Abstract
Neuroblastoma is a childhood cancer originating from embryonic neural crest cells. Amplification of the proto-oncogene N-myc, seen in ~30% of neuroblastoma tumors, is a marker for poor prognosis. Recently discovered small regulatory RNAs, microRNAs (miRNAs), are implicated in cancers, including neuroblastoma. miRNAs downregulate the expression of genes by binding to the 3-untranslated regions (3′-UTRs), thereby inhibiting translation or inducing degradation of cognate mRNAs. Our study sought to identify miRNAs that regulate N-myc expression and thereby malignancy in neuroblastoma. miRNAs whose expression negatively correlates with N-myc expression were identified from a miRNA microarray of 4 N-myc-amplified neuroblastoma cell lines. Three of these miRNAs (miR-17, miR-20a and miR-18a) belong to the miR-17-92 cluster, previously shown to be upregulated by N-myc. qPCR validation of these miRNAs in a larger panel of cell lines revealed that levels of miR-17 were inversely proportional to N-myc mRNA amounts in the N-myc-amplified cell lines. Notably, miR-17 also downregulated N-myc protein synthesis in the N-myc-amplified cells, thereby generating a negative feedback regulatory loop between the proto-oncogene and this miRNA. Moreover, the neuronal-specific RNA-binding protein HuD (ELAVL4), which regulates the processing/stability of N-myc mRNA, competes with miR-17 for a binding site in the 3′-UTR of N-myc. Thus, N-myc levels appear to be modulated by the antagonistic interactions of both miR-17, as a negative regulator, and HuD, as a positive regulator, providing further evidence of the complex cellular control mechanisms of this oncogene in N-myc-amplified neuroblastoma cells.
Keywords: neuroblastoma, N-myc, MYCN, microRNAs, miR-17, HuD
Introduction
Neuroblastoma (NB) is the most common extracranial solid tumor in children. One of the major biological factors related to poor patient survival is amplification of the N-myc gene, which is seen in ~30% of cases (1). This proto-oncogene encodes a basic helix-loop-helix transcription factor that binds to E-box elements (2) in the numerous genes it regulates. Since the majority of neuroblastoma patients with overexpression of N-myc have a lower survival rate, defining novel pathways that regulate its expression is critical for developing new therapeutic approaches to better treat this enigmatic cancer.
MicroRNAs (miRNAs) are small (~21 nucleotides) non-coding regulatory RNAs that downregulate the expression of mRNAs either by inhibiting their translation and/or facilitating the degradation of the cognate mRNA (3). Several recent studies have reported a role for miRNAs in targeting or being targets of N-myc in neuroblastoma (4–16). Of particular interest has been the relationship between N-myc expression and members of the miR-17-92 miRNA cluster, a polycistronic cluster on chromosome 13q31.32 that gives rise to 6 mature miRNAs (miR-17, miR-18a, miR-19a, miR-20a and miR-92-1). Several previous studies in human neuroblastoma cell lines have shown that the expression levels of the miR-17-92 cluster members are directly correlated with N-myc amplification and that N-myc binds to E-boxes to increase the transcription of this gene cluster (5,16). Likewise, miR-17-92 overexpression is associated with increased tumorigenicity in numerous types of cancer, including neuroblastoma (17).
Our previous studies have shown that another RNA-binding element also plays an important role in the amplification and overexpression of N-myc: the neuronal-specific RNA binding protein HuD (ELAVL4). This protein is involved in the splicing and stabilization of N-myc mRNA (18–20), and its decreased expression (consequent to loss of the short arm of chromosome #1) leads to N-myc amplification in neuroblastoma. Conversely, re-introduction of HuD genes removes the selection pressure, resulting in loss of N-myc genes (20).
In the present study, we demonstrate that miR-17, a member of the miR-17-92 cluster, is induced by N-myc and reciprocally binds to the N-myc 3-untranslated region (3′-UTR) to downregulate its expression, creating a negative-feedback loop in N-myc-amplified neuroblastoma cell lines. Moreover, the 3′-UTR region to which miR-17 binds contains an overlapping binding site for HuD and the miRNA and this RNA-binding protein compete for binding to this site. This competition may be exacerbated in N-myc-amplified neuroblastoma cells, which lack one copy of the HuD gene. A possible consequence of this imbalance is a heightened selection pressure resulting in the extremely high levels of N-myc gene amplification seen in neuroblastoma as well as the increased malignancy and decreased differentiation in N-myc-amplified neuroblastoma cells.
Materials and methods
Cell culture
All cell lines were maintained as previously described (21). Thirteen human neuroblastoma clonal cell lines or enriched populations were used in these studies: N-myc-amplified (SK-N-BE(1)n, LA1-55n, KCN-83n, BE(2)-M17V, SK-N-LD, SK-N-HM, BE(2)-C and LA1-5s), and N-myc non-amplified (SH-SY5Y, SMS-LHN, CB-JMN, SH-EP1 and SMS-KCNs).
miRNA microarray
miRNAs were isolated using mirVana™ miRNA Isolation kits (Ambion, Austin, TX, USA). Processing and initial analysis of miRNA expression levels were carried out by LC Sciences (Houston, TX, USA). The correlation between N-myc RNA and miRNA expression levels was assessed by linear regression analysis.
Quantitative real-time RT-PCR
cDNA for miRNAs were synthesized using the TaqMan® MicroRNA Reverse Transcription kit. miRNA-specific primers and miRNAs were quantified using TaqMan assays (Applied Biosystems, Foster City, CA, USA) by the comparative ΔΔCt method. Expression levels of miRNAs were normalized to U6 and expressed as a fold-change compared to the levels of the same sample of SH-SY5Y cells.
Western blot analysis
Western blot analysis of proteins was performed as previously described (21). Primary antibodies used were rabbit anti-N-myc [(C-19), (SC-791)] (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) and mouse anti-actin [(AC-74), (076K4762)] (Invitrogen Corp., Carlsbad, CA, USA).
Transfections and infections
miRNA inhibitors for miR-17 and control oligos (100 nM) (Ambion) were transiently transfected into SK-N-LD cells using Lipofectamine 2000 (Invitrogen Corp.) according to the manufacturers instructions. Lentivirus constructs containing the miR-17-92 cluster and controls were purchased from SBI Biosciences (Mountain View, CA, USA) and used to infect SK-N-LD and CB-JMN cells at a multiplicity of infection of 10, according to the manufacturer's instructions. Resulting populations were either used directly or cloned using cloning cylinders.
Luciferase and β-galactosidase assays
The 3′-UTR of N-myc (880 bp) was PCR-amplified (primers available upon request) and cloned into pMIR-Luciferase reporter plasmid (Ambion) using T4-ligase (Promega, Madison, WI, USA). The sequences of the inserts were confirmed (GENEWIZ, South Plainfield, NJ, USA).
SK-N-LD cells were co-transfected in quadruplicate with either pMIR-Luciferase-N-myc-UTR or pMIR-REPORT-Luciferase (Luciferase vector) and pMIR-REPORT-β-gal (Ambion) using Lipofectamine 2000. After 24 h, half of each of the co-transfected populations were treated with 100 nM of either the miR-17 inhibitor or a non-specific control oligo (both from Ambion). Twenty-four hours thereafter luciferase and β-gal activities were assessed (Promega). Luciferase activity was normalized to β-gal and expressed as a fold-change in the miR-17-inhibitor-treated cells compared to the control-treated cells for both the pMIR-Luciferase-full-N-myc 3′-UTR and the vector transfectants.
Colony-forming efficiency
Colony-forming efficiencies in soft agar were assessed as previously described (21). Mean colony-forming efficiency (CFE; the number of colonies divided by the cell inoculum × 100) was determined in quadruplicate in 3 independent experiments.
Gel mobility shift assays (GMSAs)
The pCRII-SNM plasmid, containing 399 nt at the 3′-end of the N-myc 3′-UTR (19) was linearized and used to generate radioactive probes for gel shift assays using the MAXIscript T7 in vitro transcription kit (Ambion) and 32P-UTP (PerkinElmer, Inc., Waltham, MA, USA). The probes generated were treated with DNase and purified using a MEGAclear kit (both from Ambion).
HuD and HuC proteins were expressed and purified as previously described (19). Bacterial lysates containing either HuD or HuC protein were mixed with the N-myc 3′-UTR probe (~10,000 cpm) and incubated for 20 min at room temperature. In competition experiments, the probe was pre-mixed in annealing buffer (19) with either mature miR-17 RNA oligo (8, 40 or 80 pmol) or non-specific RNA oligo (40 pmol) (sequences available upon request) (Dharmacon Inc., Chicago, IL, USA), denatured at 95°C for 5 min and cooled to room temperature before use. RNA-protein complexes were resolved by electrophoresis through 3.2% non-denaturing polyacrylamide gels and exposed to Kodak XAR film overnight at −80°C.
Results
Inverse relationship between N-myc mRNA and miR-17-92 cluster expression
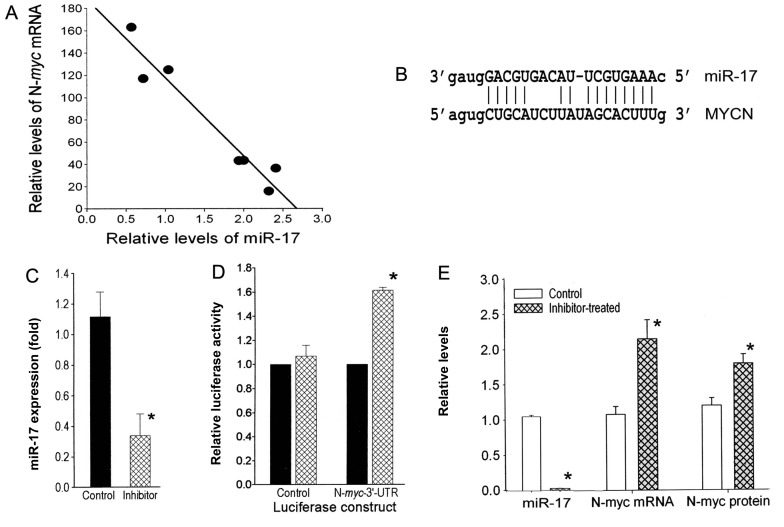
To identify miRNAs that regulate N-myc, we analyzed the expression of miRNAs in 4 human neuroblastoma cell lines with varying degrees of N-myc amplification using an miRNA microarray. The 4 cell lines had N-myc relative mRNA levels (compared to the non-amplified SH-SY5Y neuroblastoma cell line) ranging from 16- to 183-fold. The levels of each miRNA in the 4 lines were compared to the N-myc levels by linear regression analysis, and ranked in descending order according to the R2 value. Notably, from this analysis, we found a highly inverse correlation between the expression level of N-myc and most of the miR-17-92 cluster members (Table I). Additional qRT-PCR analysis for 3 of the miRNAs (miR-17, miR-20a and miR-18a) with a larger set (n=13) of neuroblastoma cell lines clearly revealed that all 3 miRNAs were significantly higher in the N-myc-amplified group (n=8) compared to the N-myc-non-amplified group (n=6) (data not shown) consistent with previous findings. Notably, as predicted by the microarray data (Table I), the qRT-PCR data ascertained that the inverse relationship between N-myc expression and miR-17 levels was highly significant (R2=0.98; P<0.001) (Fig. 1A). However, the inverse relationship was not significant for miR-20a and miR-18a in the N-myc-amplified cells. Further studies were undertaken to investigate whether the inverse correlation between miR-17 and N-myc levels is due to the downregulation of N-myc by miR-17.
Table I.
Inverse correlation between N-myc overexpression and levels of miR-17-92 cluster members.
| Cell lines | |||||
|---|---|---|---|---|---|
| mRNA/miRNA | BE(2)-M17 | BE(2)-C | SK-N-BE(1)n | SK-N-LD | R2 |
| N-myc | 183 | 117 | 47 | 16 | |
| hsa-miR-20a | 7,044 | 11,238 | 12,395 | 16,020 | 0.922 |
| hsa-miR-17 | 7,749 | 10,688 | 12,137 | 16,145 | 0.902 |
| hsa-miR-92 | 10,643 | 11,483 | 13,651 | 17,298 | 0.834 |
| hsa-miR-19b | 4,865 | 5,201 | 8,840 | 13,332 | 0.815 |
| hsa-miR-19a | 640 | 578 | 1,745 | 4,475 | 0.680 |
| hsa-miR-18a | 1,022 | 1,381 | 1,172 | 3,173 | 0.496 |
N-myc levels were determined by qRT-PCR and expressed as fold increase compared to SH-SY5Y. Values for miRNAs are fluorescent intensities from a microRNA microarray.
Figure 1.
N-myc is a target of miR-17. (A) Linear regression analysis showing an inverse correlation between miR-17 and N-myc mRNA levels in the 8 N-myc-amplified human neuroblastoma cell lines (R2=0.98; P<0.001). (B) Predicted complementary base pairing between miR-17 and the 3′-UTR of N-myc (www.microrna.org). (C) miR-17 levels in SK-N-LD cells expressing the Luciferase-N-myc-UTR reporter gene or the Luciferase vector revealed a decrease of 3.33-fold (*P<0.05) when treated with an miR-17 inhibitor compared to the control-treated cells. (D) Inhibitor treatment increased the luciferase activity 1.55-fold (*P<0.01) in cells expressing the Luciferase-N-myc-UTR construct, but had no effect on the Luciferase vector transfectants. (E) Inhibitor-treated SK-N-LD cells with a 33.3-fold (*P<0,01) decrease in the levels of miR-17 had a 2.1-fold increase in N-myc mRNA (*P<0.01) as determined by qRT-PCR and a 1.5-fold increase in N-myc protein (*P<0.02) compared to the untreated control (n=3).
N-myc 3′-UTR has miR-17 regulatory sites
Sequence analysis revealed a complementary binding site for miR-17 within the 3′-UTR of N-myc mRNA (Fig. 1B), making the oncogene a predicted target of miR-17 (www.microrna.org and www.mirbase.org). To determine whether N-myc mRNA is a target of miR-17, the N-myc full length 3′-UTR was cloned into a luciferase reporter gene and this construct (Luciferase-N-myc-UTR) or the Luciferase vector (control) was transfected into the SK-N-LD N-myc-amplified cell line. When each of these transfectants was treated with the miR-17 inhibitor, the miR-17 levels decreased significantly (3.3-fold; P<0.05) (Fig. 1C) compared to the control oligo transfectants. This decrease in miR-17 resulted in a significant 1.6-fold (P<0.01) increase in luciferase activity in cells expressing the Luciferase-N-myc-UTR construct, but had no effect on the luciferase activity in the Luciferase vector-transfectants (Fig. 1D). Thus, the binding site of miR-17 in N-myc appears to be functional.
Alteration of miR-17 expression inversely affects N-myc expression
To confirm the direct regulation of N-myc expression by miR-17, a specific inhibitor for miR-17 was transiently transfected into N-myc-amplified SK-N-LD cells. miR-17 levels were decreased 33.3-fold (P<0.01) (Fig. 1E). Consequently, N-myc mRNA and protein levels in the inhibitor-transfectants were significantly increased 2.1-fold (P<0.01) and 1.5-fold (P<0.02), respectively, compared to the control cells (Fig. 1E). Since the expression levels of the homolog miRNAs miR-18a and miR-20a remained unaltered (data not shown), these decreases in N-myc appear to be the result of the specific decrease of miR-17. However, transfection of the miR-17 inhibitor into an N-myc-non-amplified cell line SH-SY5Y did not alter the levels of N-myc mRNA or the protein, suggesting this regulation is not seen in N-myc-non-amplified cells (data not shown).
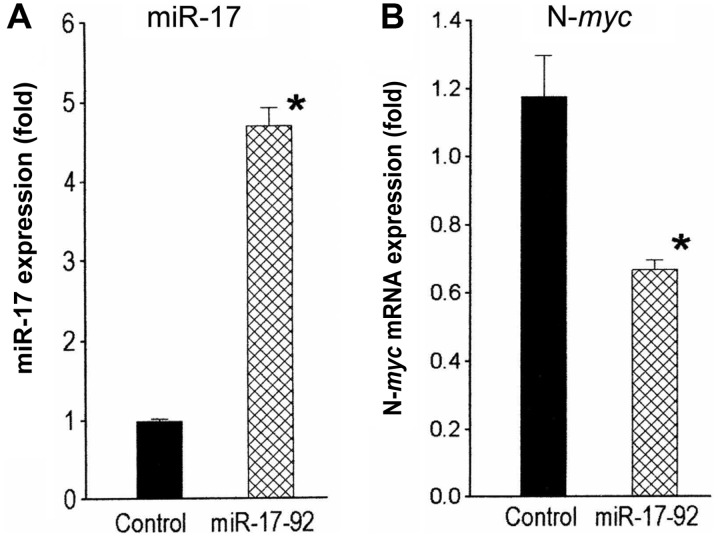
In converse experiments, levels of miR-17 were increased in SK-N-LD cells by stable infection with a miR-17-92-overexpressing cassette in a lentiviral vector. These infected cell populations had 4.8-fold (P<0.03) higher miR-17 levels compared to the vector-infected control cells (Fig. 2A). N-myc mRNA levels in the miR-17-92-overexpressing cells decreased significantly 1.7-fold (P<0.02) compared to the control (Fig. 2B). Collectively, these observations suggest that miR-17 regulates N-myc expression in N-myc-amplified human neuroblastoma cells.
Figure 2.
Increased expression of miR-17 downregulates N-myc mRNA levels. qRT-PCR analyses revealed that (A) the miR-17 levels were increased 4.8-fold (*P<0.03) and (B) N-myc mRNA was decreased 1.7-fold (*P<0.02) in the SK-N-LD cells infected with a lentiviral construct containing the miR-17-92 cluster compared to the control.
Overexpression of miR-17-92 increases tumorigenicity
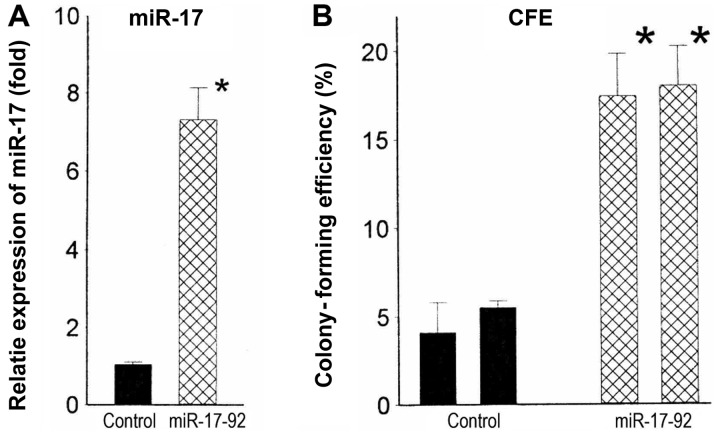
miR-17-92 overexpression is associated with increased tumorigenicity in numerous types of cancer. To assess whether elevated levels of miR-17-92 likewise contributed to increased tumorigenicity of N-myc-amplified cell lines, we stably infected the lentiviral miR-17-92 cassette into a non-amplified cell line, CB-JMN. Two miR-17-92-infected clones with average increases in miR-17 of 7.0-fold (P<0.001) (Fig. 3A) were tested for malignant potential. These infectants had colony-forming efficiencies in soft agar of 17.4 and 18.0%. By contrast, those of the control clones were 4.4 and 5.1% (Fig. 3B). Thus, increased expression of miR-17-92 appears to be responsible, at least in part, for the increased malignant potential and/or aggressiveness of N-myc-amplified tumors.
Figure 3.
Lentiviral-infected CB-JMN cells overexpressing the miR-17-92 cluster have high malignant potential. (A) Mean expression levels of miR-17 in clones of CB-JMN cells infected with the miR-17-92-constructs (n=2) or the control constructs (n=2); miR-17 levels are increased 7.0-fold (*P<0.001). (B) Colony-forming efficiency is significantly 3.3-fold higher (*P<0.001) in cells overexpressing the microRNA.
Regulation of N-myc by HuD and miR-17
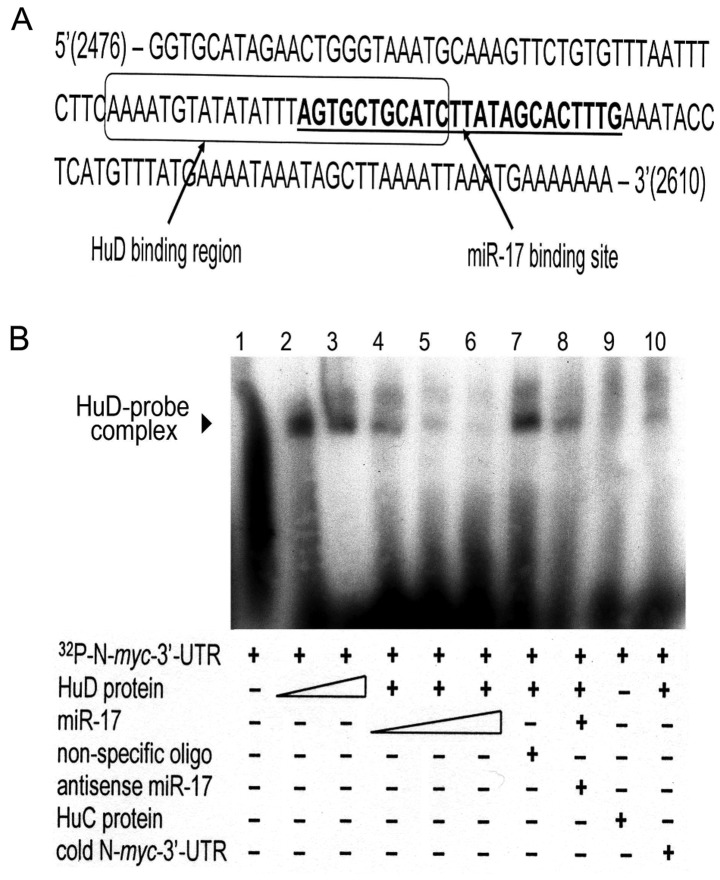
Previous studies in our laboratory and others have shown that the neuronal-specific RNA-binding protein HuD (ELAVL4) is involved in stabilizing the N-myc mRNA transcript by binding to the AU-rich elements in its 3′-UTR (18–20). Since miR-17 binds to the 3′-UTR, but destabilizes the mRNA, we examined the location of the proposed binding sites of both regulators. Notably, the binding sites overlap (Fig. 4A). To assess whether these 2 regulators compete for binding to the N-myc 3′-UTR, gel shift assays were performed. As shown in Fig. 4B, binding of the recombinant HuD protein to a radio-labeled 399-bp region from the N-myc 3′-UTR containing the 2 putative binding sites protects it from degradation by endogenous bacterial RNases, resulting in an RNA-protein complex that migrates more slowly through the gel (Fig. 4B, lanes 2 and 3). Notably, prior incubation of the N-myc probe with increasing amounts of miR-17 decreased in a dose-dependent manner the amount of HuD-N-myc complex (Fig. 4B, lanes 4–6). Formation of the HuD-mRNA complex was not abrogated by preincubation with non-specific RNA oligonucleotides (Fig. 4B, lane 7) or with miR-17 pre-annealed to its antisense sequence (Fig. 4B, lane 8) and was not noted after substitution of HuC, a closely related Hu protein, for HuD (Fig. 4B, lane 9) or pre-incubation of the HuD lysate with cold N-myc 3′-UTR prior to the addition of labeled N-myc 3′-UTR (Fig. 4B, lane 10). This experiment was repeated with a second, shorter piece (124 bp) of the N-myc 3′-UTR with the same results (data not shown). Thus, the neuronal-specific RNA-binding protein HuD and miR-17 appear to compete for adjacent overlapping binding sites in the 3′-UTR of N-myc, exerting antagonistic effects on N-myc mRNA stability and subsequent protein expression.
Figure 4.
HuD competes with miR-17 for an overlapping binding site in the N-myc 3′-UTR. (A) Sequence analysis revealed that HuD and miR-17 bind to adjacent regions in the N-myc 3′-UTR. The box indicates the AU-rich binding domain of HuD; the miR-17 region is indicated in underlined boldface type. Eleven bases are common to both. (B) A gel mobility shift assay (GMSA) revealed formation of the N-myc-HuD protein complex (lanes 2 and 3) and a concentration-dependent decrease in formation of the complex with increasing amounts of miR-17 (lanes 4–6). The complex amount was unaltered when non-specific oligos were substituted for miR-17 (lane 7), present but slightly decreased when miR-17 was pre-annealed with its antisense (lane 8), absent when HuC was substituted for HuD in the reaction (lane 9), and barely detectable when the labeled probe was diluted with saturating amounts of unlabeled probe (lane 10).
Discussion
In the present study, we identified an interaction between miR-17 and HuD, two molecules involved in the regulation of N-myc expression. Several studies have identified miRNAs that correlate with N-myc expression in neuroblastoma cell lines and tumors (4–15,22). Many of these studies revealed that the expression of the miR-17-92 cluster is: i) higher in N-myc-amplified cells (5,16,22); and ii) regulated by N-myc (5,16), as we also observed in our panel of cells. This cluster, also known as oncomiR-1, has been shown to influence tumorigenicity in many forms of cancer (17). Schulte et al (22) noted an association between high miR-17-92 levels and an unfavorable outcome in children with neuroblastoma. Moreover, in the present study, increased expression of miR-17-92 increased the tumorigenic potential of an N-myc-non-amplified cell line. Therefore, the aggressive behavior of neuroblastoma tumors with N-myc amplification could be mediated, at least in part, by downregulation of target tumor suppressor/antiproliferative/apoptotic genes (17) consequent to increased levels of miR-17-92.
The negative correlation of N-myc and miR-17 levels in N-myc-amplified neuroblastoma cell lines revealed the existence of a negative feedback loop between N-myc and miR-17. Notably, this relationship was not observed in N-myc-non-amplified cell lines. Why is this negative regulation observed only in N-myc-amplified cell lines? Our previous studies have shown that HuD binds and stabilizes the N-myc transcript (19). N-myc-amplified cells have lower levels of HuD compared to N-myc-non-amplified cells as a consequence of the loss of one HuD allele with the 1p deletion invariably accompanying N-myc amplification (20). In non-amplified cells with 2 HuD alleles, HuD protein amounts appear sufficient to block the common binding site and ensure adequate levels of N-myc translation. By contrast, lower levels of HuD may allow increased access to and promote N-myc mRNA degradation by miR-17, leading to N-myc levels too low to sustain viability. This effect may in turn select for cells that have amplified N-myc, as proposed by Grandinetti et al (20). Overcompensation, leading to excess N-myc RNA and protein, may further increase the amount of miR-17, leading subsequently to both a greater imbalance between the 2 regulators and to greater malignancy. Thus, our findings concerning the interactions between miR-17, HuD and N-myc provided one explanation for the very high levels of N-myc gene amplification found in neuroblastoma.
Increased levels of the oncogenic miR-17-92 cluster, and in particular miR-17, in N-myc-amplified tumors could be a major mediator of their aggressive nature. In addition, the negative feedback regulatory mechanism between N-myc and miR-17 could be an important regulator of cell fate (proliferation vs. apoptosis) in normal as well as in malignant neuroblasts. A similar negative feedback regulatory mechanism has been observed between c-myc and miR-17-92, in which upregulation of the miR-17-92 cluster results in downregulation of E2F (23). Greater knowledge of the complex interactions between miR-17-92, HuD and N-myc could lead to better understanding of the regulation of N-myc expression and to the discovery of additional drugs or therapeutic approaches for the more effective treatment of N-myc-amplified cancers.
Acknowledgements
The present study was supported in part by a grant (CA 77593) from the National Institutes of Health (Bethesda, MD, USA).
References
- 1.Park JR, Bagatell R, London WB, Maris JM, Cohn SL, Mattay KK, Hogarty M. COG Neuroblastoma Committee: Children's Oncology Group's 2013 blueprint for research: Neuroblastoma. Pediatr Blood Cancer. 2013;60:985–993. doi: 10.1002/pbc.24433. [DOI] [PubMed] [Google Scholar]
- 2.Bell E, Chen L, Liu T, Marshall GM, Lunec J, Tweddle DA. MYCN oncoprotein targets and their therapeutic potential. Cancer Lett. 2010;293:144–157. doi: 10.1016/j.canlet.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Schulte JH, Schowe B, Mestdagh P, Kaderali L, Kalaghatgi P, Schlierf S, Vermeulen J, Brockmeyer B, Pajtler K, Thor T, et al. Accurate prediction of neuroblastoma outcome based on miRNA expression profiles. Int J Cancer. 2010;127:2374–2385. doi: 10.1002/ijc.25436. [DOI] [PubMed] [Google Scholar]
- 5.Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V, Giacomini P, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci USA. 2010;107:1506–1511. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckley PG, Alcock L, Bryan K, Bray I, Schulte JH, Schramm A, Eggert A, Mestdagh P, De Preter K, Vandesompele J, et al. Chromosomal and microRNA expression patterns reveal biologically distinct subgroups of 11q- neuroblastoma. Clin Cancer Res. 2010;16:2971–2978. doi: 10.1158/1078-0432.CCR-09-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67:976–983. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
- 9.Shohet JM, Ghosh R, Coarfa C, Ludwig A, Benham AL, Chen Z, Patterson DM, Barbieri E, Mestdagh P, Sikorski DN, et al. A genome-wide search for promoters that respond to increased MYCN reveals both new oncogenic and tumor suppressor microRNAs associated with aggressive neuroblastoma. Cancer Res. 2011;71:3841–3851. doi: 10.1158/0008-5472.CAN-10-4391. [DOI] [PubMed] [Google Scholar]
- 10.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei JS, Song YK, Durinck S, Chen QR, Cheuk AT, Tsang P, Zhang Q, Thiele CJ, Slack A, Shohet J, et al. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27:5204–5213. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovén J, Zinin N, Wahlström T, Müller I, Brodin P, Fredlund E, Ribacke U, Pivarcsi A, Påhlman S, Henriksson M. MYCN-regulated microRNAs repress estrogen receptor-α (ESR1) expression and neuronal differentiation in human neuroblastoma. Proc Natl Acad Sci USA. 2010;107:1553–1558. doi: 10.1073/pnas.0913517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haug BH, Henriksen JR, Buechner J, Geerts D, Tømte E, Kogner P, Martinsson T, Flægstad T, Sveinbjørnsson B, Einvik C. MYCN-regulated miRNA-92 inhibits secretion of the tumor suppressor DICKKOPF-3DKK3) in neuroblastoma. Carcinogenesis. 2011;32:1005–1012. doi: 10.1093/carcin/bgr073. [DOI] [PubMed] [Google Scholar]
- 14.Buechner J, Tømte E, Haug BH, Henriksen JR, Løkke C, Flægstad T, Einvik C. Tumour-suppressor microRNAs let-7mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br J Cancer. 2011;105:296–303. doi: 10.1038/bjc.2011.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bray I, Bryan K, Prenter S, Buckley PG, Foley NH, Murphy DM, Alcock L, Mestdagh P, Vandesompele J, Speleman F, et al. Widespread dysregulation of MiRNAs by MYCN amplification and chromosomal imbalances in neuroblastoma: Association of miRNA expression with survival. PLoS One. 2009;4:e7850. doi: 10.1371/journal.pone.0007850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulte JH, Horn S, Otto T, Samans B, Heukamp LC, Eilers UC, Krause M, Astrahantseff K, Klein-Hitpass L, Buettner R, et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int J Cancer. 2008;122:699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- 17.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42:1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chagnovich D, Fayos BE, Cohn SL. Differential activity of ELAV-like RNA-binding proteins in human neuroblastoma. J Biol Chem. 1996;271:33587–33591. doi: 10.1074/jbc.271.52.33587. [DOI] [PubMed] [Google Scholar]
- 19.Lazarova DL, Spengler BA, Biedler JL, Ross RA. HuD, a neuronal-specific RNA-binding protein, is a putative regulator of N-myc pre-mRNA processing/stability in malignant human neuroblasts. Oncogene. 1999;18:2703–2710. doi: 10.1038/sj.onc.1202621. [DOI] [PubMed] [Google Scholar]
- 20.Grandinetti KB, Spengler BA, Biedler JL, Ross RA. Loss of one HuD allele on chromosome #1p selects for amplification of the N-myc proto-oncogene in human neuroblastoma cells. Oncogene. 2006;25:706–712. doi: 10.1038/sj.onc.1209095. [DOI] [PubMed] [Google Scholar]
- 21.Walton JD, Kattan DR, Thomas SK, Spengler BA, Guo HF, Biedler JL, Cheung NK, Ross RA. Characteristics of stem cells from human neuroblastoma cell lines and in tumors. Neoplasia. 2004;6:838–845. doi: 10.1593/neo.04310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulte JH, Marschall T, Martin M, Rosenstiel P, Mestdagh P, Schlierf S, Thor T, Vandesompele J, Eggert A, Schreiber S, et al. Deep sequencing reveals differential expression of microRNAs in favorable versus unfavorable neuroblastoma. Nucleic Acids Res. 2010;38:5919–5928. doi: 10.1093/nar/gkq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]