Abstract
CmeABC, a resistance-nodulation-division (RND) type of efflux pump, contributes to Campylobacter resistance to a broad spectrum of antimicrobial agents and is also essential for Campylobacter colonization of the animal intestinal tract by mediation of bile resistance. As one of the main systems for Campylobacter adaptation to different environments, CmeABC is likely subject to control by regulatory elements. We describe the identification of a transcriptional repressor for CmeABC. Insertional mutagenesis of cmeR, an open reading frame immediately upstream of the cmeABC operon, resulted in overexpression of cmeABC, as determined by transcriptional fusion (PcmeABC-lacZ) and immunoblotting with CmeABC-specific antibodies. Overexpression of the efflux pump was correlated with a moderate increase in the level of resistance of the cmeR mutant to several antimicrobials. In vitro, recombinant CmeR bound specifically to the promoter region of cmeABC, precisely, to the inverted repeat sequences in the cmeABC promoter. A single nucleotide deletion between the two half sites of the inverted repeat reduced the level of CmeR binding to the promoter sequence and resulted in overexpression of cmeABC. Together, these findings indicate that cmeR encodes a transcriptional repressor that directly interacts with the cmeABC promoter and modulates the expression of cmeABC. Mutation either in CmeR or in the inverted repeat impedes the repression and leads to enhanced production of the MDR efflux pump.
As a general and important mechanism for antimicrobial resistance, multidrug efflux systems (often named MDR pumps) contribute significantly to the intrinsic and acquired resistance to antibiotics in bacterial organisms (37, 40, 46). In addition to being key players in antibiotic resistance, MDR pumps also facilitate bacterial adaptation to deleterious environments where toxic compounds or metabolites are present. In bacteria, expression of MDR efflux pumps is usually controlled by transcriptional regulators that either repress or activate the transcription of the MDR efflux genes (14, 37, 40). Many of these regulators are local repressors that directly interact with the promoter regions of MDR efflux genes or operons. For example, repressors AcrR (Escherichia coli), QacR (Staphylococcus aureus), MtrR (Neisseria gonorrhoeae), and MexR (Pseudomonas aeruginosa) bind specifically to the promoter sequences of acrAB, qacA, mtrCDE, and mexAB, respectively, thereby inhibiting the expression of the corresponding MDR efflux gene(s) (9, 12, 17, 27). Mutations in the repressors or repressor-binding sequences impede the repression and result in overexpression of efflux pumps, which consequently increases bacterial resistance to structurally unrelated antimicrobial agents (9, 12, 16, 38, 42, 48). Recently, two-component systems were also found to be involved in the regulation of bacterial MDR pumps (3, 10, 33, 34). These examples illustrate the complexity and diversity of the regulatory mechanisms for bacterial MDR efflux pumps.
Campylobacter jejuni is the leading bacterial cause of human food-borne enteritis in many industrialized countries (11) and has become increasingly resistant to antimicrobials, compromising the effectiveness of antibiotic treatments (8, 44, 45, 53). One of the mechanisms used by Campylobacter for antimicrobial resistance is the CmeABC efflux system, a resistance-nodulation-division (RND) type of efflux pump recently identified in C. jejuni (24, 39). This efflux pump system consists of three members, including an outer membrane protein (CmeC), an inner membrane drug transporter (CmeB), and a periplasmic fusion protein (CmeA). These three proteins are encoded by a three-gene operon (cmeABC) and function together to form a membrane channel that extrudes toxic substrates directly out of Campylobacter cells (24). CmeABC contributes significantly to the intrinsic and acquired resistance of Campylobacter to structurally diverse antimicrobials (24, 26, 39). In addition, CmeABC plays a key role in bile resistance and is essential for Campylobacter growth in bile-containing media and colonization of the animal intestinal tract (25). These findings have defined the importance of CmeABC in the antimicrobial resistance and pathophysiology of Campylobacter.
Even though basal production of CmeABC in wild-type strains occurs at a level that can readily be detected with antibodies specific to the efflux pumps (24), little is known about the regulatory mechanisms that modulate the expression of cmeABC in Campylobacter cells. Understanding the regulatory system for CmeABC will provide new insights into the mechanisms by which Campylobacter contributes to multidrug resistance (MDR) and adaptation to environmental changes. In this study, we report on the identification of CmeR as a transcriptional repressor for CmeABC. The cmeR gene is located immediately upstream of cmeA and encodes a 210-amino-acid (aa) protein that shares sequence and structure similarities to the members of the TetR family of transcriptional repressors. Using various approaches, we show that CmeR represses the transcription of cmeABC by directly binding to the promoter region (specifically, to the inverted repeat [IR]) of the efflux operon. Mutations in CmeR or the CmeR-binding site impede the repression and result in the overexpression of CmeABC and enhanced resistance to multiple antibiotics.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The various Campylobacter strains, mutants, and plasmids used in this study and their sources are listed in Table 1. These isolates were routinely grown in Mueller-Hinton (MH) broth (Difco) or agar at 42°C under microaerobic conditions, which were generated with a Campypak Plus (Becton Dickinson) gas pack in an enclosed jar. When needed, MH media were supplemented with kanamycin (30 μg/ml) or chloramphenicol (4 μg/ml). E. coli cells were grown at 37°C with shaking at 200 rpm in Luria-Bertani (LB) medium. When needed, LB media were supplemented with kanamycin (30 μg/ml) or ampicillin (100 μg/ml).
TABLE 1.
Bacterial plasmids and strains used in this study
| Plasmid or strain | Description | Source or reference |
|---|---|---|
| Plasmids | ||
| pGEMT-Easy | PCR cloning vector, Ampr | Promega |
| pCMER | pGEMT-Easy containing full-length cmeR fragment, Ampr | This study |
| pCMERC | pCMER with chloramphenicol resistance cassette inserted in cmeR, Ampr Cmr | This study |
| pMW10 | E. coli-C. jejuni shuttle vector with promoterless E. coli lacZ gene, Kanr | 52 |
| pIT81 | pMW10 derivative with the cmeABC promoter of wild-type C. jejuni 81-176 inserted upstream of lacZ | This study |
| pIT3e | pMW10 derivative with the cmeA promoter of C. jejuni CR3e inserted upstream of lacZ | This study |
| pRK2013 | IncP Tra RK2+ ΔrepRK2 repE1+, Kanr | 6 |
| pQE-30 | Expression vector, Ampr | Qiagen |
| pQE-CmeR | pQE-30 derivative expressing full-length recombinant CmeR protein | This study |
| Strains | ||
| C. jejuni | ||
| NCTC 11168 | Wild type; genome sequence known | 35 |
| 81-176 | Wild type; isolated from a human | 4 |
| CR3e | 81-176 derivative; spontaneous fluoroquinolone-resistant mutant obtained after stepwise selection with ciprofloxacin | This study |
| 9B6 | 81-176 derivative; cmeB::kan | 24 |
| JL106 | 81-176 derivative; cmeC::kan | 25 |
| JL107 | 81-176 derivative; cmeR::cm | This study |
| JL108 | 81-176 containing pMW10 | This study |
| JL109 | JL107 containing pMW10 | This study |
| JL110 | 81-176 containing pIT81 | This study |
| JL111 | JL107 containing pIT81 | This study |
| JL112 | 81-176 containing pIT3e | This study |
| E. coli | ||
| JM109 | endA1 recA1 gyrA96 thi hsdR17 (rk−, mk+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | Promega |
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rk−, mk+) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
PCR.
All primers used for PCR are listed in Table 2. PCR was performed in a volume of 100 μl containing 200 μM each deoxynucleoside triphosphate, 200 nM primers, 2.5 mM MgSO4, 50 ng of Campylobacter genomic DNA, and 5 U of Taq DNA polymerase (Promega) or Pfu Turbo DNA polymerase (Stratagene). Cycling conditions varied according to the estimated annealing temperatures of the primers and the expected sizes of the products. To amplify the 0.9-kb coding sequence of cmeR from C. jejuni 81-176, primers F and R were designed from the genomic sequence of C. jejuni NCTC 11168 (35) and were used in the PCR along with the genomic DNA of strain 81-176 and Taq DNA polymerase. PCR products were purified with a QIAquick PCR purification kit (Qiagen) and subsequently sequenced. To insert the cat gene cassette into the cmeR gene, primers CHLF and CHLR (Table 2) were used in the PCR with Pfu Turbo7 DNA polymerase to amplify the entire cat gene from shuttle vector pUOA18 (49). To determine the binding of CmeR to the cmeABC promoter, primers GSF and GSR1 were used to amplify the 170-bp DNA fragment that contains the intergenic region (IT) from wild-type strain 81-176 and its mutant, strain CR3e, for gel mobility shift assays. Reverse primers GSR2, GSR3, and GSR4 were used in conjunction with primer GSF to map the specific CmeR-binding site in the IT. The locations of these PCR primers are indicated in Fig. 1A.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequencea |
|---|---|
| F | 5′-TAGAAAAGTATATTTGTATACCCT-3′ |
| R | 5′-CGCCACTAACTTGAGGCTTTA-3′ |
| R1 | 5′-AATTTTTGGCTAATTATATCTTAATTT-3′ |
| CHLF | 5′-TGCTCGGCGGTGTTCCTTT-3′ |
| CHLR | 5′-GCGCCCTTTAGTTCCTAAAG-3′ |
| RF | 5′-ATAGGATCCATGAACTCAAATAGAACACCA-3′ (BamHI) |
| RR | 5′-TTTTAAGCTTTGGAGCTATTGATT-3′ (HindIII) |
| GSF | 5′-CTAAATGGAATCAATAGCTCC-3′ |
| GSR1 | 5′-GCACAACACCTAAAGCTAAAA-3′ |
| GSR2 | 5′-TAAAAATTGTAATATTTATTACAG-3′ |
| GSR3 | 5′-ATTGTAATATTTATTACAGAAATT-3′ |
| GSR4 | 5′-GAAATTTTTGGCTAATTATAT-3′ |
| AF | 5′-AACCTCAAGTTAGCGGCGTA-3′ |
| AR | 5′-AATCCTTGCTTGCATTTTCG-3′ |
| PF | 5′-AAAAGGATCCTAAATGGAATCAATAGCTCC-3′ (BamHI) |
| PR | 5′-ATCTCGGTATGATCTAGATCA-3′ (XbaI) |
Underlining indicates restriction sites. The names of the restriction sites are listed in parentheses following the sequences.
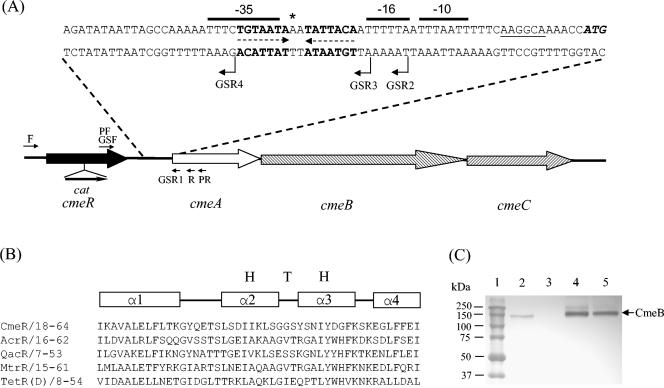
FIG. 1.
Control of cmeABC transcription by CmeR in strain 81-176. (A) Genomic organization and features of the intergenic region between cmeR-cmeABC. ORFs are indicated by boxed arrows. The start codon (ATG) of cmeA is in bold italics, and the sequences that form the IR (5′-TGTAATA-3′) are highlighted in bold and indicated by dashed arrows. The putative ribosome-binding site (AAGGCA) is underlined. The predicted −10, −16, and −35 regions of PcmeABC are overlined. The single nucleotide that was found to be deleted in CR3e is indicated by an asterisk. The locations of the key primers used for PCR are indicated by arrows. The location and orientation of the cat gene cassette (labeled Cmr) insertion in cmeR are indicated by a solid arrow. (B) Sequence alignment of the N-terminal DNA-binding domains of five repressors belonging to the TetR family. The numbers following the name of each protein indicate the amino acid numbers in each corresponding protein. The regions forming α helices are indicated by the boxes above the alignment. The HTH motif is also labeled. (C) Immunoblotting analysis of the CmeB protein in wild-type strain 81-176 and its mutants. Cell envelopes prepared from strains 81-176 (lane 2), 9B6 (lane 3; CmeB−), JL107 (lane 4; CmeR−), and CR3e (lane 5) were separated by SDS-PAGE and immunoblotted with anti-CmeB. The same amounts of total proteins were loaded in each lane. Prestained molecular mass markers (Bio-Rad) are shown in lane 1.
Sequence analysis and prediction of secondary structures.
PCR products were sequenced with an automated DNA sequencer (model 377; Applied Biosystems). Sequence analysis was performed with the Genetics Computer Group (GCG) Sequence Analysis Software Package (Oxford Molecular). The BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/) of the National Center for Biotechnology Information was used to search for homologous sequences and conserved domains in CmeR. The MOTIF program (http://motif.genome.jp) was also used to search for protein motifs in CmeR. The Peptidestructure program in GCG was used to make an initial prediction of the secondary structures of CmeR. Other programs, including SOMP and SOSUI (BCM Search Launcher Texas, Baylor College of Medicine [http://searchlauncher.bcm.tmc.edu/seq-search/struc-predict.html]), were also used to improve the prediction power.
Insertional mutation of cmeR.
An isogenic cmeR mutant was first constructed by insertional mutagenesis in strain NCTC 11168. The cmeR mutation was then introduced into strain 81-176 by natural transformation. To construct the cmeR mutant in NCTC 11168, primers F and R1, designed from the published genomic sequence (35), were used to amplify a 729-bp fragment containing the entire open reading frame (ORF) of cmeR. The PCR product was cloned into the pGEMT-Easy vector (Promega), resulting in the construction of pCMER. Since a unique BsrBRI site (which generates blunt ends) occurs in the middle of cmeR, pCMER was digested with BsrBRI to interrupt the cmeR gene. Primers CHLF and CHLR were used to amplify the 802-bp cat gene cassette from pUOA18 (49) by using Pfu Turbo DNA polymerase (Stratagene). The cat PCR product was directly ligated to BsrBRI-digested pCMER to obtain construct pCMERC. Sequencing of the construct indicated that the cat gene was inserted within the codon encoding aa 127 of CmeR in the same direction as the transcription of cmeR. The pCMERC construct, which served as a suicide vector, was electroporated into C. jejuni NCTC 11168. Transformants were selected on MH agar containing 4 μg of chloramphenicol per ml. Inactivation of the cmeR gene in the transformants by insertion of the cat gene was confirmed by PCR. To create the isogenic cmeR mutant in strain 81-176, the insertional mutation in cmeR of NCTC 11168 was transferred into strain 81-176 by natural transformation. The cmeR mutation in 81-176 was further confirmed by PCR. The cmeR mutant of 81-176 was named JL107. The levels of the CmeABC products in the cmeR mutant were determined by immunoblotting with anti-CmeB and anti-CmeC antibodies, as described previously (24). The density of each band was analyzed with the ChemilmagerIS-5500 digital imaging system (Alpha Innotech).
In vitro selection of fluoroquinolone-resistant mutants.
Fluoroquinolone-susceptible wild-type strain 81-176 was used as the parental strain for plating. Spontaneous fluoroquinolone-resistant mutants were obtained by stepwise selection on MH agar plates containing ciprofloxacin (ICN Biomedicals Inc.). At the first step, 200 μl of 2-day cultures of 81-176 containing approximately 2 × 109 CFU were plated on MH agar plates supplemented with 4 μg of ciprofloxacin per ml. Resistant colonies were selected, one of which was used for further plating with an increased ciprofloxacin concentration. The selection and plating process was repeated three times, and the final concentration of ciprofloxacin used for plating was 80 μg/ml. One clone (clone CR3e) from the final selection step was chosen for use in this study.
Susceptibility tests.
The MICs of different antimicrobials for Campylobacter were determined by a microtiter broth dilution method as described in a previous publication (24). Briefly, Campylobacter cultures were grown in MH broth to the late log phase and then diluted in MH broth to obtain an inoculum with approximately 2 × 107 CFU of bacterial cells per ml. Antimicrobial stock solutions were serially diluted twofold in 96-well microtiter plates with MH broth. The starting concentrations for the twofold dilution series were 100 mg/ml for cholic acid and choleate; 5 mg/ml for sodium dodecyl sulfate (SDS) and fusidic acid; 100 μg/ml for ciprofloxacin, norfloxacin, tetracycline, and cefotaxime; and 5 μg/ml for ampicillin, ethidium bromide, and erythromycin. The volume in each well was 120 μl. Two wells were used for each dilution of the antimicrobials. To each well, 5 μl of the bacterial inoculum was added, resulting in a final bacterial density of 8 × 105 CFU/ml. The microtiter plates were incubated for 2 days under microaerophilic conditions at 42°C. Three independent experiments were conducted to confirm the reproducibilities of the MIC patterns. The compounds used in these assays were purchased from Sigma Chemical Co. (norfloxacin, tetracycline, ampicillin, cefotaxime, erythromycin, fusidic acid, cholic acid, and choleate), ICN Biomedicals Inc. (ciprofloxacin), EM Science (SDS), and AMRESCO (ethidium bromide).
Production and purification of rCmeR.
A full-length histidine-tagged recombinant CmeR (rCmeR) was produced in E. coli by using the pQE-30 vector of the QIAexpress7 Expression System (Qiagen). The complete coding sequence of cmeR in C. jejuni 81-176 was amplified with primers RF and RR (Table 2). A restriction site (underlined in the primer sequences in Table 2) was attached to the 5′ end of each primer to facilitate the directional cloning of the amplified PCR product into the pQE-30 vector. The amplified PCR product was digested with BamHI and HindIII and was then ligated into the pQE-30 vector, which had previously been digested with BamHI and HindIII. Cloning, expression, and purification of recombinant CmeR were performed by the procedures described previously (24, 54). The plasmid in the E. coli clone producing CmeR was sequenced, with no mutations in the coding sequence of cmeR detected.
Electrophoretic mobility shift assays.
To determine the binding of CmeR to the operator region of cmeABC, electrophoretic mobility shift assays were performed by the procedure described by Alekshun et al. (2), with slight modifications. Primers GSF and GSR1 (Table 2) were used to amplify the 170-bp cmeR-cmeA IT, which was then labeled at the 3′ end with digoxigenin-11-ddUTP (DIG-11-ddUTP) by using the DIG Oligonucleotide 3′-End Labeling kit (Roche Molecular Biochemicals). An internal cmeA fragment amplified with primers AF and AR (Table 2) was labeled with DIG-11-ddUTP and was used as the control DNA for the gel shift assay. The DIG-11-ddUTP-labeled DNA fragments (0.2 pmol) were incubated with purified rCmeR in amounts ranging from 9.4 ng to 1.2 μg in 20 μl of binding buffer containing 20 mM HEPES (pH 7.6), 1 mM EDTA, 10 mM (NH4)2SO4, 5 mM dithiothreitol, 0.2% Tween 20, 30 mM KCl, and 25 ng of poly(dI-dC). The reaction mixtures were incubated at room temperature for 15 min and were then subjected to electrophoresis on a nondenaturing 6% (wt/vol) polyacrylamide gel in 0.25× TBE (22 mM Tris, 22 mM boric acid, 0.5 mM EDTA [pH 8.0]) at 200 V for 45 min. The DNA in the gels was transferred to a nylon membrane with a vacuum blotter. DIG-labeled DNA was detected and visualized by using alkaline phosphatase-conjugated anti-DIG antibody and the chemiluminescent substrate CDP-Star (Roche Molecular Biochemicals). For the competition experiments, different amounts (50-, 150-, and 300-fold molar excesses) of unlabeled DNA were added as competitors during the binding assays. To identify the specific CmeR-binding site, reverse primers GSR2, GSRR3, and GSR4 (Table 2; Fig. 1A) were used with primer GSF to generate DNA fragments spanning different portions of the IT between cmeR and cmeA. The PCR fragments were labeled with DIG and used for the DNA-binding assays, as described above.
Construction of promoter fusions.
A 640-bp DNA fragment containing PcmeABC and its flanking region was amplified from wild-type strain 81-176 with primer pair PF and PR (Table 2; Fig. 1A). The amplified PCR products were digested with BamHI and XbaI and then inserted into plasmid pMW10, a shuttle vector carrying a promoterless lacZ gene (52), to create plasmid pIT81. The same promoter region in mutant CR3e was also amplified by PCR and inserted into pMW10 to create plasmid pIT3e. Plasmids pIT81, pIT3e, and pMW10 were mobilized into various C. jejuni strains by triparental mating by using DH5α/pRK2013 (6) as the helper strain, according to the procedure described by Miller et al. (31).
β-Galactosidase assay.
The β-galactosidase (LacZ) activity in the Campylobacter strains containing the PcmeABC-lacZ transcriptional fusion was measured as described by Miller (30), with the modification that C. jejuni cultures were grown for 16 h in MH broth to log phase (absorbance at 600 nm, approximately 0.2) before they were harvested. All assays were conducted in triplicate.
Nucleotide sequence accession number.
The cmeR gene sequence of C. jejuni 81-176 determined in this study was deposited in GenBank under accession number AF466820.
RESULTS
Sequence features of cmeR and IT between cmeR and cmeABC in C. jejuni 81-176.
Analysis of the genomic sequence of C. jejuni NCTC 11168 (35) suggested that Cj0368c, an ORF immediately upstream of the cmeABC operon, likely encodes a transcription factor. The homolog of Cj0368c in strain 81-176 was amplified by PCR with primers F and R (Fig. 1A; Table 2) and subsequently sequenced and was named cmeR in this study. cmeR encodes a 210-aa protein and is transcribed in the same direction as cmeABC (Fig. 1A). The deduced amino acid sequence of CmeR in 81-176 is 99.5% identical to the encoded product of Cj0368c in strain NCTC 11168. CmeR shares sequence similarities with the members of TetR family of transcriptional repressors of efflux systems (Pfam accession number PF00440). In particular, the N-terminal region of CmeR contains a DNA-binding domain that is highly conserved among the TetR family regulators (Fig. 1B), including QacR (GenBank accession number AF053772; 50% identity in 52 aa of overlap) of S. aureus, AcrR (GenBank accession number U00734; 40% identity in 60 aa of overlap) of E. coli, and MtrR (GenBank accession number Z25797; 31% identity in 83 aa of overlap) of Neisseria gonorrhoeae. Within the domain, an α-helix-turn-α-helix (HTH) DNA-binding motif, a signature sequence of the TetR family regulators, is also present in CmeR (aa 30 to 60) (Fig. 1B). There is a 97-bp IT between cmeR and cmeA, in which an IR consisting of 7-bp half sites separated by a 2-bp spacer was identified (Fig. 1A). On the basis of the reported consensus promoter sequence of Campylobacter (52), the putative −10, −16, and −35 sequences were identified for the promoter of cmeABC in C. jejuni 81-176 (Fig. 1A). The IR is located between the predicted −10 and −35 sequences, and the −35 region partly overlaps with the half site of the IR. These sequence features suggested that CmeABC is subject to regulation and CmeR is likely a local regulator for CmeABC.
Insertional mutagenesis of cmeR increases the level of cmeABC expression.
To determine if CmeR functions as a regulator for CmeABC, CmeR was inactivated by inserting the chloramphenicol resistance gene (cat) cassette into the codon encoding aa 127 of CmeR (Fig. 1A). As shown by immunoblotting (Fig. 1C, lane 4), the level of production of CmeB in the cmeR mutant (named JL107) of strain 81-176 was substantially higher than that in the wild type. Spot densitometric analysis of the immunoblot estimated that JL107 produced approximately fivefold more CmeB than wild-type 81-176. To determine if the increased level of production of CmeB in JL107 was due to an elevated level of transcription of cmeABC, the promoter sequence (PcmeABC) of cmeABC in 81-176 was placed upstream of the promoterless lacZ gene in plasmid pMW10 to create transcriptional fusion plasmid pIT81 (Table 1), which was then transformed into wild-type strain 81-176 and JL107. As shown in Table 3, the LacZ activity in JL108 and JL109 (which carried the control plasmid pMW10) was barely detectable, indicating that the endogenous level of expression of the promoterless lacZ was low and negligible. On the basis of the measurement of β-galactosidase activity, PcmeABC was moderately active (236 Miller units) in wild-type strain 81-176 (shown for strain JL110 in Table 3), which was consistent with the fact that the CmeABC proteins were detectable by immunoblotting in the wild-type strain (Fig. 1C) (24). However, in the absence of a functional CmeR, the level of transcription of PcmeABC was elevated approximately 6.2-fold (to 1,467 Miller units; shown in strain JL111) over the wild-type level. The fold difference in the level of transcription of PcmeABC between wild-type strain 81-176 and the cmeR mutant was comparable to that from the immunoblotting results, shown in Fig. 1C. Inactivation of CmeR in strain NCTC 11168 also resulted in cmeABC overexpression (data not shown), further confirming the role of CmeR in controlling cmeABC. Together, these results indicate that CmeR represses the transcription of cmeABC and that inactivation of CmeR results in the overexpression of the efflux operon.
TABLE 3.
Effects of mutations in cmeR or the IT on cmeABC transcription
| Strain | Description | β-Galactosidase activity (Miller units)a |
|---|---|---|
| JL108 | 81-176 with pMW10 | 1.45 ± 0.25 |
| JL109 | JL107 (cmeR::cm) with pMW10 | 2.08 ± 0.30 |
| JL110 | 81-176 with pIT81 | 236 ± 2 |
| JL111 | JL107 (cmeR::cm) with pIT81 | 1,467 ± 21 |
| JL112 | 81-176 with pIT3e | 586 ± 20 |
Means of triplicate measurements ± standard deviation.
CmeR binds to the IT between cmeR and cmeA.
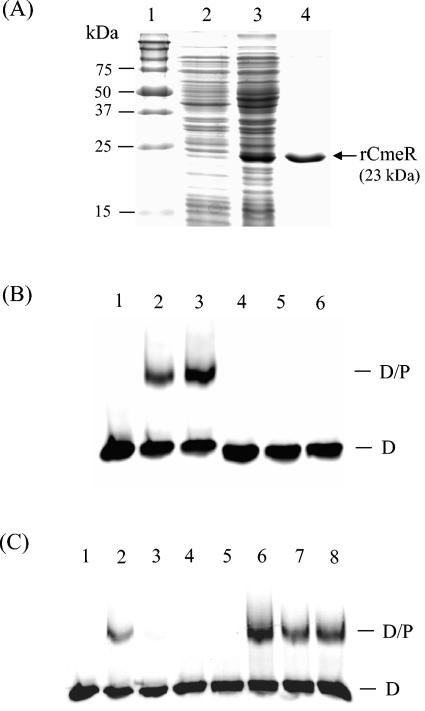
To determine if CmeR regulates the cmeABC operon via direct interaction with the promoter of cmeABC, a gel mobility shift assay was performed with rCmeR and the IT DNA amplified with primers GSF and GSR1 (Fig. 1A; Table 2). rCmeR showed a molecular mass of approximately 23 kDa on SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 2A), consistent with the calculated molecular mass from the deduced amino acid sequence of CmeR. As shown in Fig. 2B, rCmeR bound to the IT DNA but not to the control DNA, which was a 170-bp internal fragment of cmeA amplified by PCR with primers AF and AR (Table 2). The specific interaction between rCmeR and the IT DNA was further confirmed by a competition assay (Fig. 2C). The addition of a 50-fold molar excess of the unlabeled IT DNA (Fig. 2C, lane 3) completely eliminated the formation of the DNA-protein (D-P) complex, while the 170-bp control DNA did not have any effect on rCmeR binding to the IT, even at a molar excess as high as 300-fold (Fig. 2C, lanes 6 to 8). Together, these findings indicate that CmeR specifically binds to the promoter region of cmeABC. When different concentrations of rCmeR were used in the DNA-binding assay, a single retarded D-P complex was always observed on the gel (data not shown), suggesting that there is only one CmeR-binding site in the IT region.
FIG. 2.
Binding of rCmeR to the IT region between cmeR-cmeABC. (A) SDS-PAGE analysis of rCmeR produced in E. coli. Lane 1, prestained molecular mass markers (Bio-Rad); lane 2, whole-cell lysate of noninduced E. coli; lane 3, whole-cell lysate of E. coli induced with 1 mM isopropyl-β-d-thiogalactopyranoside; lane 4, rCmeR purified by Ni-nitrilotriacetic acid affinity chromatography. (B) Gel mobility shift assays with the IT DNA (lanes 1 to 3) or the control DNA (an internal cmeA fragment; lanes 4 to 6). In the DNA-binding assay, the DIG-11-dUTP-labeled DNA (0.2 pmol) was incubated with 0 ng (lanes 1 and 4), 75 ng (lanes 2 and 5), and 37.5 ng (lanes 3 and 6) of rCmeR. (C) Competition assay to determine the binding specificity of CmeR. In the DNA-binding reactions, the DIG-11-dUTP-labeled IT DNA was incubated with 0 ng (lane 1) or 37.5 ng (lanes 2 to 8) of rCmeR in the presence of the unlabeled IT DNA (lanes 3 to 5) or the control DNA (lanes 6 to 8). The unlabeled DNA was added at a 50-fold (lanes 3 and 6), 150-fold (lanes 4 and 7), or 300-fold (lanes 5 and 8) molar excess of the labeled IT DNA. The positions of the DIG-labeled IT (D) and the IT-rCmeR complex (D/P) are indicated in panels B and C.
The IR in the IT is required for specific CmeR binding.
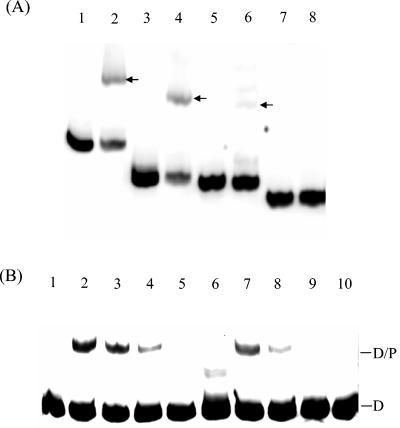
Regulators of bacterial MDR efflux systems usually bind to the IRs in their target promoter regions (14). The presence of an IR upstream of cmeABC (Fig. 1A) suggested a potential binding site for CmeR. To examine this possibility, the gel mobility shift assay was performed with a series of PCR products whose sequences span different portions of the IT region between cmeR and cmeABC (see Fig. 1A for the locations of the primers). As shown in Fig. 3A, rCmeR bound to the DNA fragments amplified by primers GSF and GSR1 or primers GSF and GSR2, and partial binding was also observed with the fragment amplified with primers GSF and GSR3. However, the PCR fragment lacking the IR derived with primers GSF and GSR4 was not bound by rCmeR, because no retarded rCmeR-DNA complex was observed on the gel (Fig. 3A, lane 8). These observations strongly indicate that the IR is the binding site for CmeR. The reduced level of binding of CmeR to the fragment obtained by PCR with primers GSF and GSR3 suggested that the immediate flanking sequence of the IR is also required for full binding by CmeR.
FIG. 3.
Localization of the specific binding site of CmeR. (A) Gel mobility shift assay with IT DNA amplified with primers GSF and GSR1 (lanes 1 and 2), GSF and GSR2 (lanes 3 and 4), GSF and GSR3 (lanes 5 and 6), or GSF and GSR4 (lanes 7 and 8). The PCR products were end labeled with DIG-11-dUTP and incubated with 0 ng (lanes 1, 3, 5, and 7) or 75 ng (lanes 2, 4, 6, and 8) of rCmeR. The positions of the retarded IT-rCmeR complex are indicated by arrows. (B) Effect of a single nucleotide deletion in the spacer of the IR on the binding of CmeR. In the gel mobility shift assay, The DIG-labeled IT DNA from wild-type strain 81-176 (lanes 1 to 5) or clone CR3e (lanes 6 to 10) was incubated with 0 ng (lanes 1 and 6), 19 ng (lanes 2 and 7), 9.5 ng (lanes 3 and 8), 4.8 ng (lanes 4 and 9), or 2.4 ng (lanes 5 and 10) of CmeR. The positions of the DIG-labeled IT (D) and the IT-rCmeR complex (D/P) are indicated on the right.
Mutation in the IR affects expression of cmeABC.
A spontaneous MDR mutant (designated CR3e) of strain 81-176, obtained by stepwise selection on ciprofloxacin-containing plates, showed high levels of resistance to fluoroquinolones and concurrently elevated levels of resistance to several other antibiotics (Table 4), although ciprofloxacin was the exclusive antimicrobial used in the stepwise selection. Immunoblotting analysis of the CmeABC proteins in CR3e indicated that the efflux proteins are overexpressed in this mutant (Fig. 1C, lane 5). To determine the mechanism responsible for the increased level of production of CmeABC in CR3e, the complete cmeR gene and the IT region between cmeR and cmeA were amplified from CR3e by PCR. Sequence analysis indicated that the cmeR gene in mutant CR3e is identical to the one in wild-type strain 81-176, indicating that mutation in CmeR was unlikely the reason for the enhanced production of CmeABC in CR3e. However, a single nucleotide deletion (marked by an asterisk in Fig. 1A) occurred between the two half sites of the IR in the promoter region of cmeABC, which was the only mutation found in the entire IT. Since the IR was the binding site for CmeR, we hypothesized that the single nucleotide deletion affected the binding by CmeR, resulting in overexpression of CmeABC in mutant CR3e. To test this hypothesis, a gel mobility shift assay was performed with the IT DNA derived from strains 81-176 and CR3e and different amounts of rCmeR. As shown in Fig. 3B, as little as 4.8 ng of rCmeR could form a detectable D-P complex with 81-176-derived IT DNA (Fig. 3B, lane 4). However, at least twice the amount of rCmeR (9.5 ng) was needed to form a visible D-P complex when the IT DNA derived from CR3e was used in the assay (Fig. 3B, lane 8). This finding was reproducible in three independent experiments and indicates that the binding of CmeR to the promoter region of cmeABC in CR3e is reduced due to the single nucleotide deletion between the half sites of the IR. To further examine the impact of the single nucleotide deletion on the transcription of cmeABC, the PcmeABC sequence bearing the single deletional mutation was transcriptionally fused to the promoterless lacZ in pMW10 to create pIT3e, which was then introduced into wild-type strain 81-176 to create strain JL112 (Table 1). As shown in Table 3, the β-galactosidase activity was approximately 2.5-fold higher in JL112 (which contained the mutated promoter-lacZ fusion) than in JL110 (which contained the wild-type promoter-lacZ fusion), indicating that the mutated PcmeABC is more active than the wild-type PcmeABC and that the single deletional mutation increases the level of transcription of cmeABC in CR3e.
TABLE 4.
Susceptibilities of C. jejuni 81-176, JL107, and CR3e to different antimicrobials
| Antimicrobial | MIC (μg/ml)a
|
||
|---|---|---|---|
| 81-176 | JL107 | CR3e | |
| Ciprofloxacin | 0.390 | 0.780 (2) | 100 (256) |
| Norfloxacin | 0.098 | 0.196 (2) | 6.25 (64) |
| Tetracycline | 0.098 | 0.098 (−) | 0.196 (2) |
| Ampicillin | 0.625 | 0.625 (−) | 2.5 (4) |
| Cefotaxime | 0.390 | 0.780 (2) | 1.56 (4) |
| Erythromycin | 0.039 | 0.156 (4) | 0.156 (4) |
| Ethidium bromide | 0.625 | 0.625 (−) | 0.625 (−) |
| Fusidic acid | 39 | 78 (2) | 78 (2) |
| Cholic acid | 3,125 | 3,125 (−) | 3,125 (−) |
| Choleate | 12,500 | 12,500 (−) | 25,000 (2) |
| SDS | 156 | 156 (−) | 156 (−) |
The numbers in parentheses indicate the fold differences in MICs between 81-176 and its mutant derivatives. −, no MIC difference was observed.
Overexpression of cmeABC correlates with enhanced resistance to multiple antibiotics.
Compared to wild-type strain 81-176, the CmeR mutant (strain JL107) showed enhanced resistance to several antibiotics (Table 4). The MICs of ciprofloxacin, norfloxacin, cefotaxime, and fusidic acid for JL107 increased twofold and the MIC of erythromycin for JL107 increased fourfold. The MIC results and the moderate differences in the MICs for the cmeR mutant and those for wild-type strain 81-176 were reproducible in three independent experiments. Seven consecutive passages (∼168 generations) of JL107 in MH broth showed the same MIC changes, indicating that the phenotype is stable. Although tetracycline, ampicillin, ethidium bromide, cholic acid, choleate, and SDS are substrates of CmeABC (24), mutant JL107 showed only a wild-type level of resistance to these substances. Overexpression of cmeABC in spontaneous mutant CR3e also appeared to be correlated with the MDR phenotype of the mutant. Although the Thr-86-Ile point mutation was present in the gyrA gene of CR3e (data not shown), the exceedingly high level of resistance to ciprofloxacin (Table 4) may be explained by the overexpression of cmeABC. In addition, the enhanced resistance of CR3e to tetracycline, ampicillin, cefotaxime, erythromycin, and fusidic acid was at least partially attributable to the overexpressed cmeABC.
DISCUSSION
This work demonstrates that CmeR functions as a transcriptional repressor for CmeABC and that the interaction of CmeR with the IR immediately upstream of cmeABC regulates the expression of this MDR operon. This conclusion is based on the following evidence. First, CmeR shares significant sequence and structural homologies with known repressors belonging to the TetR family of transcriptional regulators. Second, inactivation of CmeR by insertional mutagenesis substantially increased the level of transcription of cmeABC and, consequently, enhanced the level of production of the CmeABC proteins (Table 3 and Fig. 1C). Third, CmeR specifically bound to the unique IR upstream of cmeA, as shown by the gel mobility shift assay (Fig. 2 and Fig. 3A). Finally, a mutation in the IR (a 1-bp deletion between the two half sites) significantly reduced the level of binding by CmeR (Fig. 3B) and resulted in enhanced transcription (Table 3) and translation (Fig. 1C) of the efflux operon. Together, these findings formally define the critical role of CmeR and its specific binding site on the regulation of CmeABC in C. jejuni.
CmeR represses the transcription of cmeABC, but it also allows a moderate level of production of the efflux proteins in wild-type strains in the absence of antibiotics. This feature is different from the control of TetA by TetR, in which the basal level of expression of tetA is minimal in the absence of tetracycline (14). The difference is probably due to the fact that the TetA pump is specific for tetracycline and constitutive expression of tetA is not required in the absence of the substrate (19). In contrast, the other efflux pumps that are controlled by repressors of the TetR family and that have a broad spectrum of substrates (e.g., AcrAB, MtrCDE, and QacA) are expressed at substantial levels in wild-type strains even in the absence of specific substrates (12, 15, 48). This relatively high basal level of expression of the MDR pumps is probably required for their key roles in conferring intrinsic resistance to different antimicrobials and toxic compounds and facilitates the adaptation of bacterial organisms to environmental changes. In addition, bacterial MDR pumps are likely required for extrusion of endogenous toxic metabolites (18, 37), which also necessitates constitutive expression of the efflux pumps even in the absence of exogenous selection pressure. On the other hand, the overproduction of efflux pumps in the absence of selection pressure or substrates has been demonstrated to be deleterious to some organisms (7, 32, 43). Therefore, there is a need for regulatory systems to modulate the expression of MDR efflux pumps in bacteria. In this respect, CmeR acts as a moderator in Campylobacter to maintain balanced production of CmeABC to meet the physiological needs and facilitate the adaptation of Campylobacter to environmental changes, including antibiotic treatments.
The cmeR gene was inactivated by the insertion of an antibiotic resistance gene cassette in the middle of the ORF (Fig. 1). Although there was a possibility that the truncated N-terminal portion of CmeR was still produced in the mutant strain, it is unlikely that the truncated version of CmeR was functional in repressing the transcription of cmeABC. CmeR belongs to the TetR family, and the members of this family function as dimers. Although the DNA-binding motif is located in the N-terminal portion, the C-terminal portion is essential for dimer formation (14). Deletion of the C-terminal portion of the regulatory protein in the TetR family would prevent the formation of dimers and, consequently, would affect the binding to target DNA. Hence, the truncated CmeR, even if it were produced in the mutant strain, is not expected to perform the repressor function, as is the case with the full-length CmeR. This argument is directly supported by the findings that the CmeR mutant showed a significant increase in the level of transcription of cmeABC and the level of production of the efflux proteins (Table 3 and Fig. 1). Regardless of the expression status of the truncated CmeR in the mutant, the results from this study demonstrate that CmeR functions as a repressor for CmeABC. To determine if CmeR directly bound to the promoter sequence of cmeABC, His-tagged rCmeR was produced in E. coli and was used for the gel mobility shift assay. His-tagged recombinant proteins have commonly been used to assess the binding of MDR pump repressors to target DNA (9, 12, 42). It was unlikely that the His tag attached to rCmeR had any effect on DNA binding because rCmeR did not bind to the negative control DNA (internal cmeA fragment), while it bound specifically to the promoter DNA of cmeABC (Fig. 2). In addition, the binding specificity was further verified by competition with the nonlabeled promoter DNA and mutation of the binding site (Fig. 2 and 3).
The IR is a typical DNA motif for binding sites of regulatory proteins (14). It has been known that the correct spacing between the two half sites of an IR is critical for binding by repressors (13, 51). For example, Wissmann et al. (51) showed that a 1-bp increase or decrease in the single-base-pair spacing between the two half sites of the tet operator decreased the affinity of the operator sequence to TetR. Another study with staphylococcal QacR (13) also demonstrated that binding of QacR was dependent on correctly spaced operator half sites. In this study, we found that CmeR bound specifically to the IR in the promoter region of CmeABC (Fig. 3) and that a single nucleotide deletion between the two half sites of the IR reduced the level of CmeR binding to the promoter sequence of cmeABC (Fig. 3B) and led to a 2.5-fold increase in the level of transcription of PcmeABC (Table 3). On the basis of these findings, we can confidently link the enhanced expression of cmeABC in mutant CR3e to the single nucleotide deletion in the IR. The sequence feature of the IR upstream of cmeABC is similar to that of the IR bound by TetR, which comprises 9-bp half sites separated by a 1-bp spacer (14, 51), but is different from that of the large QacR-binding region comprising 15-bp half sites separated by a 6-bp spacer (12). On the basis of the sequence analogy of the binding sites and the known binding mechanisms of TetR and QacR (13, 20), it is speculated that CmeR may bind to its operator as a dimer in a way similar to that of TetR rather than to a pair of dimers, as is the case with QacR (13). This speculation remains to be examined in future studies.
Overexpression of MDR efflux pumps mediated by mutations in their regulatory elements is usually associated with acquired resistance to multiple antibiotics in bacteria (36, 37, 50). The results from this study also indicate that overexpression of CmeABC increases the levels of resistance of Campylobacter to several antimicrobials (Table 4). On the basis of the MIC, the enhanced resistance in the isogenic CmeR mutant (strain JL107) was moderate, but the differences were reproducible in independent experiments. At this stage it is unclear why the overexpression of CmeABC mediated by the CmeR mutation did not cause large changes in the MICs for the CmeR mutant. There is a possibility that CmeR also regulates other unidentified genes and that inactivation of CmeR may have pleiotropic effects on gene expression in Campylobacter, which potentially obscures the changes in MICs. This possibility is being examined in our laboratory. Nevertheless, the relatively small-scale increase in the resistance profiles of the CmeR mutant is not totally surprising, because it has been found in other bacteria that overexpression of a single MDR pump caused by mutations in its local repressor may not confer drug resistance to a level of clinical significance (21, 38, 47). However, overexpression of MDR pumps may allow bacteria to survive under the pressure of high antibiotic concentrations and promote the emergence of mutants with specific target gene mutations that are highly resistant to antimicrobials (46, 50). The contribution of CmeABC overexpression to the acquired antibiotic resistance in Campylobacter remains to be determined in future studies.
In mutant CR3e, which was obtained by stepwise selection on ciprofloxacin-containing plates, the high-level resistance to fluoroquinolones (Table 4) was expected because the mutant contained the specific GyrA mutation (Thr-86-Ile), which, in conjunction with the function of CmeABC, confers a high level of resistance to fluoroquinolones (26). The enhanced resistance of CR3e to other antimicrobials (Table 4) is at least partially attributable to the overexpression of CmeABC in the mutant. However, there is a possibility that other unknown mutations might also have occurred, and these might also have contributed to the increased level of antibiotic resistance in CR3e. Due to technical difficulties, our effort to introduce the single nucleotide deletion in the cmeABC promoter of CR3e into wild-type 81-176 by using natural transformation or electroporation was not successful. Thus, the involvement of unknown mutations in the enhanced resistance in CR3e cannot be totally excluded at this stage.
The expression of MDR efflux pumps can be conditionally induced by structurally diverse substrates of these pumps (1, 5, 12, 22, 23, 28, 29, 41). This induction is due to the direct interaction of the substrates with repressor molecules, which interferes with the binding of repressors to operator DNA and which results in increased levels of expression of MDR pump genes. Transcriptional regulators of the TetR family are characterized by a conserved HTH-containing DNA-binding domain at the N-terminal region and a divergent C-terminal sequence that is involved in the binding to inducing compounds (12, 19, 20). The variation in the C-terminal sequence reflects the diversity of substrates that can interact with the regulators. A conformational change occurs in the DNA-binding domain when an inducer binds to the C-terminal region of a repressor, releasing the inhibition to efflux pumps. Although CmeABC is the key pump for bile resistance in Campylobacter (25), no differences in the MICs of cholic acid and choleate were observed for the wild-type strain 81-176 and strain JL107 (an isogenic cmeR mutant of strain 81-176) (Table 4). Considering the fact that bile salts induce AcrAB expression in E. coli (41), it is possible that bile salts also induce cmeABC expression in Campylobacter. If this induction indeed occurs, it would obscure the differences in the MICs of bile salts for wild-type strain 81-176 and the CmeR mutant due to the enhanced expression of cmeABC in strain 81-176 in the presence of bile salts. At this stage, it is unclear if the expression of cmeABC is inducible and if any substrates directly interact with CmeR. Since we have constructed the transcriptional reporter system (PcmeABC-lacZ) and established the gel mobility shift assay using rCmeR, the induction of cmeABC under various conditions can now be examined in a definitive manner.
Acknowledgments
We thank W. G. Miller and R. E. Mandrell for supplying plasmids pMW10 and pRK2013 required for this study.
This work was supported by National Institutes of Health grant DK063008.
REFERENCES
- 1.Ahmed, M., C. M. Borsch, S. S. Taylor, N. Vazquez-Laslop, and A. A. Neyfakh. 1994. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 269:28506-28513. [PubMed] [Google Scholar]
- 2.Alekshun, M. N., Y. S. Kim, and S. B. Levy. 2000. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol. Microbiol. 35:1394-1404. [DOI] [PubMed] [Google Scholar]
- 3.Baranova, N., and H. Nikaido. 2002. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 184:4168-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 5.Brooun, A., J. J. Tomashek, and K. Lewis. 1999. Purification and ligand binding of EmrR, a regulator of a multidrug transporter. J. Bacteriol. 181:5131-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckert, B., and C. F. Beck. 1989. Overproduction of transposon Tn10-encoded tetracycline resistance protein results in cell death and loss of membrane potential. J. Bacteriol. 171:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engberg, J., F. M. Aarestrup, D. E. Taylor, P. Gerner-Smidt, and I. Nachamkin. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, K., L. Adewoye, and K. Poole. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fournier, B., R. Aras, and D. C. Hooper. 2000. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J. Bacteriol. 182:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. ASM Press, Washington, D.C.
- 12.Grkovic, S., M. H. Brown, N. J. Roberts, I. T. Paulsen, and R. A. Skurray. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273:18665-18673. [DOI] [PubMed] [Google Scholar]
- 13.Grkovic, S., M. H. Brown, M. A. Schumacher, R. G. Brennan, and R. A. Skurray. 2001. The staphylococcal QacR multidrug regulator binds a correctly spaced operator as a pair of dimers. J. Bacteriol. 183:7102-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grkovic, S., M. H. Brown, and R. A. Skurray. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagman, K. E., C. E. Lucas, J. T. Balthazar, L. Snyder, M. Nilles, R. C. Judd, and W. M. Shafer. 1997. The MtrD protein of Neisseria gonorrhoeae is a member of the resistance/nodulation/division protein family constituting part of an efflux system. Microbiology 143:2117-2125. [DOI] [PubMed] [Google Scholar]
- 16.Hagman, K. E., W. Pan, B. G. Spratt, J. T. Balthazar, R. C. Judd, and W. M. Shafer. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611-622. [DOI] [PubMed] [Google Scholar]
- 17.Hagman, K. E., and W. M. Shafer. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J. Bacteriol. 177:4162-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helling, R. B., B. K. Janes, H. Kimball, T. Tran, M. Bundesmann, P. Check, D. Phelan, and C. Miller. 2002. Toxic waste disposal in Escherichia coli. J. Bacteriol. 184:3699-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345-369. [DOI] [PubMed] [Google Scholar]
- 20.Hinrichs, W., C. Kisker, M. Duvel, A. Muller, K. Tovar, W. Hillen, and W. Saenger. 1994. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science 264:418-420. [DOI] [PubMed] [Google Scholar]
- 21.Jellen-Ritter, A. S., and W. V. Kern. 2001. Enhanced expression of the multidrug efflux pumps AcrAB and AcrEF associated with insertion element transposition in Escherichia coli mutants selected with a fluoroquinolone. Antimicrob. Agents Chemother. 45:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaatz, G. W., and S. M. Seo. 1995. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 39:2650-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, S., J. Nishimura, Y. Yufu, H. Ideguchi, T. Umemura, and H. Nawata. 1992. Modulation of expression of multidrug resistance gene (mdr-1) by adriamycin. FEBS Lett. 308:175-178. [DOI] [PubMed] [Google Scholar]
- 24.Lin, J., L. O. Michel, and Q. Zhang. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46:2124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, J., O. Sahin, L. Michel, and Q. Zhang. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 71:4250-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo, N., O. Sahin, J. Lin, L. O. Michel, and Q. Zhang. 2003. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob. Agents Chemother. 47:390-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 28.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45-55. [DOI] [PubMed] [Google Scholar]
- 29.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Miller, W. G., A. H. Bates, S. T. Horn, M. T. Brandl, M. R. Wachtel, and R. E. Mandrell. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen, T. N., Q. G. Phan, L. P. Duong, K. P. Bertrand, and R. E. Lenski. 1989. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol. Biol. Evol. 6:213-225. [DOI] [PubMed] [Google Scholar]
- 33.Nishino, K., and A. Yamaguchi. 2001. Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 183:1455-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishino, K., and A. Yamaguchi. 2002. EvgA of the two-component signal transduction system modulates production of the yhiUV multidrug transporter in Escherichia coli. J. Bacteriol. 184:2319-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 36.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob. Agents Chemother. 44:2233-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poole, K. 2001. Multidrug resistance in gram-negative bacteria. Curr. Opin. Microbiol. 4:500-508. [DOI] [PubMed] [Google Scholar]
- 38.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. E. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pumbwe, L., and L. J. Piddock. 2002. Identification and molecular characterisation of CmeB, a Campylobacter jejuni multidrug efflux pump. FEMS Microbiol. Lett. 206:185-189. [DOI] [PubMed] [Google Scholar]
- 40.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberg, E. Y., D. Bertenthal, M. L. Nilles, K. P. Bertrand, and H. Nikaido. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48:1609-1619. [DOI] [PubMed] [Google Scholar]
- 42.Saito, K., H. Yoneyama, and T. Nakae. 1999. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 179:67-72. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez, P., J. F. Linares, B. Ruiz-Diez, E. Campanario, A. Navas, F. Baquero, and J. L. Martinez. 2002. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother. 50:657-664. [DOI] [PubMed] [Google Scholar]
- 44.Smith, K. E., J. M. Besser, C. W. Hedberg, F. T. Leano, J. B. Bender, J. H. Wicklund, B. P. Johnson, K. A. Moore, and M. T. Osterholm. 1999. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992-1998. N. Engl. J. Med. 340:1525-1532. [DOI] [PubMed] [Google Scholar]
- 45.Trieber, C. A., and D. E. Taylor. 2000. Mechanisms of antibiotic resistance in Campylobacter, p. 441-454. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. American Society for Microbiology, Washington, D.C.
- 46.Van Bambeke, F., E. Balzi, and P. M. Tulkens. 2000. Antibiotic efflux pumps. Biochem. Pharmacol. 60:457-470. [DOI] [PubMed] [Google Scholar]
- 47.Veal, W. L., R. A. Nicholas, and W. M. Shafer. 2002. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J. Bacteriol. 184:5619-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, H., J. L. Dzink-Fox, M. Chen, and S. B. Levy. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 45:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 50.Webber, M. A., and L. J. Piddock. 2003. The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother. 51:9-11. [DOI] [PubMed] [Google Scholar]
- 51.Wissmann, A., I. Meier, and W. Hillen. 1988. Saturation mutagenesis of the Tn10-encoded tet operator O1. Identification of base-pairs involved in Tet repressor recognition. J. Mol. Biol. 202:397-406. [DOI] [PubMed] [Google Scholar]
- 52.Wosten, M. M., M. Boeve, M. G. Koot, A. C. van Nuene, and B. A. van der Zeijst. 1998. Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, Q., J. Lin, and S. Pereira. 2003. Fluoroquinolone-resistant Campylobacter in animal reservoirs: dynamics of development, resistance mechanisms, and ecological fitness. Anim. Health Res. Rev. 4:63-71. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Q., J. C. Meitzler, S. Huang, and T. Morishita. 2000. Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter major outer membrane protein. Infect. Immun. 68:5679-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]