Abstract
Purpose
Most non-small cell lung cancers (NSCLCs) are now diagnosed from small specimens, and classification using standard pathology methods can be difficult. This is of clinical relevance as many therapy regimens and clinical trials are histology dependent. The purpose of this study was to develop an mRNA expression signature as an adjunct test for routine histo-pathological classification of NSCLCs.
Experimental Design
A microarray dataset of resected adenocarcinomas (ADC) and squamous cell carcinomas (SCC) was used as the learning set for an ADC-SCC signature. The Cancer Genome Atlas (TCGA) lung RNAseq dataset was used for validation. Another microarray dataset of ADCs and matched non-malignant lung was used as the learning set for a Tumor vs. Nonmalignant signature. The classifiers were selected as the most differentially expressed genes and sample classification was determined by a nearest distance approach.
Results
We developed a 62-gene expression signature that contained many genes used in immunostains for NSCLC typing. It includes 42 genes that distinguish ADC from SCC and 20 genes differentiating non-malignant lung from lung cancer. Testing of the TCGA and other public datasets resulted in high prediction accuracies (93–95%). Additionally, a prediction score was derived that correlates both with histologic grading and prognosis. We developed a practical version of the Classifier using the HTG EdgeSeq nuclease protection-based technology in combination with next-generation sequencing that can be applied to formalin-fixed paraffin-embedded (FFPE) tissues and small biopsies.
Conclusions
Our RNA classifier provides an objective, quantitative method to aid in the pathological diagnosis of lung cancer.
Keywords: lung cancer classification, adenocarcinoma, squamous carcinoma, microarray, expression signatures
INTRODUCTION
Most cancers of the lung are carcinomas, and they may be divided into two broad categories, small cell carcinoma (SCLC, 10–20%) and non-small cell carcinoma (NSCLC, 80–90%), differing in their biology, clinical presentation and therapy (1). While there are rare types of NSCLC, the vast majority fall into three categories: adenocarcinomas (ADC), squamous cell carcinomas (SCC), and large cell carcinomas (LCC). ADCs are carcinomas that form glands, papillary structures, grow in a lepidic pattern, or secrete mucin. As the lung is a complex organ with central and peripheral compartments having different histologies and functions (2), there are multiple subtypes of ADCs (3). SCCs are believed to arise from metaplastic cells in the large airways, as there are no squamous cells in the normal respiratory tract. LCCs are undifferentiated NSCLCs that do not show morphologic or immunostain evidence of glandular or squamous differentiation. Recent studies confirmed that most or all LCCs lacking neuroendocrine features could be assigned to other NSCLC types, with only a small number of true null phenotype cases remaining (4, 5).
Originally, the clinical management of the major forms of NSCLC was similar, and the main clinical task required of the pathologist was the separation of NSCLC from SCLC (6). However during the past decade the therapy of NSCLC has undergone a paradigm shift, as we are rapidly moving from an era of empirical therapy to one of personalized therapy (“precision medicine”) based on mutational patterns and tumor classification (7). Of interest, the known oncogenic “driver” mutations for ADC and SCC are almost completely different (8) and the selection of both conventional chemotherapy and targeted therapy may be influenced by the NSCLC subtype (6, 9). While most targeted therapies for NSCLC are directed at ADCs or non-squamous histologies, the importance of the correct diagnosis of squamous tumors by pathological or molecular methods is gaining recognition (10). Hence the precise histologic classification of NSCLC is of crucial clinical importance.
Other major developments in lung cancer management are the ability to obtain computed tomography-guided needle and small core biopsies (usually allowing a pathologic diagnosis to be made irrespective of anatomic location) and imaging studies that have greatly increased the accuracy of preoperative staging. These advances have considerably reduced the number of futile lung cancer resections and staging that relied on post operative pathologic results. However, one practical disadvantage is that most initial lung cancer diagnoses (~70%) now have to be made from small biopsies or cytological specimens (6). While the pathological classification of NSCLC is relatively straightforward if there is an adequate tumor specimen and the tumor is well or moderately differentiated, the diagnosis of poorly differentiated tumors, particularly from small specimens, may be challenging. Following recommendations by the latest version of the WHO Classification (1) pathologists now use combinations of immunostains, and various algorithms have been proposed to assist in the accurate classification of NSCLC (3, 9, 11, 12). Despite these improvements, not all poorly differentiated NSCLCs in small biopsy or cytology samples can be classified into ADCs and SCCs and they are usually referred to as NSCLC-not otherwise specified (NSCLC-NOS) (6, 9). As a side note, this term should be reserved to the lung cancer specimens of small size and should not be applied to surgical resections or large biopsies; instead, resected tumors that cannot be classified as specific forms of NSCLC should be classified as LCC (13).
In order to improve the classification of lung cancer and reduce potential observer bias and variability, we developed and validated an mRNA expression-based classification of NSCLC utilizing large datasets of resected NSCLC tumors with expert pathology review. We have further developed a practical version of the Classifier based on the HTG EdgeSeq technology that, among other advantages, can be applied reproducibly to FFPE and core needle biopsies.
MATERIALS AND METHODS
Patient tumor samples
Three cohorts of lung cancer or non-malignant lung specimens were used to derive and test the RNA classifiers: A set of 275 NSCLC specimens obtained from the Pathology Core at MD Anderson Cancer Center (MDACC), consisting of 183 ADC, 80 SCC, and 12 other subtypes; a set of 83 pairs of lung ADCs and matched non-malignant lung tissues obtained from British Columbia Cancer Research Centre in collaboration with Early Detection Research Network (EDRN) and the Canary Foundation; and a set of 979 NSCLC (490 ADC, 489 SCC) and non-malignant lung tissues (n=108) from The Cancer Genome Atlas (TCGA). Reference pathologists for the tumor specimen diagnoses were Ignacio Wistuba and Jaime Rodriguez for the MDACC set, Adi Gazdar for the EDRN/Canary set, and the TCGA Lung Cancer Pathology Panel (chaired by William Travis and including Natasha Rekhtman) for the TCGA set. For most of the tumors analyzed in this study, immunostaining was not utilized for pathologic classification. Histologic typing of the TCGA set was performed by light microscopy according to the previous WHO classification (14).
Expression Profiling Datasets
MDACC set
Frozen tissues from NSCLC tumors resected at MDACC were used to generate multiple 5-micron thick sections. Representative tissue sections were hematoxylin and eosin (H&E)-stained and reviewed to estimate the percentage of tumor and non-malignant cells. About 5–10 sections were processed to extract RNA, whose quality was assessed on Nano Series II RNA LAB-chips using Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA). Cases were selected with the following defined characteristics: tumor (vs. non-malignant) ≥ 70%, malignant cells (vs. stromal cells) ≥ 30%, RNA Integrity Number (RIN) ≥ 4 (range 0–10). RNA samples were shipped to UT Southwestern for expression profiling. Five hundred nanograms of RNA were labeled and hybridized to the Illumina Beadchip array HumanWG-6 V3 (San Diego, CA). Array data were pre-processed using the R package mbcb (15) for background correction. The arrays were then log-transformed and quantile-normalized. This dataset was submitted to Gene Expression Omnibus (GEO) under the accession GSE41271.
EDRN/Canary set
RNA was prepared with TRIzol (Invitrogen) as previously described from ADCs resected in British Columbia, Canada and collected by Dr. Stephen Lam (16, 17). Profiling was done on Illumina HumanWG-6 V3 at UT Southwestern and processed similarly to the MDACC set. This dataset was submitted to GEO under the accession GSE75037.
TCGA Set
RNAseq data were downloaded from the TCGA portal (18). The archive filenames were unc.edu_LUAD.IlluminaHiSeq_RNASeqV2.1.13.0 for ADC and unc.edu_LUSC.IlluminaHiSeq_RNASeqV2.1.10.0 for SCC. The extracted files consisted of 548 and 539 samples respectively. Using the barcode key provided (19) we found that the first set had 490 ADCs and 58 non-malignant lung samples and the second set had 489 SCCs and 50 non-malignant samples.
FFPE Material
FFPE specimens from resected NSCLC (n=35), non-malignant lung tissue (n=11) or core needle biopsies (n=36) were collected at MDACC. They were then cut in a microtome at 4-micron tick sections, mounted on glass slides using nuclease-free conditions, and sent to HTG labs for EdgeSeq profiling (see below).
Statistical Analysis
A Sweave report documenting all statistical steps (written in R code) pertaining to this manuscript is available from the authors on request. In brief, classifiers were generated from the training set’s two classes by generating a volcano plot and selecting the n most significantly overexpressed genes in the two classes, where n is optimized by 5-fold stratified cross validation with 100 iterations (which resulted in n=21 for the ADC-SCC classifier, and n=10 for the Tumor-Nonmalignant classifier, see Supplementary Figure 1). We thus obtained a 2n-gene classifier (42 genes for ADC-SCC classification, 20 genes for Tumor-Nonmalignant classification). Classification was determined by a nearest distance approach that compares each sample’s expression values for the classifier genes to the mean expression values in the training set’s two classes (called class centroids). Pearson correlation was used as a similarity measure. The relative magnitude of this measure determined the class prediction. i.e. if the Pearson correlation was greater with the ADC centroid than with the SCC centroid, then the sample was predicted to be ADC, and vice versa. A correlation plot was generated from each sample’s correlation pair (correlation with ADC, correlation with SCC). A score for each sample was calculated as the signed distance from its plotted location to the diagonal where the correlations are equal. After normalization, this score ranged from −1 to +1. A positive score indicates ADC prediction while a negative score indicates SCC prediction. The magnitude of these scores can be viewed as an estimate of the prediction’s confidence.
HTG EdgeSeq assay
The assay couples quantitative nuclease protection (qNPA) with Next-Generation Sequencing (NGS) to measure gene expression in small FFPE or frozen samples without RNA extraction (20). A more detailed description of the assay is available in the Supplementary Material. Briefly, the FFPE specimens from MDACC were scraped into tubes and lysed in HTG’s lysis buffer, followed by the introduction of gene-specific DNA nuclease protection probes (NPPs). After allowing the NPPs to hybridize to their target RNAs, which can be both soluble or cross-linked in the biological matrix, S1 nuclease is added which removes excess unhybridized NPPs and RNAs, leaving behind only NPPs hybridized to their target RNAs. Thus, a stoichiometric conversion of the target RNA to the DNA nuclease protection probes is achieved, producing a virtual 1:1 ratio of NPP to RNA. The qNPA steps are automated on the HTG EdgeSeq processor, which is followed by PCR to add sequencing adaptors and tags. The labeled samples are pooled, cleaned, and sequenced on an NGS platform using standard protocols. Data from the NGS instrument are processed and reported by the HTG EdgeSeq parser software. Supplementary Fig. 6 shows an example of the assay results for 25 FFPE samples used in this study. Good dynamic range and reproducibility were obtained, which indicates that the assay is both sensitive and specific.
RESULTS
SEER-based classification of NSCLC
We examined the Surveillance, Epidemiology and End Results (SEER) database for lung cancer classifications which covers the years 2008–2012 (21) and identified 227,000 lung cancer cases, of which 83.4% were classified as NSCLC, 13.3% as SCLC, and 3.1% were unclassified (Carcinoma-NOS). Of the NSCLCs, 51.9% were ADCs, 27.1% were SCCs, 2.5% were LCCs, 5.9% were other forms of NSCLC, and 12.6% were not classified (NSCLC-NOS). From these figures, a total of 13.6% of lung cancer cases, which after sampling adjustment represent about 22,000 patients yearly, were not histologically classified. These patients therefore were not eligible for histology type-based therapies.
ADC-SCC Signature: Training on the MDACC set
To build a classifier distinguishing ADC and SCC, we used the MDACC dataset which contains mRNA profiles for 183 ADCs and 80 SCCs from surgically resected frozen specimens. Forty-two genes differentially expressed between the two subtypes were selected from a volcano plot (Fig. 1A and B, Supplementary Table 1). These genes were highly significant with expression fold differences > 2.6 and T test p-value < 10−18 (FDR < 10−16). Some of the proteins encoded by the significant genes are already used as immunostains in clinical diagnostic procedures and include high molecular weight keratins (KRTs), NKX2-1 (TITF1), TP63 and DSG3 (desmoglein 3).
Figure 1.
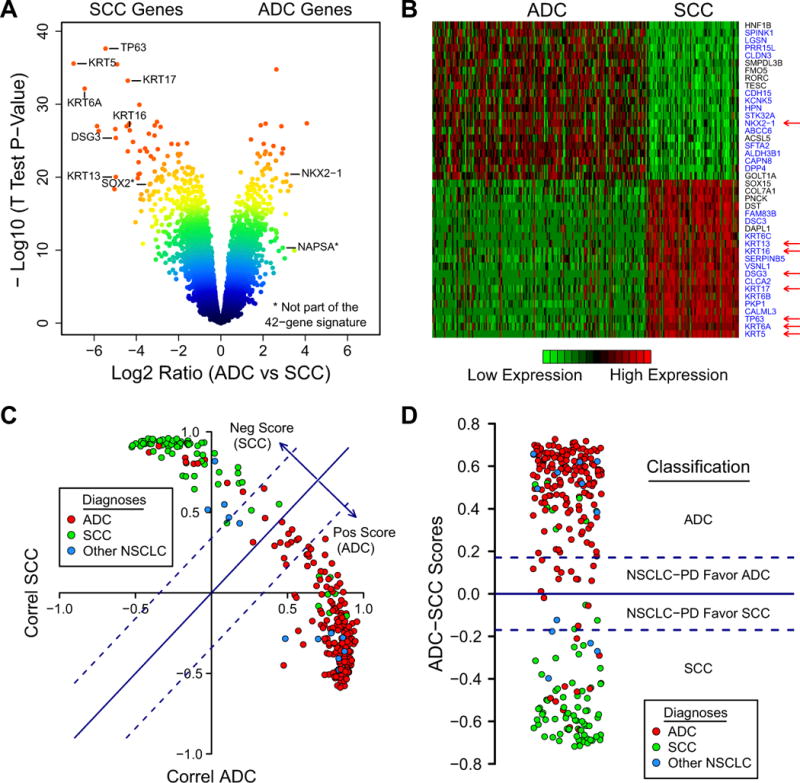
(A) A volcano plot shows that many genes are significantly different between ADC and SCC. Among these are several genes for immunostains typically used by pathologists, including high molecular weight keratins (KRTs), TP63, DSG3, and TITF1 (NKX2-1). Color-coding is related to the distance of each point to the plot’s origin and shows significance levels (red: highly significant). (B) Heatmap of ADC-SCC signature in the MDACC dataset. Red, high mRNA expression; green, low mRNA expression. Twenty-one genes overexpressed in ADC and twenty-one genes overexpressed in SCC were selected for this signature. Blue labels, genes known to be relevant to lung cancer pathogenesis. Arrows, commonly used immunostains; (C) “Correlation plot” where each point represents the Pearson correlation values between the 42-gene signature expression of individual samples and the mean expression of ADCs (x-axis) and SCCs (y-axis). The scores shown as arrows are defined as (Correl ADC − Correl SCC)/2. They are proportional to the distances from each point to the line y = x. Dotted lines represent cutoff score values below which the samples are thought to be less well-differentiated. Color-coding in this and similar plots represents pathological diagnoses. (D) “Score plot” where the y-axis represents the scores calculated from (C). PD, Poorly Differentiated. Dotted lines are the score cutoffs.
We defined the Classifier using this training set as follows: We first calculated the Pearson correlations between each sample’s 42-gene signature expression values and the mean expression values of the same 42 genes in the ADC group and in the SCC group (the class centroids). A “correlation plot” was generated (Fig. 1C) where each point represents the pair of correlation values associated with each sample. On the straight line y = x the two correlations are equal. Below this line, the correlation with the ADC group is higher than with the SCC group, and vice-versa. For each point we defined a score as
This score is proportional to the distance from each point to the dividing line (arrows in Fig 1C; also plotted vertically in Fig 1D). We interpret positive scores as predicting ADC histology while negative scores predict SCC histology. The two dotted lines are cutoff scores set at
where sd is the standard deviation and abs is the absolute value. This cutoff is equal to +/− 0.17 in the MDACC set and can be viewed as a prediction threshold: values above 0.17 are predicted to be ADC while values below −0.17 are predicted to be SCC. Intermediate values are predicted as poorly differentiated (see below). Using these definitions 170 of 183 ADCs (93%; original histologic review) had positive scores and were thus correctly classified, while 72 of 80 SCCs (90%) had negative score and were corrected classified. Overall the accuracy within this training set was 92% (Table 1).
Table 1.
Prediction accuracies of ADC-SCC and Tumor-Nonmalignant Signatures
| Signature Type | Dataset | Expression Platform | Training or Testing | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|---|---|
| ADC-SCC | SPORE/MDACCa | Illumina WG-6 V3 | Training | 170/183 (93%)c | 72/80 (90%)d | 242/263 (92%) |
| ADC-SCC | SPORE/MDACCb | Illumina WG-6 V3 | Training | 170/178 (96%) | 78/83 (94%) | 248/261 (95%) |
| ADC-SCC | TCGAa | RNAseq | Testing | 475/490 (97%) | 456/489 (93%) | 931/979 (95%) |
| ADC-SCC | TCGAb | RNAseq | Testing | 423/437 (97%) | 437/453 (96%) | 860/890 (97%) |
| ADC-SCC | EDRN/Canary | Illumina WG-6 V3 | Testing | 82/83 (99%) | NA | 82/83 (99%) |
|
| ||||||
| Tumor-Nonmalignant | EDRN/Canary | Illumina WG-6 V3 | Training | 83/83 (100%)e | 83/83 (100%)f | 166/166 (100%) |
| Tumor-Nonmalignant | TCGA | RNAseq | Testing | 959/979 (98%) | 108/108 (100%) | 1067/1087 (98%) |
| Tumor-Nonmalignant | SPORE/MDACC | Illumina WG-6 V3 | Testing | 252/275 (92%) | NA | 252/275 (92%) |
|
| ||||||
| ADC-SCC | FFPE Resected | HTG EdgeSeq | Testing | 16/17 (94%) | 18/18 (100%) | 34/35 (97%) |
| ADC-SCC | FFPE CNB | HTG EdgeSeq | Testing | 15/19 (79%) | 17/17 (100%) | 32/36 (89%) |
| Tumor-Nonmalignant | FFPE Resected | HTG EdgeSeq | Testing | 33/35 (94%) | 11/11 (100%) | 44/46 (96%) |
| Tumor-Nonmalignant | FFPE CNB | HTG EdgeSeq | Testing | 35/36 (97%) | NA | 35/36 (97%) |
Original histopathological diagnosis
Revised histopathological diagnosis
Predicted ADC (score > 0)/Diagnostic ADC
Predicted SCC (score < 0)/Diagnostic SCC
Predicted Tumor (score > 0)/Diagnostic Tumor
Predicted Non-Malignant (score < 0)/Diagnostic Non-Malignant
Abbreviations: FFPE, Formalin-Fixed Paraffin-Embedded; CNB, Core Needle Biopsy
The discrepancies and borderline classified cases (with scores lower than the specified cutoffs) were re-evaluated by the MD Anderson pathologists using immunostains when appropriate: these pathologists on re-review felt that about half of the discrepant diagnoses should be changed, with an improved accuracy of 95%. However, we stress that no change was done to the Classifier itself, which was based on the original histologic diagnosis.
ADC-SCC Signature: Testing on the TCGA Set
As a validation set we used the TCGA-lung RNAseq data. Scores were calculated as before and values larger than 0.17 predicted ADC while values lower than −0.17 predicted SCC (Fig. 2). Intermediate values predicted poorly differentiated tumors favoring ADC (positive low scores) or SCC (negative low scores) (see below). Including these lower scores, we thus obtained 97% correct prediction for the ADC set of samples (475 out of 490), and 93% correct prediction for the SCC set of samples (456 of 489; overall: 95%; Table 1). Interestingly, the non-malignant lung tissues largely fell into the same classification group as the ADCs.
Figure 2.
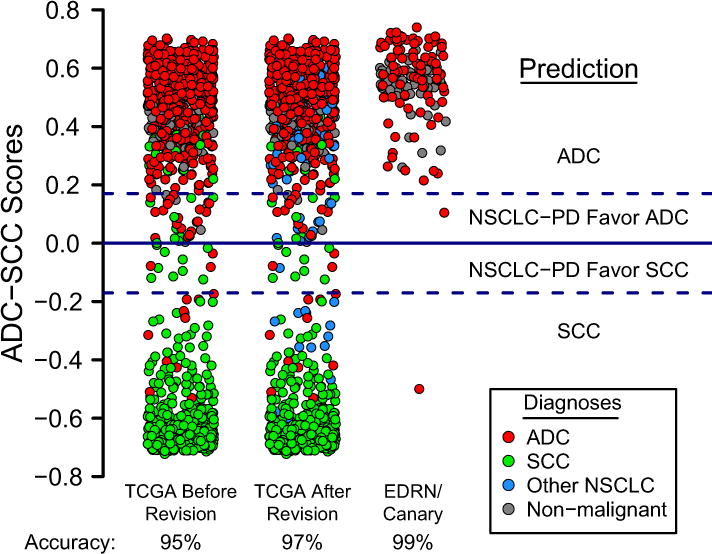
The ADC-SCC signature is validated on the TCGA and EDRN/Canary datasets and shown as score plots as in Fig. 1D. The prediction accuracies for TCGA (before revision) were 97% (ADC) and 93% (SCC). Overall accuracy: 95% (Table 1). These accuracies were 97% and 96% respectively (overall: 97%) after revision of diagnosis of selected TCGA cases which included many NSCLC-NOS (blue). The prediction accuracy for the EDRN/Canary dataset which consists of ADCs only was 99%.
Based on these class prediction results, selected cases were re-examined by pathologists from the TCGA studies. Many of the SCCs that were scored as ADCs turned out on re-examination to be called NSCLC-NOS or other subtypes such as undifferentiated LCCs (Fig. 2). Using these revised diagnoses, the prediction accuracy for the ADC group was unchanged at 97%, while the accuracy for the SCC group increased from 93% to 96% (overall accuracy: 97%, Table 1).
Tumor-Nonmalignant Signature
As observed above, most of the non-malignant lung samples in the TCGA set were classified as “ADC”. We thus generated an additional signature to distinguish tumor from non-malignant lung. As a training set, we chose the EDRN/Canary set consisting of 83 pairs of ADC specimens and matched non-malignant lung controls. Using the same method as for the ADC-SCC signature, we generated a volcano plot (Fig. 3A) and ranked differentially expressed genes between tumor and non-malignant using their distance to the plot’s origin. Twenty genes were selected as most differentially expressed (10 overexpressed in the tumor group, 10 overexpressed in the non-malignant group; Fig. 3B, Supplementary Table 1). A correlation plot (Fig. 3C) shows that the two groups are clearly separated in the training set (100% correct classification; Table 1), and a score ranging from −1 (non-malignant) to +1 (tumor) can be computed (Fig. 3C and D).
Figure 3.
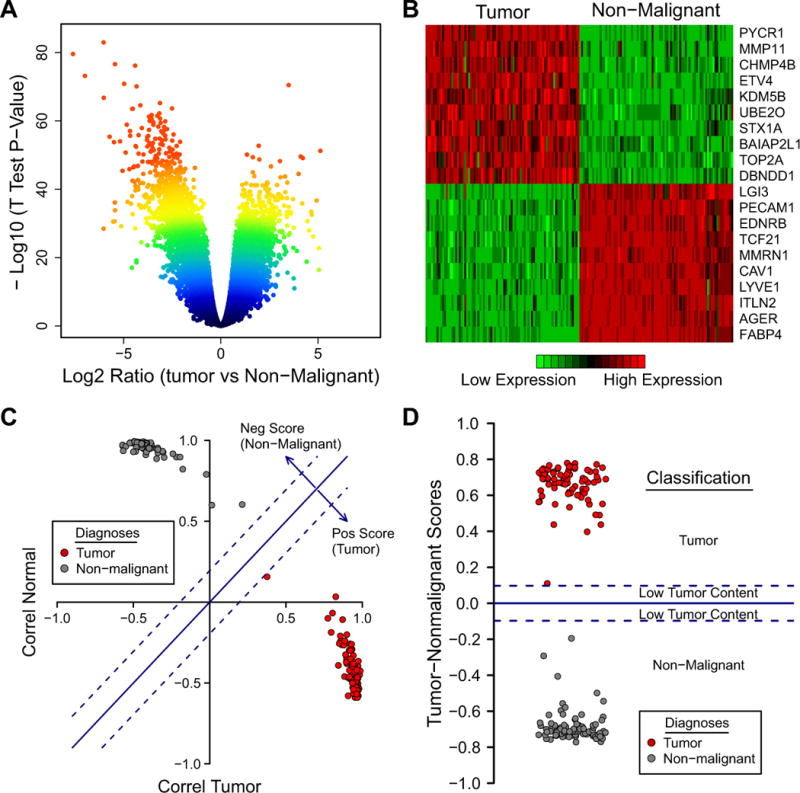
(A) A volcano plot shows that many genes are significantly different between tumor and non-malignant. (B) Ten genes overexpressed in tumors and ten genes overexpressed in non-malignant lung tissue were selected for the Tumor vs Nonmalignant signature. (C) This signature applied to the training set can distinguish tumor and non-malignant with complete accuracy (100%). (D) A score plot with the cutoff scores (+/− 0.10) shows a range of possible tumor content (tumor, low-tumor content, non-malignant).
As a validation set for the Tumor-Nonmalignant signature, we again used the TCGA dataset. Fig. 4 shows the score values with 100% prediction accuracy for the non-malignant group and 98% prediction accuracy for the tumor group (overall: 98%; Table 1). This shows that a 20-gene signature is sufficient to differentiate NSCLCs from non-malignant lung with high accuracy.
Figure 4.
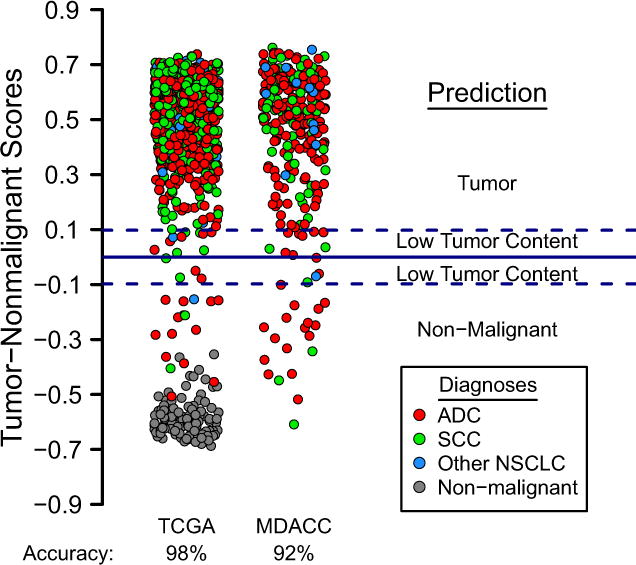
The Tumor-Nonmalignant signature is validated on the TCGA dataset, which consists of 979 lung tumors and 108 non-malignant lung tissues. The prediction accuracies are 98% for tumors and 100% for non-malignant lung (overall: 98%, Table 1). Many tumor samples have negative scores which could be due to larger stromal infiltration. A score plot for the MDACC dataset, which has tumor samples only, shows an accuracy of 92% (Table 1).
The ADC-SCC and Tumor-Nonmalignant prediction scores can be combined in a 2-D plot that clearly segregates the three groups (Supplementary Fig. 2).
Validation in public datasets
To further validate the ADC-SCC and Tumor-Nonmalignant signatures we looked at several public mRNA expression datasets containing sufficiently large numbers of NSCLC samples (n > 20 each) and deposited in GEO. The selected 22 datasets contained 1,560 ADCs, 732 SCCs, and 340 non-malignant lung tissues.
Both signatures gave highly accurate predictions (Supplementary Table 2). For the ADC-SCC signature, the average sensitivity (ADC prediction) and specificity (SCC prediction) were 95% and 89% respectively (overall: 93%), similar to the TCGA test set prediction values (95%). For the Tumor-Nonmalignant signature, the sensitivity (tumor prediction) and the specificity (non-malignant prediction) were 83% and 100% respectively (overall: 86%). The tumor prediction was significantly lower than the corresponding TCGA test set prediction (98%), which probably reflects larger stromal contents in the various GEO cohorts. However the non-malignant prediction was the same at 100%. Together, these results provide strong validation of our histology signatures in essentially all of the deposited datasets currently available.
ADC-SCC score as an estimate of differentiation
We hypothesized that ADC-SCC scores are related to the degree of differentiation, i.e. the higher the score (in absolute value), the better the differentiation. To support this, we first looked at ADC subtypes as reported by TCGA. We found that the lepidic subtype had higher scores than both solid (P = 0.026) and invasive mucinous (P = 0.071) subtypes (Supplementary Fig. 3A), consistent with the belief that tumors having extensive lepidic (non-invasive) components are better differentiated than those with large solid component (22). Further, we looked at known genes with different mutational spectra in ADCs and SCCs to see whether their mutation (or amplification) was associated with higher score. For ADC we indeed observed significantly higher scores for cases with mutation in EGFR, CTNNB1, HER2, BRAF, or KRAS (Supplementary Fig. 3B). For SCC we found significant differences for SOX2 amplification and for NFE2L2 or PIK3CA mutation (Supplementary Fig. 3C). Hence lung tumors with histology-specific mutations (which in some cases may be more differentiated) tended to have higher ADC-SCC scores.
Finally, one of us (AFG) looked at a random subset of the TCGA pathology slides and graded them using a modification of the standard grading system (23). Specifically we selected 50 ADC slides and 50 SCC slides randomly but with a uniform score distribution, and their degree of differentiation (grading) was assessed in a blinded fashion, i.e. without knowledge of their prediction score. Grading was then compared to prediction scores using two statistical tests. Significant associations with ADC and SCC scores were found (P < 0.03 for ADC and P < 10−4 for SCC; Supplementary Fig. 4). Together these data strongly suggest that the signature scores are correlated with the degree of differentiation.
ADC-SCC score and prognosis prediction
Since tumor grading is also correlated with patient survival (high grade tumors, i.e. poorly differentiated tumors, having worse prognosis (23, 24)) we hypothesized that a similar relationship between ADC-SCC score and survival would exist. We therefore looked at three cohorts with available clinical information: the MDACC set (non-neoadjuvant cases only: 145 ADCs and 64 SCCs), the Director’s Challenge set (ADC only; n=423) (25), and a SCC study (GSE4573; SCC only; n=106) (26). In most of these cases, high ADC or SCC scores were indeed associated with better prognosis (Supplementary Fig. 5).
Application of the Classifier to FFPE resected tumor specimens and small biopsies
In preliminary studies, the 62-gene signature was adapted to the HTG EdgeSeq technology (HTG Molecular Diagnostics) and was used to classify ADC from SCC and tumor from non-malignant lung using 4-micron sections from FFPE surgically resected tumors and core needle biopsy specimens. We applied our Classifier to these data and found that 34 of 35 FFPE resected samples (97%) were correctly classified as ADC or SCC and 32 of 36 FFPE core needle biopsies (89%) were also correctly classified (Table 1). Interestingly almost all discrepant cases had low scores (< 0.1 in absolute value; Supplementary Table 3) and had been independently diagnosed as poorly differentiated, thus explaining the majority of the discrepancies. Additionally, the ADC-SCC score correlation between FFPE resected tumors and matched CNB samples from the same patients was r=0.98, demonstrating the ability to use small, clinically relevant samples with this assay. Taken together, these results indicate that our Classifier, adapted to the HTG EdgeSeq platform, can also determine NSCLC subtype in fixed tissue, both from resected and CNB specimens.
DISCUSSION
The advent of “precision medicine” has made accurate classification of NSCLC a necessity for the clinical management of these tumors. However, currently the majority of diagnostic specimens (~70%) are small biopsies or cytological specimens (27), greatly increasing the difficulty of accurately diagnosing poorly differentiated tumors. Based on the anticipated 225,000 new cases of lung cancer for 2016 (28), of which an estimated 85% will be NSCLC, this amounts to over 130,000 NSCLC cases per year in the USA that will be diagnosed from small biopsies or cytology specimens. Cases without definitive diagnoses, and those wrongly classified, may not receive optimal therapy or may not be eligible for histology classification-restricted clinical trials. While the use of small panels of immunostains has greatly aided this task, about 5–10% of small biopsy cases at major medical centers will still be signed out as NSCLC-NOS. Examination of the SEER data registry suggests that the high diagnostic standards present at major medical centers may not extend to the medical community as a whole. Thus 14% of the lung cancer cases in the SEER registry were not further classified, amounting to an estimated 22,000 cases per year in the USA. In addition, an unknown percentage of cases will be misclassified or subject to arbitrary diagnosis by pathologists using varying pathological criteria or interpretation. A recent European interobserver study examined the diagnostic accuracy on lung cancer small biopsies for the distinction between ADC and SCC and related these to immunostaining and mutation analysis (29). The study was performed on prospectively collected biopsies obtained by bronchoscopy or transthoracic needle biopsy of patients with NSCLC. Eleven experienced pulmonary pathologists independently read H&E-stained slides of 110 cases, resulting in a kappa value of 0.55 +/− 0.10 and the diagnosis of NSCLC-NOS was given on average to 29.5% of the biopsies. This indicates that even experienced pathologists at major medical centers may disagree on interpretation or may not be able to fully classify a relatively high percentage of small biopsy specimens without the use of immunostains or other adjunct tests.
The widespread use of immunostains for the classification of NSCLC has greatly reduced the number of cases in the NOS category (30) and most of these tumors can now be classified with a single SCC and a single ADC marker (1). These findings led the new WHO Classification to recommend using immunostaining for SCC markers such as TP63 or its isoform p40 (deltaNp63) and high molecular weight keratins as well as ADC markers such as NKX2-1 (TTF-1) and Napsin A to classify poorly differentiated lung cancers including NSCLC-NOS (1, 31). However, interpretation of immunostains is not uniform and alternative approaches to lung cancer classification are being explored as adjunct tools to aid the pathologic diagnosis of lung cancers. These methods include digital nuclear imaging, mutation analysis, copy number variations, and various other molecular methods, either singly or in combination (32–34).
In this report, we developed and validated a gene expression classifier from a training set consisting of 263 surgically resected tumors in order to accurately and non-subjectively separate ADC from SCC. The list of top differentially expressed genes heavily favored SCC, possibly reflecting the greater pathologic heterogeneity and molecular complexity of ADCs and their multiple subtypes (3, 11). Thus, we selected an equal number of top genes significantly overexpressed in ADCs (n=21) and SCCs (n=21) so as not to bias the selection in favor of one NSCLC type. Not surprisingly, many of the selected genes are among the most frequently used and reliable immunostains in routine pathological practice (Fig. 1B, red arrows) or are known to play a role in lung cancer or in one of the main subtypes (Fig. 1B, blue labels). We validated the Classifier using the TCGA lung cancer datasets, which were available in a different platform (RNAseq) than our training set (Illumina BeadArray). We obtained very high prediction accuracies (95%) in spite of the fact that a fraction of the TCGA diagnostic materials were found to be of less than optimal quality (e.g. frozen sections instead of permanently fixed H&E slides) and in spite of the partially subjective nature of pathological diagnosis (29). In fact a significant limitation to the TCGA project was that the materials for immunostaining were not always available. Nevertheless, two of us (WDT, NR), who are the TCGA reference pathologists, reviewed the discrepancies, and this resulted in even better classification accuracy (Table 1, “Revised histopathological diagnosis”).
Interestingly, several non malignant lung TCGA specimens were classified as ADC by the signature, so we used the EDRN/Canary dataset to develop another classifier, containing 20 genes, that separated tumor cells from non-malignant lung with high accuracy. The combined 62-gene signature could now segregate ADC, SCC and non-malignant lung in this TCGA test set.
There are no squamous cells in the normal lung. Squamous metaplasia arises as the result of noxious stimuli such as tobacco exposure, mechanical trauma, inflammation or infection. Many of the SCC-associated classifier genes are involved in squamous differentiation, including basal (stem) cell proliferation, expression of high molecular weight keratins, desmosome formation, calcium regulation or cornified envelope formation (35–37). ADCs demonstrate considerable heterogeneity of morphological and biological subtypes (31). However, most of the ADC genes had relevance to lung cancer or were known to be ADC-specific. Both NKX2-1 and NAPSA are routinely used in many pathology classification schemes, however the latter, with a rank of 177, was not part of the top 21 genes overexpressed in the ADC group, and could not be used in the Classifier. Our data indicate that other genes in the Classifier, such as the trypsin inhibitor SPINK1 which is already known to be overexpressed in lung ADCs (38), may be good candidates for new immunostains in pathologic diagnosis, provided sensitive and specific antibodies are available. For SCC identification, the Classifier selected several high molecular weight keratins (KRTs) as well as TP63 among the top genes, but excluded SOX2, a gene frequently amplified in SCCs, although it was also significantly overexpressed in SCC (rank = 46) (9, 39, 40).
The Classifier can also provide a score that reflects the degree of differentiation. In support of this, we observed that NSCLCs with mutations that are specific for ADCs (EGFR, KRAS and others) or SCCs (SOX2 amplification, NFE2L2 mutation) tended to have higher magnitude scores than tumors that were wild-type for these mutations or amplifications (Supplementary Fig. 3B and C). In addition, the lepidic subtype of ADCs which is believed to be more differentiated also had a relatively higher score (Supplementary Fig. 3A). Finally, evaluation of tumor grade from TCGA histopathology slides revealed a strong concordance between prediction score and histologic grading (high score was associated with better differentiation). Thus our Classifier can be interpreted both qualitatively and quantitatively.
Consistent with the association between histologic grading and survival, our signature turned out to have prognostic value as well (high score associated with better survival). Thus this Classifier has the additional advantage of being of prognostic importance and may be useful in selecting the subpopulation of curative resected lung cancer patients that will benefit from adjuvant therapy.
Previous ADC-SCC gene signatures have been reported (41–46) and about 10–45% of the genes in these signatures overlap with ours. Two of these signatures were formally developed as classifiers, with external tumor set validation. The first, from Hou et al (44), comprises 50 unique genes (15 of which overlapped with our signature) and were validated in one external dataset with a prediction accuracy of 84%. In order to directly compare this classifier with our own, we tested it in TCGA RNAseq data using the class centroids provided by the study and Pearson correlation to predict the class. The resulting prediction had an accuracy of 92% (sensitivity: 99%, specificity: 84%) while our Classifier showed 95% accuracy (97% sensitivity, 93% specificity). The second study, from Wilkerson et al (46), had 15 genes (4 overlapping with our signature) and a reported prediction accuracy of 81% in external validation. Using the TCGA validation test, this corresponded to an accuracy of 92% (sensitivity: 90%, specificity 95%). Our current study thus offers the following advantages over prior ones: a) a slightly better overall accuracy; b) a balance between sensitivity and specificity; c) the ability to distinguish non-malignant from lung cancer; d) validation in a larger number of public NSCLC expression datasets with high prediction accuracies (93% for the ADC-SCC classification); e) the quantitative aspect of our Classifier and its correlation with differentiation and prognosis (this point also supports removing the term LCC and replacing it with poorly differentiated NSCLC); f) the ability, as mentioned below, to classify small biopsy samples and FFPE materials, using technology that can be transferred to a CLIA-certified environment.
While some pathologists may question the necessity for a molecular classification of NSCLC, the large number of non-classified cases, and the potential lack of diagnosis reproducibility even among experienced lung cancer pathologists, point to the value of a non-subjective test. This may even be a necessity in institutions or countries where immunostains are not routinely used and where staff pathologists may apply highly variable diagnostic criteria. An especially relevant use of a molecular classification would be for large multinational clinical trials where no central pathology review is available. We will also need further evaluation in a set of cases that have been diagnosed utilizing established immunohistochemical methods recommended by the 2015 WHO classification. Unfortunately these new criteria could not be applied to the datasets evaluated in this study.
Finally, to demonstrate the potential clinical applicability of our Classifier, we have developed an extraction‐free, highly sensitive, automated and cost‐effective NGS version based on the HTG EdgeSeq technology and have shown that its accuracy is similar to the original microarray-based Classifier (Table 1). In fact, the NGS classifier can be reproducibly applied to clinically challenging sample types, such as FFPE materials and core-needle biopsies.
In summary we have developed and validated a sensitive and specific gene expression classifier for NSCLC that distinguishes ADC from SCC, and lung cancer from normal lung. The Classifier was shown to be largely independent of the major gene expression platforms in common usage. Most of the genes in the Classifier are relevant to lung cancer or are known to be differentially expressed in NSCLC. The development and further validation of a practical and cost effective FFPE-based CLIA-certified version has the potential to lead to a widespread clinical application of the Classifier.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Personalized therapy and entry into clinical trials for non-small lung cancer (NSCLC) are heavily dependent on accurate histological classification. While most cases can be classified using routine pathology review of H&E and immunostains, examination of the US cancer registries indicates that many cases of NSCLC remain unclassified. To address this important clinical problem, we developed and validated a quantitative gene expression signature for the highly accurate classification of NSCLC. As the signature score reflects differentiation, it also provides prognostic information for early stage resected NSCLC, and thus may help identify patients who would benefit from adjuvant chemotherapy. Additionally we developed a next-generation sequencing (NGS) laboratory assay utilizing HTG Molecular Diagnostic’s HTG EdgeSeq chemistry and demonstrated that its performance is similar to the original microarray-based classifier. Importantly, the NGS classifier can be applied reproducibly to clinically challenging sample types, such as formalin-fixed paraffin-embedded (FFPE) materials and core-needle biopsies.
Acknowledgments
We wish to thank HTG Molecular Diagnostics Vice Presidents John Wineman and Patrick Roche for their support and contribution to this work.
Financial Support: This work was generously supported by the NCI Specialized Program in Research Excellence (SPORE) in Lung Cancer, P50CA70907, the Lungevity Foundation, the NCI Early Detection Research Network (EDRN), U01CA086402, and the Canary Foundation. The HTG EdgeSeq work was supported by NIH grant R44HG005949.
Footnotes
Disclosure of Potential Conflicts of Interest: Ihab Botros and Debrah Thompson are both employees and stockholders of HTG Molecular Diagnostics, Inc. (HTG). Ignacio Wistuba is a consultant for HTG and an occasional participant of its advisory board. He also received financial support from HTG as part of a research agreement. No other authors of this article have financial ties to HTG.
References
- 1.Travis W, Brambilla E, Burke A, Marx A, Nicholson A, editors. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. 4th. Lyon: International Agency for Research on Cancer; 2015. [DOI] [PubMed] [Google Scholar]
- 2.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers–a different disease. Nat Rev Cancer. 2007;7:778–90. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 3.Travis W, Brambilla E, Noguchi M, Geisinger K, Beer D, Powell C, et al. The new IASLC/ATS/ERS international multidisciplinary lung adenocarcinoma classification. J Thorac Oncol. 2009;4:244–85. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rekhtman N, Tafe LJ, Chaft JE, Wang L, Arcila ME, Colanta A, et al. Distinct profile of driver mutations and clinical features in immunomarker-defined subsets of pulmonary large-cell carcinoma. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26:511–22. doi: 10.1038/modpathol.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Clinical Lung Cancer Genome Project (CLCGP) and Network Genomic Medicine (NGM) A genomics-based classification of human lung tumors. Science translational medicine. 2013;5:209ra153. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gazdar AF. Should we continue to use the term non-small-cell lung cancer? Ann Oncol. 2010;21(Suppl 7):vii225–vii9. doi: 10.1093/annonc/mdq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gazdar AF, Minna JD. Precision medicine for cancer patients: lessons learned and the path forward. J Natl Cancer Inst. 2013;105:1262–3. doi: 10.1093/jnci/djt219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shames DS, Wistuba II. The evolving genomic classification of lung cancer. J Pathol. 2014;232:121–33. doi: 10.1002/path.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cagle PT, Allen TC, Dacic S, Beasley MB, Borczuk AC, Chirieac LR, et al. Revolution in lung cancer: new challenges for the surgical pathologist. Archives of pathology & laboratory medicine. 2011;135:110–6. doi: 10.5858/2010-0567-RA.1. [DOI] [PubMed] [Google Scholar]
- 10.Oliver TG, Patel J, Akerley W. Squamous non-small cell lung cancer as a distinct clinical entity. American journal of clinical oncology. 2015;38:220–6. doi: 10.1097/COC.0b013e3182a0e850. [DOI] [PubMed] [Google Scholar]
- 11.Kerr KM. Pathologist and molecular biologist, ever the twain shall meet? Lung Cancer. 2009;63:161–3. doi: 10.1016/j.lungcan.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Rekhtman N, Ang DC, Sima CS, Travis WD, Moreira AL. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24:1348–59. doi: 10.1038/modpathol.2011.92. [DOI] [PubMed] [Google Scholar]
- 13.Loo PS, Thomas SC, Nicolson MC, Fyfe MN, Kerr KM. Subtyping of undifferentiated non-small cell carcinomas in bronchial biopsy specimens. J Thorac Oncol. 2010;5:442–7. doi: 10.1097/JTO.0b013e3181d40fac. [DOI] [PubMed] [Google Scholar]
- 14.Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC, editors. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Lyon: International Agency for Research on Cancer; 2004. [Google Scholar]
- 15.Ding LH, Xie Y, Park S, Xiao G, Story MD. Enhanced identification and biological validation of differential gene expression via Illumina whole-genome expression arrays through the use of the model-based background correction methodology. Nucleic Acids Res. 2008;36:e58. doi: 10.1093/nar/gkn234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22:1197–211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tam KW, Zhang W, Soh J, Stastny V, Chen M, Sun H, et al. CDKN2A/p16 inactivation mechanisms and their relationship to smoke exposure and molecular features in non-small-cell lung cancer. J Thorac Oncol. 2013;8:1378–88. doi: 10.1097/JTO.0b013e3182a46c0c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Cancer Genome Atlas - Data Portal [cited 2016 April 12] Available from: https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp.
- 19.TCGA barcode - TCGA - National Cancer Institute - Confluence Wiki [cited 2016 April 12] Available from: https://wiki.nci.nih.gov/display/TCGA/TCGA+barcode.
- 20.HTG EdgeSeq Chemistry [cited 2016 April 12] Available from: https://www.htgmolecular.com/science/htg-edgeseq-chemistry.
- 21.Cancer Statistics Review, 1975–2012 - SEER Statistics [cited 2016 April 12] Available from: http://seer.cancer.gov/csr/1975_2012/
- 22.Travis WD. Classification of lung cancer. Seminars in roentgenology. 2011;46:178–86. doi: 10.1053/j.ro.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Cancer Grading Manual. 2nd. Springer; 2013. [Google Scholar]
- 24.Tumor Grade Fact Sheet - National Cancer Institute [cited 2016 April 12] Available from: http://www.cancer.gov/about-cancer/diagnosis-staging/prognosis/tumor-grade-fact-sheet.
- 25.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–7. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raponi M, Zhang Y, Yu J, Chen G, Lee G, Taylor JM, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–72. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 27.Travis WD, Rekhtman N. Pathological diagnosis and classification of lung cancer in small biopsies and cytology: strategic management of tissue for molecular testing. Semin Respir Crit Care Med. 2011;32:22–31. doi: 10.1055/s-0031-1272866. [DOI] [PubMed] [Google Scholar]
- 28.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 29.Thunnissen E, Noguchi M, Aisner S, Beasley MB, Brambilla E, Chirieac LR, et al. Reproducibility of histopathological diagnosis in poorly differentiated NSCLC: an international multiobserver study. J Thorac Oncol. 2014;9:1354–62. doi: 10.1097/JTO.0000000000000264. [DOI] [PubMed] [Google Scholar]
- 30.Lewis DR, Check DP, Caporaso NE, Travis WD, Devesa SS. US lung cancer trends by histologic type. Cancer. 2014;120:2883–92. doi: 10.1002/cncr.28749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–85. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thunnissen EB, Diegenbach PC. Classification of lung carcinoma by means of digital nuclear image analysis. Analytical and quantitative cytology and histology/the International Academy of Cytology [and] American Society of Cytology. 1986;8:301–4. [PubMed] [Google Scholar]
- 33.Bishop JA, Benjamin H, Cholakh H, Chajut A, Clark DP, Westra WH. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res. 2010;16:610–9. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 34.Ramani RG, Jacob SG. Improved classification of lung cancer tumors based on structural and physicochemical properties of proteins using data mining models. PLoS One. 2013;8:e58772. doi: 10.1371/journal.pone.0058772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratushny V, Gober MD, Hick R, Ridky TW, Seykora JT. From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J Clin Invest. 2012;122:464–72. doi: 10.1172/JCI57415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baroni A, Buommino E, De Gregorio V, Ruocco E, Ruocco V, Wolf R. Structure and function of the epidermis related to barrier properties. Clinics in dermatology. 2012;30:257–62. doi: 10.1016/j.clindermatol.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Fuchs E. Scratching the surface of skin development. Nature. 2007;445:834–42. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazar V, Suo C, Orear C, van den Oord J, Balogh Z, Guegan J, et al. Integrated molecular portrait of non-small cell lung cancers. BMC Med Genomics. 2013;6:53. doi: 10.1186/1755-8794-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger K, Yatabe Y, et al. Diagnosis of lung cancer in small biopsies and cytology: implications of the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification. Archives of pathology & laboratory medicine. 2013;137:668–84. doi: 10.5858/arpa.2012-0263-RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger K, Yatabe Y, et al. Diagnosis of lung adenocarcinoma in resected specimens: implications of the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification. Archives of pathology & laboratory medicine. 2013;137:685–705. doi: 10.5858/arpa.2012-0264-RA. [DOI] [PubMed] [Google Scholar]
- 41.Hofmann HS, Bartling B, Simm A, Murray R, Aziz N, Hansen G, et al. Identification and classification of differentially expressed genes in non-small cell lung cancer by expression profiling on a global human 59.620-element oligonucleotide array. Oncology reports. 2006;16:587–95. [PubMed] [Google Scholar]
- 42.Rohrbeck A, Neukirchen J, Rosskopf M, Pardillos GG, Geddert H, Schwalen A, et al. Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers. J Transl Med. 2008;6:69. doi: 10.1186/1479-5876-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuner R, Muley T, Meister M, Ruschhaupt M, Buness A, Xu EC, et al. Global gene expression analysis reveals specific patterns of cell junctions in non-small cell lung cancer subtypes. Lung Cancer. 2009;63:32–8. doi: 10.1016/j.lungcan.2008.03.033. [DOI] [PubMed] [Google Scholar]
- 44.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez-Palencia A, Gomez-Morales M, Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R, et al. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer. 2011;129:355–64. doi: 10.1002/ijc.25704. [DOI] [PubMed] [Google Scholar]
- 46.Wilkerson MD, Schallheim JM, Hayes DN, Roberts PJ, Bastien RR, Mullins M, et al. Prediction of lung cancer histological types by RT-qPCR gene expression in FFPE specimens. The Journal of molecular diagnostics : JMD. 2013;15:485–97. doi: 10.1016/j.jmoldx.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


