Abstract
The airway epithelium forms a barrier between the internal and external environments. Epithelial dysfunction is critical in the pathology of many respiratory diseases, including cystic fibrosis. Ets homologous factor (EHF) is a key member of the transcription factor network that regulates gene expression in the airway epithelium in response to endogenous and exogenous stimuli. EHF, which has altered expression in inflammatory states, maps to the 5′ end of an intergenic region on Chr11p13 that is implicated as a modifier of cystic fibrosis airway disease. Here we determine the functions of EHF in primary human bronchial epithelial (HBE) cells and relevant airway cell lines. Using EHF ChIP followed by deep sequencing (ChIP-seq) and RNA sequencing after EHF depletion, we show that EHF targets in HBE cells are enriched for genes involved in inflammation and wound repair. Furthermore, changes in gene expression impact cell phenotype because EHF depletion alters epithelial secretion of a neutrophil chemokine and slows wound closure in HBE cells. EHF activates expression of the SAM pointed domain-containing ETS transcription factor, which contributes to goblet cell hyperplasia. Our data reveal a critical role for EHF in regulating epithelial function in lung disease.
Keywords: ChIP sequencing (ChIP-seq), epithelium, ETS transcription factor family, lung injury, siRNA, transcription factor, transcriptomics, Ets homologous factor (EHF)
Introduction
The airway epithelium plays a critical role in lung function in both normal and pathological states. It forms an active barrier between the external and internal environments, responding to a wide range of stimuli. Because this cell layer is constantly exposed to the outside atmosphere, it has developed mechanisms to respond rapidly to external insults. Following mechanical damage, epithelial cells proliferate and migrate to close the resulting wound (1). In response to foreign pathogens and particles, the cells release immune factors (2). The airway epithelium also drives the mucociliary escalator, which physically removes debris from the lungs (3, 4). In the presence of chronic damage or infection, these pathways can malfunction and contribute to lung pathology in diseases such as cystic fibrosis (CF)2 (3, 5, 6).
CF is a common autosomal recessive disorder caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. The main cause of morbidity and mortality in CF is lung disease (7). Airway dysfunction in CF patients is complex, but altered ion transport leads to a decreased ability to clear microorganisms, chronic infection, and inflammation. Neutrophil-dominant inflammation is present early in the disease (8–10). There is increased secretion of cytokines, including IL-13 and IL-17a, in the lungs, which is exacerbated in patients infected with Pseudomonas aeruginosa, the predominant pathogen in the CF airway (11–13). IL-13 augments mucin production by the lung epithelium and triggers goblet cell hyperplasia (14). IL-17a induces neutrophil chemokine production by epithelial cells (15). The amplified mucus and cytokine levels cause destruction of lung tissue by initiating remodeling and fibrosis, which correlate with pulmonary dysfunction (16–19). Similar pathways are activated in other airway diseases, including chronic obstructive pulmonary disease (COPD) and asthma (20, 21). Modulation of gene expression underlies the response of the lung epithelium to the environment, both in normal and disease states, so it is critical to understand the causative mechanisms. Pivotal to these processes is the transcription factor (TF) network that regulates development, normal function, and response to disease in respiratory epithelial cells. TFs respond to normal and aberrant stimuli by binding to the genome at regulatory sites, thereby altering gene expression. Ets homologous factor (EHF) is a member of the epithelium-specific Ets transcription factor subfamily that is expressed in the bronchial epithelium (22–24). In response to inflammatory mediators, including IL-1β and TNF-α, bronchial EHF expression is increased in an NF-κB-dependent manner, suggesting a role for EHF in the epithelial response to inflammation (25). In prostate cancer, loss of EHF leads to epithelial-to-mesenchymal transition (EMT) and increased metastatic potential (26). In EMT, epithelial cells lose contact with adjacent cells and become more motile (27). Similar pathways may be involved in lung fibrosis (28).
Recent genome-wide association studies led to an interest in EHF as a potentially important factor in CF lung pathology. These studies identified intergenic SNPs at Chr11p13 that strongly associate with lung disease severity in CF patients carrying the most common disease-causing mutation, F508del (29, 30). These SNPs are likely within or close to cis-regulatory elements for nearby genes. EHF maps immediately adjacent (5′) to this intergenic region, suggesting a role as a modifier of lung disease in CF patients. We hypothesize that EHF regulates genes involved in cellular processes that impact the severity of epithelial pathology in airway disease, including inflammation and wound repair.
This study investigates the role of EHF in primary human bronchial epithelial (HBE) cells. Using ChIP followed by sequencing (ChIP-seq) and RNA sequencing (RNA-seq) after EHF depletion, we identified putative direct targets of EHF in HBE cells. These genes are enriched in processes that are involved in lung pathology. Cellular assays determined that EHF-modulated changes in gene expression alter epithelial phenotypes. Our results suggest that EHF is involved in multiple processes that influence lung function in disease. Of particular relevance to CF are the release of neutrophil chemokines and wound repair.
Results
ChIP-seq generates a genome-wide binding signature for EHF in primary HBE cells
To identify genome-wide binding sites for EHF, ChIP-seq was performed on cultures from two tissue donors. The antibody specific for EHF was used previously for ChIP-seq in Calu-3 lung adenocarcinoma cells (31). The HOMER irreproducible discovery rate pipeline (32) was used to call a total of 11,326 peaks with an irreproducible discovery rate (IDR) < 0.05 (supplemental Table S1). Sonicated input DNA was used as background. The normalized tag counts at each called peak were significantly correlated between the two biological replicates (r = 0.29, p < 0.0001).
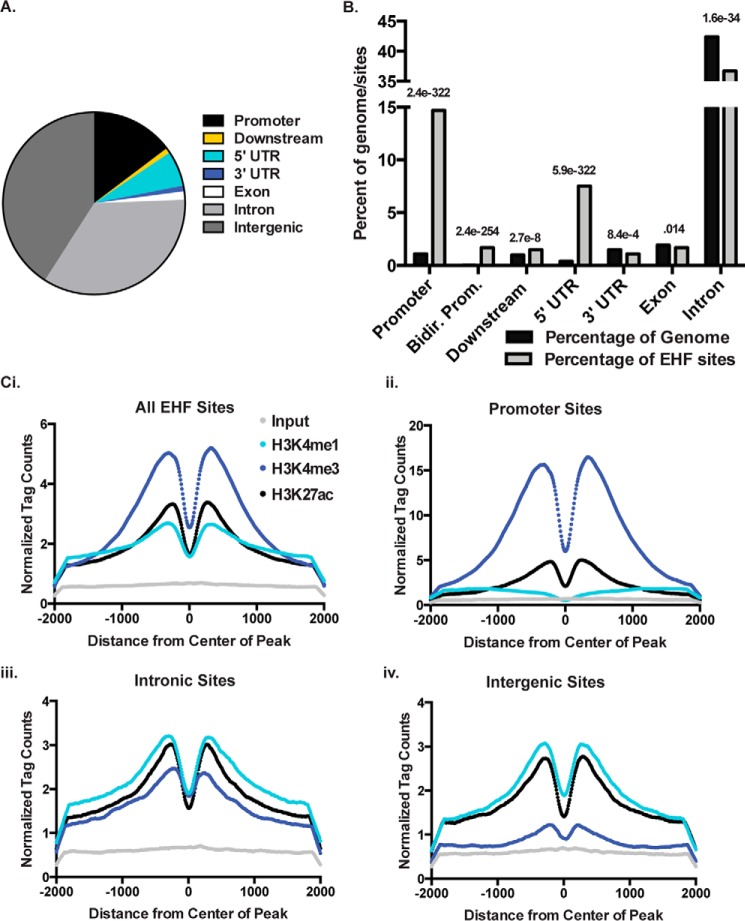
The Cis-regulatory Element Annotation System (CEAS) software (33) was used to annotate peaks based on genomic location (Fig. 1A) and determined that EHF sites were enriched at promoters (−1 kb relative to the transcription start site) and 5′ UTRs of genes (Fig. 1B). Consistent with its role as a TF controlling cell type-specific gene expression, peaks of EHF occupancy were also found in intergenic and intronic regions, which may represent cis-regulatory elements such as enhancers. These regulatory sites are marked by specific histone modifications. Active or poised promoters and enhancers are enriched for histone 3 lysine 4 trimethylation (H3K4me3) and H3K4me1, respectively, whereas active regions are specifically marked by histone 3 lysine 27 acetylation (H3K27ac) (34, 35). To determine whether EHF peaks are found in regions with these histone modifications, as predicted by their genomic locations, ChIP-seq was performed using antibodies against H3K4me1, H3K4me3, and H3K27ac in HBE cells. EHF sites were enriched for all marks with a bimodal distribution around the center of the peak, in accordance with a nucleosome-depleted center region where transcription factors bind (Fig. 1C). Consistent with an overrepresentation of EHF sites at promoters, the H3K4me3 mark was the most enriched.
Figure 1.
EHF ChIP-seq sites are overrepresented at promoters and enriched for activate histone marks. EHF ChIP-seq was performed in undifferentiated HBE cells from two donor cultures. A, percentage of EHF-binding sites found at promoters (1 kb upstream of TSSs) and downstream of genes (1 kb), 5′ UTRs, 3′ UTRs, exons, introns, and intergenic sites. B, percentage of EHF ChIP-seq peaks found at specific sites compared with genomic background. Bidirectional promoters (Bidir. prom.) are considered 2.5 kb upstream of TSSs. The p values, derived using CEAS (one-sided binomial test), are shown. C, normalized tag counts from H3K4me1, H3K4me3, and H3K27ac ChIP-seq in undifferentiated HBE cells as measured from the center of all EHF-binding sites (i), promoter-binding sites (ii), intronic binding sites (iii), and intergenic binding sites (iv). Tag counts for input sonicated HBE DNA provided a control.
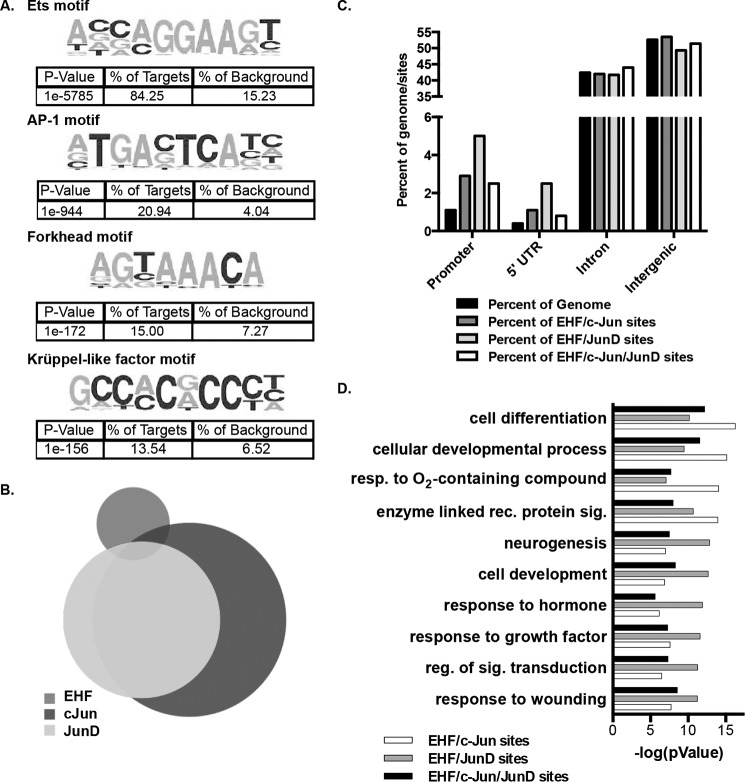
De novo motif analysis was performed on the significant EHF ChIP-seq peaks, which identified the epithelium-specific Ets motif as the most enriched, found in 84.25% of sites within 200 bp of the center of the peak (Fig. 2A). The motif analysis also identified secondary enriched motifs. The binding motifs for activator protein 1 (AP-1), Forkhead domain, and Krüppel-like factor (KLF) transcription factor families were among the most enriched secondary motifs, found in 20.94%, 15%, and 13.54% of peaks, respectively (Fig. 2A).
Figure 2.
EHF and the AP-1 transcription factor complex bind at overlapping sites genome-wide. A, de novo motif analysis within 200 bp of the center of EHF ChIP-seq peaks in undifferentiated HBE cells identified enriched motifs. The p values, derived by HOMER using cumulative binomial distributions, and the percentage of EHF sites and of background containing each motif are shown below the sequence. B, intersection of EHF, c-Jun, and JunD ChIP-seq binding sites in Calu-3 cells. C, annotation of EHF/c-Jun, EHF/JunD, and EHF/c- Jun/JunD overlapping sites compared with the percentage of the genome. The p values were derived using CEAS (one-sided binomial test). D, gene ontology process enrichment analysis was performed on the nearest gene to each EHF/c-Jun, EHF/JunD, and EHF/c-Jun/JunD intersecting region. Shown are the negative logs of the p values, derived using MetaCore. Resp., response; rec., receptor; sig., signaling; reg., regulation.
To confirm that EHF and AP-1 co-localize in the genome, ChIP-seq for c-Jun and JunD, members of the AP-1 protein complex, was performed in Calu-3 lung adenocarcinoma cells. ENCODE (Encyclopedia of DNA Elements) consortium-validated antibodies were used to perform two biological replicates for each protein, and the HOMER IDR method was used to call significant peaks of transcription factor binding. A total of 11,517 and 7524 sites were identified for c-Jun and JunD, respectively. In addition, the HOMER IDR protocol was used to call peaks in two biological replicates of EHF ChIP performed in Calu-3 cells, which we published previously (31). These three data sets were then intersected (Fig. 2B). Twenty-five percent of the EHF peaks overlapped at least one c-Jun or JunD peak, with 59% of these containing both c-Jun and JunD sites, 32% having only c-Jun, and 9% containing only JunD. EHF sites that overlap JunD binding sites are enriched at the promoters (Fig. 2C). The nearest gene annotation method was used to determine potential genes regulated by the overlapping binding sites, and these were subjected to a gene ontology process enrichment analysis using MetaCore from Thomson Reuters version 6.26 (Fig. 2D). The overlapping sites are found near loci involved in differentiation, development, and wound repair, suggesting that they may have a combined role in regulating phenotypes important for lung pathology.
EHF regulates genes involved in lung pathology
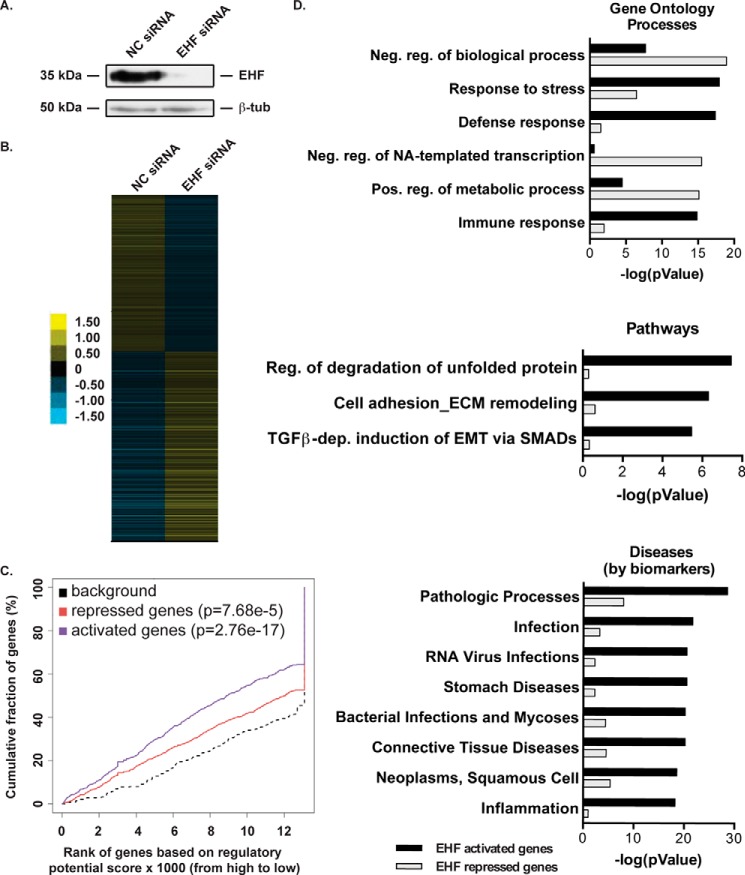
To detect genes that are directly and indirectly regulated by EHF, RNA-seq was performed on EHF-depleted (using a targeted siRNA) and negative control siRNA-treated HBE cells (Fig. 3A). Cufflinks (36) was used to identify a total of 1145 genes that were differentially regulated (DEGs) following EHF depletion (FDR < 0.05), with 625 increasing and 520 decreasing in expression (Fig. 3B and supplemental Table S2). A multidimensional scaling analysis was performed, which confirmed that four of five samples within each treatment clustered together and away from the second group (supplemental Fig. S1).
Figure 3.
EHF depletion followed by RNA-seq identifies targets involved in the immune response and gene regulation. A, siRNA depletion of EHF in undifferentiated HBE cells was measured by Western blot with β-tubulin (β-tub) as a loading control. B, relative expression of 1145 genes that show a significant change in expression following EHF depletion in undifferentiated HBE cells (FDR < 0.05). Each row signifies a different gene. The scale is log2 -fold change relative to average gene expression. C, activating and repressive function prediction of EHF in undifferentiated primary HBE cells as determined by BETA software. The red, purple, and dashed lines indicate the down-regulated, up-regulated, and non-differentially expressed genes, respectively. The p values of activated and repressed genes compared with non-regulated genes were determined by a Kolmogorov-Smirnov test. D, MetaCore enrichment analysis of direct targets of EHF. Analysis was performed on EHF-activated (black) and -repressed (gray) genes and divided into gene ontology processes (top panel), pathways (center panel), and diseases (bottom panel). Shown are the negative logs of the p values, derived using MetaCore. Neg., negative; NA, nucleic acid; pos., positive; dep., dependent.
To identify putative direct targets of EHF, the EHF ChIP-seq data and the DEGs identified by RNA-seq after EHF depletion were intersected using Binding and Expression Target Analysis (BETA) (37). EHF was found to have significant activating and repressive function (Fig. 3C), consistent with gene expression changes in the positive and negative direction following EHF knockdown. This analysis revealed 425 putative direct targets that are repressed by EHF and 434 that are activated (supplemental Tables S3 and S4). The forkhead box A1 (FOXA1) motif was enriched in EHF sites near repressed genes compared with non-regulated genes (p = 2.85e-13). The EHF-repressed genes included the following, which encode transcription factors: GATA-binding protein 6 (GATA6), v-myc avian myelocytomatosis viral oncogene homolog (MYC), CCAAT/enhancer binding protein (C/EBP) γ and β (CEBPG and CEBPB), HOP homeobox (HOPX), and KLF5. Genes activated by EHF include the SAM pointed domain-containing ETS transcription factor (SPDEF), which is involved in airway goblet cell development (38, 39), and the inflammatory S100 calcium binding proteins S100A8, S100A9, and S100A12.
An enrichment analysis was performed on up-regulated and down-regulated predicted direct targets using MetaCore (Fig. 3D). EHF-activated genes were found in pathways including regulation of degradation of misfolded proteins, extracellular matrix remodeling, and EMT; diseases comprising infections and inflammation; and gene ontology processes including response to stress, immune response, and response to wounding. EHF-repressed genes were enriched in gene ontologies involving negative regulation of RNA synthesis, transcription, and metabolic processes.
EHF and FOXA1 may co-regulate transcription factors important for lung epithelial development
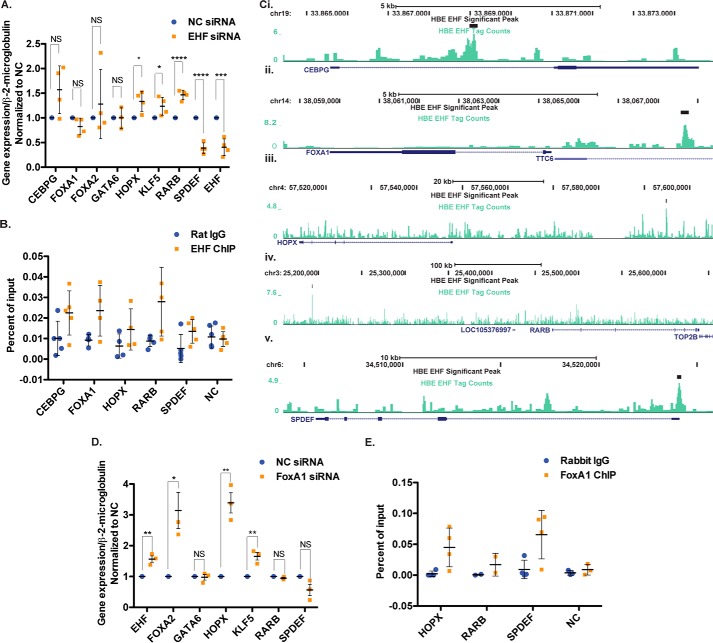
Lung organogenesis and airway epithelial differentiation are complex processes coordinated by a network of transcription factors. Multiple TFs within this network were identified as putative EHF targets in our genome-wide data. These include genes with nearby peaks of EHF occupancy and/or DEGs after EHF depletion, among which are GATA6, HOPX, KLF5, SPDEF, retinoic acid receptor β (RARB), FOXA1, and FOXA2. To validate direct or indirect targets, EHF was depleted in primary HBE cell cultures derived from four HBE donors (supplemental Fig. S2). Expression of key TFs was determined using RT-qPCR (Fig. 4A). HOPX, KLF5, and RARB abundance significantly increased following EHF depletion, and SPDEF levels significantly decreased. CEBPG and FOXA1 showed an increase and decrease, respectively, that did not reach statistical significance (CEBPG, p = 0.056; FOXA1, p = 0.08). Because of the substantial variation between primary cultures, these non-significant changes may be biologically relevant but would require additional validation. GATA6 showed no change, and FOXA2 levels were variable between donors. Nearby peaks of EHF occupancy evident in the ChIP-seq data suggest that most of these genes may be direct targets (Fig. 4C and supplemental Fig. S3). To confirm this prediction, four biological (donor) replicates of EHF ChIP qPCR were performed in primary HBE cultures (Fig. 4B). EHF binding was confirmed at intron 1 of CEBPG, intergenic sites 5′ of FOXA1, HOPX, and RARB, and the promoter of SPDEF.
Figure 4.
EHF and FOXA1 regulate other transcription factors involved in airway epithelial differentiation in HBE cells. A, EHF depletion by siRNA followed by RT-qPCR identified changes in TF gene expression (n = 4). B, EHF ChIP-qPCR confirmed binding at specific sites identified in EHF ChIP-seq (n = 4). C, Genome Browser tracks show EHF peaks identified by IDR in HBE cells at the top (black), below are the EHF ChIP-seq tag counts (teal) and the gene (blue) for CEPBG (i), FOXA1 (ii), HOPX (iii), RARB (iv), and SPDEF (v). D, alterations in EHF levels and EHF-regulated TF gene expression after FOXA1 siRNA knockdown, shown by RT-qPCR (n = 3). E, FOXA1 ChIP qPCR confirmed its binding to a subset of EHF sites (n = 4). All experiments were performed in undifferentiated HBE cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; NS, not significant; unpaired two-tailed Student's t test. Bars show the average of all experiments, with error bars representing standard deviation.
The sites of EHF occupancy near CEBPG, HOPX, RARB, and SPDEF also contain sequences with the Forkhead domain motif. Because EHF represses all but one of these genes (SPDEF), and the FOXA1 motif is enriched among EHF binding sites near repressed genes genome-wide, we next asked whether FOXA1 co-regulates gene expression with EHF at these loci. FOXA1 was depleted in HBE primary cultures from three donors using a specific siRNA and a non-targeting control, and gene expression was measured by RT-qPCR (supplemental Fig. S4 and Fig. 4D). Depletion of FOXA1 significantly elevated expression of EHF, FOXA2, HOPX, and KLF5 and decreased SPDEF levels (p = 0.008). RARB did not change. Furthermore, the EHF sites near HOPX and SPDEF were bound by FOXA1 as shown by four biological replicates of ChIP qPCR in primary HBE cells from two donors (Fig. 4E). The FOXA1 enrichment near RARB was lower than the other sites, which may explain why FOXA1 knockdown does not alter RARB expression.
EHF depletion alters the expression of cytokines important for neutrophil migration and survival
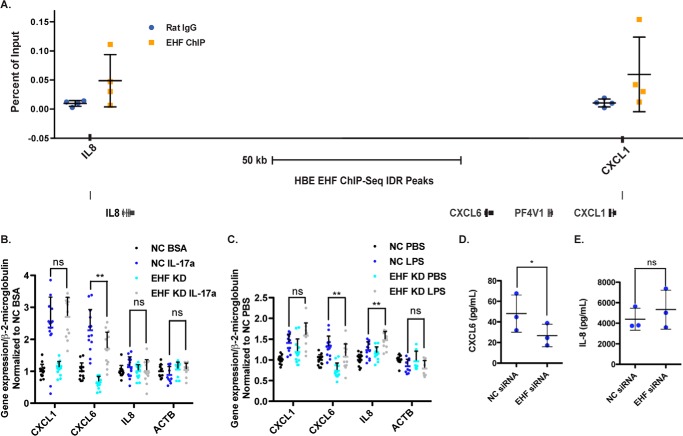
Genes involved in inflammation and infection are enriched among putative direct targets of EHF identified by ChIP-seq and RNA-seq. Two EHF binding sites were observed at the Chr4q13.3 region encompassing the genes for the neutrophil chemokines CXCL1, CXCL6, and IL8, one 5′ to IL8 and the other 3′ to CXCL1 (Fig. 5A). The sites identified by EHF ChIP-seq were confirmed by ChIP qPCR in HBE cell chromatin from four donors (Fig. 5A). The bronchial epithelium releases these cytokines in response to stimuli, including IL-17a, a pro-inflammatory cytokine involved in multiple lung pathologies, including CF (12, 15), and LPS, a component of the Gram-negative cell wall that induces a strong immune response in the lung (40). The role of EHF in regulating these immune mediators was studied in Calu-3 lung adenocarcinoma cells, which express CXCL1, CXCL6, and IL8. To determine whether EHF regulates secretion of the three cytokines encoded by these genes, both at a basal level and in an inflammatory milieu, EHF was depleted in Calu-3 cells using a specific siRNA, with a non-targeting siRNA as a control (supplemental Figs. S5 and S6). Cells were then treated with vehicle (BSA) or IL-17a for 4 h or vehicle (PBS) or LPS from P. aeruginosa for 24 h, and RNA was extracted. Cytokine expression was then quantified using RT-qPCR (Fig. 5, B and C). CXCL6 expression was repressed upon EHF depletion under normal conditions and following LPS and IL-17a treatment. EHF knockdown was associated with increased IL8 expression following LPS treatment, but there was no change in basal levels. CXCL1 was not significantly regulated by EHF under either condition. To determine whether the changes in CXCL6 transcript abundance altered CXCL6 cytokine secretion, colorimetric sandwich ELISAs were utilized to quantify its release into conditioned medium. EHF depletion decreased CXCL6 secretion (Fig. 5D) but did not significantly alter IL-8 secretion (Fig. 5E), consistent with no change in IL8 expression at basal levels.
Figure 5.
EHF depletion in Calu-3 cells alters secretion of a neutrophil chemokine. A, EHF ChIP qPCR confirmed its binding to sites near IL8, CXCL6, and CXCL1 in HBE cells. Shown is the average of all experiments with standard deviation (n = 4) (top panel). Also shown is a graphic of the Chr4q13.3 region with EHF ChIP-seq peaks (bottom panel). B and C, EHF was depleted (siRNA) in Calu-3 cells, followed by their exposure to carrier, IL-17a (B), or LPS (C). Gene expression was measured by RT-qPCR. β-actin (ACTB) was included as a negative control. **, p < 0.01 by two-way analysis of variance plus multiple comparisons test; ns, not significant (average with standard deviation). D and E, secretion of CXCL6 (D) and IL-8 (E) into medium conditioned by NC- and EHF-depleted Calu-3 quantified by colorimetric sandwich ELISA (n = 3). *, p < 0.05; paired two-tailed Student's t test. Bars show the average of all experiments, with error bars representing standard deviation.
Depletion of EHF slows wound closure in wild-type HBE cells and HBE cells from cystic fibrosis patients
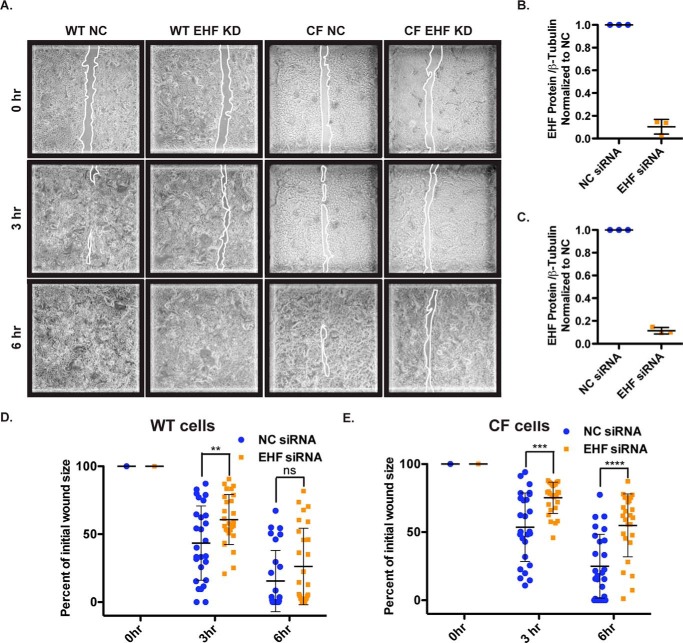
Among genes with nearby peaks of EHF occupancy that are also activated by EHF are many that are involved in EMT and response to wounding. When the lung epithelial barrier is damaged, cells at the edge of the wound migrate to close it. This mechanism may be impaired in airway epithelial cells that carry a CFTR mutation, suggesting that it contributes to CF lung pathology (5, 41). Wound repair can be measured in vitro with a wound scratch assay, which was performed in primary HBE cells from three wild-type and three CF donors. EHF was depleted using siRNA, and the cells were grown to a confluent layer on plastic (undifferentiated). The layer was scratched, and the wounds were imaged 0, 3, and 6 h post-injury (Fig. 6A). By 6 h, many of the scratches had closed. The cells were lysed, and EHF depletion was confirmed by quantification of Western blots by gel densitometry (Fig. 6, B and C). The initial wound size was not significantly different between the negative control (NC) and EHF siRNA-treated cells. The area was measured at subsequent time points, and the percentage of initial injury size was calculated (Fig. 6, D and E). In WT HBE cells at the 3-h time point, the scratches in EHF-depleted cells remained significantly larger than those in NC cells (p < 0.01). In CF cells, EHF knockdown significantly delayed the reduction in wound size at both 3 and 6 h. This corresponds to a slowing of wound closure in EHF-depleted cells compared with the NC in both WT and CF cells.
Figure 6.
EHF depletion slows wound closure in WT and CF HBE cells. A, images of NC and EHF siRNA-treated WT and CF undifferentiated HBE cells 0, 3, and 6 h after initial wounding. Outlines of wounds are traced in white. B and C, siRNA depletion of EHF was confirmed by Western blot and quantified using gel densitometry in WT (B) and CF cells (C) (average with standard deviation). D and E, percent of initial wound size was measured at all time points. Shown is the average with standard deviation for WT (D) and CF HBE cells (E) (three donor cultures). **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; NS, not significant; unpaired two-tailed Student's t test. Bars show the average of all experiments, with error bars representing standard deviation.
Discussion
Our data show that EHF is a critical regulator of multiple lung epithelial functions that are integral to pulmonary diseases, including cystic fibrosis. Among CF patients, there is a wide variation in airway phenotypes, which are under strong genetic control (42). However, lung disease severity does not correlate with specific mutations in CFTR (43), implicating other loci (gene modifiers) in modulating lung disease in this population. Our data suggest that EHF may be one of those modifiers based on its functions described here and its genomic location at Chr11p13, adjacent to SNPs that strongly associate with lung disease severity in CF patients (29, 30). Previous genome-wide studies of EHF demonstrated its contribution to the maintenance of epithelial barrier integrity and wound repair in Calu-3 lung adenocarcinoma cells (31) and in the mouse cornea (44). Here we investigated the role of EHF in primary HBE cells with a focus on understanding how it may contribute to lung pathology. The outcomes not only confirm our previous observations but also reveal substantial novel findings on how EHF participates in lung epithelial differentiation and inflammation.
The intersection of ChIP-seq data sets gives an overall indication of the function of EHF with respect to gene regulation. EHF sites are overrepresented at promoters (1 kb upstream of the TSS). A role for EHF in both activating and repressing gene expression is confirmed by the BETA output, which shows significant activity in both directions. This is consistent with previous studies at specific loci that demonstrate both active and repressive actions for EHF, depending on the cellular context (22, 45, 46). In our data, the fraction of genes activated by EHF is greater than the repressed component, consistent with the presence of active histone marks at EHF sites.
Because EHF occupies the relatively common Ets motif, and it can increase or decrease gene expression, its interactions with other TFs are likely critical to specify binding location and mechanisms of action. The motif analysis of EHF binding sites in HBE cells shows that the most enriched secondary motif is for AP-1. These data are consistent with our previous findings that the AP-1 motif is overrepresented in sites of EHF occupancy in Calu-3 cells (31). The AP-1 TF complex is made up of hetero- and homodimers of Jun, Fos, and other family members. Regulation of transcription by AP-1 has an important role in differentiation, cell injury, inflammation, and apoptosis in the lung (47, 48). AP-1 activation may be elevated in CF tracheal cells compared with their wild-type counterparts (49). EHF represses genes that contain both EHF and AP-1 motifs at their promoters (22), which suggests that the two factors can co-regulate gene expression. To determine whether EHF and Jun proteins co-localize genome-wide, we performed ChIP-seq for c-Jun and JunD in Calu-3 cells and intersected these data with our EHF occupancy profile published previously (31). There is significant coincidence between binding sites for EHF and both AP-1 complex proteins. The overlapping sites are found near genes involved in multiple pathways important for lung epithelial function in disease, including differentiation and wound repair. Further investigation of genes co-regulated by EHF and AP-1 could elucidate potential targets to modulate inflammation and wound repair in the airway.
Normal homeostasis in the lung epithelium depends on appropriate responses to external stimuli. An abnormal or exaggerated reaction can initiate or exacerbate lung disease. The pivotal role of several TFs in airway development and differentiation is well documented, particularly through studies in rodents. Nkx2–1 is the first marker of anterior foregut cells that will become respiratory epithelial progenitors (50). FoxA1 and FoxA2 are pioneer transcription factors required for branching morphogenesis early in lung development and epithelial differentiation at later stages (51, 52). Retinoic acid receptor (RAR) signaling is also necessary for proper branching morphogenesis (53). GATA6 is involved in lung epithelial differentiation throughout development but is also active in post-natal epithelial regeneration (54–56). Hopx acts downstream of Nkx2–1 to repress GATA6/Nkx2–1-induced expression of type II pneumocyte genes and is critical for alveolar development (57), whereas Klf5 is required for maturation of the alveolar respiratory epithelium (58). SPDEF is involved in airway goblet cell differentiation and mucus production and antagonizes the action of FoxA2 (59). Although these TFs are best studied in airway epithelial development, they are also likely to be involved in epithelial regeneration and dysplasia during disease. Here we demonstrate that EHF contributes to the transcriptional network controlling pulmonary gene expression.
By integrating the results of EHF ChIP-seq and its depletion followed by RNA-seq, we identified putative direct targets of the protein. TFs involved in lung development are among these identified targets. EHF represses HOPX, KLF5, and RARB, likely through binding to the promoter or a nearby cis-regulatory element. EHF binding sites are enriched for the Forkhead motif, which is bound by both FOXA1 and FOXA2. Interestingly, FOXA1 and EHF regulate each other and impact the expression of downstream TF targets similarly. Hence, these two factors may cooperate and, perhaps, participate together in a feedback loop.
Although it represses most TFs involved in lung development, EHF enhances the expression of SPDEF, which participates in goblet cell differentiation in the airway and is induced by IL-13 (38, 39). Given the known role of SPDEF in goblet cells (38, 39, 60), these data implicate EHF in activating goblet cell differentiation and, perhaps, perturbing that of other lineages. This suggests that EHF contributes to an IL-13-mediated cell differentiation pathway by activating SPDEF and repressing other transcription factors. IL-13 is a Th2 cytokine that induces goblet cell hyperplasia in the airway through SPDEF, with known roles in CF, asthma, and COPD (11, 13, 38, 39). Previously, immunohistochemical analysis showed that the most intense staining for EHF in the airway was in mucous gland epithelial cells, with a smaller fraction of bronchial epithelial cells being positive for EHF (22, 23). The pivotal role of submucosal glands in CF airway pathology (61) is particularly relevant to these observations.
In airway diseases such as CF, both IL-13 and IL-17a play a prominent role in promoting inflammation. Having observed that EHF influences the expression of genes downstream of IL-13, we chose to also investigate a potential function downstream of IL-17a. IL-17a induces bronchial epithelial cells to release neutrophil chemokines, including CXCL1, CXCL6, and IL-8 (15). These cytokines promote the migration and survival of neutrophils. This is critical for CF lung disease, as neutrophil-dominant inflammation is present early in the illness (8–10). Release of proteases by neutrophils contributes to lung damage (62, 63). EHF depletion in lung adenocarcinoma cells increases IL8 expression and decreases CXCL6 expression. This correlates with changes in secretion of CXCL6. By increasing CXCL6 secretion, EHF could increase neutrophil recruitment and survival, thereby exacerbating inflammation in the CF lung.
Another important function of lung epithelial cells that may be impaired in CF is response to wounding (5, 41). This is problematic in a disease characterized by inflammation and resulting damage to the lung epithelium. We showed previously that EHF depletion slows scratch wound closure in Calu-3 lung adenocarcinoma cells (31). Consistent with these earlier observations are current genome-wide studies in HBE cells that show an enrichment of EHF-activated genes in EMT and response to wounding. Moreover, we demonstrate here that EHF depletion slows wound scratch closure in both WT and CF HBE cells. By further impairing a phenotype that may already be compromised in CFTR mutant epithelial cells, EHF could modify lung disease severity in CF patients. There are certain limitations to the scratch assays performed here because the cells are not differentiated on permeable supports. Hence, some epithelial cells types are absent and cannot contribute to normal wound closure. Differences in the transcriptome of differentiated and undifferentiated cells may also alter their interactions with the environment.
Overall, this study makes a strong case for EHF as a potential modifier of lung pathology in cystic fibrosis and other diseases in which the immune system and epithelial repair mechanisms play a critical role, including COPD and asthma. Although we have not yet established a correlation between the SNPs associated with lung disease severity in CF and EHF transcription, we predict, based on our EHF functional data in HBE cells, that regulatory variants may impact its expression. A detailed investigation of control mechanisms for EHF is required to reveal their contribution to airway disease.
Experimental procedures
Cell culture
Calu-3 cells were obtained from the ATCC and cultured by standard methods. Passage 1 (P1) primary bronchial epithelial cells were obtained under protocol 03-1396 approved by the University of North Carolina at Chapel Hill Biomedical Institutional Review Board. Informed consent was obtained from authorized representatives of all organ donors. Cells were grown on collagen-coated plastic in bronchial epithelial growth medium (64). Passage 2 or 3 primary cells were used for the study.
ChIP-seq and ChIP qPCR
Antibodies used in ChIP-seq were against EHF (clone 5A.5) (22), H3K4me1 (ab8895, Abcam), H3K4me3 (07-0473, Millipore), H3K27ac (ab4729), c-Jun (sc-1694, Santa Cruz Biotechnology), and JunD (sc-74). ChIP-seq was performed as described previously (31) (ChIP-seq experiment information is shown in supplemental Table S5). For EHF ChIP-seq, replicates were performed on HBE cells from two donors. One donor was used for histone modification ChIP-seq. For c-Jun and JunD ChIP-seq, two biological replicates were performed on Calu-3 cells. Peaks were called using the HOMER-IDR method (IDR < 0.05) (32). Histone tag directories were generated using HOMER (65). Downstream analysis was performed using HOMER and CEAS (33). To confirm the EHF results, ChIP was performed as described, followed by qPCR using primer pairs specific for each target of interest and SYBR Green (Life Technologies). FoxA1 ChIP was performed as described previously (66) with an antibody against FoxA1 (ab23738). For primers, see supplemental Table S6.
RNA-seq
HBE cells were treated with either Silencer Select negative control siRNA 2 (4390846) or EHF siRNA (s25399) (both from Ambion, 30 nm) using RNAiMax transfection reagent (Life Technologies). 48 h after transfection, RNA was isolated from five samples of each treatment using TRIzol (Life Technologies). RNA-seq was performed as described previously (31) (see supplemental Table S7 for replicate information). Differentially expressed genes were identified using TopHat and Cufflinks (36). Direct targets of EHF were identified using BETA (37). This software assigns transcription factor-binding peaks to genes using a model that decreases the regulatory potential of the TF binding site as the distance to the TSS increases and then sums the contribution of multiple sites near a gene (in this analysis, within 10 kb or a CCCTC-binding factor (CTCF) site, whichever is closer). It also takes into account the significance of gene expression changes following modulation of the transcription factor (with an FDR < 0.1).
EHF and FoxA1 knockdown followed by RT-qPCR
TF depletion was done in three (FoxA1) or four (EHF) HBE donor cultures. EHF depletion was performed as described above. For FOXA1, HBE cells were treated with 20 nm control siRNA-A (sc-37007, Santa Cruz Biotechnology) or HNF-3α siRNA (sc-37930, Santa Cruz Biotechnology) using RNAiMax. RNA was isolated for 48 (FOXA1) or 72 (EHF) h as above. RT-qPCR was performed using the TaqMan reverse transcription reagent kit (Life Technologies), primer pairs specific to each gene, and SYBR Green (for primers, see supplemental Table S6).
Western blot
HBE and Calu-3 cells were lysed in NET buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 1× Sigma Protease Inhibitor), and proteins were analyzed by standard methods. The antibodies used were specific for EHF (clone 5A.5), β-tubulin (T4026, Sigma-Aldrich), and GAPDH (Cell Signaling Technology).
LPS and IL-17a treatment of Calu-3 cells
Cells were forward transfected with NC or EHF siRNA as above. Forty-eight hours later, cells were serum-starved for 24 h and then treated with PBS or 1 μg/ml P. aeruginosa LPS (Sigma, L9134) for 4 h or BSA or 10 ng/ml IL-17a (R&D Systems) for 24 h. RNA was isolated using TRIzol, and RT-qPCR (SYBR Green) was performed as above (for primers, see supplemental Table S6).
ELISAs
Cells were forward-transfected with NC or EHF siRNA as described. 48 h post-transfection, the medium was replaced with serum-free medium and conditioned for 24 h. The medium was removed and diluted. Quantikine colorimetric sandwich ELISA for IL-8 (D800C) and CXCL6 (DGC00) (both from R&D Systems) was performed according to the protocol of the manufacturer.
Wound repair assay
The wound closure assay was performed as described previously (31) on three donor wild-type HBE cultures and three donor CF HBE cultures. Images were obtained at room temperature using a Leica DMi1 microscope with a PH1 ×10 objective using an Olympus DP22 camera and cellSens Entry software.
Author contributions
S. L. F. and A. H. conceived and coordinated the study and wrote the manuscript. M. J. M. optimized and performed the EHF ChIP-seq experiments shown in Fig. 1. S. G. performed the H3K4me3 ChIP-seq experiments shown in Figs. 1 and 2. L. C. J. generated the RNA-seq libraries for the experiments shown in Fig. 3. S. H. R. isolated and provided the primary HBE cells. S. H. L. grew the primary HBE cells, provided expert advice on these cells, and assisted with the wound closure experiments shown in Fig. 6. S. L. F. performed and analyzed the remaining experiments shown. A. T. generated and provided the EHF antibody necessary to complete these studies and gave valuable insights into the biology of EHF. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Drs. H. Dang and M. Schipma for helpful bioinformatics discussions, and we also thank Drs. M. R. Knowles and W. K. O'Neal for their intellectual contributions.
This work was supported by National Institutes of Health Grants R01HL117843, R01HL094585, and R01HD068901 (to A. H.); F30HL124925 (to S. F.); T32GM008061; and P30DK065988. This work was also supported by Cystic Fibrosis Foundation Grants Harris14P0 (to A. H) and CFF-Bouch15P0). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Tables S1–S6 and Tables S1–S7.
All data were deposited into Gene Expression Omnibus (GEO) GSE with accession number GSE85403.
- CF
- cystic fibrosis
- COPD
- chronic obstructive pulmonary disease
- TF
- transcription factor
- EHF
- Ets homologous factor
- EMT
- epithelial-to-mesenchymal transition
- HBE
- human bronchial epithelial
- RNA-seq
- RNA sequencing
- ChIP-seq
- ChIP followed by deep sequencing
- IDR
- irreproducible discovery rate
- CEAS
- Cis-regulatory Element Annotation System
- H3K4me1
- histone 3 lysine 4 monomethylation
- H3K4me3
- histone 3 lysine 4 trimethylation
- H3K27ac
- histone 3 lysine 27 acetylation
- KLF
- Krüppel-like factor
- DEG
- differentially expressed gene
- FDR
- false discovery rate
- BETA
- Binding and Expression Target Analysis
- NC
- negative control
- TSS
- transcription start site
- RAR
- retinoic acid receptor
- SPDEF
- SAM pointed domain-containing ETS transcription factor
- qPCR
- quantitative PCR.
References
- 1. Crosby L. M., and Waters C. M. (2010) Epithelial repair mechanisms in the lung. Am. J. Physiol. Lung. Cell Mol. Physiol. 298, L715–L731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hallstrand T. S., Hackett T. L., Altemeier W. A., Matute-Bello G., Hansbro P. M., and Knight D. A. (2014) Airway epithelial regulation of pulmonary immune homeostasis and inflammation. Clin. Immunol. 151, 1–15 [DOI] [PubMed] [Google Scholar]
- 3. Knowles M. R., and Boucher R. C. (2002) Mucus clearance as a primary innate defense mechanism for mammalian airways. J. Clin. Invest. 109, 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fahy J. V., and Dickey B. F. (2010) Airway mucus function and dysfunction. N. Engl. J. Med. 363, 2233–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schiller K. R., Maniak P. J., and O'Grady S. M. (2010) Cystic fibrosis transmembrane conductance regulator is involved in airway epithelial wound repair. Am. J. Physiol. Cell Physiol. 299, C912–C921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cohen T. S., and Prince A. (2012) Cystic fibrosis: a mucosal immunodeficiency syndrome. Nat. Med. 18, 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grosse S. D., Boyle C. A., Botkin J. R., Comeau A. M., Kharrazi M., Rosenfeld M., and Wilfond B. S. (2004) Newborn screening for cystic fibrosis: evaluation of benefits and risks and recommendations for state newborn screening programs. National Center on Birth Defects and Developmental Disabilities. C.D.C. M.M.W.R. 53, 1–36 [PubMed] [Google Scholar]
- 8. Khan T. Z., Wagener J. S., Bost T., Martinez J., Accurso F. J., and Riches D. W. (1995) Early pulmonary inflammation in infants with cystic fibrosis. Am. J. Respir. Crit. Care Med. 151, 1075–1082 [DOI] [PubMed] [Google Scholar]
- 9. Konstan M. W., Hilliard K. A., Norvell T. M., and Berger M. (1994) Bronchoalveolar lavage findings in cystic fibrosis patients with stable, clinically mild lung disease suggest ongoing infection and inflammation. Am. J. Respir. Crit. Care Med. 150, 448–454 [DOI] [PubMed] [Google Scholar]
- 10. Rosenfeld M., Gibson R. L., McNamara S., Emerson J., Burns J. L., Castile R., Hiatt P., McCoy K., Wilson C. B., Inglis A., Smith A., Martin T. R., and Ramsey B. W. (2001) Early pulmonary infection, inflammation, and clinical outcomes in infants with cystic fibrosis. Pediatr. Pulmonol. 32, 356–366 [DOI] [PubMed] [Google Scholar]
- 11. Hartl D., Griese M., Kappler M., Zissel G., Reinhardt D., Rebhan C., Schendel D. J., and Krauss-Etschmann S. (2006) Pulmonary T(H)2 response in Pseudomonas aeruginosa-infected patients with cystic fibrosis. J. Allergy Clin. Immunol. 117, 204–211 [DOI] [PubMed] [Google Scholar]
- 12. Tan H. L., Regamey N., Brown S., Bush A., Lloyd C. M., and Davies J. C. (2011) The Th17 pathway in cystic fibrosis lung disease. Am. J. Respir. Crit. Care Med. 184, 252–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tiringer K., Treis A., Fucik P., Gona M., Gruber S., Renner S., Dehlink E., Nachbaur E., Horak F., Jaksch P., Döring G., Crameri R., Jung A., Rochat M. K., Hörmann M., et al. (2013) A Th17- and Th2-skewed cytokine profile in cystic fibrosis lungs represents a potential risk factor for Pseudomonas aeruginosa infection. Am. J. Respir. Crit. Care Med. 187, 621–629 [DOI] [PubMed] [Google Scholar]
- 14. Kuperman D. A., Huang X., Koth L. L., Chang G. H., Dolganov G. M., Zhu Z., Elias J. A., Sheppard D., and Erle D. J. (2002) Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat. Med. 8, 885–889 [DOI] [PubMed] [Google Scholar]
- 15. Prause O., Laan M., Lötvall J., and Lindén A. (2003) Pharmacological modulation of interleukin-17-induced GCP-2-, GRO-α- and interleukin-8 release in human bronchial epithelial cells. Eur. J. Pharmacol. 462, 193–198 [DOI] [PubMed] [Google Scholar]
- 16. Durieu I., Peyrol S., Gindre D., Bellon G., Durand D. V., and Pacheco Y. (1998) Subepithelial fibrosis and degradation of the bronchial extracellular matrix in cystic fibrosis. Am. J. Respir. Crit. Care Med. 158, 580–588 [DOI] [PubMed] [Google Scholar]
- 17. Hamutcu R., Rowland J. M., Horn M. V., Kaminsky C., MacLaughlin E. F., Starnes V. A., and Woo M. S. (2002) Clinical findings and lung pathology in children with cystic fibrosis. Am. J. Respir. Crit. Care Med. 165, 1172–1175 [DOI] [PubMed] [Google Scholar]
- 18. Hilliard T. N., Regamey N., Shute J. K., Nicholson A. G., Alton E. W., Bush A., and Davies J. C. (2007) Airway remodelling in children with cystic fibrosis. Thorax 62, 1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Regamey N., Jeffery P. K., Alton E. W., Bush A., and Davies J. C. (2011) Airway remodelling and its relationship to inflammation in cystic fibrosis. Thorax 66, 624–629 [DOI] [PubMed] [Google Scholar]
- 20. Kim V., Rogers T. J., and Criner G. J. (2008) New concepts in the pathobiology of chronic obstructive pulmonary disease. Proc. Am. Thorac. Soc. 5, 478–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holgate S. T. (2011) The sentinel role of the airway epithelium in asthma pathogenesis. Immunol. Rev. 242, 205–219 [DOI] [PubMed] [Google Scholar]
- 22. Tugores A., Le J., Sorokina I., Snijders A. J., Duyao M., Reddy P. S., Carlee L., Ronshaugen M., Mushegian A., Watanaskul T., Chu S., Buckler A., Emtage S., and McCormick M. K. (2001) The epithelium-specific ETS protein EHF/ESE-3 is a context-dependent transcriptional repressor downstream of MAPK signaling cascades. J. Biol. Chem. 276, 20397–20406 [DOI] [PubMed] [Google Scholar]
- 23. Silverman E. S., Baron R. M., Palmer L. J., Le L., Hallock A., Subramaniam V., Riese R. J., McKenna M. D., Gu X., Libermann T. A., Tugores A., Haley K. J., Shore S., Drazen J. M., and Weiss S. T. (2002) Constitutive and cytokine-induced expression of the ETS transcription factor ESE-3 in the lung. Am. J. Respir. Cell Mol. Biol. 27, 697–704 [DOI] [PubMed] [Google Scholar]
- 24. Kas K., Finger E., Grall F., Gu X., Akbarali Y., Boltax J., Weiss A., Oettgen P., Kapeller R., and Libermann T. A. (2000) ESE-3, a novel member of an epithelium-specific Ets transcription factor subfamily, demonstrates different target gene specificity from ESE-1. J. Biol. Chem. 275, 2986–2998 [DOI] [PubMed] [Google Scholar]
- 25. Wu J., Duan R., Cao H., Field D., Newnham C. M., Koehler D. R., Zamel N., Pritchard M. A., Hertzog P., Post M., Tanswell A. K., and Hu J. (2008) Regulation of epithelium-specific Ets-like factors ESE-1 and ESE-3 in airway epithelial cells: potential roles in airway inflammation. Cell Res. 18, 649–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Albino D., Longoni N., Curti L., Mello-Grand M., Pinton S., Civenni G., Thalmann G., D'Ambrosio G., Sarti M., Sessa F., Chiorino G., Catapano C. V., and Carbone G. M. (2012) ESE3/EHF controls epithelial cell differentiation and its loss leads to prostate tumors with mesenchymal and stem-like features. Cancer Res. 72, 2889–2900 [DOI] [PubMed] [Google Scholar]
- 27. Thiery J. P., and Sleeman J. P. (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 28. Kim K. K., Kugler M. C., Wolters P. J., Robillard L., Galvez M. G., Brumwell A. N., Sheppard D., and Chapman H. A. (2006) Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc. Natl. Acad. Sci. U.S.A. 103, 13180–13185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wright F. A., Strug L. J., Doshi V. K., Commander C. W., Blackman S. M., Sun L., Berthiaume Y., Cutler D., Cojocaru A., Collaco J. M., Corey M., Dorfman R., Goddard K., Green D., Kent J. W. Jr., et al. (2011) Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat. Genet. 43, 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Corvol H., Blackman S. M., Boëlle P. Y., Gallins P. J., Pace R. G., Stonebraker J. R., Accurso F. J., Clement A., Collaco J. M., Dang H., Dang A. T., Franca A., Gong J., Guillot L., Keenan K., et al. (2015) Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat. Commun. 6, 8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fossum S. L., Mutolo M. J., Yang R., Dang H., O'Neal W. K., Knowles M. R., Leir S. H., and Harris A. (2014) Ets homologous factor regulates pathways controlling response to injury in airway epithelial cells. Nucleic Acids Res. 42, 13588–13598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karmel A. (2014) Homer-idr: second pass updated. 10.5281/zenodo.11619 [DOI] [Google Scholar]
- 33. Ji X., Li W., Song J., Wei L., and Liu X. S. (2006) CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 34, W551–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., and Ren B. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 35. Zhou V. W., Goren A., and Bernstein B. E. (2011) Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 12, 7–18 [DOI] [PubMed] [Google Scholar]
- 36. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., and Pachter L. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang S., Sun H., Ma J., Zang C., Wang C., Wang J., Tang Q., Meyer C. A., Zhang Y., and Liu X. S. (2013) Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc. 8, 2502–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park K. S., Korfhagen T. R., Bruno M. D., Kitzmiller J. A., Wan H., Wert S. E., Khurana Hershey G. K., Chen G., and Whitsett J. A. (2007) SPDEF regulates goblet cell hyperplasia in the airway epithelium. J. Clin. Invest. 117, 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen G., Korfhagen T. R., Xu Y., Kitzmiller J., Wert S. E., Maeda Y., Gregorieff A., Clevers H., and Whitsett J. A. (2009) SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J. Clin. Invest. 119, 2914–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pace E., Ferraro M., Siena L., Melis M., Montalbano A. M., Johnson M., Bonsignore M. R., Bonsignore G., and Gjomarkaj M. (2008) Cigarette smoke increases Toll-like receptor 4 and modifies lipopolysaccharide-mediated responses in airway epithelial cells. Immunology 124, 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Itokazu Y., Pagano R. E., Schroeder A. S., O'Grady S. M., Limper A. H., and Marks D. L. (2014) Reduced GM1 ganglioside in CFTR-deficient human airway cells results in decreased β1-integrin signaling and delayed wound repair. Am. J. Physiol. Cell Physiol. 306, C819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vanscoy L. L., Blackman S. M., Collaco J. M., Bowers A., Lai T., Naughton K., Algire M., McWilliams R., Beck S., Hoover-Fong J., Hamosh A., Cutler D., and Cutting G. R. (2007) Heritability of lung disease severity in cystic fibrosis. Am. J. Respir. Crit. Care Med. 175, 1036–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cystic Fibrosis Genotype-Phenotype Consortium (1993) Correlation between genotype and phenotype in patients with cystic fibrosis. N. Engl. J. Med. 329, 1308–1313 [DOI] [PubMed] [Google Scholar]
- 44. Stephens D. N., Klein R. H., Salmans M. L., Gordon W., Ho H., and Andersen B. (2013) The Ets transcription factor EHF as a regulator of cornea epithelial cell identity. J. Biol. Chem. 288, 34304–34324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taniue K., Oda T., Hayashi T., Okuno M., and Akiyama T. (2011) A member of the ETS family, EHF, and the ATPase RUVBL1 inhibit p53-mediated apoptosis. EMBO Rep. 12, 682–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kunderfranco P., Mello-Grand M., Cangemi R., Pellini S., Mensah A., Albertini V., Malek A., Chiorino G., Catapano C. V., and Carbone G. M. (2010) ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PLoS ONE 5, e10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reddy S. P., and Mossman B. T. (2002) Role and regulation of activator protein-1 in toxicant-induced responses of the lung. Am. J. Physiol. Lung Cell Mol. Physiol. 283, L1161–L1178 [DOI] [PubMed] [Google Scholar]
- 48. Mossman B. T., Lounsbury K. M., and Reddy S. P. (2006) Oxidants and signaling by mitogen-activated protein kinases in lung epithelium. Am. J. Respir. Cell Mol. Biol. 34, 666–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Verhaeghe C., Remouchamps C., Hennuy B., Vanderplasschen A., Chariot A., Tabruyn S. P., Oury C., and Bours V. (2007) Role of IKK and ERK pathways in intrinsic inflammation of cystic fibrosis airways. Biochem. Pharmacol. 73, 1982–1994 [DOI] [PubMed] [Google Scholar]
- 50. Herriges M., and Morrisey E. E. (2014) Lung development: orchestrating the generation and regeneration of a complex organ. Development 141, 502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wan H., Kaestner K. H., Ang S. L., Ikegami M., Finkelman F. D., Stahlman M. T., Fulkerson P. C., Rothenberg M. E., and Whitsett J. A. (2004) Foxa2 regulates alveolarization and goblet cell hyperplasia. Development 131, 953–964 [DOI] [PubMed] [Google Scholar]
- 52. Wan H., Dingle S., Xu Y., Besnard V., Kaestner K. H., Ang S. L., Wert S., Stahlman M. T., and Whitsett J. A. (2005) Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J. Biol. Chem. 280, 13809–13816 [DOI] [PubMed] [Google Scholar]
- 53. Mollard R., Ghyselinck N. B., Wendling O., Chambon P., and Mark M. (2000) Stage-dependent responses of the developing lung to retinoic acid signaling. Int. J. Dev. Biol. 44, 457–462 [PubMed] [Google Scholar]
- 54. Yang H., Lu M. M., Zhang L., Whitsett J. A., and Morrisey E. E. (2002) GATA6 regulates differentiation of the distal lung epithelium. Development 129, 2233–2246 [DOI] [PubMed] [Google Scholar]
- 55. Zhang Y., Rath N., Hannenhalli S., Wang Z., Cappola T., Kimura S., Atochina-Vasserman E., Lu M. M., Beers M. F., and Morrisey E. E. (2007) GATA and Nkx factors synergistically regulate tissue-specific gene expression and development in vivo. Development 134, 189–198 [DOI] [PubMed] [Google Scholar]
- 56. Zhang Y., Goss A. M., Cohen E. D., Kadzik R., Lepore J. J., Muthukumaraswamy K., Yang J., DeMayo F. J., Whitsett J. A., Parmacek M. S., and Morrisey E. E. (2008) A Gata6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nat. Genet. 40, 862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yin Z., Gonzales L., Kolla V., Rath N., Zhang Y., Lu M. M., Kimura S., Ballard P. L., Beers M. F., Epstein J. A., and Morrisey E. E. (2006) Hop functions downstream of Nkx2.1 and GATA6 to mediate HDAC- dependent negative regulation of pulmonary gene expression. Am. J. Physiol. Lung Cell. Mol. Physiol. 291, L191–L199 [DOI] [PubMed] [Google Scholar]
- 58. Wan H., Luo F., Wert S. E., Zhang L., Xu Y., Ikegami M., Maeda Y., Bell S. M., and Whitsett J. A. (2008) Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development 135, 2563–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Korfhagen T. R., Kitzmiller J., Chen G., Sridharan A., Haitchi H.-M., Hegde R. S., Divanovic S., Karp C. L., and Whitsett J. A. (2012) SAM-pointed domain ETS factor mediates epithelial cell-intrinsic innate immune signaling during airway mucous metaplasia. Proc. Natl. Acad. Sci. U.S.A. 109, 16630–16635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rajavelu P., Chen G., Xu Y., Kitzmiller J. A., Korfhagen T. R., and Whitsett J. A. (2015) Airway epithelial SPDEF integrates goblet cell differentiation and pulmonary Th2 inflammation. J. Clin. Invest. 125, 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Widdicombe J. H., and Wine J. J. (2015) Airway gland structure and function. Physiol. Rev. 95, 1241–1319 [DOI] [PubMed] [Google Scholar]
- 62. Birrer P., McElvaney N. G., Rüdeberg A., Sommer C. W., Liechti-Gallati S., Kraemer R., Hubbard R., and Crystal R. G. (1994) Protease-antiprotease imbalance in the lungs of children with cystic fibrosis. Am. J. Respir. Crit. Care Med. 150, 207–213 [DOI] [PubMed] [Google Scholar]
- 63. Voynow J. A., Fischer B. M., and Zheng S. (2008) Proteases and cystic fibrosis. Int. J. Biochem. Cell Biol. 40, 1238–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fulcher M. L., Gabriel S., Burns K. A., Yankaskas J. R., and Randell S. H. (2005) Well differentiated human airway epithelial cell cultures. Methods Mol. Med. 107, 183–206 [DOI] [PubMed] [Google Scholar]
- 65. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kerschner J. L., Gosalia N., Leir S. H., and Harris A. (2014) Chromatin remodeling mediated by the FOXA1/A2 transcription factors activates CFTR expression in intestinal epithelial cells. Epigenetics 9, 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.