ABSTRACT
Hutchinson-Gilford progeria syndrome (HGPS) is caused by a mutation in LMNA that produces an aberrant lamin A protein, progerin. The accumulation of progerin in HGPS cells leads to an aberrant nuclear morphology, genetic instability, and p53-dependent premature senescence. How p53 is activated in response to progerin production is unknown. Here we show that young cycling HGPS fibroblasts exhibit chronic DNA damage, primarily in S phase, as well as delayed replication fork progression. We demonstrate that progerin binds to PCNA, altering its distribution away from replicating DNA in HGPS cells, leading to γH2AX formation, ATR activation, and RPA Ser33 phosphorylation. Unlike normal human cells that can be immortalized by enforced expression of telomerase alone, immortalization of HGPS cells requires telomerase expression and p53 repression. In addition, we show that the DNA damage response in HGPS cells does not originate from eroded telomeres. Together, these results establish that progerin interferes with the coordination of essential DNA replication factors, causing replication stress, and is the primary signal for p53 activation leading to premature senescence in HGPS. Furthermore, this damage response is shown to be independent of progerin farnesylation, implying that unprocessed lamin A alone causes replication stress.
KEYWORDS: HGPS, progerin, senescence, aging, p53, telomere
INTRODUCTION
The study of children with rare accelerated aging (progeria) syndromes has provided insight into the normal process of aging. Hutchinson-Gilford progeria syndrome (HGPS) is caused by a heterozygous, autosomal dominant mutation in LMNA (1), the gene that encodes lamin A, a key structural protein in the nuclear lamina. The lamina, a protein network that lines the inner nuclear membrane, provides structural support for the nucleus and is important for chromatin attachment, DNA replication, nuclear organization, and gene transcription in ways that are not completely understood. Farnesylation of prelamin A at the C terminus allows targeting to the inner nuclear membrane, where it is subsequently cleaved by the zinc metalloproteinase Zmpste24 to release mature lamin A into the lamina and nucleoplasm. In HGPS, the most common LMNA mutation (G608G) activates a cryptic splice donor site in exon 11, resulting in prelamin A mRNA with a 150-bp internal deletion, leading to a 50-amino-acid truncation in a region that contains the Zmpste24 cleavage site (reviewed in reference 2). The mutant lamin, termed progerin, retains the farnesyl lipid anchor, interferes with the integrity of the nuclear lamina, and causes the formation of misshapen nuclei. The retention of progerin on the inner nuclear membrane interferes with the function and normal distribution of lamin A, causing a loss of peripheral heterochromatin, increased DNA damage, impaired recruitment of DNA repair proteins to sites of DNA damage (3), and persistent activation of DNA damage response proteins (4). In addition to its structural role, the colocalization of nucleoplasmic lamin A with the replication proteins PCNA and DNA polymerase δ within nuclear foci suggests an involvement of lamin A in DNA replication (5, 6). Additionally, the interaction of lamin A with LAP2Aα (7) and RB (8) is important for regulation of cell cycle entry. In support of the role of lamins in replication, it has been shown recently that the accumulation of prelamin A in U2OS cells leads to replication stress by interfering with PCNA (9). Hence, it is possible that the accumulation of unprocessed progerin in HGPS cells interferes with these functions of lamin A, leading to genome instability.
Varela and colleagues (10) reported a marked upregulation of p53-responsive genes, including BTG2, Gadd45α, and p21WAF1 in Zmpste24−/− mouse cells, and proposed that Zmpste24 deficiency, which results in prelamin A accumulation, elicits a stress response that activates p53. The p53 protein plays a critical role in coordinating the cellular response to diverse stress signals by regulating the expression of coding genes and noncoding RNA genes that collectively contribute to apoptosis, cell cycle control, and cellular senescence (11). The activation of cellular senescence, an irreversible cell cycle arrest in G1 accompanied by distinct morphological and biochemical changes, is well established (12). Replicative senescence is triggered by critically short telomeres that activate a DNA damage response that is mediated primarily by the ATM kinase and p53 (13). In addition to telomere shortening, other stress signals, including oncogene activation (14), replication stress (15), oxidative stress (16), and sublethal DNA damage (17), lead to stress-induced (premature) senescence mediated through the p53 and/or the p16-pRB pathway. Compelling evidence supports the idea that both replicative senescence (telomere dependent) and stress-induced senescence (telomere independent) occur in vivo (18, 19) and that both forms of senescence play a role in organismal aging and serve as important mechanisms of tumor suppression (20, 21).
The integrity of telomeres in HGPS is controversial, and it remains unclear if shortened or dysfunctional telomeres contribute to DNA damage and premature senescence in HGPS cells (22). In this study, we investigated the mechanism that leads to p53-dependent premature senescence in HGPS cells. We find that chronic DNA damage resulting from DNA replication stress leads to p53 activation and premature senescence. Progerin induces this response independently of eroded telomeres and retention of its lipid anchor, implying that unprocessed nucleoplasmic lamin A itself is causal in HGPS premature senescence.
RESULTS
p53-dependent premature senescence in HGPS cells is telomere independent.
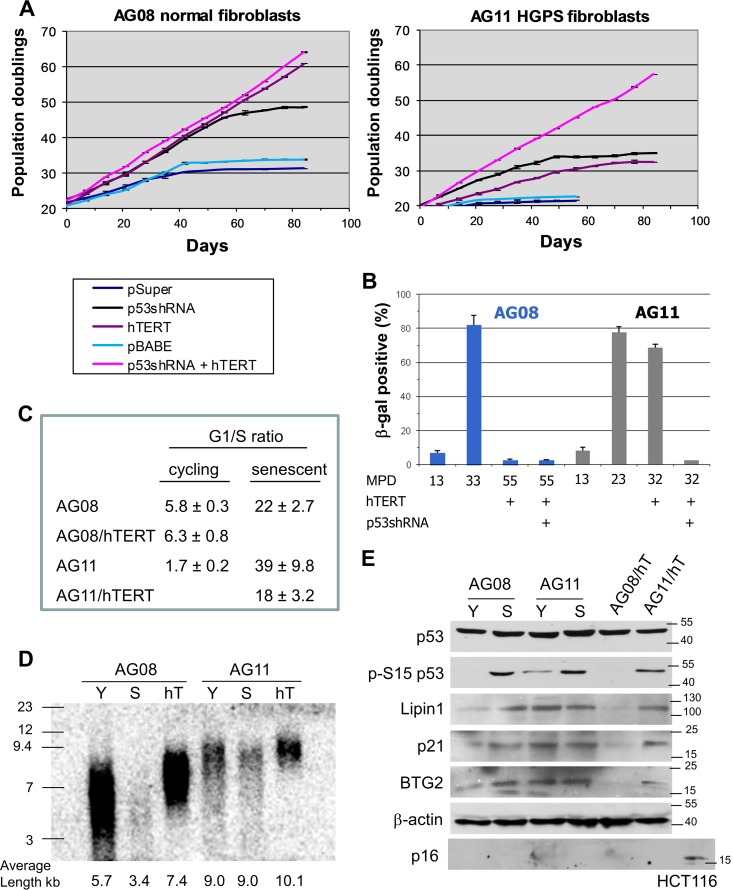
To investigate the determinants that govern cellular senescence in HGPS cells, we compared the growth of HGPS fibroblasts (AG11513) to that of age- and gender-matched normal cells (AG08470). AG08 cells undergo senescence at a population doubling (PD) of 32 to 34, and short hairpin RNA (shRNA)-mediated inhibition of p53 extended replicative life span by approximately 15 PDs (Fig. 1A). AG11 HGPS cells senesce prematurely at a PD of 22, and knockdown of p53 extended life span by approximately 13 PDs. This is consistent with previous observations showing that p53 promotes cellular senescence and that p53 inactivation extends cellular life span. Ectopic expression of the telomerase catalytic subunit (hTERT) revealed a striking difference between AG08 and AG11 cells. We found that hTERT expression immortalized the normal AG08 cells as expected (23, 24); however, in AG11 cells, hTERT expression extended life span by approximately 10 PDs but did not lead to immortalization (Fig. 1A). hTERT-expressing AG11 cells acquired a senescence phenotype on the basis of morphology, β-galactosidase staining, failure to divide, and cell cycle analysis (Fig. 1A to C). Notably, AG11 HGPS cells could only be immortalized upon p53 repression and ectopic hTERT expression (Fig. 1A and B).
FIG 1.
Premature senescence in HGPS cells is dependent on p53 and not dependent on telomere shortening. (A) Measurement of the number of mean population doublings over the life span of normal AG08 fibroblasts (left) and HGPS AG11 fibroblasts (right) expressing p53 shRNA, hTERT, or p53shRNA plus hTERT. Data represent averages from 3 biological replicates counted with 4 technical replicates each week. Standard error of the mean (SEM) values are shown as error bars. (B) β-Galactosidase staining of normal AG08 and HGPS AG11 fibroblasts expressing hTERT alone or p53shRNA plus hTERT at the indicated mean population doubling (MPD). (C) The G1/S ratios were determined from cell cycle profiles obtained by flow cytometry after propidium iodide staining. Cellular senescence is reflected by an increase in the G1/S ratio. (D) TRF assay of AG08 and AG11 cells showing overall telomere length in young (Y), senescent (S), and hTERT (hT)-expressing cells. This assay was repeated 3 times, and a representative result is shown. The average telomere length, determined by pixel density maxima, is shown. (E) Western blot analysis of young cycling and senescent AG08 and AG11 cells; AGO8 and AG11 cells expressing hTERT at population doubling 35 were also compared. Protein extracts were immunoblotted with the indicated antibodies. HCT116 lysates served as a positive control for p16 expression.
To determine if the inability of hTERT to immortalize HGPS cells was the result of an inability to elongate telomeres, telomere length was measured using the terminal restriction fragment (TRF) length assay. hTERT extended telomere length in both AG08 and AG11 cells; hence, AG11 cells undergo senescence despite having long telomeres (Fig. 1D). Moreover, unlike normal AG08 cells that enter senescence with short telomeres (3.4 kb compared to 5.7 kb), AG11 HGPS cells showed no significant attrition of telomeres (remained at 9 kb) at the onset of premature senescence (Fig. 1D).
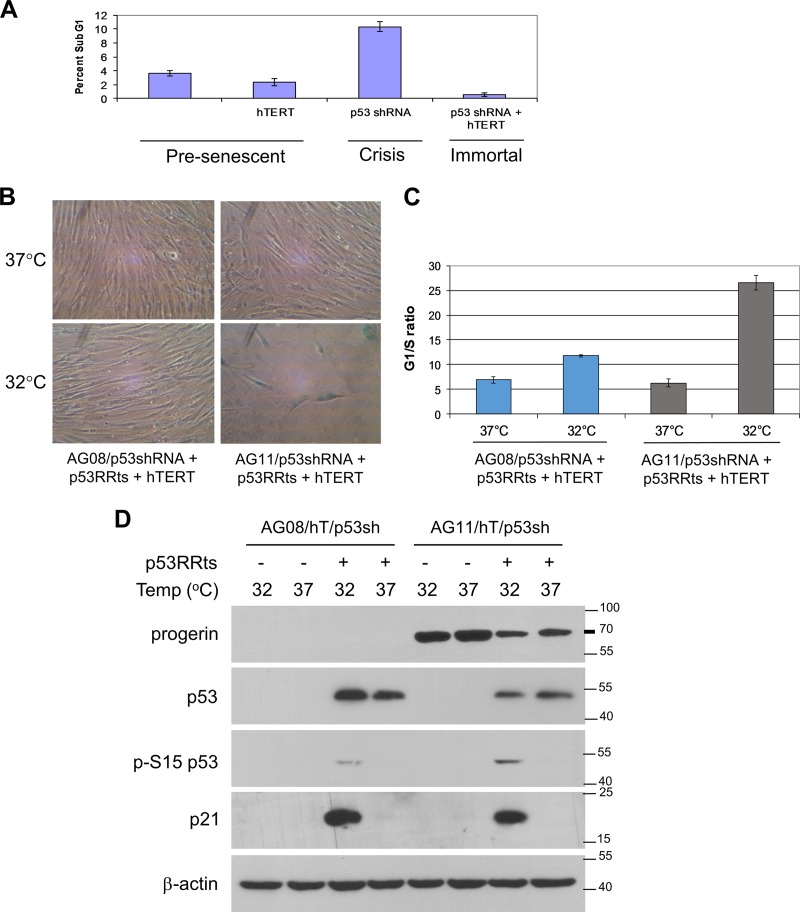
We detected p53 activation in senescent AG08 and high- and low-passage-number AG11 cells as well as in senescent hTERT-expressing AG11 cells, as measured by p53 phosphorylation at Ser15 and by elevated expression of p53 target genes p21, BTG2, and Lpin1 (Fig. 1E). p53 activation and p53 target gene expression were also seen in young, cycling AG11 cells (Fig. 1E). p16 expression, which can lead to senescence independently of p53 (25), was not detected in senescent AG08 and AG11 cells (Fig. 1E). Taken together, these results indicate that premature senescence in HGPS cells is mediated by p53 and that p53 activation is likely the result of a telomere-independent signal. Inhibition of p53 allows HGPS cells to bypass the telomere-independent senescence barrier, but as telomeres shorten with increasing passage number, the cells encounter a second barrier to proliferation (crisis) that is dependent on critically short telomeres (Fig. 2A). In HGPS cells with downregulated p53, hTERT expression restores telomere length and bypasses the second barrier, leading to immortalization.
FIG 2.
Progerin-induced premature senescence is dependent on p53. (A) Determination of crisis (apoptosis) in AG11 HGPS cells expressing pBABE vector control (MPD 25), hTERT (MPD 45), p53 shRNA (MPD 49), or both hTERT and p53shRNA (MPD 45). Apoptosis was determined by flow cytometry on the basis of sub-G1 content after propidium iodide staining. At the time of analysis, MPD 49 p53 shRNA cells were in a state of decline in which overall cell numbers were decreasing, while all other cell lines were actively growing. (B to D) p53RRts and hTERT were coexpressed in normal AG08/p53 shRNA and HGPS AG11/p53 shRNA. The cells were assessed at an MPD of 60 (AG08/p53sh/p53RRts/hTERT) or MPD of 50 (AG11/p53sh/p53RRts/hTERT). hTERT was expressed so that the contribution of progerin could be assessed independently of telomere erosion during premature senescence. (B) Cell growth was assessed at 37°C and at 32°C. Cells were visualized after β-galactosidase staining. hTERT-p5RRts-expressing AG11 cells at 32°C were 82% ± 5.6% positive β-galactosidase, demonstrating telomere-independent senescence. (C) The cells used for panel B were stained with propidium iodide, and the G1/S ratios were obtained from the cell cycle profiles using flow cytometry. Values shown represent the means and SEM (n = 3). (D) Western blot analysis confirming p53 knockdown and p53RRts and progerin expression in the cells used for panel B. The activation of p53 at 32°C is demonstrated by the upregulation of p21 and by the phosphorylation of p53 at Ser15.
To further investigate the dependency of progerin-induced senescence on p53, we introduced a temperature-sensitive (ts) mutation (A138V) into a p53 retroviral expression vector that is resistant to p53 shRNA (p53RR). The p53ts protein is inactive at 37°C and undergoes a conformational change at 32°C so that it resembles the wild-type p53 protein. Previously, we reported that p53RR induces cellular senescence when expressed in late-passage p53 shRNA-expressing cells (26). p53RRts and hTERT were coexpressed in AG08/p53shRNA and AG11/p53shRNA cells, and cell growth was assessed at 37°C and at 32°C. Ectopic hTERT was included in these experiments to ensure that telomeres remained capped and did not contribute to the senescence program. At 37°C, these cells grew normally. Upon p53 activation at 32°C, only the HGPS AG11/p53shRNA cells entered senescence, as determined by β-galactosidase staining (Fig. 2B) and cell cycle analysis (Fig. 2C); the AG08/p53shRNA cells continued to proliferate. We confirmed ectopic expression of p53RRts in the AG08 and AG11 cells and confirmed that progerin was expressed in the AG11 cells (Fig. 2D). We detected strong activation of p53RRts at 32°C in the AG11 cells as assessed by p53 phosphorylation at Ser15. A much smaller proportion of the total p53RRts was activated in the AG08 cells at 32°C (Fig. 2D). Interestingly, p21 was induced in both AG08 and AG11 cells at 32°C, and yet this was insufficient to induce senescence in AG08 cells, suggesting that p21 is insufficient to induce cellular senescence and/or that other p53 effector genes are involved in this process, as noted previously (26). However, senescence is a process that requires irreversible DNA damage (13), and the p53 activation in AG08 likely represents transient stress such as the temperature shift. In contrast, the AG11 cells must be continually signaling p53 and activating the senescence program. Since the AG11 cells also are expressing hTERT, this stress signal does not originate from uncapped telomeres. Together, these data confirm that premature senescence in HGPS cells is dependent on p53 and independent of telomere erosion.
Cycling progeria cells exhibit chronic DNA damage resulting from DNA replication stress.
An increased amount of unrepaired DNA damage is considered a hallmark of HGPS cells (3, 27). In addition, Varela et al. (10) reported the presence of a DNA damage response in Zmpste24−/− mice and activation of a p53-dependent transcriptional pathway. Zmpste24 encodes the metalloproteinase involved in the maturation of lamin A, and as a result, prelamin A accumulates in Zmpste24−/− mice, similar to HGPS patients with mutations in the LMNA gene. These reports prompted us to investigate DNA damage in HGPS cells. Cycling HGPS fibroblasts, but not normal fibroblasts, exhibit DNA damage, as reflected by immunostaining and colocalization of γH2AX and 53BP1, two markers used to detect DNA double-strand breaks (28) and stalled replication forks (29, 30) (Fig. 3A). Increased expression of γH2AX in cycling AG11 cells was confirmed by Western blotting (Fig. 3B). Interestingly, serum-starved HGPS cells that arrest in the G0/G1 phase of the cell cycle do not exhibit γH2AX and 53BP1 staining (Fig. 3A) and show diminished activation of p53 and reduced expression of p21 and γH2AX (Fig. 3B), suggesting that the DNA damage accumulates during cell cycling. To determine if only cells in S phase form DNA damage-induced foci, cells were exposed to bromodeoxyuridine (BrdU) for 8 h and then immunostained for 53BP1. In HGPS AG11 cells, 47.7% of BrdU-positive cells contained 53BP1 foci, and only 8.1% of the BrdU-negative cells contained 53BP1 foci. In normal AG08 cells, 53BP1 foci were found in 5.8% of BrdU-positive cells and in 4.0% of BrdU-negative cells (Fig. 3C). This indicates that DNA damage occurs predominantly during S phase. We then examined the cell cycle profiles of AG08 and AG11 cells by flow cytometry and found that cycling (young) HGPS fibroblasts exhibit a greater proportion of cells in S phase (Fig. 3D) and grow more slowly in culture than normal fibroblasts (data not shown), suggesting a delayed passage through S phase. hTERT-expressing AG11 HGPS cells similarly exhibit a greater proportion of cells in S phase than hTERT-expressing AG08 normal cells (Fig. 3D). In addition, immunofluorescent detection of γH2AX combined with propidium iodide (PI) staining of DNA to define the cell cycle phase in which the double-strand breaks occur, confirmed that DNA damage occurs predominantly during S phase (Fig. 3E).
FIG 3.
Cycling HGPS cells exhibit chronic DNA damage associated with DNA replication stress. (A) Immunostaining for γH2AX and 53BP1 in cycling or noncycling (serum-starved) HGPS AG11 or cycling-normal AG08 cells. Draq5 was used to stain nuclei. (B) Western blot analysis of γH2AX, H2AX, p53, phospho-Ser15 p53, and p21 in cycling (Cyc) and serum-starved (SS) AG08 and AG11 cells. Cells were serum starved for 5 days. (C) Cells were labeled with BrdU (10 nM) for 8 h and immunostained with antibodies against 53BP1. The proportion of BrdU-positive and BrdU-negative cells showing ≥5 53BP1 foci is indicated by the black shading. Data presented are means and SEM from 3 independent experiments; 150 to 200 cells were counted in each experiment. (D) Young cycling AG08 (MPD 22) and AG11 (MPD 13) cells were subjected to cell cycle analysis by flow cytometry after propidium iodide staining. Also shown are hTERT-expressing AG08 and AG11 cells. The following percentages of cells were in S phase: AG08, 13% ± 0.7%; AG11, 35% ± 2.7%; AG08 hT, 12.5% ± 1.4%; and AG11, 18.6% ± 1%. Shown are the mean values and SEM (n = 3). (E) The proportion of cells in different phases of the cell cycle that were expressing γH2AX was measured by flow cytometry using propidium iodide to determine DNA content and antibodies to γH2AX to determine the extent of DNA damage. The values shown represent the means and SEM from 3 independent experiments.
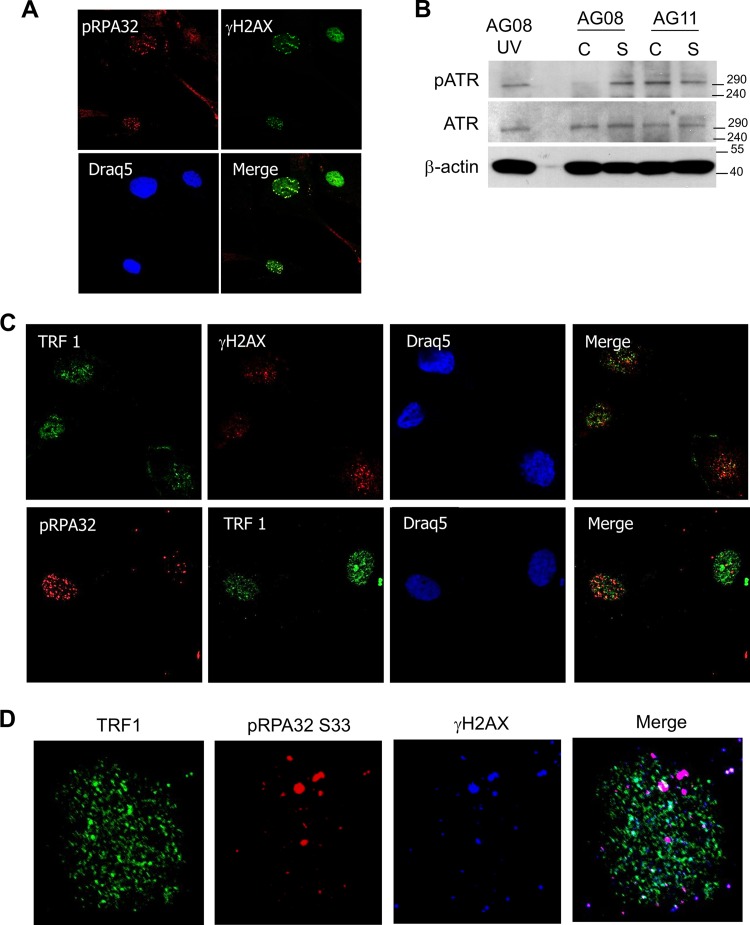
To investigate the possibility that replication stress in AG11 cells leads to DNA damage during S phase, we examined the localization of phospho-RPA32 (Ser33) in HGPS cells and found colocalization with γH2AX (Fig. 4A). Phospho-RPA32 was not detected in cycling AG08 cells (results not shown). RPA binds to single-stranded DNA and accumulates at stalled replication forks (31). Importantly, we find that ATR is activated in cycling and senescent AG11 cells (Fig. 4B). ATR kinase initiates the signaling cascade in response to the collapse of stalled replication forks and promotes phosphorylation of RPA at Ser 33 (32, 33).
FIG 4.
DNA damage from replication stress in HGPS cells colocalizes with pRPA32 and not with telomeres (TRFI). (A) Immunostaining for γH2AX and pRPA32 in cycling AG11 cells. Phospho-RPA32 (Ser33) serves as a marker for single-stranded DNA that is associated with stalled DNA replication forks. (B) Western blot analysis of phospho-ATR and total ATR in cycling (C) and senescent (S) AG08 and AG11 cells. β-Actin served as a loading control. UV-treated normal cells (AG08) served as a positive control for pATR expression. (C) Immunostaining for γH2AX, TRF1, and pRPA32 in AG11 HGPS cells. pRPA is a marker for single-stranded DNA that accumulates at stalled DNA replication forks, and TRF1 is a marker for telomeres. AG11 cells were doubly stained with TRF1 and γH2AX or pRPA32 and TRF2. (D) Nucleus of an AG11 cell that was triply stained with pRPA (Dylight 649), TRF1 (Alexa Fluor 488), and γH2AX (Cy3).
While our findings suggest that telomere integrity is maintained in HGPS cells, other laboratories have reported that DNA damage occurs at telomeres during normal cellular senescence and during progerin-induced premature senescence (34, 35). These findings are based on the presence of telomere aggregates that are associated with the nuclear lamina and with γH2AX foci. To determine if DNA damage colocalizes to telomeres, HGPS AG11 cells were immunostained with γH2AX, pRPA32, and TRF1, a marker for telomeres (36). While γH2AX was found to colocalize with pRPA32 (Fig. 4A), little colocalization with TRF1 was observed in progeria fibroblasts (Fig. 4C). Colocalization of TRF1 and γH2AX was detected in normal AG08 cells undergoing replicative senescence (data not shown). Simultaneous costaining for TRF1, γH2AX, and pRPA32 confirmed that the DNA damage in HGPS is distinct from telomeric regions (Fig. 4D).
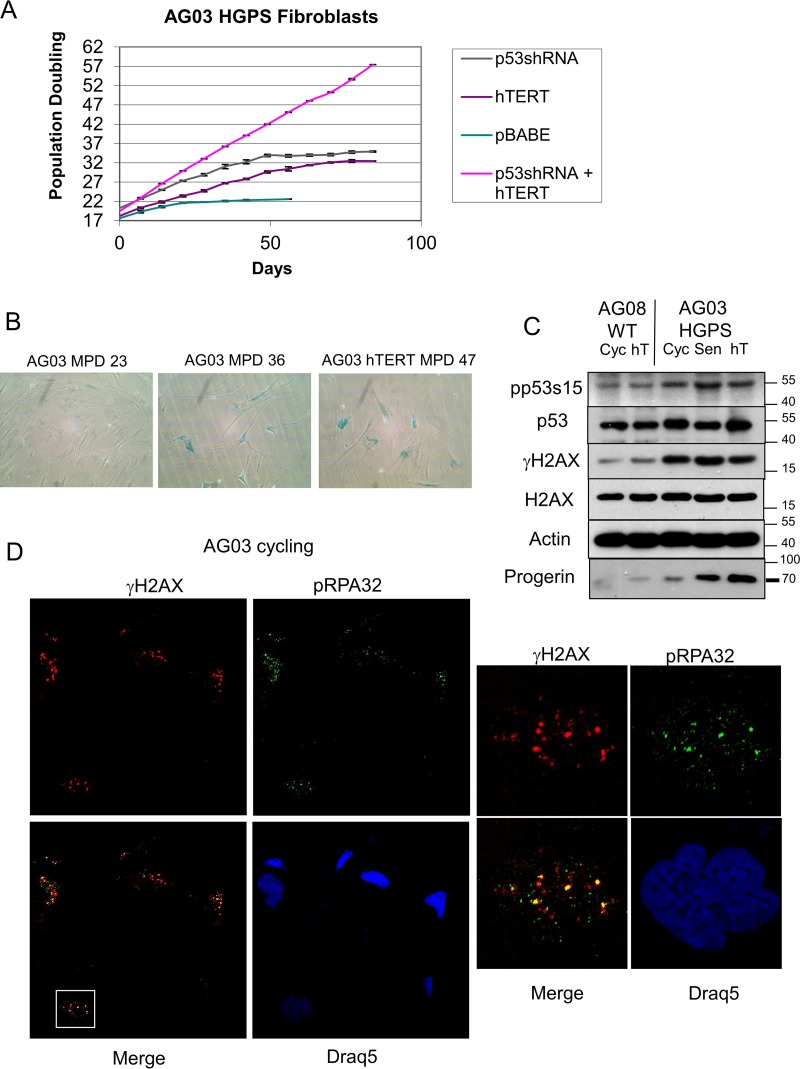
We next extended our findings to an additional HGPS cell strain, AG03198 (AG03). We ectopically expressed p53 shRNA and hTERT in AG03 cells. As expected, p53 shRNA extended the life span of the AG03 cells, but hTERT alone was unable to immortalize the cells (Fig. 5A), and they ultimately entered senescence (Fig. 5B). As was seen in AG11 cells, only the combination of p53 knockdown and ectopic hTERT expression allowed the immortalization of AG03 cells (Fig. 5A). We confirmed p53 activation and the accumulation of γH2AX and progerin in cycling and senescent AG03 cells (Fig. 5C). In addition, we provide support for replication stress in AG03 cells by visualization of pRPA32 colocalization with γH2AX (Fig. 5D).
FIG 5.
AG03 HGPS fibroblasts show p53-dependent and telomere-independent premature senescence and replication stress. (A) Measurement of the number of mean population doublings over the life span of HGPS AG03 fibroblasts expressing pBABE vector control, p53 shRNA, hTERT, or p53shRNA plus hTERT. Data represent averages from 3 biological replicates counted with 4 technical replicates each week. SEM values are shown as error bars. (B) β-Galactosidase staining of cycling low-passage-number HGPS AG03 cells (MPD 23), senescent high-passage-number AG03 cells (MPD 36), and senescent AG03/hTERT cells (MPD 47). (C) Western blot analysis of young cycling normal AG08 and HGPS AG03 cells, immortal AG08/hTERT cells at MPD 44, and senescent AG03/hTERT cells at MPD 46. Protein extracts were immunoblotted with the indicated antibodies. Progerin levels, as expected, accumulate with population doubling. (D) Immunostaining for γH2AX (Cy3) and pRPA32 (Alexa Fluor 488) in cycling low-passage-number HGPS AG03 cells. Phospho-RPA32 (Ser33) serves as a marker for single-stranded DNA that is associated with stalled DNA replication forks. Draq5 was used to stain nuclei. The white box indicates the magnified view of a single nucleus to demonstrate colocalization of γH2AX with pRPA32.
The distribution of the DNA replication elongation factor PCNA is altered in HGPS cells.
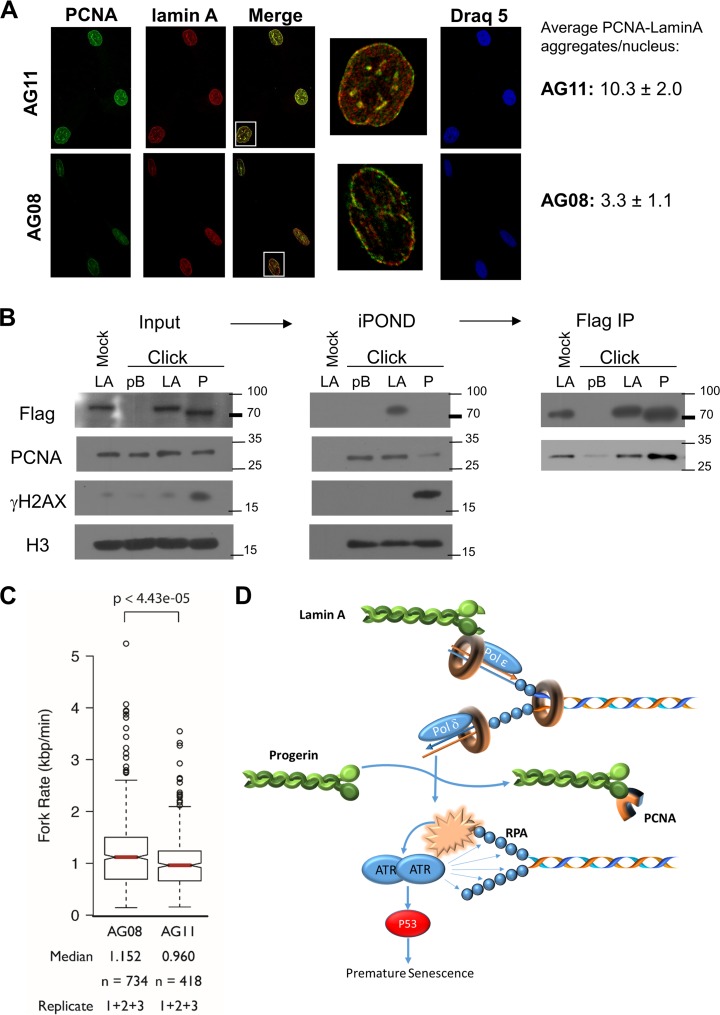
Human lamins may regulate DNA replication by binding PCNA and positioning PCNA on chromatin (6). Previous studies demonstrated that disruption of lamin organization by a dominant-negative lamin A mutant (ΔNLA), missing the first 33 amino acids of human lamin A, repressed DNA replication. This was associated with the redistribution of DNA replication proteins RFC and PCNA away from chromatin to form nuclear aggregates along with the nuclear lamins (37). In addition, accumulation of prelamin A has been shown to interfere with PCNA function and cause replication stress (9). Similarly, ectopic expression of progerin (LAΔ50) was conjectured to interfere with S phase progression and DNA replication (38). As a result of these findings, we examined the distribution of PCNA in early-passage AG08 cells and AG11 cells. Normal AG08 cells show diffuse staining of PCNA and lamin A throughout the nucleoplasm. In contrast, PCNA is present in nuclear aggregates along with lamin A in HGPS cells (Fig. 6A). This finding supports a model in which progerin sequesters PCNA, making it unavailable to associate with DNA and promote chain elongation at replication forks (39).
FIG 6.
Progerin sequesters PCNA. (A) Immunostaining for lamin A (Cy3) and PCNA (Alexa Fluor 488) in normal AG08 and HGPS AG11 cells. The white boxes indicate the magnified view of a single nucleus to demonstrate colocalization and PCNA aggregation. These aggregates were quantitated by analysis of the maxima of merged images using ImageJ. Numbers represent the averages from 100 to 200 cells of each type from two biological replicates. (B) The iPOND procedure was carried out in BJ cells (MPD 58-60) expressing vector control (pB), Flag-lamin A (LA), or Flag-progerin (P). The click reaction catalyzes the addition of biotin-azide to EdU-labeled DNA. A mock reaction without biotin-azide was carried out on lamin A input sample as a control. Antistreptavidin beads were used to pull down biotin-labeled chromatin and associated proteins. (Middle) This was followed by immunoblotting to detect PCNA, Flag, γH2AX, and H3 histone. (Right) After iPOND, the depleted extracts were subjected to immunoprecipitation (IP) with FLAG antibody, followed by Western blotting with FLAG or PCNA antibodies. (C) The rates of DNA replication fork progression in normal AG08 cells and HGPS AG11 cells (both MPD of 19) were measured using DNA combing. The distributions of rates from three replicates are presented as a box plot where the median is indicated by the red horizontal bar, the box spans the first through third quartiles, the whiskers extend to the last data points within 1.5 times the interquartile range, and outliers are plotted as circles. The distributions were compared using the Mann-Whitney U test, and the P value is indicated. (D) Accumulation of progerin leads to PCNA aggregation, replication stress, and p53 activation. The data are consistent with a model in which the accumulation of progerin causes the sequestration of PCNA, leading to its aggregation away from replication. The absence and miscoordination of PCNA leads to replication stress and the subsequent activation of p53. p53 activation in turn leads to premature senescence in HGPS cells.
We used iPOND capture technology to monitor the presence of lamin A, progerin, and PCNA on replicating DNA in fibroblasts ectopically expressing FLAG-lamin A, FLAG-progerin, or empty vector (pBABE). EdU (5-ethyyl-2′-deoxyuridine)-labeled DNA was covalently linked to biotin in the presence of copper (click reaction), and proteins bound to EdU-labeled DNA were purified on streptavidin beads, eluted, and visualized by Western blotting (Fig. 6B). BJ fibroblasts and lamin A-expressing BJ fibroblasts proliferate with a doubling time of approximately 41 to 42 h (MPD of 56); proliferation is slower in progerin-expressing BJ fibroblasts, and these cells undergo premature senescence (described in the next section). As a result, a long EdU incubation time is required to label replicating DNA in human fibroblasts and progerin-expressing fibroblasts. While this precludes identification of active replisomes, the iPOND procedure is useful to detect proteins associated with newly replicated DNA. Lamin A and PCNA, but not progerin, are purified with EdU-labeled DNA. γH2AX is detected only in progerin-expressing cells and is bound to EdU-labeled DNA. We detect similar amounts of PCNA bound to EdU-labeled DNA in the pBABE- and HA-lamin A-expressing cells, and far less PCNA bound to EdU-labeled DNA in the progerin-expressing cells. Histone H3 serves as a control to ensure equivalent amounts of chromatin in the input samples and in the iPOND samples. Importantly, we find that PCNA coimmunoprecipitates with FLAG-lamin A as well as with FLAG-progerin in the extracts used to perform iPOND (Fig. 6B). These results show that progerin binds PCNA and suggest that progerin promotes the redistribution of PCNA away from replicating DNA, resulting in replication stress and γH2AX formation.
To determine if DNA replication fork progression is compromised in HGPS cells, replication fork velocity was measured using DNA combing. Normal AG08 and HGPS AG11 cells at the same population doubling were pulse-labeled with CldU (5′-chlorodeoxyuridine) and IdU (5′-iododeoxyuridine) consecutively for 30 min each. Individual DNA fibers were stretched onto salinized glass coverslips, and the replication fork rate was calculated by expressing the length of IdU tracks as a function of time (Fig. 6C). We found that replication forks moved at 1.15 kbp/min in AG08 cells and slowed to 0.96 kbp/min in AG11 cells (P = 0.00004). The decreased rate of replication fork progression in AG11 suggests that progerin causes DNA replication stress (40), leading to DNA damage, p53 activation, and premature senescence in progeria (Fig. 6D).
Progerin-induced premature senescence is dependent on p53 and is associated with replication stress.
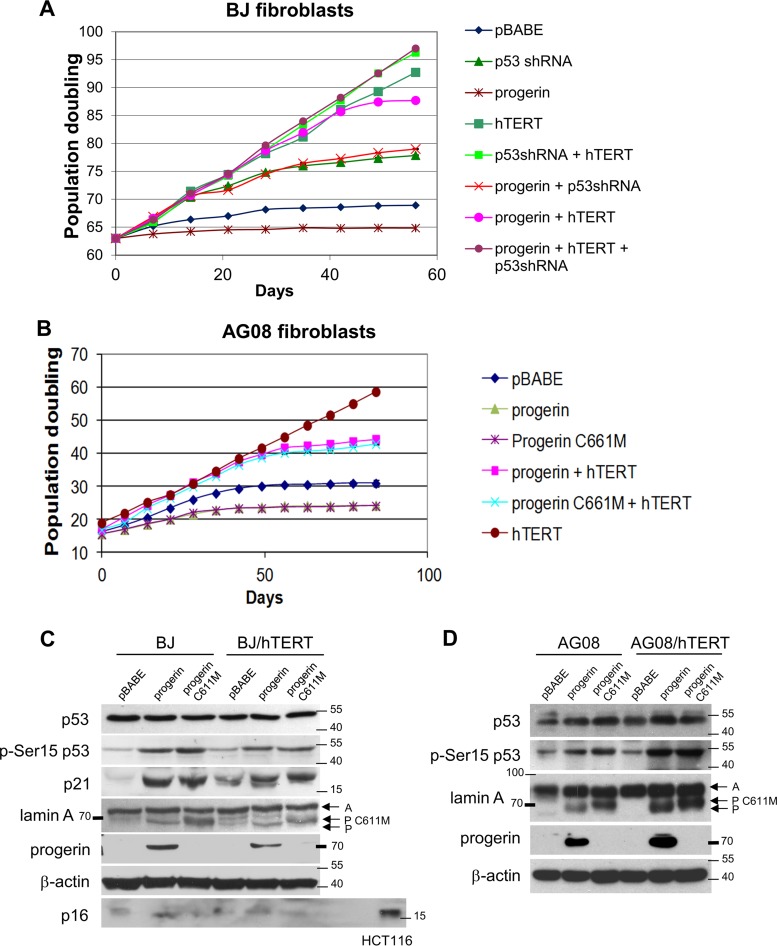
To investigate the dependency of progerin-induced cellular senescence on p53, we expressed progerin ectopically in (i) normal BJ and AG08 fibroblasts, (ii) cells where p53 was repressed (BJ/p53shRNA), (iii) cells where hTERT was ectopically expressed (BJ/hTERT, AG08/hTERT), and (iv) cells where p53 was repressed and hTERT was expressed (BJ/p53shRNA/hTERT). The timing of cellular senescence was determined by measuring population doublings (Fig. 7A and B), and progerin expression was confirmed by Western blotting (Fig. 7C and D). Progerin expression led to p53 activation as shown by increased p53 phosphorylation at Ser15 and by increased expression of p21 (Fig. 7C and D). Progerin induced senescence in all cells except in those where p53 was depleted (BJ/p53shRNA and BJ/p53shRNA/hTERT). hTERT expression delayed but did not prevent progerin-induced cellular senescence in both BJ and AG08 cells (Fig. 7A and B), similar to what was seen previously in AG11 and AG03 HGPS cells expressing endogenous progerin (Fig. 1A and 5A). Notably, ectopic progerin expression in normal AG08 cells resulted in the accumulation of γH2AX foci that colocalized with 53BP1 and pRPA32 (Fig. 8A), consistent with our earlier findings in AG11 cells expressing endogenous progerin.
FIG 7.
Progerin and progerin C661M promote premature senescence and replication stress. (A and B) Measurement of the number of mean population doublings in normal BJ fibroblasts (A) or normal AG08 fibroblasts (B) expressing progerin, progerin C661M, hTERT, p53 shRNA, or empty vector (pBABE) as indicated. Data represent averages from 3 biological replicates counted with 4 technical replicates each week. SEM values are shown as error bars. Senescent arrest was confirmed by acidic β-galactosidase, with 80% ± 4.4% in progerin and 82% ± 3.9% in progerin C661M staining blue. (C) Western blot analysis of BJ and BJ/hTERT cells expressing empty vector (pBABE), progerin, or progerin C661M. Protein extracts were immunoblotted with the indicated antibodies. (D) Western blot analysis of AG08 and AG08/hTERT cells expressing empty vector (pBABE), progerin, or progerin C661M. Protein extracts were immunoblotted with the indicated antibodies. A, processed lamin A; P C611M, progerin C611M; P, progerin. Note that the progerin antibody used on the Western blot does not detect the progerin C611M mutant. p53 activation is reflected by phospho-Ser15 p53 and p21 expression. β-Actin served as a loading control. HCT116 cells served as a positive control for p16 expression.
FIG 8.
Accumulation of progerin induces DNA damage and replication stress. (A) Immunostaining of γH2AX, 53BP1, and pRPA32 in normal AG08 fibroblasts expressing ectopic progerin. (B) Immunostaining of γH2AX, 53BP1, and pRPA32 in normal AG08 fibroblasts expressing ectopic progerin C661M. (C) Immunostaining using antiprogerin (Cy3-secondary antibody) and anti-53BP1 (Alexa Fluor 488-secondary antibody) and Draq5 in HGPS AG11 cells. Arrows indicate cells that lack endogenous progerin staining. (D) Cell counts of cells depicted in panel C showing the percentage of AG11 HGPS cells that immunostain (+) for progerin (red bars) with increasing mean population doublings (MPD). The proportion of cells with 5 or more 53BP1 foci is also shown. Data represent 100 to 200 cells in 3 individual experiments.
Progerin-induced senescence does not require farnesylation of progerin.
To investigate the requirement of progerin farnesylation to induce premature senescence, we generated a farnesyl site mutant in progerin (C611M) at the C-terminal CAAX motif that directs farnesylation of the cysteine residue in progerin and lamin A by farnesyltransferase. Mutation at this site is expected to interfere with the recruitment of progerin and lamin A to the nuclear membrane due to loss of the lipid anchor. Unexpectedly, we found that progerin C611M completely mimics progerin in its ability to promote premature senescence in normal AG08 cells (Fig. 7B), activate p53 (Fig. 7D), and promote γH2AX foci that colocalize with pRPA32 (Fig. 8B). These data indicate that blocking the farnesylation of progerin does not influence its role in promoting replication stress. They suggest that a population of mature lamin A in the nucleoplasm is critical in the coordination of the replication fork. When the unprocessed form of lamin A accumulates, it causes S-phase delay and replication stress, leading to DNA damage. The probability of this DNA damage not being repaired increases with the duration of replication stress. Such a model is supported by examining progerin accumulation in individual HGPS cells. We find that progerin expression and DNA damage increases as cells grow in culture (Fig. 8C and D).
DISCUSSION
The accumulation of progerin in HGPS cells is associated with altered nuclear architecture, chronic DNA damage, defective DNA repair mechanisms, and genomic instability (41). Using a Zmpste24−/− mouse model that results in the accumulation of farnesylated and carboxymethylated prelamin A, Varela et al. (10) reported that Zmpste24 deficiency elicits a stress response that activates p53. Although activation of the p53 signaling pathway in response to stress is well studied, the cellular response to progerin that triggers p53 activation remains uncharacterized. In this study, we report that progerin expression leads to replication stress. This is supported by our findings showing ATR activation, colocalization of γH2AX with 53BP1 and with pRPA32, PCNA redistribution, and decreased replication fork progression in cycling HGPS cells. This is unique to HGPS cells and is not seen in cycling normal cells. Moreover, we show that ectopically expressed progerin binds PCNA and sequesters it away from replicating DNA. We propose that DNA damage resulting from the collapse of stalled replication forks serves as the primary signal for p53 activation and premature senescence in HGPS cells. Consistent with this model, we show that DNA damage occurs predominantly during S phase and is not detected when HGPS cells are placed under serum-free conditions to prevent proliferation. The inability of ectopic hTERT expression to immortalize HGPS cells is also consistent with this model, since stalled replication forks would not be rescued by telomere elongation. Replication stress would continue to activate p53 even in the presence of protected telomeres. The association of progerin with PCNA, the accumulation of lamin A/PCNA aggregates in HGPS cells, and decreased replication rate support this model, suggesting that progerin directly interferes with the proper coordination of essential DNA replication factors such as PCNA (Fig. 6D).
The role of telomeres in HGPS is controversial, with some reports describing that HGPS cells contain shorter telomeres than age-matched controls (42, 43) and another finding that there is no difference in telomere length (22). Some reports indicate that hTERT-mediated cellular immortalization is possible in certain HGPS strains (34, 44, 45), while others have found that HGPS fibroblasts are resistant to hTERT immortalization (46, 47). Moreover, hTERT does not protect against progerin-induced aberrant nuclear shape and DNA damage (48). DNA damage at telomeres has been reported in cells undergoing replicative senescence and in cells undergoing progerin-induced premature senescence (34, 35). In these studies, telomere dysfunction is reflected by the presence of telomere aggregates that are associated with the nuclear lamina and with γH2AX foci. In our study, we find no evidence of telomere erosion or telomere dysfunction in HGPS cells. HGPS cells enter senescence with long telomeres, and ectopic hTERT expression is unable to immortalize HGPS cells in the absence of p53 repression. In addition, we find that hTERT expression does not prevent premature senescence induced by ectopically expressed progerin. Moreover, we do not detect DNA damage at telomeres in HGPS cells. We do find that hTERT expression extends the proliferative capacity in HGPS cells and delays the onset of senescence in HGPS cell strains as well as in normal cells that express ectopic progerin or progerin C661M. We are unable to provide an explanation for this. A recent study reports that ectopic telomerase expression prevents progerin-induced proliferative defects, progerin-induced premature senescence, and progerin-induced DNA damage (49), consistent with previous reports (34, 50). Telomerase, however, was unable to rescue two other progerin-induced defects, namely, repression of the heterochromatic marker H3K27me3 and nuclear abnormalities. Hence, telomerase expression rescues some but not all progerin-induced phenotypes. The extent of rescue may depend on the levels of progerin and telomerase expression. These studies are not inconsistent with the idea that telomerase has functions beyond telomere stabilization. There is evidence for a diverse range of noncanonical telomerase functions in proliferation, stem cells, apoptosis, and gene expression (51). For example, Masutomi et al. reported that hTERT participates in chromatin maintenance and that hTERT affects the response to DNA double-strand breaks. They demonstrated that suppression of hTERT expression impaired DNA damage responses and diminished the capacity for DNA repair in response to genotoxic damage (52). In addition, the impact on proliferation may be a key feature of hTERT expression, which could explain why S phase is not delayed as significantly in hTERT AG11 cells (Fig. 3D). Based on these data, we suggest that telomerase extends the life span of HGPS cells through its reparative function, regulation of proliferation, and its role in chromatin maintenance.
A nucleoplasmic population of lamin A was reported to colocalize within chromatin foci that contain the replication proteins PCNA and DNA polymerase δ; these foci are believed to represent sites of DNA replication in early S phase (5, 6). Interestingly, lamin A/C deficiency enhances replication stress in cells treated with interstrand cross-linking agents, suggesting a role for lamin A/C in the recovery of stalled replication forks (53). Unprocessed prelamin A likely interferes with DNA replication by sequestering replication and repair factors (9, 39, 54). Progerin differs from mature lamin A in several ways, including (i) having a 50-amino-acid internal deletion, (ii) remaining bound to the inner nuclear membrane through a farnesyl lipid anchor, and (iii) retention of a methylated farnesylcysteine at the carboxy terminus of the protein. Progerin, unlike processed lamin A, may fail to interact functionally with proteins that maintain replication fork integrity, and this could be due to the 50-amino-acid deletion in progerin or because of mislocalization to the inner nuclear membrane. Notably, methylation of the terminal farnesylcysteine of progerin is important for progerin-induced premature senescence and not for progerin-induced nuclear abnormalities (55). This finding could explain why farnesyltransferase inhibitors improve nuclear shape but do not alleviate DNA damage (4). Thus, methylation of the terminal cysteine may contribute to progerin's ability to disrupt DNA replication and replication fork integrity.
Farnesyltransferase inhibitors have been used to reverse the aberrant nuclear morphology present in HGPS fibroblasts by interfering with the recruitment of progerin to the nuclear membrane (56). However, a mouse model with a knock-in nonfarnesylated progerin was shown to still develop phenotypes of progeria, implying that the lipid anchor is not required to induce the disease (57). We generated a farnesyl site mutant in progerin (C611M) at the C-terminal CAAX motif that directs farnesylation to the cysteine residue in progerin. Mutation at this site interferes with the recruitment of progerin to the nuclear membrane, promoting nucleoplasmic localization. Progerin C611M behaves identically to progerin in promoting premature senescence in normal AG08 and BJ cells, activating p53 and promoting DNA damage foci that colocalize with pRPA32. These data suggest that blocking the farnesylation of progerin and its recruitment to the inner nuclear membrane does not influence its role in replication stress and premature senescence. This implies that progerin has an effect on coordinating DNA replication in the nucleoplasm rather than having a significant impact on HGPS through disruption of the nuclear lamina. Thus, we propose that progerin, unlike processed lamin A, fails to interact functionally with proteins (such as PCNA) that regulate DNA replication. This leads to replication stress, causes DNA damage, activates p53, and induces premature senescence. These events contrast with the paradigm of telomere erosion and represent an alternative pathway to p53-mediated senescence.
MATERIALS AND METHODS
Plasmid constructs.
The p53 shRNA construct was created using hairpin inserts with the sequence GACTCCAGTGGTAATCTAC into BglII and XhoI sites that were directionally ligated into the pSuper.retro.puro backbone (58). The cDNAs for lamin A and progerin were generated by reverse transcription-PCR (RT-PCR) amplification using RNA isolated from human normal or HGPS fibroblasts and cloned into pBABEhygro or pBABEpuro using the BamHI and EcoRI sites. The following primers were used (lowercase letters indicate BamHI and EcoRI restriction sites): lamin A and progerin, cgggatccATGGAGACCCCGTCC and ggaattcTTACATGATGCTGCAGTTCTG. The progerin mutant C661M cDNA was generated by PCR amplification using progerin cDNA as a template with the 5′ primer described above and the 3′ primer replaced with ggaattcTTACATGATGCTCATGTTCTG (boldface indicates which bases differ from the wild type). A11 constructs were verified by DNA sequencing. The hTERT and p53 RNA interference-resistant (p53RR) expression constructs were described previously (24, 26). The temperature-sensitive mutation (A138V) was generated by PCR-based mutagenesis of p53RR using GTTTTGCCAACTGGTCAAGACCTGCCCTG and its complementary strand.
Cell culture and retroviral infection.
The BJ human fibroblast cell strain was maintained as previously described (59). The progeria strains AG03198 and AG11513 and its age-matched control, AG08470, were obtained from Coriell Institute and were maintained in minimal essential medium-Earle's balance salts supplemented with 10% fetal bovine serum (FBS). All experiments were conducted using cells grown under 2 to 5% oxygen and 5% carbon dioxide, using a sealed chamber. These cells were modified to express the ecotropic receptor to allow infections with ecotropic viral supernatants. The fibroblasts were infected with amphotropic isotyped virus (Phoenix-A packaging cells) containing the ecotropic receptor (pm5-Eco) and then selected in G418 (1 mg/ml) for 2 weeks to generate BJ, AG08, and AG11 Eco strains. Ecotropic retroviral supernatants were produced by transfection of Phoenix-E packaging cells with the various pBABE or pSuper retroviral constructs using Fugene. After 48 or 72 h, the medium was collected, supplemented with 0.8 μg/ml Polybrene, and used to infect fibroblast strains. Selection was performed 48 to 72 h after viral infection using 1 μg/ml puromycin for 3 days (pSuper or pBABEpuro) or 150 μg/ml hygromycin for 4 days (pBABEhygro). The acidic β-galactosidase assay was performed as described previously (60).
Cell counting.
All cell lines were generated by viral infection simultaneously from the same parent cells at an identical population doubling for a given experiment. Every 7 days, cells were counted by Coulter Counter and replated at 1:10 or 1:20 dilution. The mean population doubling (MPD) was calculated by the formula MPD = log (Nf/Ni)/log 2, where Nf is the number of cells counted and Ni is the number of cells seeded. Long-term counting studies (usually 100 days) involved generating 3 biological replicates at the start of the experiment from the same parent line. Each of these was used to generate 4 technical replicate cell counts. The averages of all replicate counts were used to generate the growth curves. Short-term growth rate experiments were performed by seeding 50,000 cells in triplicate for each time point and were harvested for counting daily.
Western blotting and antibodies.
Cells were lysed directly in 2× Laemmli protein sample buffer (4% SDS, 25 mM Tris-HCl [pH 6.8], 20% glycerol, 0.1 M dithiothreitol [DTT]). Protein concentrations were quantified by bicinchoninic acid (BCA) assay (Pierce) using the protocol from the supplier. Equal amounts of protein were resolved by PAGE, transferred onto nitrocellulose membranes, and blocked in 5% milk and 2% BSA in TBS solution containing 0.5% Tween 20. Primary or secondary antibodies were diluted in TBS containing 2% BSA and 0.5% Tween 20. Antibodies against the following proteins were used: H2AX (07-627) and progerin (13A4; cannot detect progerin C611M, since this change alters the epitope) (Millipore); γH2AX (NB100-78356) (Novus); ATR (sc-1887), phospho-S428 ATR (sc-109912), β-actin (sc-47778), BTG2 (sc-33775), lamin A (Sc-20680), lipin (sc-98450), p16 (sc-759), p21 (sc-397), and p53 (DO-1) (Santa Cruz); and phospho(S-15)-p53 (9284) (Cell Signaling Technologies). Anti-rabbit and anti-mouse secondary antibodies were conjugated with horseradish peroxidase (HRP) (Jackson IR).
Southern blot analysis.
Cell pellets were resuspended in lysis buffer (10 mM NaCl, 10 mM Tris, pH 8.0, 25 mM EDTA, 0.5% SDS), digested overnight with proteinase K (100 μg/ml), and RNase (50 μg/ml) treated for 30 min. The samples then were phenol-chloroform extracted and ethanol precipitated, followed by digestion overnight with HinfI and RsaI (New England BioLabs). The concentration of digested DNA samples was assessed by NanoDrop and visualized after electrophoresis on 0.7% agarose to confirm equal loading. The 0.7% gels were transferred overnight through capillary action to Hybond-plus membranes followed by UV cross-linking. The probe was generated by using a 1-kb fragment of human telomere sequence in a Klenow reaction with random primers and [32P]dCTP. Hybridization was carried out overnight at 42°C in Hyb solution (5% SDS, 6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], and 50% formamide) followed by extensive washing. Blots were visualized by autoradiography using a PhosphorImager (Typhoon Trio). Average telomere length was determined as described by Kimura et al. (61) using Gel Analyzer software.
Cell cycle analysis.
Cells were fixed on ice in 70% ethanol, washed with phosphate-buffered saline (PBS) containing 1% BSA, incubated with 100 μg/ml RNase A for 10 min at 37°C, and finally resuspended in PBS containing propidium iodide (50 μg/ml). Cell cycle distribution was examined by flow cytometry using a FACSCalibur flow cytometer (Becton Dickinson). Costaining with γH2AX involved staining with anti-γH2AX antibody followed by anti-rabbit fluorescein isothiocyanate-conjugated secondary antibody. Cells were also stained with PI as described above. Cells treated with doxorubicin were used as controls to determine the threshold for γH2AX-positive staining.
Immunofluorescence microscopy.
Cells on glass coverslips were fixed with 1% paraformaldehyde in PBS for 10 min and permeabilized with 0.5% Triton X-100 for 10 min. Antibodies were diluted in TBS–0.5% Tween 20 containing 1% BSA and 1/50 donkey serum (Jackson). Specific protein localization was visualized after staining with antibodies to 53BP1 (1/400; 4937; Cell Signaling), γH2AX (1/400; NB100-78356; Novus), phospho-S33 RPA32 (1/500; NBP1-23016; Novus), TRF1 (1/400; sc-56807; Santa Cruz), lamin A (1/1,000; Sc-20680; Santa Cruz), or PCNA (1/50; sc-56; Santa Cruz) for 1 h. Secondary antibodies included goat anti-mouse antibody–Alexa Fluor 488 (Invitrogen), goat anti-rabbit antibody–Alexa Fluor 488 (Invitrogen), goat anti-mouse antibody–Cy3 (Jackson), and goat anti-rabbit antibody–Cy3 (Jackson). Secondary antibodies were incubated for 40 min at a concentration of 1/1,000 in TBS–0.5% Tween 20 containing 1% BSA and 1/50 donkey serum. All samples were washed 3 times with TBS containing 0.5% Tween 20 between antibody incubations. The nuclei were stained with the fluorescent DNA dye Draq5 (Alexis) according to the manufacturer's instructions. Triple-stained images required that the RPA32 antibody be conjugated to biotin using EZ-link sulfo-NHS-LC-biotin ((sulfosuccinimidyl 6-[biotinamido] hexanoate) Thermo). Visualization of biotin-RPA32 antibody staining used streptavidin-Dylight 649 (Abcam). Images were obtained using an Olympus FluoView 300 confocal laser-scanning microscopic system (Carsen group) with a single optical section at ×400 magnification. Images were analyzed and superimposed using ImageJ software.
Isolation of proteins on nascent DNA (iPOND).
Cultured human fibroblasts (MPD of 58 to 60) were synchronized by serum starvation (48 h) followed by serum stimulation (16 h). Cells were grown in 10 μM EdU (5-ethyyl-2′-deoxyuridine; Sigma-Aldrich) for 8 h. After labeling, cells were cross-linked in 1% formaldehyde–PBS for 20 min at room temperature, quenched using 0.125 M glycine, and washed in PBS containing 1% BSA. Cell pellets were resuspended in 0.1% Triton-X–PBS to permeabilize. Cells were washed once in 1% PBS prior to the click reaction. The click reaction was performed by suspending cells in a reaction buffer consisting of 4 mM CuSO4, 1 mM Tris(2-carboxyethyl)-phosophine, and 1 μM biotin-azide (Sigma-Aldrich) in PBS for 1 h at room temperature. Cell lysis and protein capture on streptavidin-agarose beads was performed as described previously by Sirbu et al. (29).
Molecular combing.
Primary AG08 (MPD of 19) and AG11 (MPD of 19) fibroblasts were synchronized through contact inhibition and 48 h of serum starvation before being plated to 25% confluence for the DNA combing experiment. Growing cells were sequentially labeled with CldU and IdU, harvested following trypsinization, and cast into agarose plugs as described previously (62). The processing of the plugs, DNA combing, immunofluorescence, imaging, and analysis of replication progression followed established procedures (62). Replication rates for individual forks were determined by measuring the length of IdU tracks that were adjacent to a CldU track (to eliminate forks that initiated during the labeling period) and dividing by the labeling time. The distributions of fork rates for three independent replicates are presented as box plots, and the data were aggregated to produce the box plot shown in Fig. 5C. The two-sided Mann-Whitney U test was used to compare the distributions of DNA replication fork rates between AG08 cells and AG11 cells.
ACKNOWLEDGMENTS
Funding was provided through grants from the Progeria Research Foundation (S.B.), the Canadian Cancer Society Research Institute (Impact grant 702310 to G.W.B.), and the Canadian Institutes of Health Research (grant MOP-79368 to G.W.B.).
We thank David Miller for reviewing the manuscript.
S.B. and K.W. designed experiments, prepared figures, and wrote the manuscript. K.W., D.C., W.M., and M.S. performed the experiments. B.H. and G.W.B. designed, performed, and analyzed the DNA combing experiments and edited the manuscript.
REFERENCES
- 1.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS. 2003. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burtner CR, Kennedy BK. 2010. Progeria syndromes and ageing: what is the connection? Nat Rev Mol Cell Biol 11:567–578. doi: 10.1038/nrm2944. [DOI] [PubMed] [Google Scholar]
- 3.Liu B, Wang J, Chan KM, Tjia WM, Deng W, Guan X, Huang JD, Li KM, Chau PY, Chen DJ, Pei D, Pendas AM, Cadinanos J, Lopez-Otin C, Tse HF, Hutchison C, Chen J, Cao Y, Cheah KS, Tryggvason K, Zhou Z. 2005. Genomic instability in laminopathy-based premature aging. Nat Med 11:780–785. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Rusinol A, Sinensky M, Wang Y, Zou Y. 2006. DNA damage responses in progeroid syndromes arise from defective maturation of prelamin A. J Cell Sci 119:4644–4649. doi: 10.1242/jcs.03263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kennedy BK, Barbie DA, Classon M, Dyson N, Harlow E. 2000. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev 14:2855–2868. doi: 10.1101/gad.842600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shumaker DK, Solimando L, Sengupta K, Shimi T, Adam SA, Grunwald A, Strelkov SV, Aebi U, Cardoso MC, Goldman RD. 2008. The highly conserved nuclear lamin Ig-fold binds to PCNA: its role in DNA replication. J Cell Biol 181:269–280. doi: 10.1083/jcb.200708155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pekovic V, Harborth J, Broers JL, Ramaekers FC, van Engelen B, Lammens M, von Zglinicki T, Foisner R, Hutchison C, Markiewicz E. 2007. Nucleoplasmic LAP2alpha-lamin A complexes are required to maintain a proliferative state in human fibroblasts. J Cell Biol 176:163–172. doi: 10.1083/jcb.200606139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez J, Calvo F, Gonzalez JM, Casar B, Andres V, Crespo P. 2010. ERK1/2 MAP kinases promote cell cycle entry by rapid, kinase-independent disruption of retinoblastoma-lamin A complexes. J Cell Biol 191:967–979. doi: 10.1083/jcb.201004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobb AM, Murray TV, Warren DT, Liu Y, Shanahan CM. 2016. Disruption of PCNA-lamins A/C interactions by prelamin A induces DNA replication fork stalling. Nucleus 7:498–511. doi: 10.1080/19491034.2016.1239685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varela I, Cadinanos J, Pendas AM, Gutierrez-Fernandez A, Folgueras AR, Sanchez LM, Zhou Z, Rodriguez FJ, Stewart CL, Vega JA, Tryggvason K, Freije JM, Lopez-Otin C. 2005. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature 437:564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- 11.Vousden KH, Prives C. 2009. Blinded by the light: the growing complexity of p53. Cell 137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Porath I, Weinberg RA. 2005. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol 37:961–976. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. 2004. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol Cell 14:501–513. doi: 10.1016/S1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 14.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593–602. doi: 10.1016/S0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 15.Bartek J, Bartkova J, Lukas J. 2007. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 16.Chen Q, Ames BN. 1994. Senescence-like growth arrest induced by hydrogen peroxide in human diploid fibroblast F65 cells. Proc Natl Acad Sci U S A 91:4130–4134. doi: 10.1073/pnas.91.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Leonardo A, Linke SP, Clarkin K, Wahl GM. 1994. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev 8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 18.Collado M, Blasco MA, Serrano M. 2007. Cellular senescence in cancer and aging. Cell 130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Sedivy JM. 2007. Telomeres limit cancer growth by inducing senescence: long-sought in vivo evidence obtained. Cancer Cell 11:389–391. doi: 10.1016/j.ccr.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Collado M, Serrano M. 2010. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer 10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahin E, Depinho RA. 2010. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature 464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao K, Blair CD, Faddah DA, Kieckhaefer JE, Olive M, Erdos MR, Nabel EG, Collins FS. 2011. Progerin and telomere dysfunction collaborate to trigger cellular senescence in normal human fibroblasts. J Clin Investig 121:2833–2844. doi: 10.1172/JCI43578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 24.Vaziri H, Benchimol S. 1998. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol 8:279–282. doi: 10.1016/S0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 25.Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, Campisi J. 2003. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J 22:4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheaton K, Muir J, Ma W, Benchimol S. 2010. BTG2 antagonizes Pin1 in response to mitogens and telomere disruption during replicative senescence. Aging Cell 9:747–760. doi: 10.1111/j.1474-9726.2010.00601.x. [DOI] [PubMed] [Google Scholar]
- 27.Scaffidi P, Misteli T. 2006. Lamin A-dependent nuclear defects in human aging. Science 312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10:886–895. doi: 10.1016/S0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 29.Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev 25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward IM, Chen J. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 276:47759–47762. doi: 10.1074/jbc.M009785200. [DOI] [PubMed] [Google Scholar]
- 31.Cimprich KA, Cortez D. 2008. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson E, Nievera CJ, Klimovich V, Fanning E, Wu X. 2006. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J Biol Chem 281:39517–39533. doi: 10.1074/jbc.M605121200. [DOI] [PubMed] [Google Scholar]
- 33.Vassin VM, Anantha RW, Sokolova E, Kanner S, Borowiec JA. 2009. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J Cell Sci 122:4070–4080. doi: 10.1242/jcs.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benson EK, Lee SW, Aaronson SA. 2010. Role of progerin-induced telomere dysfunction in HGPS premature cellular senescence. J Cell Sci 123:2605–2612. doi: 10.1242/jcs.067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raz V, Vermolen BJ, Garini Y, Onderwater JJ, Mommaas-Kienhuis MA, Koster AJ, Young IT, Tanke H, Dirks RW. 2008. The nuclear lamina promotes telomere aggregation and centromere peripheral localization during senescence of human mesenchymal stem cells. J Cell Sci 121:4018–4028. doi: 10.1242/jcs.034876. [DOI] [PubMed] [Google Scholar]
- 36.de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 37.Spann TP, Moir RD, Goldman AE, Stick R, Goldman RD. 1997. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J Cell Biol 136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, Collins FS. 2004. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A 101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Musich PR, Zou Y. 2009. Genomic instability and DNA damage responses in progeria arising from defective maturation of prelamin A. Aging (Albany NY) 1:28–37. doi: 10.18632/aging.100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat Cell Biol 16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musich PR, Zou Y. 2011. DNA-damage accumulation and replicative arrest in Hutchinson-Gilford progeria syndrome. Biochem Soc Trans 39:1764–1769. doi: 10.1042/BST20110687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB. 1992. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci U S A 89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Decker ML, Chavez E, Vulto I, Lansdorp PM. 2009. Telomere length in Hutchinson-Gilford progeria syndrome. Mech Ageing Dev 130:377–383. doi: 10.1016/j.mad.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Huang S, Risques RA, Martin GM, Rabinovitch PS, Oshima J. 2008. Accelerated telomere shortening and replicative senescence in human fibroblasts overexpressing mutant and wild-type lamin A. Exp Cell Res 314:82–91. doi: 10.1016/j.yexcr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouellette MM, McDaniel LD, Wright WE, Shay JW, Schultz RA. 2000. The establishment of telomerase-immortalized cell lines representing human chromosome instability syndromes. Hum Mol Genet 9:403–411. doi: 10.1093/hmg/9.3.403. [DOI] [PubMed] [Google Scholar]
- 46.Corso C, Parry EM, Faragher RG, Seager A, Green MH, Parry JM. 2005. Molecular cytogenetic insights into the ageing syndrome Hutchinson-Gilford Progeria (HGPS). Cytogenet Genome Res 111:27–33. doi: 10.1159/000085666. [DOI] [PubMed] [Google Scholar]
- 47.Wallis CV, Sheerin AN, Green MH, Jones CJ, Kipling D, Faragher RG. 2004. Fibroblast clones from patients with Hutchinson-Gilford progeria can senesce despite the presence of telomerase. Exp Gerontol 39:461–467. doi: 10.1016/j.exger.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 48.Scaffidi P, Misteli T. 2008. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol 10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chojnowski A, Ong PF, Wong ES, Lim JS, Mutalif RA, Navasankari R, Dutta B, Yang H, Liow YY, Sze SK, Boudier T, Wright GD, Colman A, Burke B, Stewart CL, Dreesen O. 27 August 2015. Progerin reduces LAP2alpha-telomere association in Hutchinson-Gilford progeria. eLife 4. doi: 10.7554/eLife.07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kudlow BA, Stanfel MN, Burtner CR, Johnston ED, Kennedy BK. 2008. Suppression of proliferative defects associated with processing-defective lamin A mutants by hTERT or inactivation of p53. Mol Biol Cell 19:5238–5248. doi: 10.1091/mbc.E08-05-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cong Y, Shay JW. 2008. Actions of human telomerase beyond telomeres. Cell Res 18:725–732. doi: 10.1038/cr.2008.74. [DOI] [PubMed] [Google Scholar]
- 52.Masutomi K, Possemato R, Wong JM, Currier JL, Tothova Z, Manola JB, Ganesan S, Lansdorp PM, Collins K, Hahn WC. 2005. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc Natl Acad Sci U S A 102:8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh M, Hunt CR, Pandita RK, Kumar R, Yang CR, Horikoshi N, Bachoo R, Serag S, Story MD, Shay JW, Powell SN, Gupta A, Jeffery J, Pandita S, Chen BP, Deckbar D, Lobrich M, Yang Q, Khanna KK, Worman HJ, Pandita TK. 2013. Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest. Mol Cell Biol 33:1210–1222. doi: 10.1128/MCB.01676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Misteli T, Scaffidi P. 2005. Genome instability in progeria: when repair gets old. Nat Med 11:718–719. doi: 10.1038/nm0705-718. [DOI] [PubMed] [Google Scholar]
- 55.Ibrahim MX, Sayin VI, Akula MK, Liu M, Fong LG, Young SG, Bergo MO. 2013. Targeting isoprenylcysteine methylation ameliorates disease in a mouse model of progeria. Science 340:1330–1333. doi: 10.1126/science.1238880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toth JI, Yang SH, Qiao X, Beigneux AP, Gelb MH, Moulson CL, Miner JH, Young SG, Fong LG. 2005. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc Natl Acad Sci U S A 102:12873–12878. doi: 10.1073/pnas.0505767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang SH, Andres DA, Spielmann HP, Young SG, Fong LG. 2008. Progerin elicits disease phenotypes of progeria in mice whether or not it is farnesylated. J Clin Investig 118:3291–3300. doi: 10.1172/JCI35876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brummelkamp TR, Bernards R, Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 59.Wheaton K, Riabowol K. 2004. Protein kinase C delta blocks immediate-early gene expression in senescent cells by inactivating serum response factor. Mol Cell Biol 24:7298–7311. doi: 10.1128/MCB.24.16.7298-7311.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A 92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kimura M, Stone RC, Hunt SC, Skurnick J, Lu X, Cao X, Harley CB, Aviv A. 2010. Measurement of telomere length by the Southern blot analysis of terminal restriction fragment lengths. Nat Protoc 5:1596–1607. doi: 10.1038/nprot.2010.124. [DOI] [PubMed] [Google Scholar]
- 62.Yang J, O'Donnell L, Durocher D, Brown GW. 2012. RMI1 promotes DNA replication fork progression and recovery from replication fork stress. Mol Cell Biol 32:3054–3064. doi: 10.1128/MCB.00255-12. [DOI] [PMC free article] [PubMed] [Google Scholar]