Abstract
Mutations affecting components of the mitochondrial electron transport chain have been shown to increase lifespan in multiple species including the worm Caenorhabditis elegans. While it was originally proposed that decreased generation of reactive oxygen species (ROS) resulting from lower rates of electron transport could account for the observed increase in lifespan, recent evidence indicates that ROS levels are increased in at least some of these long-lived mitochondrial mutants. Here, we show that the long-lived mitochondrial mutant isp-1 worms have increased resistance to oxidative stress. Our results suggest that elevated ROS levels in isp-1 worms cause the activation of multiple stress-response pathways including the mitochondrial unfolded protein response, the SKN-1-mediated stress response, and the hypoxia response. In addition, these worms have increased expression of specific antioxidant enzymes, including a marked upregulation of the inducible superoxide dismutase genes sod-3 and sod-5. Examining the contribution of sod-3 and sod-5 to the oxidative stress resistance in isp-1 worms revealed that loss of either of these genes increased resistance to oxidative stress, but not other forms of stress. Deletion of sod-3 or sod-5 decreased the lifespan of isp-1 worms and further exacerbated their slow physiologic rates. Thus, while deletion of sod-3 and sod-5 genes has little impact on stress resistance, physiologic rates or lifespan in wild-type worms, these genes are required for the longevity of isp-1 worms. Overall, this work shows that the increased resistance to oxidative stress in isp-1 worms does not account for their longevity, and that resistance to oxidative stress can be experimentally dissociated from lifespan.
Keywords: Aging, oxidative stress, Caenorhabditis elegans, superoxide dismutase, isp-1, mitochondria, reactive oxygen species, genetics, lifespan
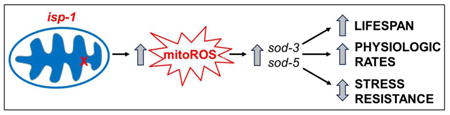
Graphical Abstract
Introduction
Reactive oxygen species (ROS) are generated as a bi-product of normal metabolism as electrons being passed down the electron transport chain (ETC) are leaked directly to oxygen to form superoxide. These ROS have been shown to cause oxidative damage to functional units of the cell, such as DNA, protein and lipids. Since excessive oxidative damage can impair function, and oxidative damage has been shown to increase with age [1], it has been proposed that reactive oxygen species are one of the primary causes of aging [2]. As mitochondria are thought to be the primary site of ROS generation in a cell, the Mitochondrial Free Radical Theory of Aging further proposes that ROS-induced damage to the mitochondria is the main contributor to aging [3]. Based on this theory, decreasing the rate of electrons flowing through the ETC could be predicted to increase longevity by decreasing the production of ROS (assuming equivalent efficiency). Consistent with this idea, a number of mutations that affect proteins in the ETC have been shown to increase lifespan in the worm C. elegans [4-8], as well as in other species, including flies [9] and mice [10, 11].
isp-1 encodes the Rieske iron sulfur protein of complex III in the electron transport chain [6]. While the complete absence of ISP-1 protein results in lethality, a point mutation (qm150) in the exon 6 of 8 exons in the isp-1 gene has been shown to markedly increase lifespan [6]. These mutants have been shown to have a decreased rate of oxidative phosphorylation [6, 12, 13] (note that while oxidative phosphorylation is measured by oxygen consumption, which takes place at complex V, ROS are produced at earlier points in the ETC), an increase in the levels of reduced CoenzymeQ9 resulting from decreased oxidation by complex III [14], and decreased levels of complex I [13]. Based on these observations, it was originally proposed that isp-1 worms live long because of decreased production of ROS (see Figure 5 [6]). However, more recent findings cast doubt on this conclusion, suggesting that ROS levels are actually increased in isp-1 worms despite their decreased rate of oxidative phosphorylation. Whole isp-1 worms exhibit increased staining with two ROS-sensitive fluorescent dyes: DCF (dichlorofluorescein) and DHE (dihydroethidium)[15]. Further, it was also shown that mitochondrial ROS are elevated in isp-1 worms as MitoSox staining is increased in mitochondria isolated from isp-1 worms compared to WT [16]. While measurements of oxidative damage in whole isp-1 worms showed no difference in the level of protein carbonyls [17], the levels of 4-HNE modified proteins were found to be increased in mitochondria isolated from isp-1 worms compared to WT [18]. Combined these data indicate that ROS levels and oxidative damage are not decreased in isp-1 worms.
While unchecked ROS can cause damage to various components of a cell, there are a number of antioxidant enzymes that are present to detoxify ROS [19]. Superoxide dismutase (sod genes) converts superoxide, the primary form of ROS generated in the mitochondria, into hydrogen peroxide, which can then be converted to water by catalase (ctl genes), glutathione peroxidase (gpx genes) or peroxiredoxin (prdx genes). Glutaredoxins (glrx) and thioredoxin (trx) can reduce peroxiredoxins and other targets to restore their function after oxidation. The activity of thioredoxin can then be restored by thioredoxin reductase (trxr genes). Glutathione-S-transferase (gst genes) can act to detoxify products of oxidative stress.
In C. elegans there are five sod genes: sod-1, sod-2 and sod-4 are the primary cytoplasmic, mitochondrial, and extracellular sod genes respectively, and correspond to SOD1, SOD2 and SOD3 in humans. These three sod genes account for 99% of the sod mRNA present in the worm [20]. C. elegans also express two additional sod genes that are not found in most organisms. sod-3 and sod-5 are inducible sod genes that are normally expressed at low levels (less than 1% of total sod mRNA) in the mitochondria and cytoplasm, respectively. While it is known that the expression of these sod genes is induced under stress [21], the function of these genes is poorly understood. Deletion of either sod-3 or sod-5 has no effect on lifespan, sensitivity to juglone-induced oxidative stress, development time, fertility, defecation cycle length or rate of movement in WT worms [22, 23].
In this paper we study the relationship between oxidative stress resistance and longevity in the long-lived isp-1 mitochondrial mutant. We find that although these worms have increased ROS, they have increased resistance to oxidative stress, which is associated with an upregulation of antioxidant defense genes, and the activation of various stress response pathways. Examining the role of the inducible sod genes in the stress resistance and longevity of isp-1 worms, reveals that deletion of these genes increases resistance to acute oxidative stress but decreases lifespan in isp-1 worms. This indicates that having increased levels of ROS does not necessarily result in increased sensitivity to oxidative stress, and that increasing resistance to oxidative stress does not necessarily increase lifespan.
Materials and Methods
Strains
C. elegans strains were cultured on nematode growth medium (NGM) agar plates, which were seeded with OP50, a slow-growing mutant of Escherichia coli. All strains were maintained at 20°C. Wild-type animals were N2 Bristol strain. The following strains were used in these experiments:
| Strain | Description |
|---|---|
| WT(N2) | Wild-type control strain |
| isp-1(qm150) | Long-lived mitochondrial mutant with point mutation in exon 6 of 8 of gene encoding Rieske iron sulfur protein of complex III |
| sod-3(tm760) | Inducible mitochondrial superoxide dismutase mutant with deletion removing exons 2 and 3 of 5 exons. These exons contain part of the active site of the enzyme |
| sod-5(tm1146) | Inducible cytoplasmic superoxide dismutase mutant with deletion affecting exons 2 and 3 of 5 exons. These exons contain part of the active site of the enzyme |
| MQ1378 qmEx413[Psod-3::sod-3:GFP] | Transgenic worms expressing SOD-3:GFP for visualization of SOD-3 expression levels |
| CL2166 dvIs19[Pgst-4::GFP] | Transgenic worms expressing GFP under the promoter of the SKN-1 target gene gst-4 for monitoring activation of SKN-1 oxidative stress response |
| SJ4100 zcIs13[Phsp-6::GFP] | Transgenic worms expressing GFP under the hsp-6 promoter for monitoring activation of the mitochondrial unfolded protein response |
We also generated the following double mutant strains: isp-1(qm150);sod-3(tm760), isp-1(qm150);sod-5(tm1146), isp-1(qm150);Psod-3::sod-3:GFP, isp-1(qm150);Pgst-4::GFP, and isp-1(qm150);Phsp-6::GFP.
mRNA expression levels
Isolation of mRNA
One to two plates of well fed, synchronized, pre-fertile young adult worms were collected in M9 buffer [Na2HPO4 (42.3 uM), KH2PO4 (22uM), NaCl (85 uM), MgSO4 (1mM)]. Worms were washed three times in M9 buffer and flash frozen in Trizol (ThermoFisher Scientific). Once samples for all of the independent replicates had been collected, RNA from all of the samples was extracted at the same time. RNA isolation was done as previously described [24]. Note that the RNA samples used for RNA sequencing were completely independent of those used for quantitative real-time RT-PCR.
RNA sequencing and analysis
For the RNA sequencing (RNA-seq) experiments, we collected six independent samples for WT and isp-1 worms. RNA was isolated independently for all twelve samples. Subsequently, the isp-1 samples were pooled prior to library preparation. Sequencing libraries were prepared using the Kapa Biosystems stranded mRNA-Seq kit for the Illumina platform. Libraries were sequenced using 1×75 bp sequencing on the Illumina NextSeq 500 platform at the Van Andel Research Institute to a depth of ∼30M reads per sample. Read quality was assessed using FASTQC v. 0.11.3 (www.bioinformatics.babraham.ac.uk/projects/fastqc/) and aligned first to the E. coli K-12 genome [25] to assess contamination, then aligned to the WBcel235 C. elegans genome assembly [26] using Subread v. 1.5.0 [27] with default parameters. Transcript abundances were quantified using featureCounts v. 1.4.6 [28] with strand specific read counting. Differential gene expression analysis was performed using the edgeR package v. 3.14.0 [29] in R v. 3.3.0.
To compare gene expression in WT, isp-1, isp-1;sod-3 and isp-1;sod-5 worms, we collected 9 biological replicates per strain and isolated mRNA separately. Sets of three mRNA isolates were pooled at equal concentrations to generate three samples for library preparation and sequencing. Sequencing was performed as described above at a depth of ∼10M reads per sample. RNAseq data is available at NCBI Gene Expression Omnibus (GEO): GSE95240. Heatmaps were generated using the pheatmap package v. 1.0.8 in R v. 3.3.0 (https://CRAN.R-project.org/package=pheatmap). Pathway and gene ontology enrichment analyses were performed using the commonly differentially expressed genes (q < 0.05) between isp-1, isp-1;sod-3, and isp-1;sod-5 relative to WT unless stated otherwise. KEGG and Reactome pathway enrichment analysis was performed using the BACA package v. 1.3 [30] in R v. 3.3.0. Enriched pathways were required to have a minimum of five genes and q < 0.05. Gene ontology enrichment was performed with the GOseq package [31] v. 1.26.0 in R v. 3.3.0 and geneontology.org. Plots were generated using ggplot2 v. 2.2.1 (http://www.springer.com/us/book/9780387981413) and cowplot v. 0.7.0 in R v. 3.3.0 (https://CRAN.R-project.org/package=cowplot).
Quantitative real-time RT-PCR
Quantitative Real-Time RT-PCR (qPCR) was performed in two steps as previously described [24]. mRNA was converted to cDNA using a High-Capacity cDNA Reverse Transcription kit (Life Technologies/Invitrogen). qPCR was performed using a FastStart Universal SYBR Green kit (Roche) in an AP Biosystems RT-PCR machine.
Fluorescent reporter strains
Fluorescent reporter intensity quantification in adult worms was performed by mounting randomly selected adult worms onto an unseeded NGM plate (nematode growth medium) and imaging using a fluorescence dissecting microscope (Nikon SMZ1500) as reported previously [21]. Fluorescent reporter intensity quantification in larval stage worms was performed using a Cellomics Arrayscan high content imager as previously described [32].
Oxidative stress assays
Sensitivity to oxidative stress was assessed using multiple paradigms involving two superoxide-generating compounds: paraquat (PQ) and juglone. PQ produces superoxide through redox cycling: it receives an electron from a donor such as NADPH oxidase, then passes the electron on to dioxygen to generate superoxide, thereby regenerating PQ ion in the process[33]. PQ is thought to increase superoxide levels primarily in the mitochondria [34, 35]. Juglone (5-hydroxy-1,4-napthalenedione) is a natural extract from black walnut trees. It is also believed to generate superoxide through redox cycling [36].
Paraquat development assay
To test oxidative stress during development, a minimum of 40 eggs were transferred to plates containing low concentrations of paraquat ranging from 0.2 mM to 0.35 mM. The developing worms were then monitored for the latest developmental stage attained. The stage to which the worms developed appeared to be sensitive to the exact concentration of paraquat as some variation was observed with different batches of paraquat or different ages of plates. To eliminate this variability, all of the strains were always tested at the same time on the same batch of plates.
Acute oxidative stress during development
Worms that were hatched on NGM plates were transferred to plates containing 200 mM paraquat at either the L2 or L4 stage of development. The survival of these worms was then checked hourly for the 12 hour duration of the assay. At each time point, dead worms were counted and removed. 25 worms per strain per trial were tested.
Acute oxidative stress during adulthood
Sensitivity to oxidative stress was tested acutely during adulthood by placing day 1 or day 8 adult worms on plates containing various concentrations of juglone ranging from 180-300 μM. Survival was monitored at 1, 2, 3 and 4 hours. For this assay, plates were made fresh on the day of the assay as the activity of juglone in plates declines over time. Each trial utilized 25 worms per strain for each concentration of juglone.
Chronic oxidative stress during adulthood
To test sensitivity to oxidative stress throughout adulthood, day 1 adult worms were transferred to plates containing 4 mM paraquat. 100 μM FUdR was also added to these plates to prevent internal hatching of progeny (bagging). Survival was monitored daily until all of the worms had died. The results represent 115 death events for both WT and isp-1 worms.
Heat stress assay
Worms grown on NGM plates at 20°C were transferred to a 37°C incubator on day 1 of adulthood. Survival was monitored hourly until all of the worms had died. Each replicate used 20 worms per strain.
Osmotic stress assay
Worms grown on regular NGM plates (50 mM NaCl) were transferred to plates containing 500 mM NaCl on day 1 of adulthood. Survival was checked at 24 and 48 hours. Each replicate used 20 worms per strain.
Bacterial pathogen stress assay
Worms were exposed to Pseudomonas aeruginosa to induce bacterial pathogen stress following the slow kill assay outlined by Kirienko et. al. [37]. Each replicate used 20 worms per strain.
Lifespan
Lifespan studies were completed at 20°C on plates containing 100 μM FUdR (fluorodeoxyuridine; Sigma). Although this concentration of FUdR can increase the lifespan of certain strains [38], we have found that isp-1 worms have increased lifespan with or without FUdR and that the lifespan of sod-3 and sod-5 mutants is equivalent to WT with or without FUdR. Worms that died from internal hatching of progeny, expulsion of internal contents or desiccation on the side of the dish were removed from the study. Four independent biological replicates were performed with more than 240 death events recorded for each strain.
Post-embryonic development
Eggs were collected and allowed to hatch over a period of 3 hours. After 3 hours, 25 L1 worms per strain per trial were transferred to a new plate. Beginning at 35 hours worms were checked every 4 hours. Worms that had reached adulthood were counted and removed.
Self-brood size
To determine the average number of progeny produced by each strain, L4 worms were placed on individual NGM plates. Worms were transferred daily until egg laying ceased and the total number of live progeny produced was counted. Each replicate consisted of 5 worms. Progeny were allowed to grow to the pre-fertile young adult stage to facilitate counting and were immobilized in a 4°C cold room prior to counting.
Defecation cycle length
Defecation cycle length in young adult worms was measured as the average time between consecutive pBoc contractions. Results represent 10 worms per replicate.
Thrashing rate
Thrashing rate was measured manually. Twenty day 1 adult worms were transferred to an unseeded NGM plate and 1 ml of M9 buffer was added. The total number of body bends was counted for 30 seconds for a randomly selected 10 worms.
Detecting ROS using dihydroethidium (DHE)
ROS levels were measured using DHE (ThermoFisher Scientific, D1168) as previously described [39] with the following modifications. 30mM DHE stock in DMSO was aliquoted and stored at -80°C. 5 μl of stock DHE was diluted in 5 mL of PBS (30μM concentration). Age matched day 1 adult worms (approximately 100 individuals) were picked into a 1.5 tube and washed 3 times in 1 mL PBS. On the final wash, all but 100 μl PBS was removed, and 100 ul of 30μM DHE was added to a final concentration of 15uM DHE in 200 μl. Worms were incubated for 1 hour on a shaker at room temperature, then washed 3 times in PBS and mounted on a 1.5% agarose pad and immobilized with 5mM levamisole. Worms were imaged at 40× using an upright Leica compound microscope (DM5500B). 30-40 worms were imaged for 3 biological replicates. Fluorescence intensity of ethidium labeled ROS was quantified in the anterior pharynx using a ROI (region of interest) method and ImageJ.
Measuring oxidative damage
Carbonyl groups were detected using the Protein Oxidation Detection Kit (Millipore, S7150). Briefly, pellets of day 1 adult worms (1-2 60 mm plates per sample) were collected and flash frozen in lysis Buffer (150 mM KCl, 1mM EDTA, 0.25% SDS, 1% NP-40, 50 mM Tris/HCL pH 7.4), with DTT added to a final concentration of 50mM to prevent oxidation occurring after cell lysis. Samples were freeze-thawed on ice, and sonication was performed using a Bioruptor (Diagenode). Protein concentration of diluted samples (5mM DTT) was determined using the reducing agent compatible BCA kit (Pierce/Thermoscientfic 23250). 5 μg total protein was put into each DNPH and control reaction, and separated on a 10% gel, transferred, and developed according to standard methods. Quantification was performed using ImageJ by collecting total pixel intensity of equally thresholded ROI, with the negative control background subtracted from each sample. Intensity was normalized to WT, and 3 biological replicates were averaged.
Statistical analysis
For all assays, a minimum of three independent biological replicates were performed. Statistical significance of differences was determined using a one-way ANOVA, two-way ANOVA, or repeated measures ANOVA, and a Bonferroni posttest. For lifespan and chronic paraquat data, differences between the survival curves were assessed using a log-rank test. Error bars indicate standard error of the mean.
Results
Increased resistance to oxidative stress in isp-1 worms
During the original characterization of isp-1 worms, it was reported that isp-1 worms fail to develop on plates containing the superoxide-generating compound paraquat at concentrations that did not prevent WT worms from reaching adulthood [6]. We confirmed this result by showing that WT worms can develop to adulthood at concentrations up to at least 0.35 mM paraquat, while isp-1 worms fail to develop to adulthood even at 0.2 mM paraquat (Fig. 1A). While this result is consistent with isp-1 worms having increased sensitivity to oxidative stress, it is also possible that the difference results from the slower development time of isp-1 worms, thereby giving them a more prolonged exposure to paraquat in order to reach adulthood.
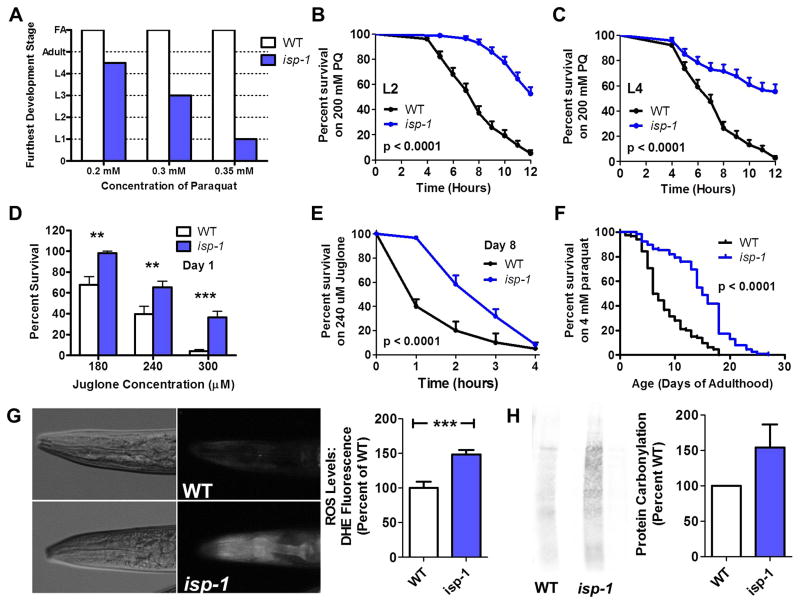
Fig. 1. isp-1 worms have increased resistance to oxidative stress.
(A) isp-1 eggs exposed to paraquat fail to develop to fertile adults at concentrations (0.2 mM – 0.35 mM) that do not prevent wild-type N2 worms from reaching adulthood. However, in an acute oxidative stress assay where worms are exposed to 200 mM paraquat, isp-1 worms show increased survival compared to wild-type worms at the L2 (B) and L4 stages (C) of development. Increased resistance to oxidative stress is also observed during adulthood in isp-1 worms as day 1 adult isp-1 worms (D) and day 8 adult isp-1 worms (E) both survive juglone-induced oxidative stress (180-300 μM) better than wild-type worms. Similarly, in a chronic oxidative stress assay where worms are exposed to a low concentration of paraquat (4 mM) beginning on day 1 of adulthood, isp-1 worms survive significantly longer than wild-type worms. This indicates that isp-1 worms have increased resistance to oxidative stress that begins before the L2 stage of development. Error bars indicate SEM. FA = fertile adult. **p<0.01, ***p<0.001.
To determine whether the long development time of isp-1 worms was responsible for their increased sensitivity to oxidative stress in the paraquat development assay, we performed an acute oxidative stress assay on developing isp-1 worms at the L2 and L4 stages of development. In this assay, we exposed isp-1 and WT worms to 200 mM paraquat and monitored survival. In contrast to the paraquat development assay, we found that isp-1 worms exhibited increased survival compared to WT worms at both the L2 and L4 stage of development (Fig. 1 B,C). This suggests either that isp-1 worms have a differential ability to handle acute high doses of oxidative stress, compared to chronic low doses, or that the increased sensitivity to oxidative stress in the paraquat development assay results from the long development time of isp-1 worms.
To gain more insight into oxidative stress sensitivity in isp-1 worms, we performed both acute and chronic assays of oxidative stress in adult isp-1 worms. In an acute assay of oxidative stress resistance in which worms are exposed to another superoxide-generating compound juglone, isp-1 worms were found to be more resistant to oxidative stress than WT worms at day 1 and day 8 of adulthood (Fig. 1 D,E). Finally, we tested resistance to oxidative stress during adulthood using a chronic exposure to 4 mM paraquat beginning at day 1 of adulthood. Again, we found that isp-1 worms have markedly increased survival compared to WT worms (Fig. 1F).
Since isp-1 worms have been reported to have elevated levels of ROS [16, 39], it is counterintuitive that these worms would have increased resistance to oxidative stress. As such, we sought to confirm that isp-1 worms have increased ROS. We stained isp-1 and WT worms with dihydroethidium (DHE), a compound that emits red fluorescence when oxidized and has been used as an in vivo sensor of ROS [39, 40]. We found that isp-1 worms exhibited increased DHE fluorescence compared to WT worms, thereby confirming that these worms have elevated ROS (Fig. 1G). Similarly, we measured oxidative damage to proteins by measuring protein carbonylation and found an increase in isp-1 worms compared to WT worms (Fig. 1H). Combined our results suggest that isp-1 worms have increased resistance to oxidative stress throughout development and adulthood despite having elevated levels of ROS.
Upregulation of inducible sod genes in isp-1 worms
Based on our observation that isp-1 worms exhibit increased resistance to oxidative stress, we hypothesized that increased ROS production in isp-1 worms might trigger the upregulation of antioxidant defence genes, and activation of stress response pathways leading to oxidative stress resistance. To determine whether antioxidant defenses and stress response pathways are activated in isp-1 worms, we initially used RNAseq to obtain a global view of the transcriptional changes present in isp-1 worms. We focussed specifically on genes encoding antioxidant defense enzymes and those involved in known stress response pathways including the cytoplasmic unfolded protein response, the mitochondrial unfolded protein response, the endoplasmic reticulum unfolded protein response, the hypoxia response, and the SKN-1-mediated oxidative stress response.
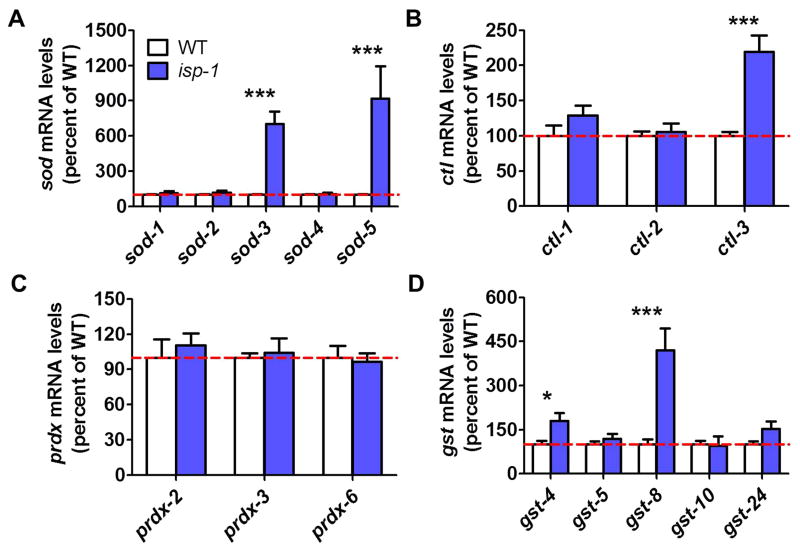
Among the superoxide dismutase genes (SOD), we found that only sod-3 and sod-5 were upregulated (Supplementary Fig. S1A). Among the catalase genes, we observed upregulation of ctl-3 (Supplementary Fig. S1B), while there was no upregulation of any of the peroxiredoxin (prdx), glutathione peroxidase (gpx) or glutaredoxin (glrx) genes (Supplementary Fig. S1C-E). We found that one thioredoxin gene (trx-2) was upregulated, as was the thioredoxin reductase trxr-2 (Supplementary Fig. 1F). Finally, we observed upregulation of multiple glutathione-S-transferase genes (gst-3, gst-4, gst-8, gst-12, gst-13, gst-14, gst-15, gst-16, gst-19, gst-20, gst-21, gst-24, gst-25, gst-29, gst-31, gst-33, gst-34, gst-37, gst-41, gst-44, gsto-1, and gsto-2) (Supplementary Fig. 1G).
To confirm the results of the RNAseq experiment, we performed quantitative real-time RT-PCR for selected antioxidant genes to ensure that the results are reproducible and no significant differences were missed. As with the RNAseq data, we found that sod-3 and sod-5 are significantly upregulated in isp-1 worms, while the expression of other sod genes are unchanged (Fig. 2A). Similarly, we found that the expression of ctl-3 is increased in isp-1 worms, while ctl-1, ctl-2, and all three prdx genes are equivalent to wild-type (Fig. 2B,C). Finally, we observed a significant upregulation of specific glutathione-S-transferase genes (Fig. 2D).
Fig. 2. Inducible sod genes are upregulated in isp-1 worms.
The expression of antioxidant enzymes was assessed using quantitative real-time RT-PCR A. sod-3 and sod-5 were markedly upregulated in isp-1 worms (A). Among the catalase genes ctl-3 expression was increased in isp-1 worms (B), but none of the peroxiredoxins (C) showed increased expression. Specific glutathione-S-transferase genes (gst-4, gst-8) were also significantly upregulated in isp-1 worms. Error bars indicate SEM. *p<0.05, ***p<0.001.
Activation of stress response pathways in isp-1 worms
Next, we sought to determine the extent to which stress response pathways are activated in isp-1 worms. The RNAseq data indicated no activation of the cytosolic unfolded protein response (hsp-16.11, hsp-16.2) or the ER unfolded protein response (hsp-4) (Supplementary Fig. S2A). With respect to the mitochondrial unfolded protein response, we observed a 50% increase in hsp-6 levels, but no difference in the expression of hsp-60 (Supplementary Fig. S2A). Examining the expression of the top genes shown to be induced during short term hypoxia [41] revealed that almost all of these genes are also upregulated in isp-1 worms, suggesting that the hypoxia response is activated (Supplementary Fig. S2B). Finally, we found that the expression of SKN-1 target genes [42] were not consistently upregulated in isp-1 worms but we did see a marked increase in gst-4 (Supplementary Fig. S2C).
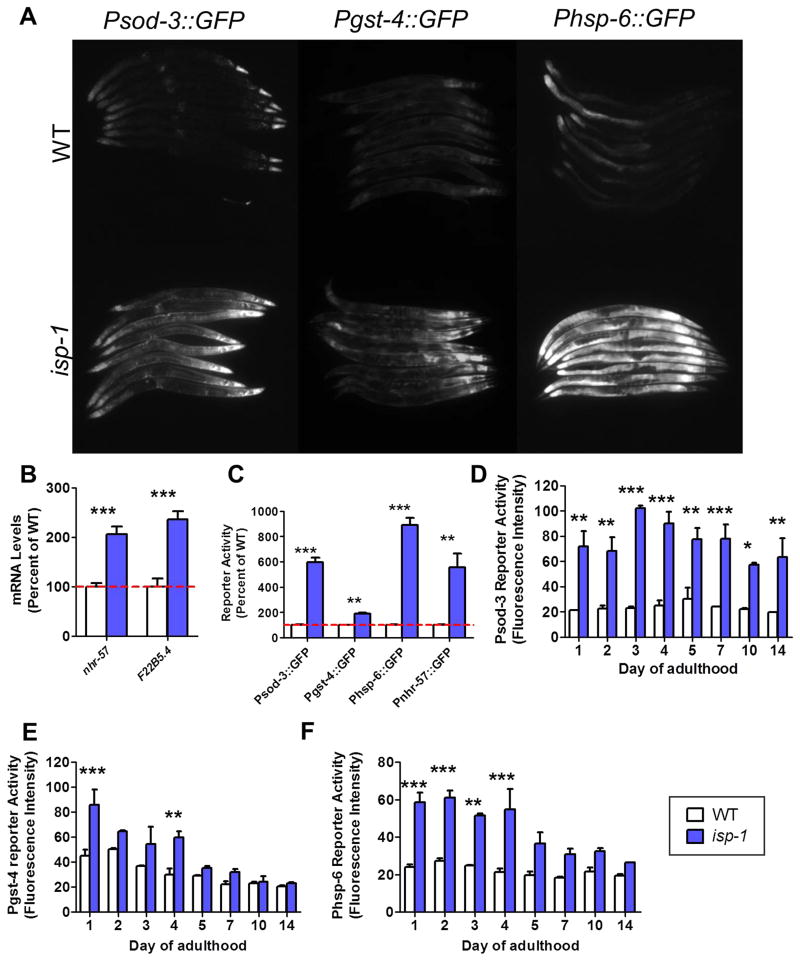
In order to confirm the results from the RNAseq data, we used fluorescent reporter strains or quantitative real-time RT-PCR. We found that reporter strains for the SKN-1-mediated oxidative stress response (Pgst-4::GFP) and the mitochondrial unfolded protein response (Phsp-6::GFP) were both upregulated in isp-1 worms (Fig. 3A). Similarly, we found that target genes for the hypoxia response (nhr-57, F22B5.4) were upregulated in isp-1 worms by quantitative real-time RT-PCR (Fig. 3B).
Fig. 3. Multiple stress response pathways are activated in isp-1 worms.
Fluorescent reporter strains and quantitative real-time RT-PCR were used to monitor the activation of stress response pathways in isp-1 worms. A. Fluorescent reporter strains for an antioxidant stress response (Psod-3::sod-3:GFP), the SKN-1-mediated oxidative stress response (Pgst-4::GFP), and the mitochondrial unfolded protein response (Phsp-6::GFP) were all activated in isp-1 worms. B. Two representative targets of the HIF-1-mediated hypoxia response (nhr-57, F22B5.4) were upregulated in isp-1 worms. C. Examining the time course of stress response activation revealed that all four stress response reporters (Psod-3::GFP, Pgst-4::GFP, Phsp-6::GFP, and Pnhr-57::GFP) already showed increased activation compared to WT worms in developing worms that were 1 day post-hatching. D. The Psod-3::sod-3:GFP reporter shows increased activation throughout adulthood in isp-1 worms. E. The increased activity of the Pgst-4::GFP reporter in isp-1 worms decreases with age, but this stress response pathway has been shown to lose function with age. F. The mitochondrial unfolded protein response exhibits increased activation in isp-1 worms throughout adulthood but the magnitude of increase decreases with age (the ability of this pathway to respond to stress is known to decline with age. Error bars indicate SEM. *p<0.05,**p<0.01, ***p<0.001.
Having shown that antioxidant genes and stress response pathways are activated in isp-1 worms at day 1 of adulthood, we sought to determine if these pathways are also activated during development and whether their activation persists throughout adulthood. We found that at 1 day post-hatching fluorescent reporter strains for sod-3 (antioxidant gene expression), gst-4 (SKN-1-mediated oxidative stress response), hsp-6 (mitochondrial unfolded protein response) and nhr-57 (hypoxia response) were already activated in isp-1 worms (Fig. 3C). This result is consistent with the increased resistance to oxidative stress observed in developing isp-1 worms. The upregulation of sod-3 persisted until at least day 14 of adulthood (Fig. 3D), while the increased expression of gst-4 and hsp-6 diminished with age (Fig. 3E,F). Note that the ability to activate the gst-4 and hsp-6 reporters is lost with age [21].
Paradoxical increase in resistance to oxidative stress with deletion of inducible sod genes
Based on the marked upregulation of sod-3 and sod-5 in isp-1 worms, we sought to determine whether the increased expression of these genes contributed to the increased resistance to oxidative stress of isp-1 worms. Since SOD acts to detoxify ROS, deletion of sod genes should result in a decreased ability to detoxify ROS resulting in increased sensitivity to oxidative stress. However, both sod-3 and sod-5 are normally expressed at very low levels each accounting for less than 1% of the total sod mRNA [20]. According to our RNAseq data, sod-3 makes up 1.6% of the total sod mRNA in WT worms, while sod-5 account for just 0.1% (Supplementary Fig. S3). In isp-1 worms, sod-3 mRNA accounts for 16.6% of total sod mRNA and sod-5 accounts for 2.1% (Supplementary Fig. S3). Nonetheless, the expression of sod-3 and sod-5 in isp-1 worms is still less than sod-2 and sod-1, respectively. To define the contribution of sod-3 and sod-5 to oxidative stress resistance in isp-1 worms, we generated isp-1;sod-3 and isp-1;sod-5 double mutants and compared their stress resistance to isp-1 worms.
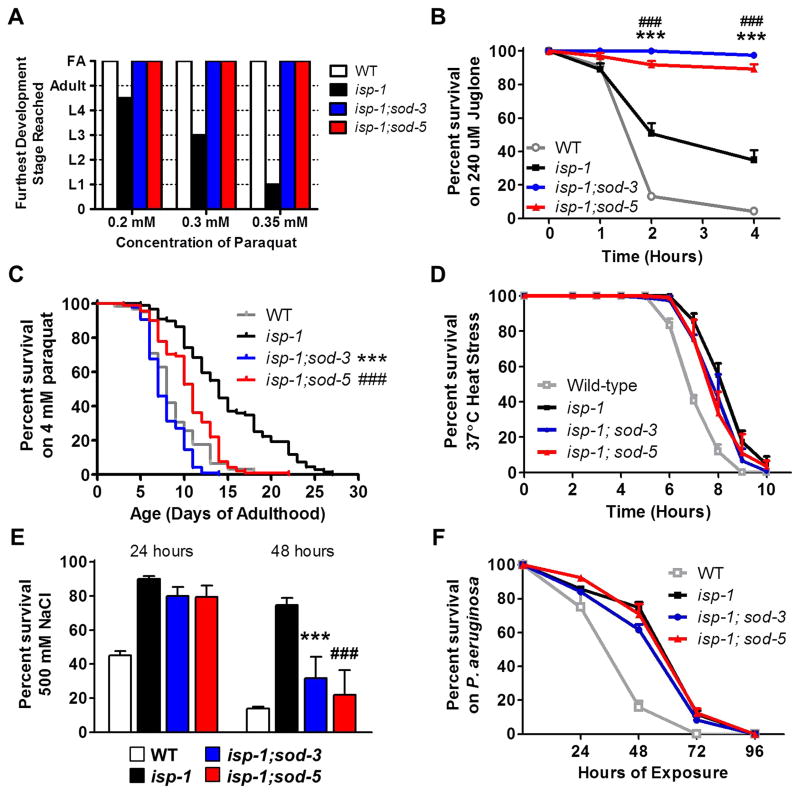
Surprisingly, we found that the deletion of sod-3 or sod-5 resulted in increased resistance to oxidative stress of isp-1 worms in the paraquat development assay. As with WT worms, isp-1;sod-3 and isp-1;sod-5 worms were able to develop to adulthood at concentrations up to at least 0.35 mM paraquat, a concentration at which isp-1 worms fail to develop past the L1 larval stage (Fig. 4A). Similarly, we found that deletion of sod-3 or sod-5 increased resistance to oxidative stress in isp-1 worms in an acute juglone oxidative stress assay on day 1 of adulthood (Fig. 4B). In contrast, isp-1;sod-3 and isp-1;sod-5 worms were found to have decreased survival in a chronic oxidative stress survival assay compared to isp-1 worms (Fig. 4C).
Fig. 4. Deletion of inducible sod genes increases resistance to oxidative stress.
Deletion of either of the inducible sod genes, sod-3 or sod-5, resulted in increased resistance to oxidative stress during development in the paraquat development assay (A) and on day 1 of adulthood in the juglone assay (240 μM juglone) (B). Conversely, isp-1;sod-3 and isp-1;sod-5 double mutants were more sensitive to a chronic exposure to oxidative stress (4 mM paraquat) beginning at day 1 of adulthood (C). The increased resistance to stress was specific to oxidative stress as isp-1;sod-3 and isp-1;sod-5 worms have stress sensitivity equivalent to isp-1 worms in assays of heat stress (37°C) (D), osmotic stress (500 mM NaCl) (E), and bacterial pathogen stress (Pseudomonas aeruginosa) (F). Error bars indicate SEM. Significant differences between isp-1 and isp-1;sod-3 are indicated by ***, those between isp-1 and isp-1;sod-5 are indicated by ###.
To determine if the deletion of sod-3 or sod-5 resulted in increased resistance to other stresses, we compared isp-1 worms to isp-1;sod-3 and isp-1;sod-5 worms in a heat stress assay (exposure to 37°C heat)(Fig. 4D), an osmotic stress assay (exposure to 500 mM NaCl) (Fig. 4E), and a bacterial pathogen stress assay (exposure to Pseudomonas aeruginosa) (Fig. 4F). In each case, we found that isp-1 worms are more resistant to stress than WT worms, but that deletion of sod-3 or sod-5 did not further increase resistance to stress. Combined, this indicates that the loss of either of the inducible sod genes specifically affects sensitivity to oxidative stress.
The inducible sod genes are required for isp-1 longevity and mitigating the slowing of physiologic rates
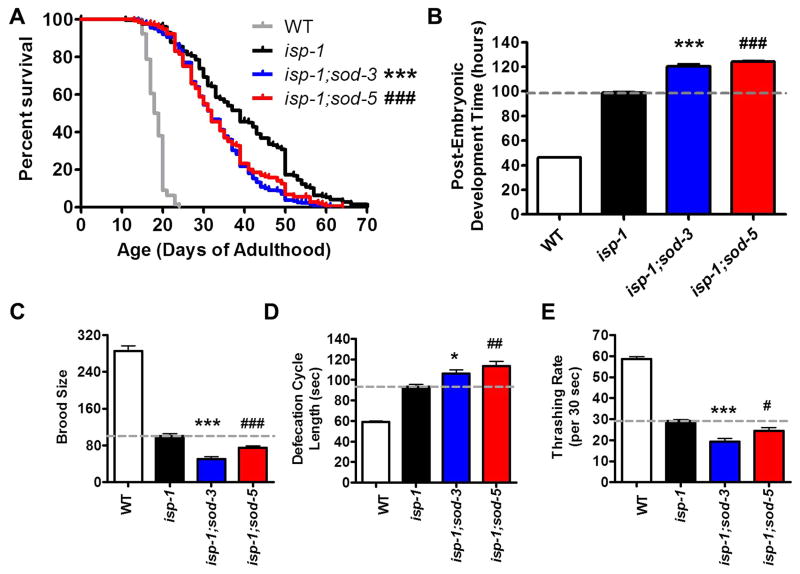
To determine whether the increased expression of sod-3 and sod-5 in isp-1 worms contributes to their long lifespan, we examined the lifespan of isp-1;sod-3 and isp-1;sod-5 double mutants. While deletion of sod-3 or sod-5 has no impact on lifespan in wild-type worms [22], deletion of either of the inducible sod genes in isp-1 worms resulted in a significant decrease in lifespan (Fig. 5A, Supplementary Table S1).
Fig 5. Deletion of inducible superoxide dismutase genes reduces lifespan and exacerbates abnormal physiological rates in isp-1 worms.
A. isp-1 worms have increased lifespan compared to wild-type worms. Deletion of either sod-3 or sod-5 significantly decreases the lifespan of isp-1 worms. (B) isp-1 worms develop slower than wild-type worms. The post-embryonic development time of isp-1 worms was further slowed by deletion of sod-3 or sod-5. (C) isp-1 worms have decreased brood size compared to wild-type worms. isp-1 brood size is further decreased by deletion of sod-3 or sod-5. (D) The defecation cycle length of isp-1 worms is slower than wild-type worms. This phenotype is exacerbated by deletion of sod-3 or sod-5. (E) The thrashing rate of isp-1 worms is slower than wild-type worms and is further decreased by deletion of sod-3 or sod-5. Note that the deletion of sod-3 or sod-5 has no effect on any of these phenotypes in wild-type worms. Error bars indicate SEM. *p<0.05, **p<0.01, ***p<0.001.
Since the presence of the inducible sod genes contributes to the increased longevity of isp-1 worms, we sought to determine whether these genes impacted other physiologic rates, which have been shown to be altered in isp-1 worms [6, 17]. Unlike lifespan where the deletion of the inducible sod genes resulted in a reversion towards WT, we found that deletion of sod-3 or sod-5 resulted in exacerbation of the slow development, decreased brood size, slow defecation, and slow thrashing phenotypes of isp-1 worms (Fig. 5B-E). It is important to note that the development time of isp-1;sod-3 and isp-1;sod-5 worms is significantly longer than isp-1 and thus cannot account for their increased ability to develop to adulthood in the paraquat development assay. Overall, while deletion of sod-3 or sod-5 has little impact on the physiology, stress resistance and lifespan of wild-type worms, these genes are required for isp-1 longevity, and their absence significantly worsens the slow physiology of isp-1 worms.
Deletion of inducible sod genes does not affect ROS levels or oxidative damage in isp-1 worms
To gain further insight into the effect of deleting inducible sod genes on lifespan and resistance to oxidative stress in isp-1 worms, we compared the levels of ROS and oxidative damage in isp-1;sod-3 and isp-1;sod-5 worms to isp-1 worms. Using DHE fluorescence to measure ROS levels, we found that while ROS levels are increased in isp-1;sod-3 and isp-1;sod-5 worms compared to WT, there was no difference from isp-1 worms (Supplementary Figure 4A). Similarly, measuring oxidative damage, we found no difference in the levels of protein carbonylation between isp-1 worms and isp-1;sod-3 or isp-1;sod-5 worms (Supplementary Figure 4B,C). Thus, the differences in oxidative stress resistance and lifespan caused by deletion of the inducible sod genes cannot be attributed to changes in ROS levels or oxidative damage. This result is consistent with the fact that sod-3 and sod-5 are not the primary sod gene in the mitochondria or cytoplasm respectively.
Deletion of inducible sod genes does not increase expression of antioxidant or stress response genes in isp-1 worms
Based on our observation of increased resistance to oxidative stress in isp-1;sod-3 and isp-1;sod-5 worms, we used quantitative real-time RT-PCR to determine how the deletion of the inducible sod genes affected antioxidant genes expression and the expression of genes involved in specific stress response pathways. Among the antioxidant genes tested (sod-1, sod-2, sod-4, ctl-1, ctl-2, ctl-3, prdx-2, prdx-3, prdx-6, gst-8), we did not observe any differences between isp-1 worms and isp-1;sod-3 or isp-1;sod-5 worms (Supplementary Fig. S5). Similarly, most of the stress response genes we tested (nhr-57, F22B5.3, sodh-1, gst-4, hsp-16.2, hsp-6) showed similar expression between isp-1 and isp-1;sod-3 or isp-1;sod-5 worms (Supplementary Fig. S5). We did find that two HIF-dependent hypoxia response genes, mtl-1 and comt-4, exhibit decreased expression in the isp-1;sod double mutant strains compared to isp-1 worms (Supplementary Fig. S5).
Deletion of inducible sod genes alters gene expression in isp-1 worms
To gain a more comprehensive view of the changes in gene expression induced by deletion of sod-3 or sod-5 in isp-1 worms, we performed RNAseq. Overall, we found that isp-1;sod-3 and isp-1;sod-5 worms exhibit similar changes in gene expression to isp-1 worms (Supplementary Fig. S6, S7). Of the genes that are upregulated in isp-1 worms, 39.9% and 40.7% are upregulated in isp-1;sod-3 and isp-1;sod-5 worms, respectively (Supplementary Fig. S8). Of genes that are downregulated in isp-1 worms, 29.6% and 45.7% are also downregulated in isp-1;sod-3 and isp-1;sod-5 worms respectively (Supplementary Fig. S8). The top GO terms for the genes that are modulated in all three mutants are indicated in Supplementary Fig. S9.
Careful analysis of the gene expression heat map (Supplementary Fig. S6) revealed specific genes that show opposite patterns of gene expression in isp-1 worms from isp-1;sod-3 and isp-1;sod-5 worms (Supplementary Fig. S10). This includes genes that are significantly upregulated in isp-1 worms but significantly downregulated in isp-1;sod-3 and isp-1;sod-5 mutants (dod-24, T22B7.7, B1086.3) and genes that are significantly downregulated in isp-1 worms but significantly upregulated in isp-1;sod-3 and isp-1;sod-5 worms (clec-43, Y38F1A.6, C09B8.4, B0511.11). Taken together, the inverted expression profiles for these genes in isp-1 and isp-1;sod-3 and isp-1;sod-5 double mutants demonstrates that changes in transcriptional and metabolic profiles exist, which may begin to explain the differences we observed in lifespan, stress resistance and physiologic rates.
To further explore what genes or groups of genes might account for the phenotypic differences between isp-1 worms and isp-1;sod-3 and isp-1;sod-5 worms, we performed KEGG enrichment analysis and Reactome enrichment analysis on the RNAseq gene expression results to identify functional groups of changes that show a different pattern of gene expression in isp-1 worms compared to isp-1;sod-3 and isp-1;sod-5 worms. In the KEGG analysis, we found that genes in the “ribosome” category are upregulated in isp-1 worms but not in the double mutants (Supplementary Fig. S11). Interestingly, we found that genes involved in “oxidative phosphorylation” and the “citrate cycle” are downregulated in isp-1;sod-3 and isp-1;sod-5 worms but not in isp-1 worms (Supplementary Fig. S11). This result is consistent with our observation that isp-1;sod-3 and isp-1;sod-5 worms have slower physiologic rates than isp-1 worms. In the reactome analysis, we found that there are seven functional categories of genes that are upregulated in isp-1 worms but not in isp-1;sod-3 or isp-1;sod-5 worms (Supplementary Fig. S12). Future studies will be required to determine the extent to which the changes in gene expression between isp-1 and the isp-1;sod double mutants that we have identified contribute to the phenotypic differences between these strains.
Discussion
Lack of correlation between levels of ROS and resistance to oxidative stress
In this work, we show that isp-1 worms exhibit increased sensitivity to oxidative stress in a paraquat development assay, but increased resistance to oxidative stress in an acute juglone assay at the L2 and L4 stages of development. These results are consistent with previous studies showing that fewer isp-1 worms develop to adulthood on plates containing 0.2-0.8 mM paraquat compared to WT worms [6], but that isp-1 worms at the L4 developmental stage have increased survival on 16 mM paraquat plates compared to WT worms [43]. We further go on to show using two different assays that isp-1 worms are also resistant to oxidative stress during adulthood. Combined, our data supports the interpretation that the increased sensitivity to oxidative stress in isp-1 worms in the paraquat development assay is an artifact of their extended development time (100 hours for isp-1 versus 45 hours for WT, [6]) and that isp-1 worms have increased resistance to oxidative stress throughout development and adulthood. This demonstrates that when performing assays involving externally-generated oxidative stress (e.g. exposure to ROS-generating compounds such as paraquat) it is important to consider the duration of exposure and uptake of the compound in the interpretation of the results, as these factors can also influence survival.
We also observed differing results in chronic versus acute assays of oxidative stress sensitivity during adulthood. We found that, in comparison to isp-1 worms, isp-1;sod-3 and isp-1;sod-5 worms show increased resistance to acute oxidative stress during adulthood, but increased sensitivity to chronic oxidative stress. The fact that isp-1;sod-3 and isp-1;sod-5 worms have decreased lifespan compared to isp-1 worms likely contributes to the fact that these worms also have decreased survival in the chronic paraquat assay. Combined, these results suggest that it is important to perform multiple different assays of sensitivity to oxidative stress to obtain a more complete understanding of stress resistance in a given strain.
Previous studies have demonstrated that isp-1 worms have increased levels of ROS as measured by staining with three different ROS-sensitive dyes: DCF, DHE and MitoSox [16, 39] and either normal or increased oxidative damage [17, 18]. Because isp-1 worms demonstrated increased resistance to oxidative stress, we sought to confirm the elevation of ROS levels and oxidative damage in isp-1 worms by staining with a ROS-sensitive fluorescent dye, DHE, and measuring protein carbonylation, respectively. Our results showing increased ROS in isp-1 worms clearly indicate that it is possible to have increased resistance to oxidative stress and elevated levels of ROS. Furthermore, this result suggests that measuring sensitivity to oxidative stress cannot be used to infer ROS levels.
Upregulation of stress response pathways in isp-1 mutants
To explain the mechanism by which isp-1 worms have increased resistance to oxidative stress despite elevated ROS, we hypothesized that these worms activate antioxidant defenses and stress response pathways to compensate for the increased levels of ROS. This hypothesis is supported by our data that demonstrates increased expression of multiple antioxidant genes and genes associated with stress response pathways. These findings are supported by previous studies showing increased expression of sod-3 [6, 18], and the HIF-1-mediated hypoxia response genes nhr-57 and F22B5.4 [39] in isp-1 worms, as well as the activation of SKN-1-mediated oxidative stress reporter Pgst-4::GFP [44], and mitoUPR reporter Phsp-6::GFP [45-47].
Of the stress pathways examined, we found that the majority of the top genes that are activated by short term hypoxia are upregulated in isp-1 worms. Previous work has demonstrated that the hypoxia-induced HIF-1 target genes nhr-57 and F22B5.4 are upregulated in isp-1 worms [15]. The fact that these genes are activated by treatment with paraquat, and that HIF-1 is required for both the upregulation of those genes and for the long lifespan on isp-1 worms suggests that a ROS-activated, HIF-dependent hypoxia response contributes to the longevity of isp-1 worms. Since it has been shown that hypoxia can cause increased generation of mitochondrial ROS [48, 49], it is likely that the overlap in gene expression changes between short term hypoxia and isp-1 mutation results from the fact that both interventions increase mitochondrial ROS.
Two previous studies have examined gene expression changes in isp-1 worms using microarray technologies [50, 51]. The study by Cristina et al. used an Illumina microarray, while the study by Yee et al. used an Affymetrix microarray. We compared the results of the two previous experiments with our RNAseq data using genes in our dataset that exhibited a two-fold change. We found that there were only 39 genes that were found to be upregulated in all three studies (Supplementary Fig. S13, Supplementary Table S2). This represents a small fraction of the 1428, 703 and 1797 genes that were found to be upregulated in our study, the study by Cristina et al., and the study by Yee et al. Similarly, there were only 3 genes out of 376, 105 or 1765 downregulated genes that were found to be in common between all three studies (Supplementary Fig. S13, Supplementary Table S2). While different technologies were utilized in each study, it is still uncertain why the degree of overlap between the three studies represents such a small percentage of the total genes identified in each experiment. Of the genes that did show overlap between all three studies were sod-3, the focus of our current study, and cdr-2 the focus of the study by Cristina et al.[50].
Role of inducible superoxide dismutase genes in longevity, stress resistance and physiologic rates
While humans and many other organisms have three SOD genes – one for each compartment of the cell (cytoplasm, mitochondria and extra-cellular) – C. elegans have five sod genes. In addition to the primary cytoplasmic (sod-1), mitochondrial (sod-2), and extracellular (sod-4) sod genes, the additional sod genes include sod-3, which is localized to the mitochondrial matrix [52, 53], and sod-5 in the cytoplasm [54]. The function of these two additional sod genes is not known. Moreover, our previous work suggested that these sod genes may be expendable as the deletion of sod-3 or sod-5 had no effect on lifespan, development, fertility or movement [22, 23]. While we did observe a small decrease in survival in sod-3 worms exposed to 4 mM paraquat, there was no difference when exposed to 240 μM juglone, and sod-5 mutants exhibited normal sensitivity to oxidative stress in both assays [22].
The fact that we observed little or no phenotypic abnormalities in sod-3 and sod-5 deletion mutants is consistent with the fact that these genes are normally expressed at very low levels. Previous results relying on quantitative real-time RT-PCR indicated that sod-3 and sod-5 account for less than 1% of the total sod mRNA in a worm [20]. Since this result could have been influenced by the efficiency of the primers used for the qPCR reaction, we looked at the distribution of sod mRNA in our RNAseq data. We obtained similar results with sod-3 accounting for 1.6% of the total sod mRNA and sod-5 accounting for just 0.1% of the total sod mRNA, compared to 34.5% and 57.9% for sod-2 and sod-1 respectively.
In isp-1 worms, we found that the large increase in sod-3 and sod-5 expression resulted in these genes making up a much larger proportion of total sod mRNA (16.6% and 2.1% respectively). Because of the marked upregulation of the inducible sod genes in isp-1 worms, we examined their contribution to stress resistance, lifespan and physiologic rates in isp-1 worms by deleting sod-3 or sod-5 in an isp-1 mutant background. Since SOD acts to detoxify superoxide, it would be predicted that deletion of sod genes would lead to increased levels of ROS, due to less ROS detoxification. Thus, it is a surprising result that deletion of either sod-3 or sod-5 resulted in increased resistance to oxidative stress in isp-1 worms in both the paraquat development assay and the acute juglone assay on day 1 of adulthood. It is also surprising that deletion of either of the inducible sod genes has the opposite effect on survival in the chronic paraquat assay. This indicates that the outcome of oxidative stress assays needs to be carefully interpreted, and highlights the importance of using multiple paradigms to assess sensitivity to oxidative stress.
Despite the fact that deletion of the inducible sod genes increased resistance to oxidative stress (except in the chronic paraquat assay), both isp-1;sod-3 and isp-1;sod-5 double mutants had decreased lifespan compared to isp-1 worms. This indicates that resistance to oxidative stress can be experimentally dissociated from longevity. Furthermore, since deletion of sod-3 or sod-5 decreased isp-1 lifespan without affecting resistance to heat stress, osmotic stress or bacterial pathogen stress, this indicates that the increased resistance to stress in isp-1 worms does not account for their longevity.
Conclusions
In this work we show that isp-1 worms have increased resistance to oxidative stress despite having elevated levels of ROS, and that resistance to oxidative stress can be further increased through the deletion of either of the inducible superoxide dismutase genes. The fact that deletion of sod-3 or sod-5 increases resistance to oxidative stress but decreases lifespan demonstrates that resistance to oxidative stress can be experimentally dissociated from longevity. Importantly, while the function of the inducible sod genes in WT worms is unknown, we show that these genes impact stress resistance, lifespan and physiologic rates in the isp-1 mutant background (Supplementary Table S3).
Supplementary Material
Highlights.
isp-1 worms have increased resistance to oxidative stress despite having elevated levels of ROS
Deletion of inducible superoxide dismutase genes paradoxically increases resistance to oxidative stress
Inducible superoxide dismutase genes can impact longevity, stress resistance and physiologic rates
Resistance to oxidative stress can be experimentally dissociated from longevity
Acknowledgments
This work was supported by the National Institute of General Medical Sciences (NIGMS) by grant number RO1 GM121756 (PI: Jeremy Van Raamsdonk) and the Van Andel Research Institute (VARI). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We would like to thank Eric Collins, Julie Koeman, Mary Rhodes and Marie Adams from the VARI Genomics Core Facility for library preparation and RNA sequencing. We would also like to acknowledge the C. elegans knockout consortium and the National Bioresource Project of Japan for providing strains used in this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and dis-ease. Drug metabolism reviews. 1998;30(2):225–43. doi: 10.3109/03602539808996310. [DOI] [PubMed] [Google Scholar]
- 2.Harman D. Aging: a theory based on free radical and radiation chemistry. Journal of gerontology. 1956;11(3):298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 3.Harman D. The biologic clock: the mitochondria? Journal of the American Geriatrics Society. 1972;20(4):145–7. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 4.Wong A, Boutis P, Hekimi S. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics. 1995;139(3):1247–59. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272(5264):1010–3. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 6.Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Developmental cell. 2001;1(5):633–44. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 7.Yang W, Hekimi S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging cell. 2010;9(3):433–47. doi: 10.1111/j.1474-9726.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature genetics. 2003;33(1):40–8. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 9.Copeland JM, Cho J, Lo T, Jr, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Current biology : CB. 2009;19(19):1591–8. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes & development. 2005;19(20):2424–34. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dell'agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, Prelle A, Roubertoux P, Rizzuto R, Zeviani M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Human molecular genetics. 2007;16(4):431–44. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- 12.Falk MJ, Zhang Z, Rosenjack JR, Nissim I, Daikhin E, Sedensky MM, Yudkoff M, Morgan PG. Metabolic pathway profiling of mitochondrial respiratory chain mutants in C. elegans. Mol Genet Metab. 2008;93(4):388–97. doi: 10.1016/j.ymgme.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suthammarak W, Morgan PG, Sedensky MM. Mutations in mitochondrial complex III uniquely affect complex I in Caenorhabditis elegans. The Journal of biological chemistry. 2010;285(52):40724–31. doi: 10.1074/jbc.M110.159608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasta V, Sedensky M, Morgan P, Hahn SH. Altered redox status of coenzyme Q9 reflects mitochondrial electron transport chain deficiencies in Caenorhabditis elegans. Mitochondrion. 2011;11(1):136–8. doi: 10.1016/j.mito.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Current biology : CB. 2010;20(23):2131–6. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS biology. 2010;8(12):e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang W, Li J, Hekimi S. A Measurable increase in oxidative damage due to reduction in superoxide detoxification fails to shorten the life span of long-lived mitochondrial mutants of Caeno-rhabditis elegans. Genetics. 2007;177(4):2063–74. doi: 10.1534/genetics.107.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dingley S, Polyak E, Lightfoot R, Ostrovsky J, Rao M, Greco T, Ischiropoulos H, Falk MJ. Mitochondrial respiratory chain dysfunction variably increases oxidant stress in Caenorhabditis elegans. Mitochondrion. 2010;10(2):125–36. doi: 10.1016/j.mito.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Raamsdonk JM, Hekimi S. Reactive Oxygen Species and Aging in Caenorhabditis elegans: Causal or Casual Relationship? Antioxidants & redox signaling. 2010;13(12):1911–53. doi: 10.1089/ars.2010.3215. [DOI] [PubMed] [Google Scholar]
- 20.Doonan R, McElwee JJ, Matthijssens F, Walker GA, Houthoofd K, Back P, Matscheski A, Vanfleteren JR, Gems D. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes & development. 2008;22(23):3236–41. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dues DJ, Andrews EK, Schaar CE, Bergsma AL, Senchuk MM, Van Raamsdonk JM. Aging causes decreased resistance to multiple stresses and a failure to activate specific stress re-sponse pathways. Aging. 2016;8(4):777–95. doi: 10.18632/aging.100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS genetics. 2009;5(2):e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Raamsdonk JM, Hekimi S. Superoxide dismutase is dispensable for normal animal lifespan. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(15):5785–90. doi: 10.1073/pnas.1116158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machiela E, Dues DJ, Senchuk MM, Van Raamsdonk JM. Oxidative stress is increased in C. elegans models of Huntington's disease but does not contribute to polyglutamine toxicity phenotypes. Neurobiology of disease. 2016;96:1–11. doi: 10.1016/j.nbd.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–62. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 26.C.e.S. Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282(5396):2012–8. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 27.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic acids research. 2013;41(10):e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for as-signing sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 29.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differen-tial expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fortino V, Alenius H, Greco D. BACA: bubble chArt to compare annotations. BMC bioin-formatics. 2015;16:37. doi: 10.1186/s12859-015-0477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome biology. 2010;11(2):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaar CE, Dues DJ, Spielbauer KK, Machiela E, Cooper JF, Senchuk M, Hekimi S, Van Raamsdonk JM. Mitochondrial and cytoplasmic ROS have opposing effects on lifespan. PLoS genetics. 2015;11(2):e1004972. doi: 10.1371/journal.pgen.1004972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinis-Oliveira RJ, Duarte JA, Sanchez-Navarro A, Remiao F, Bastos ML, Carvalho F. Paraquat poisonings: mechanisms of lung toxicity, clinical features, and treatment. Critical reviews in toxicology. 2008;38(1):13–71. doi: 10.1080/10408440701669959. [DOI] [PubMed] [Google Scholar]
- 34.Castello PR, Drechsel DA, Patel M. Mitochondria are a major source of paraquat-induced reactive oxygen species production in the brain. The Journal of biological chemistry. 2007;282(19):14186–93. doi: 10.1074/jbc.M700827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cocheme HM, Murphy MP. Complex I is the major site of mitochondrial superoxide production by paraquat. The Journal of biological chemistry. 2008;283(4):1786–98. doi: 10.1074/jbc.M708597200. [DOI] [PubMed] [Google Scholar]
- 36.Inbaraj JJ, Chignell CF. Cytotoxic action of juglone and plumbagin: a mechanistic study using HaCaT keratinocytes. Chemical research in toxicology. 2004;17(1):55–62. doi: 10.1021/tx034132s. [DOI] [PubMed] [Google Scholar]
- 37.Kirienko NV, Cezairliyan BO, Ausubel FM, Powell JR. Pseudomonas aeruginosa PA14 pathogenesis in Caenorhabditis elegans. Methods in molecular biology. 2014;1149:653–69. doi: 10.1007/978-1-4939-0473-0_50. [DOI] [PubMed] [Google Scholar]
- 38.Van Raamsdonk JM, Hekimi S. FUdR causes a twofold increase in the lifespan of the mitochondrial mutant gas-1. Mechanisms of ageing and development. 2011;132(10):519–21. doi: 10.1016/j.mad.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol. 2010;20(23):2131–6. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei Y, Kenyon C. Roles for ROS and hydrogen sulfide in the longevity response to germline loss in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2016;113(20):E2832-41. doi: 10.1073/pnas.1524727113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng D. Genome-wide analyses of expression data and HIF-1 binding sites provide insights to the HIF-1 hypoxia-inducible factor in Caenorhabditis elegans. Ph D thesis Iowa State University. 2013 [Google Scholar]
- 42.Oliveira RP, Porter Abate J, Dilks K, Landis J, Ashraf J, Murphy CT, Blackwell TK. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging cell. 2009;8(5):524–41. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walter L, Baruah A, Chang HW, Pace HM, Lee SS. The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLoS biology. 2011;9(6):e1001084. doi: 10.1371/journal.pbio.1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khan MH, Ligon M, Hussey LR, Hufnal B, Farber R, 2nd, Munkacsy E, Rodriguez A, Dillow A, Kahlig E, Rea SL. TAF-4 is required for the life extension of isp-1, clk-1 and tpk-1 Mit mutants. Aging (Albany NY) 2013 doi: 10.18632/aging.100604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett CF, Vander Wende H, Simko M, Klum S, Barfield S, Choi H, Pineda VV, Kaeberlein M. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nature communications. 2014;5:3483. doi: 10.1038/ncomms4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suthammarak W, Somerlot BH, Opheim E, Sedensky M, Morgan PG. Novel Interac-tions between Mitochondrial Superoxide Dismutases and the Electron Transport Chain. Aging cell. 2013 doi: 10.1111/acel.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dancy BM, Brockway N, Ramadasan-Nair R, Yang Y, Sedensky MM, Morgan PG. Glutathione S-transferase mediates an ageing response to mitochondrial dysfunction. Mech Ageing Dev. 2015 doi: 10.1016/j.mad.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schieber M, Chandel NS. TOR signaling couples oxygen sensing to lifespan in C. elegans. Cell reports. 2014;9(1):9–15. doi: 10.1016/j.celrep.2014.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate hypoxic signaling. Current opinion in cell biology. 2009;21(6):894–9. doi: 10.1016/j.ceb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cristina D, Cary M, Lunceford A, Clarke C, Kenyon C. A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5(4):e1000450. doi: 10.1371/journal.pgen.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yee C, Yang W, Hekimi S. The Intrinsic Apoptosis Pathway Mediates the Pro-Longevity Response to Mitochondrial ROS in C. elegans. Cell. 2014;157(4):897–909. doi: 10.1016/j.cell.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giglio MP, Hunter T, Bannister JV, Bannister WH, Hunter GJ. The manganese superoxide dismutase gene of Caenorhabditis elegans. Biochemistry and molecular biology international. 1994;33(1):37–40. [PubMed] [Google Scholar]
- 53.Hunter T, Bannister WH, Hunter GJ. Cloning, expression, and characterization of two manganese superoxide dismutases from Caenorhabditis elegans. The Journal of biological chemistry. 1997;272(45):28652–9. doi: 10.1074/jbc.272.45.28652. [DOI] [PubMed] [Google Scholar]
- 54.Jensen LT, Culotta VC. Activation of CuZn superoxide dismutases from Caenorhabditis elegans does not require the copper chaperone CCS. The Journal of biological chemistry. 2005;280(50):41373–9. doi: 10.1074/jbc.M509142200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.