Lowman HB et al. VEGF and the Fab fragment of a humanized neutralizing antibody: crystal structure of the complex at 2.4 A resolution and mutational analysis of the interface. Structure 1998; 6: 1153–1167. [DOI] [PubMed] [Google Scholar]
- Jenkins SJ, Ruckerl D, Cook PC, Jones LH, Finkelman FD, van Rooijen N et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 2011; 332: 1284–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redente EF, Higgins DM, Dwyer-Nield LD, Orme IM, Gonzalez-Juarrero M, Malkinson AM. Differential polarization of alveolar macrophages and bone marrow-derived monocytes following chemically and pathogen-induced chronic lung inflammation. J Leukocyte Biol 2010; 88: 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoger JL, Gijbels MJ, van der Velden S, Manca M, van der Loos CM, Biessen EA et al. Distribution of macrophage polarization markers in human atherosclerosis. Atherosclerosis 2012; 225: 461–468. [DOI] [PubMed] [Google Scholar]
- Casazza A, Laoui D, Wenes M, Rizzolio S, Bassani N, Mambretti M et al. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell 2013; 24: 695–709. [DOI] [PubMed] [Google Scholar]
- Mittelbronn M, Platten M, Zeiner P, Dombrowski Y, Frank B, Zachskorn C et al. Macrophage migration inhibitory factor (MIF) expression in human malignant gliomas contributes to immune escape and tumour progression. Acta Neuropathol 2011; 122: 353–365. [DOI] [PubMed] [Google Scholar]
- Verschuren L, Kooistra T, Bernhagen J, Voshol PJ, Ouwens DM, van Erk M et al. MIF deficiency reduces chronic inflammation in white adipose tissue and impairs the development of insulin resistance, glucose intolerance, and associated atherosclerotic disease. Circ Res 2009; 105: 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiner PS, Preusse C, Blank AE, Zachskorn C, Baumgarten P, Caspary L et al. MIF Receptor CD74 is Restricted to Microglia/Macrophages, Associated with a M1-Polarized Immune Milieu and Prolonged Patient Survival in Gliomas. Brain Pathol 2015; 25: 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoochani A, Schwarz MA, Yakubov E, Engelhorn T, Doerfler A, Buchfelder M et al. MIF-CD74 signaling impedes microglial M1 polarization and facilitates brain tumorigenesis. Oncogene 2016; 35: 6246–6261. [DOI] [PubMed] [Google Scholar]
- Girard E, Strathdee C, Trueblood E, Queva C. Macrophage migration inhibitory factor produced by the tumour stroma but not by tumour cells regulates angiogenesis in the B16-F10 melanoma model. Br J Cancer 2012; 107: 1498–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen T, Leng L, Fan J, Cao K, Duan Z et al. MIF produced by bone marrow-derived macrophages contributes to teratoma progression after embryonic stem cell transplantation. Cancer Res 2012; 72: 2867–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KV, Chang JP, Parachoniak CA, Pandika MM, Aghi MK, Meyronet D et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer cell 2012; 22: 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot JF. High-dose antiangiogenic therapy for glioblastoma: less may be more? Clin Cancer Res 2011; 17: 6109–6111. [DOI] [PubMed] [Google Scholar]
- Thiele M, Kerschbaumer RJ, Tam FW, Volkel D, Douillard P, Schinagl A et al. Selective targeting of a disease-related conformational isoform of macrophage migration inhibitory factor ameliorates inflammatory conditions. J Immunol 2015; 195: 2343–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Kim J, Ozawa T, Zhang M, Westphal M, Deen DF et al. Anti-VEGF antibody treatment of glioblastoma prolongs survival but results in increased vascular cooption. Neoplasia 2000; 2: 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentzas S, Simoneau E, Bridgeman VL, Vermeulen PB, Foo S, Kostaras E et al. Vessel co-option mediates resistance to anti-angiogenic therapy in liver metastases. Nat Med 2016; 22: 1294–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao Y, Park SY, Henry V, Smith BD, Tiao N, Flynn DL et al. Novel MET/TIE2/VEGFR2 inhibitor altiratinib inhibits tumor growth and invasiveness in bevacizumab-resistant glioblastoma mouse models. Neuro Oncol 2016; 18: 1230–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahbari NN, Kedrin D, Incio J, Liu H, Ho WW, Nia HT et al. Anti-VEGF therapy induces ECM remodeling and mechanical barriers to therapy in colorectal cancer liver metastases. Sci Transl Med 2016; 8: 360ra135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz A, Harter PN, Cremer S, Yalcin BH, Gurnik S, Yamaji M et al. Endothelial cell-derived angiopoietin-2 is a therapeutic target in treatment-naive and bevacizumab-resistant glioblastoma. EMBO Mol Med 2015; 8: 39–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JL, Sainson RC, Oon CE, Turley H, Leek R, Sheldon H et al. DLL4-Notch signaling mediates tumor resistance to anti-VEGF therapy in vivo. Cancer Res 2011; 71: 6073–6083. [DOI] [PubMed] [Google Scholar]
- Kessler T, Sahm F, Blaes J, Osswald M, Rubmann P, Milford D et al. Glioma cell VEGFR-2 confers resistance to chemotherapeutic and antiangiogenic treatments in PTEN-deficient glioblastoma. Oncotarget 2015; 6: 31050–31068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HR, Hong B, Kim H, Park CK, Park SH, Park S et al. Assessment of bevacizumab resistance increased by expression of BCAT1 in IDH1 wild-type glioblastoma: application of DSC perfusion MR imaging. Oncotarget 2016; 7: 69606–69615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao Y, Liang J, Holmes L, Henry V, Sulman E, de Groot JF. Acquired resistance to anti-VEGF therapy in glioblastoma is associated with a mesenchymal transition. Clin Cancer Res 2013; 19: 4392–4403. [DOI] [PubMed] [Google Scholar]
- Mao XG, Wang C, Liu DY, Zhang X, Wang L, Yan M et al. Hypoxia upregulates HIG2 expression and contributes to bevacizumab resistance in glioblastoma. Oncotarget 2016; 7: 47808–47820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi FS, Lawrence D, Lezcano C, Wu X, Zhou J, Sasada T et al. Bevacizumab plus ipilimumab in patients with metastatic melanoma. Cancer Immunol Res 2014; 2: 632–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moughon DL, He H, Schokrpur S, Jiang ZK, Yaqoob M, David J et al. Macrophage blockade using CSF1R inhibitors reverses the vascular leakage underlying malignant ascites in late-stage epithelial ovarian cancer. Cancer Res 2015; 75: 4742–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.