Abstract
In the smut fungus Ustilago maydis, the pheromone signal is transmitted via a mitogen-activated protein kinase module to the high-mobility-group (HMG) domain transcription factor Prf1, leading to its activation. This triggers sexual and pathogenic development since Prf1 binds to the PRE boxes located in the promoters of the a and b mating type genes. Here, we present the characterization of rop1 and hmg3, encoding two additional sequence-specific HMG domain proteins. While hmg3 mutants are slightly impaired in mating and do form conjugation hyphae, rop1 deletion strains display a severe mating and filamentation defect and do not respond to pheromone stimulation. In particular, rop1 is essential for pheromone-induced gene expression in axenic culture. Constitutive expression of prf1 fully complements the mating defect of rop1 mutants, indicating that rop1 is required for prf1 gene expression. Indeed, we could show that Rop1 binds directly to specific elements in the prf1 promoter. Surprisingly, on the plant surface, rop1 deletion strains do form conjugation hyphae and express sufficient amounts of prf1 to cause full pathogenicity. This indicates the involvement of additional components in the regulation of prf1 gene expression during pathogenic growth.
The high-mobility-group (HMG) box superfamily of transcription factors is defined by conserved DNA-binding domains. Based on primary sequence and DNA-binding characteristics, this class of proteins is divided into two major subfamilies. One of these subfamilies includes proteins with multiple HMG boxes exemplified by the HMG-1 and HMG-2 proteins, which have a rather nonspecific affinity for DNA (11, 17, 30). The second subfamily consists of proteins with a single HMG domain, which binds sequence-specifically to variants with the consensus motif (A/T)(A/T)CAAAG (52). Members of this subfamily are important regulators of diverse differentiation processes, including the mammalian sex-determining factor SRY (44), the related SOX proteins (40), and the lymphoid-specific transcription factors LEF-1 and TCF-1 (49). In yeasts and fungi, HMG box proteins of this class are especially involved in sexual differentiation. Examples are Mat-Mc and Ste11 from Schizosaccharomyces pombe, which determine mating type and control conjugation as well as sporulation, respectively (27, 46), Pcc1 in Coprinus cinereus, which functions as a repressor of A-regulated sexual morphogenesis (38), and Prf1 in Ustilago maydis, an essential factor for sexual and pathogenic development (18).
In U. maydis, the causative agent of corn smut disease, fusion of compatible partners is genetically controlled by the biallelic a locus, encoding pheromone (mfa1/2) and receptor (pra1/2) genes (6). Subsequent filamentous growth and pathogenic development are regulated by the multiallelic b locus (4). This locus codes for two homeodomain proteins, bE and bW, which dimerize to generate an active transcription factor when derived from different alleles (16, 24). Basal as well as pheromone-induced transcription of the genes in the a and b loci is regulated by the HMG box protein Prf1, which binds to pheromone response elements (PREs) present in the regulatory regions of the a and b mating type genes (18, 51). Consequently, prf1 deletion mutants are sterile and nonpathogenic.
To perform its function, Prf1 needs to be activated through both a conserved mitogen-activated protein (MAP) kinase module consisting of Kpp4/Ubc4 (MAPKKK), Fuz7/Ubc5 (MAPKK), and Kpp2/Ubc3 (MAP kinase) (1, 4, 22, 35-37) and a conserved cyclic AMP (cAMP) signaling pathway (29, 41). Activation of the MAP kinase cascade results in prf1-dependent gene expression, and among the genes whose transcription is activated are prf1 itself, mfa, pra, bE, and bW (18, 51). In addition, the cAMP pathway triggers pheromone-responsive expression of these genes (29, 41). Cross talk between protein kinase A and MAP kinase signaling during mating is mediated by the posttranscriptional modification of Prf1 at protein kinase A as well as MAP kinase phosphorylation sites (22).
Recent experiments have indicated that the signaling pathway bifurcates downstream of the MAP kinase Kpp2. One branch leads to the described transcriptional responses, and all of these require Prf1 (18, 37). The other branch triggers conjugation tube formation, and this morphological transition is independent of prf1 (37). In addition to posttranscriptional regulation, prf1 is regulated on the transcriptional level by an upstream activating sequence (UAS) located in its promoter (19). The UAS determines expression through environmental signals (e.g., nutrients). Furthermore, two PRE boxes in the prf1 promoter are probably involved in the autoregulation of prf1 transcription.
In this study, we have investigated the roles of additional HMG domain proteins in U. maydis development. We describe the characterization of rop1 in detail, which turned out to be a transcriptional regulator of prf1 exclusively during axenic growth.
MATERIALS AND METHODS
Strains and growth conditions.
The Escherichia coli K-12 derivatives DH5α (Bethesda Research Laboratories) and Top10 (Invitrogen) were used for cloning purposes, and E. coli Rosetta(DE3)(pLysS) (Novagen) was used for protein expression. The U. maydis strains used in this study are listed in Table 1. U. maydis strains were grown as indicated at 28°C in liquid complete medium (CM) (21), nitrate minimal medium (NM) supplemented with either 1% glucose or 1% maltose (19), YEPSL (0.4% yeast extract, 0.4% peptone, 2% sucrose), or potato dextrose (PD) (2.4% PD broth [Difco]) medium or solid PD agar. For induction of the crg1 promoter, strains were grown in CM medium containing 1% glucose to an optical density at 600 nm (OD600) of 0.5, washed twice with water, resuspended in CM medium with 1% arabinose as a carbon source, and grown for the time indicated in each experiment.
TABLE 1.
U. maydis strains used in this study
| Strain | Reference | Plasmid transformed | Integration locus | Progenitor strain |
|---|---|---|---|---|
| FB1 (a1 b1) | 3 | |||
| FB2 (a2 b2) | 3 | |||
| SG200 (a1:mfa2 bE1 bW2) | 5 | |||
| CL13 (a1 bE1 bW2) | 5 | |||
| FB1Pcrg1:fuz7DD | 37 | |||
| FB1Δkpp2-1Pcrg1:fuz7DD | 37 | |||
| FB1Δprf1Pcrg1:fuz7DD | 37 | |||
| HA232 (a2 b2 B-UAS3prf1:ip) | 19 | |||
| FB1Δrop1 | This study | pΔrop1-hyg | rop1 | FB1 |
| FB2Δrop1 | This study | pΔrop1-hyg | rop1 | FB2 |
| SG200Δrop1 | This study | pΔrop1-hyg | rop1 | SG200 |
| HA232Δrop1 | This study | pΔrop1-hyg | rop1 | HA232 |
| FB1Δhmg3 | This study | pΔhmg3-nat | hmg3 | FB1 |
| FB2Δhmg3 | This study | pΔhmg3-nat | hmg3 | FB2 |
| SG200Δhmg3 | This study | pΔhmg3-nat | hmg3 | SG200 |
| FB1Δrop1Pcrg1:fuz7DD | This study | pΔrop1-hyg | rop1 | FB1Pcrg1:fuz7DD |
| FB1Δrop1Δprf1Pcrg1:fuz7DD | This study | pΔrop1-hyg | rop1 | FB1Δprf1Pcrg1:fuz7DD |
| FB1Δhmg3Pcrg1:fuz7DD | This study | pΔhmg3-nat | hmg3 | FB1Pcrg1:fuz7DD |
| FB1Δhmg3Δrop1Pcrg1:fuz7DD | This study | pΔhmg3-nat | hmg3 | FB1Δrop1Pcrg1:fuz7DD |
| FB1Δrop1prf1con | This study | pPRF1con | prf1 | FB1Δrop1 |
| FB2Δrop1prf1con | This study | pPRF1con | prf1 | FB2Δrop1 |
| FB2Δrop1Potef:rop1 | This study | p123Potef:rop1 | ip | FB2Δrop1 |
| SG200Δrop1Potef:rop1 | This study | p123Potef:rop1 | ip | SG200Δrop1 |
| FB1Pprf1:gfp | This study | pPprf1-1746:gfp | ip | FB1 |
| FB2Pprf1:gfp | This study | pPprf1-1746:gfp | ip | FB2 |
| FB1Δrop1Pprf1:gfp | This study | pPprf1-1746:gfp | ip | FB1Δrop1 |
| FB2Δrop1Pprf1:gfp | This study | pPprf1-1746:gfp | ip | FB2Δrop1 |
Hygromycin B was from Roche (Pensberg, Germany), nourseothricin (ClonNAT) was obtained from the Hans Knöll Institute (Jena, Germany), and carboxin was from Riedel de Haen (Seelze, Germany). All other chemicals were of analytical grade and were obtained from Sigma or Merck.
Plasmids and plasmid construction.
Plasmids pTZ19R (Pharmacia), pSP72 (Promega), pSL1180 (Pharmacia), and pBS(+)SKII (Stratagene) were used for cloning, subcloning, and sequencing of genomic fragments, and pCR2.1TOPO (Invitrogen) was used for cloning and sequencing of fragments generated by PCR. pET15b (Novagen) was used for protein expression in E. coli. Sequence analysis of genomic fragments and fragments generated by PCR was performed with an automated sequencer (ABI 377) and standard bioinformatic tools.
pMF1h contains a hygromycin resistance cassette as an SfiI fragment, and pMF1n contains a nourseothricin resistance cassette as an SfiI fragment (8). p123 is a pSP72 derivative containing the enhanced green fluorescent protein (EGFP) gene egfp (Clontech) fused to the otef promoter, an nos terminator, and a carboxin resistance cassette (53). pCU4 codes for carboxin resistance and carries the gfp gene under the control of the constitutive otef promoter (33); pAH298HMG is a pET15b derivative for the heterologous expression of the HMG domain of Prf1 comprising amino acids 1 to 289 as an N-terminally 6xHis-tagged fusion protein (18).
To isolate the hmg3 gene, a 0.77-kb PCR fragment was generated with primers hmg3uni1 (GCGGCATCAGAGCATCG) and hmg3rev2 (GGACTCAGAAGCATCGGC). The amplified hmg3 fragment was used to screen a genomic cosmid library (43). A 3.3-kb region of the hybridizing cosmid 20C9, comprising the hmg3 open reading frame, was sequenced.
The rop1 gene was subcloned from a genomic bacterial artificial chromosome library (LionBioscience) as a 7.7-kb SacII fragment into the SacII site of pBS(+)SKII, and the resulting plasmid was designated pRop1-7.7S.
cDNA of rop1 was isolated from a λgt10 cDNA library (6) by PCR. The PCR fragments were cloned in pCR2.1-TOPO to yield pTB20 to pTB25 and sequenced.
Deletion constructs were generated according to Kämper (23). In particular, for pΔrop1-hyg, a 1.1-kb fragment comprising the 5′ flank and a 1.0-kb fragment comprising the 3′ flank of the rop1 open reading frame were generated by PCR on U. maydis FB1 DNA with primer combinations OTB27L (CATTCCCTTTCGTCGTCCTTGATC)-OTB28L2 (CACGGCTGAGTGGCCTGAAGCAGTCAAATCACGCCAGG) and OTB15R1 (GTGGGCCATCTAGGCCGCACTGAAGTCTTGACACAGATGC)-OTB16R2 (GGTCAGTACTTGATACTAAGCGCC), respectively. These fragments were then digested with SfiI and ligated to the 2.7-kb SfiI hygromycin resistance cassette from pMF1-h (8). The resulting ligation products were cloned into pCR2.1-TOPO. This plasmid, pΔrop1-hyg, was subsequently used as a template to amplify the rop1 deletion construct with primers OTB27L and OTB16R2.
To generate pΔhmg3-nat, a 1.0-kb fragment comprising the 5′ flank and a 1.0-kb fragment comprising the hmg3 3′ region from bp 1713 to 2663 were generated by PCR on U. maydis FB1 DNA with primer combinations L152uni (CGACTTGGAGAAGTGCGCG)-L152SfiIrev (ACACGGCCTGAGTGGCCTAGCGAGATGGAGTTGGGGC) and R152SfiIuni (TGTGGGCCATCTAGGCCCGGCGCGAATCAAGTACAGAGC)-R152rev (GCGATGAGTTTGGCGAGACGG), respectively. These fragments were then digested with SfiI and ligated to the 1.4-kb SfiI nourseothricin resistance cassette from pMF1n (8). The resulting ligation products were cloned into pCR2.1-TOPO to yield pΔhmg3-nat. This plasmid was subsequently used as a template to amplify the hmg3 deletion construct with primers L152uni and R152rev.
Plasmid p123Potef:rop1 was constructed by replacing the 0.7-kb NcoI-NotI egfp gene fragment of p123 with a 2.2-kb NcoI-HindIII fragment from pET15bRop1 (see below) coding for the N-terminally 6xHis-tagged version of Rop1. p123Potef:rop1 carries the 6xHis::rop1 allele under the control of the constitutive otef promoter.
In pPprf1−1746:gfp, the prf1 gene promoter up to position −1746 is ligated to the gfp gene. To yield this plasmid, the otef promoter in pCU4 (33) was replaced by the prf1 promoter.
pET15bRop1 was constructed by ligating a 1.8-kb SalI-BglII-digested PCR product generated with primers Rop1-5′SalI (GTCGACATGGCGCAACAGGGCTATGGC) and Rop1-3′BglII (AGATCTTCAGTGCCCAGAGGCGC) encompassing the complete rop1 open reading frame into the XhoI and BamHI sites of pET15b to yield a translational 6xHis::rop1 fusion at the N terminus of rop1.
pET15bHMGRop1 was constructed by ligating a 0.91-kb SalI- and BglII-digested PCR product generated with primers Rop1HMG-5′SalI (GTCGACCTGTCCAATCATCCGACC) and Rop1HMG-3′BglII (AGATCTGTGTTGCAAGTTCGCCACG) encoding the HMG domain of Rop1 (amino acids 100 to 401 of the protein) into the XhoI and BamHI sites of pET15b to yield a translational 6xHis::rop1100-401 fusion.
For transformation into U. maydis, linear deletion constructs were generated from the deletion plasmids by PCR; pPRF1con (19) was digested with DraI, and plasmids p123Potef:rop1 and pPprf1−1746:gfp were linearized with SspI prior to transformation. In all cases, single homologous integration events into the respective loci were verified by Southern analysis.
DNA and RNA procedures.
Standard molecular techniques were used for DNA and RNA procedures (42). Transformation of U. maydis was performed as published previously (43). U. maydis DNA was isolated as described (20). RNA from strains grown in liquid culture was prepared as described (29) or following the Trizol reagent protocol (Invitrogen). The following probes were used for Northern analyses: a 0.67-kb EcoRV fragment and a 1.3-kb EcoRI-EcoRV fragment from pSP4.2EcoRV (6) for mfa1 and pra1, respectively; a 2.6-kb PvuII fragment from pbW2-Nde-bE1 (10) for bE and bW; and a 1.6-kb EcoRV fragment from pRF-6.0B (19) for prf1. For rop1, a 0.2-kb fragment was generated with primers OTB9 (ACCTCGGCCACCTAATGC) and OTB12 (GGCGATATCGGTAGGTGG) by PCR on FB1 DNA. For hmg3, a 0.77-kb PCR product generated with primers hmg3uni1 and hmg3rev2 (see above) was used. Radioactive labeling was performed with the NEBlot kit (New England Biolabs). A 5′-end-labeled oligonucleotide complementary to the U. maydis 18S rRNA was hybridized as a loading control in Northern analyses (7). For visualization and quantification of radioactive signals, a PhosphorImager (Storm 840; Molecular Dynamics) and the program ImageQuant (Molecular Dynamics) were used.
Mating, pheromone stimulation, and pathogenicity assay.
To test for mating, compatible strains were cospotted on charcoal-containing PD plates (21), and the plates were sealed with Parafilm and incubated at 28°C for 48 h. For pheromone stimulation, strains were grown in CM with 1% glucose to an OD600 of 0.6. Synthetic pheromone (kindly provided by M. Tönnies and H. Kessler) dissolved in dimethyl sulfoxide was added to a final concentration of 2.5 μg/ml, and cells were harvested for microscopic observations and RNA preparations after 5 h of incubation in a 15-ml plastic tube on a tissue culture roller at 28°C.
Plant infections of the corn variety Early Golden Bantam (Olds Seeds, Madison, Wis.) were performed as described previously (36). Fungal structures on the plant surface were visualized by Calcofluor staining as described previously (9).
Fluorimetric measurement of GFP.
Cells grown in NM plus 1% glucose or 1% maltose to an OD600 of 0.5 to 0.8 were pelleted and resuspended in sterile H2O to an OD600 of 1.0; 200 μl of cell suspension was transferred to a microtiter plate, and fluorescence was measured in a Tecan Saphire fluorescence reader. GFP fluorescence was measured at a wavelength of 485 nm for excitation and 520 nm for emission, with a bandwidth of 7.5 nm in both cases. Fluorescence was normalized to the OD600 (22). At least two independent cultures were scored, and each was measured in triplicate to yield mean values.
Protein expression and purification and electrophoretic mobility shift assays.
Rosetta(DE3)(pLysS) cells containing plasmid pET15bHMGRop1 or pAH289HMG (18) were grown in dYT (1.6% trypto-peptone, 1% yeast extract, 0.5% NaCl) containing 1% glucose, ampicillin (100 μg/ml), and chloramphenicol (34 μg/ml) at 37°C. At an OD600 of ≈0.5, cells were shifted to 20°C, and expression was induced by the addition of 0.1 mM isopropylthiogalactopyranoside (IPTG). After overnight incubation, cells were harvested and resuspended in buffer A (20 mM Tris, pH 7.9, 0.5 M NaCl, 5 mM imidazole, 0.1 mM Triton X-100, and complete protease inhibitor cocktail [Roche]). All subsequent steps were performed on ice or at 4°C. Cells were disrupted by five passages through a French pressure cell (Minicell, 20,000 lb/in2). After centrifugation (13,000 × g, 1 h, 4°C) the cleared lysates were subjected to Ni-NTA affinity chromatography (Ni-nitrilotriacetic acid resin, Qiagen) according to the manufacturer's protocol. For washing steps, buffer A containing 60 mM imidazole was used, and elution was performed with buffer A containing 1 M imidazole. The purified His-tagged HMG domains of Rop1 and Prf1 (Rop1100-401 and Prf11-289 proteins, respectively) were desalted on PD10 columns (Pharmacia) with retention buffer (25 mM HEPES, pH 7.9, 100 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, and complete protease inhibitor cocktail; see above) for elution. Fractions (0.5 ml) were collected and assayed for protein content and purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and staining with Coomassie blue.
Electrophoretic mobility shift assays were performed with 100 to 200 ng of purified Rop1100-401 or Prf11-289 protein in retention buffer (see above) supplemented with 1 μg of bovine serum albumin and 0.4 μg of poly(dI-dC) in a total volume of 10 μl. Samples were incubated for 20 min at room temperature, 32P-labeled probe (10,000 cpm) was added, and incubation was continued for 30 min. For competition experiments, unlabeled probes were added after 10 min of incubation, and labeled probes were introduced after further incubation for 10 min.
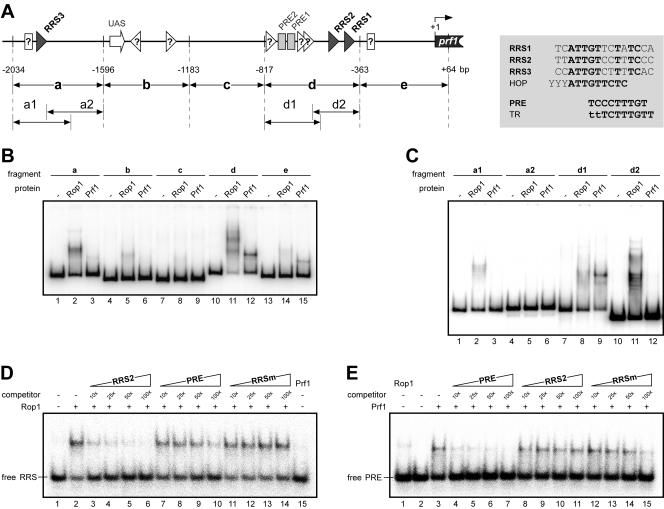
The probes used for the experiments were consecutive (fragments a to e in Fig. 6) and overlapping restriction fragments (a1 and a2 and d1 and d2, Fig. 6) of the prf1 promoter generated by digests of plasmid pRF-6.0B (19). Fragment a is a 438-bp SphI-XhoI fragment; fragment b is a 413-bp XhoI-NdeI fragment; fragment c is a 366-bp NdeI-NspI fragment; fragment d is a 454-bp NspI-XbaI fragment; fragment e is a 427-bp XbaI-SalI fragment; fragment a1 is a 282-bp SphI-NcoI fragment; fragment a2 is a 276-bp NspI-XhoI fragment; fragment d1 is a 268-bp NspI-BpmI fragment; and fragment d2 is a 228-bp SacII-XbaI fragment. In addition, three pairs of complementary oligonucleotides were annealed to yield three double-stranded 33-bp probes with single-base 5′ overhangs: PREuni (CAAATCTGTGAATCCCTTTGTGCCAGTTGACTG) annealed to PRErev (GCAGTCAACTGGCACAAAGGGATTCACAGATTT); RRS2uni (TGCAACCGACTTATTGTCCTTTCCCGAACTCCA) annealed to RRS2rev (TTGGAGTTCGGGAAAGGACAATAAGTCGGTTGC); and RRSm-uni (ATCACCATCACCCGGTGAGGGGCACCAGCGCGC) annealed to RRSm-rev (AGCGCGCTGGTGCCCCTCACCGGGTGATGGTGA). Equimolar amounts of the DNA fragments were 5′-end labeled by T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (6,000 Ci/mmol) according to the manufacturer's instructions.
FIG. 6.
Rop1 binds to specific sequences in the prf1 promoter in vitro. (A) Schematic representation of the prf1 promoter. The UAS is indicated as an open arrow, RRS sequences are depicted as black triangles, and open triangles denote putative additional RRS motifs as suggested by sequence similarity and electrophoretic mobility shift assays. PRE elements are indicated as light grey rectangles, open rectangles denote putative additional PRE elements as suggested by sequence similarity and electrophoretic mobility shift assays. Numbers below indicate positions relative to the ATG. Promoter fragments used in electrophoretic mobility shift assays are indicated as arrows below the scheme. The inset to the right gives the sequences of the three RRS sites identified. Bases present in all three RRS motifs are given in bold. For comparison, the PRE box, a half site of the hypoxic operator (HOP) recognized by Rox1p in S. cerevisiae, and the TR element recognized by Ste11 in S. pombe are shown. (B to E) Electrophoretic mobility shift assays with His-tagged HMG domains of Rop1 and Prf1 protein with (B) prf1 promoter fragments a to e and (C) prf1 promoter fragments a1 and a2 and d1 and d2. (D) Interaction of the Rop1 HMG domain with labeled RRS2 oligonucleotide was analyzed in the presence of increasing amounts (given as fold molar excesses) of the different unlabeled competitors indicated above the lanes. In lane 15, binding of the Prf1 HMG domain to the RRS2 probe was tested in the absence of competitor. (E) Interaction of the Prf1 HMG domain with labeled PRE oligonucleotide was analyzed in the presence of increasing amounts (given as fold molar excess) of the different unlabeled competitors indicated above the lanes. Binding of the Rop1 HMG domain to the PRE probe was tested in the absence of any competitor (lane 1).
Protein-DNA complexes were separated in polyacrylamide gels (4%) in 0.5x Tris-borate-EDTA buffer at 9 mA per gel for 1 to 3 h, depending on probe size. Gels were dried, and radioactive probes and complexes were visualized by PhosphorImager analysis (Molecular Dynamics).
Microscopic observation.
For microscopic observation, a Zeiss Axiophot microscope with differential interference contrast optics was used. Calcofluor fluorescence was observed with a standard DAPI (4′,6′-diamidino-2-phenylindole) filter set. GFP fluorescence was detected with a specific filter set (BP 470/20, FT493, BP 505 to 530; Zeiss, Jena, Germany). Pictures were taken with a charge-coupled device camera (Hamamatsu, Herrsching, Germany). Image processing was done with Image Pro (Media Cybernetics), Adobe Photoshop 6.0, and Canvas 6.0 (Deneba Systems).
Nucleotide sequence accession numbers.
The GenBank accession numbers for rop1 and hmg3 are AY677184 and AY677183, respectively.
RESULTS
Identification of additional HMG domain proteins encoded by U. maydis.
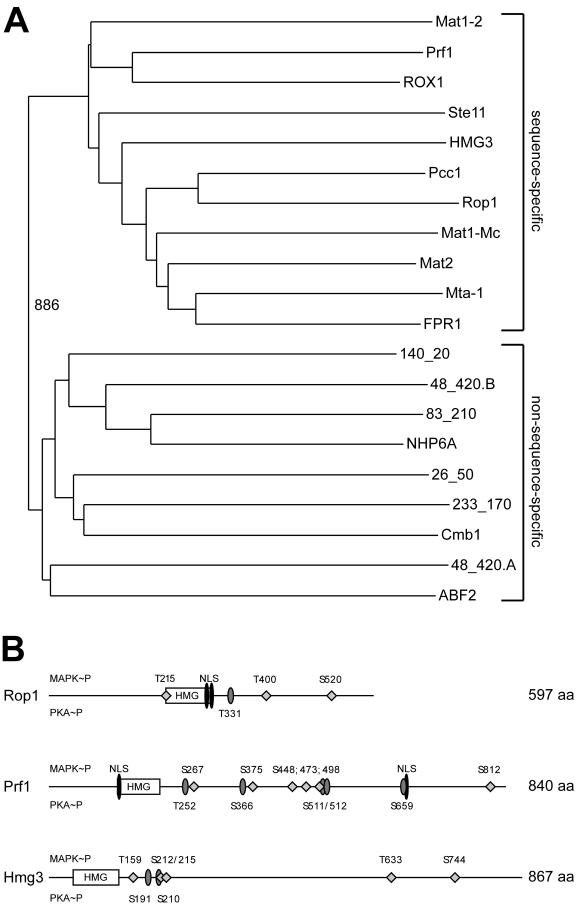
As the HMG box transcription factor Prf1 is essential for mating but not required for the process of conjugation tube formation (37), we were interested in identifying additional transcription factors encoded by the U. maydis genome which may control this morphogenetic process. In the human pathogens Cryptococcus neoformans and Candida albicans as well as in the phytopathogen Magnaporthe grisea Ste12-like homeodomain proteins are important for filamentous growth and virulence (12, 31, 32, 39, 55, 56). However, we were not able to identify an open reading frame coding for an Ste12-like protein in the U. maydis genome. Therefore, we concentrated on the HMG box family of transcriptional regulators as such factors are known to be involved in the regulation of sexual development in fungi. When highly conserved HMG DNA-binding domains were used to screen the U. maydis genome sequence, seven predicted open reading frames coding for potential HMG domain proteins were identified besides Prf1. One of these predicted open reading frames contains two HMG boxes (Fig. 1A, 48_420.A, and 48_420.B). While the HMG domains of two proteins, termed Rop1 and Hmg3, clustered with the sequence-specific class (Fig. 1A), the other six belonged to the non-sequence-specific class (Fig. 1A).
FIG.1.
U. maydis genome contains three sequence-specific HMG domain proteins. (A) Bootstrap analysis (with Clustal X [48] and neighbor-joining plot programs) of the nine HMG domains encoded by the U. maydis genome compared to sequence-specific and non-sequence-specific HMG domains of several fungal species. Amino acid positions in the proteins are given as lowercase numbers. ABF243-111 (S. cerevisiae, accession no. sp|Q02486), Cmb1141-220 (S. pombe, accession no. sp|Q10241), NHP6A21-89 (S. cerevisiae, accession no. sp|P11632), FPR1169-237 (Podospora anserina, accession no. sp|P35693) Mta-1116-184 (Neurospora crassa, accession no. sp|P36981), Mat2 (Gibberella fujikuroi; accession no. dbj|BAA75905.1), Mat1-Mc103-171 (S. pombe, accession no. sp|P10840), Pcc124-98 (C. cinereus, dbj BAA33018.1), Ste1116-80 (S. pombe, accession no. sp|P36631), ROX110-83 (S. cerevisiae, accession no. sp|P25042), and Mat1-2129-203 (C. heterostrophus, accession no. pir|S34811). The HMG domains of Prf1126-197 (accession no. gb|AAC32736), Rop1215-295 (accession no. gb|AY677184), and Hmg357-140 (accession no. gb|AY677183) cluster with the sequence-specific class, whereas the other six Ustilago HMG domains given as annotation codes (http://mips.gsf.de/genre/proj/ustilago) appear in the non-sequence-specific class. (B) Schematic representation of U. maydis Rop1, Prf1, and Hmg3 domain structure. Features were identified with the PrositeScan and PSORT engines. Putative MAP kinase (diamonds) and protein kinase A (grey ellipses) phosphorylation sites are shown; numbers above the line indicate MAP kinase phosphorylation sites, and numbers below protein kinase A phosphorylation sites. The HMG domains are represented by open rectangles, and the potential nuclear localization sequences (NLS) are depicted as black ellipsoids. The numbers on the right indicate the sizes of the proteins in amino acids.
We focused on the two putative sequence-specific HMG domain proteins Rop1 and Hmg3. The rop1 open reading frame comprises 2,778 bp. cDNA analysis revealed two introns of 462 bp and 441 bp. The HMG domain encoded by rop1 (amino acids 216 to 296) shows closest homology to Pcc1 of Coprinus cinereus, a negative regulator of pseudoclamp formation (38) (Fig. 1A). The deduced amino acid sequence of Rop1 is predicted to contain two putative nuclear localization signals (http://psort.nibb.jp) located between amino acids 282 and 296 and three potential MAP kinase sites of the consensus L/PXS/TP (13) (Fig. 1B). In addition, one potential protein kinase A phosphorylation site is predicted at amino acid position 331 (http://www.expasy.org/cgi-bin/scanprosite) (Fig. 1B).
The HMG domain of Hmg3 shows closest homology to Mat2 from Gibberella fujikuroi. The intronless hmg3 gene comprises 2,604 bp and codes for a protein of 867 amino acids that is predicted to be localized in the nucleus (http://psort.nibb.jp) but does not contain an obvious nuclear localization sequence. Hmg3 carries four potential MAP kinase (Fig. 1B) and two potential protein kinase A phosphorylation sites (http://www.expasy.org/cgi-bin/scanprosite) (Fig. 1B), respectively.
rop1 is required for mating.
To analyze the cellular functions of these putative transcription factors, we constructed rop1 and hmg3 deletion strains. To this end the genes in strains FB1 (a1 b1), FB2 (a2 b2), and SG200 (a1::mfa2 bE1 bW2) were replaced with either a hygromycin or nourseothricin resistance cassette (see Materials and Methods for details). The resulting deletion strains were viable, showed no apparent growth defect (not shown), and displayed a budding pattern indistinguishable from that of wild-type strains (see below).
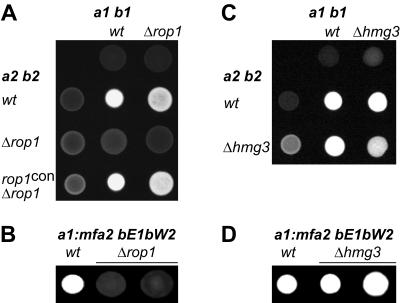
To test for mating, compatible deletion strains were cospotted on PD-charcoal plates. On these plates compatible wild-type strains fuse and form dikaryotic hyphae, which appear as white fuzziness (Fig. 2A). When cospotted with an a2 b2 wild-type partner, compatible FB1Δrop1 strains displayed slightly attenuated filamentation, while compatible mixtures of an a1 b1 wild-type strain and FB2Δrop1 strains showed significant reduction in filament formation (Fig. 2A). This might be due to different genetic backgrounds of the a1 b1 and a2 b2 strains. However, mixtures of compatible Δrop1 strains were unable to develop dikaryotic hyphae (Fig. 2A), indicating that rop1 is required for successful recognition or fusion with a compatible partner. The Δrop1 phenotype could be complemented by introducing the rop1 gene under the control of the constitutive otef promoter into a rop1 deletion strain (Fig. 2A). In addition, the effects of the rop1 deletion were studied in the solopathogenic SG200 strain. The SG200 wild-type strain shows filamentous growth on PD-charcoal plates because of autocrine pheromone stimulation and the presence of an active bE/bW heterodimer (Fig. 2B) (5). SG200Δrop1 strains were nonfilamentous (Fig. 2B), which suggests a postfusion role for Rop1.
FIG. 2.
Mating and filamentation defects of rop1 and hmg3 deletion strains. The strains indicated on top are FB1 (a1 b1) and SG200 (a1::mfa2 bE1 bW2) derivatives. Strains indicated to the left are FB2 (a2 b2) derivatives. Indicated wild-type (wt) and mutant strains were spotted alone or in combinations on charcoal-containing PD plates. Dikaryotic and b-dependent filaments appear as white fuzziness.
The plate mating assay with compatible hmg3 deletion strains revealed a reduction in dikaryotic filament formation which was only apparent in crosses of compatible Δhmg3 strains (Fig. 2C). Filamentation of the solopathogenic SG200Δhmg3 strains was similar to that of the respective wild type (Fig. 2D). This indicates minor defects in cell fusion of hmg3 deletion strains.
rop1 is essential for conjugation tube formation upon pheromone stimulation, while hmg3 is not.
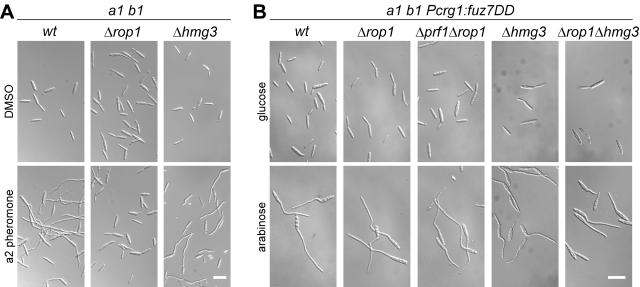
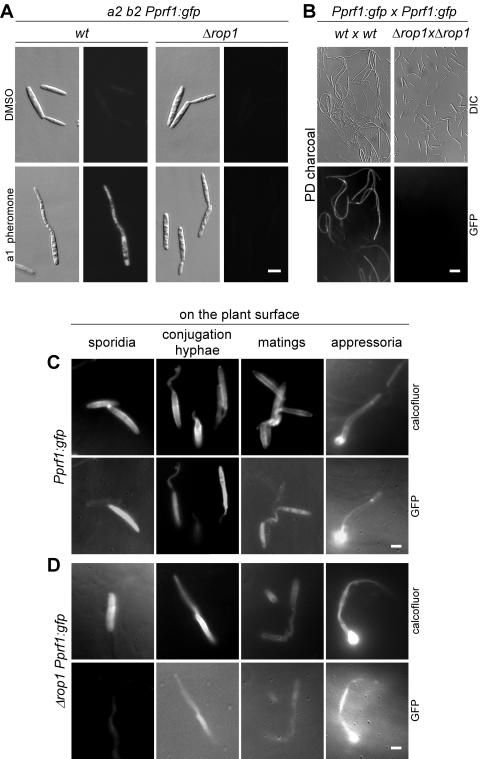
To examine the roles of these two HMG box proteins in the pheromone signaling pathway, we tested the deletion strains for their ability to form conjugation hyphae upon stimulation with synthetic pheromone (47). When stimulated with pheromone, wild-type cells react with the formation of conjugation hyphae, and this morphological transition could also be observed with hmg3 mutant cells (Fig. 3A). In contrast, pheromone-stimulated Δrop1 cells were unable to form conjugation hyphae (Fig. 3A). On these grounds, we concentrated on further characterization of rop1, which is essential for conjugation tube formation.
FIG. 3.
Influence of rop1 and hmg3 deletions on conjugation tube formation. (A) The FB1 (a1 b1) wild-type (wt) and mutant strains indicated on top were stimulated with a2 pheromone for 5 h (lower panel) or treated with dimethyl sulfoxide for the same period of time (upper panel). All pictures were taken at the same magnification. Bar, 20 μm. (B) All strains indicated on top are derivatives of FB1Pcrg1::fuz7DD, encoding an arabinose-inducible, constitutively active allele of the MAPKK Fuz7. Cell morphology after 5 h in arabinose-containing medium (lower panels) was compared to that of cells grown with glucose as the carbon source for the same period of time (upper panels). All pictures were taken at the same magnification. Bar, 20 μm.
Overexpression of the constitutively active MAPKK Fuz7DD leads to the formation of conjugation tubes in rop1 deletion strains.
The observed phenotypes of rop1 mutants could be due to defects upstream or downstream of the MAP kinase cascade, which is involved in transmitting the pheromone signal. To analyze this, we deleted rop1 in a strain coding for a constitutively active allele of the MAPKK fuz7 (fuz7DD) under the control of the arabinose-inducible crg1 promoter. The arabinose-induced FB1Pcrg1::fuz7DDΔrop1 strain developed conjugation tubes as efficiently as the progenitor strain (Fig. 3B). This makes it unlikely that Rop1 is the transcription factor regulating conjugation tube formation downstream of the MAP kinase cascade.
To check for redundant functions of Rop1, Hmg3, and Prf1 in this process, we examined Δprf1 Δrop1 and Δrop1 Δhmg3 double mutants. Again, arabinose induction of fuz7DD resulted in efficient formation of conjugation hyphae in these strains (Fig. 3B), making it unlikely that these transcription factors can substitute for each other.
Rop1 is essential for pheromone-responsive gene expression.
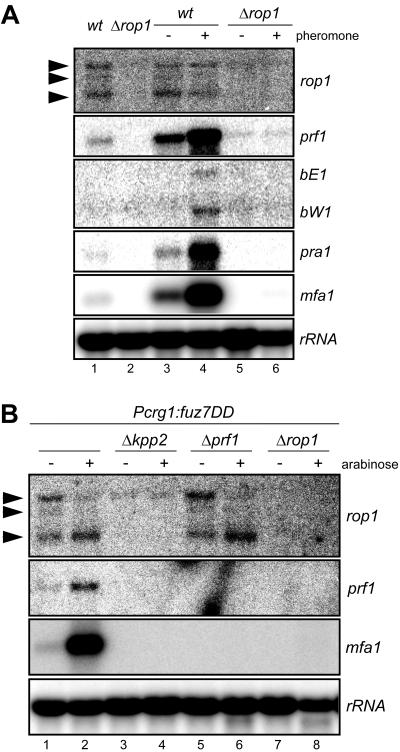
To further dissect the role of rop1 during mating, we performed Northern analyses of prf1, pra1, mfa1, bE, and bW after pheromone treatment of rop1 mutant and wild-type cells as controls (Fig. 4). As shown before (51) the pheromone-responsive genes prf1, bE1, W1, pra1, and mfa1 showed a basal level of expression (Fig. 4A, lanes 1 and 3) which was strongly induced upon stimulation with compatible pheromone (Fig. 4A, lane 4). In contrast, in rop1 mutants, neither basal nor pheromone-induced transcription of these genes could be detected (Fig. 4A, lanes 5 and 6).
FIG. 4.
rop1 is required for pheromone-responsive gene expression. RNA was prepared from the strains listed on top and subjected to Northern analysis. (A) All strains indicated on top are FB1 (a1 b1) derivatives. Cells were either untreated (lanes 1 and 2) or treated for 5 h with synthetic a2 pheromone dissolved in dimethyl sulfoxide (+) or with the same volume of dimethyl sulfoxide (−) (lanes 3 to 6); 10 μg of total RNA was loaded per lane. The blot was probed successively with the gene probes indicated on the right. (B) All strains used are derivatives of strain FB1Pcrg1::fuz7DD. Strains were grown with glucose (−) or arabinose (+) as the carbon source. RNA was isolated, and 12 μg of total RNA was loaded per lane. The filter was hybridized in succession with the gene probes indicated on the right. The rRNA probe served as a loading control. Arrowheads mark the three rop1 transcripts.
We also analyzed expression of these genes in a strain carrying the arabinose-inducible constitutively active allele of fuz7 (fuz7DD). Again, rop1 was required for basal as well as induced expression of mfa1 and prf1 under these conditions (Fig. 4B, lanes 7 and 8). Since deletion of rop1 affects the transcription of several genes regulated by Prf1, rop1 is most likely essential for the expression of the pheromone response factor prf1. This observation also suggests that rop1 acts downstream of the Kpp2 MAP kinase cascade. Alternatively, Rop1 could lie in a parallel pathway feeding into prf1 gene transcription.
Regulation of rop1 gene expression.
Northern analyses of rop1 gene expression in haploid cells revealed the presence of three rop1 transcripts of different sizes. Whereas the amounts and ratios of these transcripts remained unaffected by pheromone stimulation (Fig. 4A, lanes 3 and 4), the expression of the constitutively active MAPKK Fuz7DD induced the disappearance of the larger transcript (Fig. 4B, lanes 1 and 2) and enhanced the amounts of the smallest rop1 transcript (Fig. 4B, lanes 1 and 2). Moreover, the MAP kinase Kpp2 was required for the accumulation of all rop1 transcripts (Fig. 4B, lanes 3 and 4), while deletion of prf1 did not affect the amounts and ratios of the rop1 transcripts (Fig. 4B, lanes 6 and 7). This demonstrates that kpp2 but not prf1 is involved in the transcriptional regulation of rop1.
To rule out the possibility that the MAP kinase Kpp6 (9) represses rop1 gene expression in the absence of Kpp2 protein, we have analyzed rop1 transcript levels under conditions where the MAP kinase module was genetically activated in the presence of either wild-type Kpp2 or the unphosphorylatable Kpp2AEF version (37). In the strain expressing Kpp2AEF, significantly lower levels of rop1 transcripts were detected (not shown; H. Eichhorn and R. Kahmann, unpublished). This illustrates that a functional Kpp2 protein is required for rop1 expression. Since the rop1 gene contains two introns of 462 and 441 bp in its 5′ region, we considered the larger transcripts to be unspliced variants. Some hints that would support this notion came from the sequencing of 5′- and 3′-rapid amplification of cDNA ends (RACE) products (data not shown). 5′-RACE experiments revealed the existence of four RNA species corresponding to unspliced, spliced for either the first or second intron, or spliced at both introns (not shown). 3′-RACE with an oligo(dT) primer amplified only the rop1-specific product without introns, indicating that the unspliced or partially spliced rop1 RNAs are not polyadenylated (not shown). Taken together, these data indicate that the MAP kinase module is required for rop1 expression as well as for correct processing of the rop1 message.
Δrop1 defects are complemented by constitutive expression of prf1.
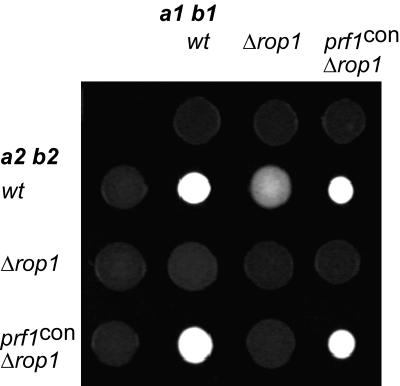
To substantiate the assertion that the observed phenotypes of rop1 mutants are due to the insufficient expression of prf1 we replaced the native prf1 promoter with the constitutive tef promoter in the Δrop1 background. The constitutive expression of prf1 in compatible rop1 deletion strains restored mating on charcoal (Fig. 5), indicating that Rop1 is an important regulator of prf1.
FIG. 5.
Constitutive expression of prf1 restores mating of rop1 deletion strains. The strains indicated on top are FB1 (a1 b1) derivatives. Strains indicated to the left are FB2 (a2 b2) derivatives. The indicated wild-type (wt) and mutant strains were spotted alone or in combinations on charcoal-containing PD plates. Dikaryotic filaments appear as white fuzziness.
The prf1 promoter is subject to complex transcriptional regulation. On the one hand it is autoregulated by Prf1 itself via the pheromone response elements (PREs) in the proximal part of the promoter (18). On the other hand prf1 gene expression is subject to nutritional signaling via the upstream activating sequence (UAS) in the distal part of the promoter (19). To assay whether rop1 acts via the UAS, we deleted rop1 in HA232. This strain carries a gfp reporter gene under the control of a triple repeat of the UAS (19). As shown before, UAS reporter gene activity is highly induced in cells grown in NM with glucose compared to cells grown in NM with maltose (19) (Table 2). Since the expression levels of the reporter gene were comparable in the rop1 deletion strain (Table 2), rop1 does not regulate prf1 transcription via the UAS.
TABLE 2.
Influence of rop1 deletion on UAS activity and pheromone induction of the prf1 promoter
| Strain | Relative fluorescencea (±SEM)
|
|||
|---|---|---|---|---|
| NM-glucose | NM-maltose | Control | Pheromone | |
| HA232 | 1.00 (±0.09) | 0.13 (±0.02) | ND | ND |
| HA232Δrop1 | 0.98 (±0.07) | 0.15 (±0.02) | ND | ND |
| FB1Pprf1:gfp | ND | ND | 1.00 (±0.03) | 36.02 (±0.34) |
| FB1Δrop1Pprf1:gfp | ND | ND | 0.19 (±0.03) | 0.11 (±0.02) |
| FB2Pprf1:gfp | ND | ND | 1.00 (±0.10) | 13.28 (±0.09) |
| FB2Δrop1Pprf1:gfp | ND | ND | 0.45 (±0.19) | 0.40 (±0.06) |
Relative fluorescence was calculated from measured GFP fluorescence per OD600. The average relative fluorescence of the wild-type strains (HA232, FB1Pprf1:gfp, and FB2Pprf1:gfp) grown in glucose-containing medium or treated with dimethyl sulfoxide (control) was set to 1.
Rop1 protein binds the prf1 promoter specifically.
To analyze whether Rop1 directly regulates prf1 gene expression via elements in the prf1 promoter, we examined heterologously expressed His-tagged versions of Rop1 and Prf1 proteins comprising the HMG domains (Rop1100-401 and Prf11-289 respectively) in electrophoretic mobility shift assays (Fig. 6). Under the binding conditions used, Prf11-289 binds to the PRE-containing fragment d (Fig. 6B, lane 12) and shows weak interactions with fragments a and e (Fig. 6B, lanes 3 and 15). The latter can be explained by the presence of PRE-related elements in these fragments.
To map the binding sites more precisely, fragments a and d were digested into two overlapping fragments and used in electrophoretic mobility shift assays (Fig. 6C). Again, Prf11-289 only showed significant interaction with the PRE-containing fragment d1 (Fig. 6C, lane 9). Under the same conditions, purified Rop1100-401 showed strong binding to prf1 promoter fragments a and d (Fig. 6B, lanes 2 and 11), whereas for fragments b, c, and e only very weak interactions could be observed (Fig. 6B, lanes 5, 8, and 14). To delineate the Rop1 recognition site, the high-affinity-binding fragments a and d were digested into two overlapping fragments and used in electrophoretic mobility shift assays (Fig. 6C). This experiment revealed a strong specific interaction of Rop1100-401 with fragments a1 and d2 (Fig. 6C, lanes 2 and 11), while the interaction with fragment d1 was only weak (Fig. 6C, lane 8).
A comparison of the DNA sequences of fragments a1 (without the a2 overlap) and d2 led to the identification of three nearly identical sequence stretches of 11 bp with the consensus ATTGT(T/C)(C/T)T(A/T)TC with significant similarities to binding sites of other sequence-specific HMG domain proteins (Fig. 6A). This motif is exclusively present in the Rop1100-401 binding fragments of the prf1 promoter (Fig. 6A, fragments a and d), which made it a good candidate for a Rop1 recognition site (RRS). To test this directly, synthetic double-stranded oligonucleotides comprising one of the three putative RRS sequences were tested in electrophoretic mobility shift assays. Indeed, Rop1100-401 showed a high affinity for all three oligonucleotides, whereas Prf1 did not (Fig. 6D, data shown for RRS2, lanes 2 and 15).
To demonstrate specificity of the DNA-protein interactions, we performed competition experiments by adding increasing amounts of unlabeled oligonucleotides comprising either the PRE box (TCCCTTTGT), the RRS2 element (ATTGTCCTTTC), or a mutagenized RRS motif (RRSm; CGGTGGAGCGA). The retarded Rop1 RRS2 complex was competed efficiently with a 10-fold molar excess of RRS2 oligonucleotide (Fig. 6D, lanes 3 to 6), while a 100-fold molar excess of the PRE oligonucleotide was required for significant competition (Fig. 6D, lanes 7 to 10). As expected for a sequence-specific interaction, the RRSm oligonucleotide containing the mutagenized RRS motif showed no competition at all (Fig. 6D, lanes 11 to 14). This demonstrates that Rop1100-401 protein binds sequence-specifically to the RRS motif in vitro. In addition, Rop1100-401 showed a very weak affinity for the related PRE box (Fig. 6E, lane 1). This serves to explain the weak interaction of Rop1100-401 protein with the prf1 promoter fragment d1 containing two PRE boxes (Fig. 6C, lane 8). On the other hand, only the PRE (Fig. 6E, lanes 4 to 7) but not the RRS2 (Fig. 6E, lanes 8 to 11) or the RRSm oligonucleotide (Fig. 6E, lanes 12 to 15) was efficient in competing with Prf1 binding to the labeled PRE oligonucleotide (Fig. 6E). Taken together, these results demonstrate that the HMG domains of Rop1 and Prf1 recognize distinct elements in the prf1 promoter.
rop1 deletion strains are fully pathogenic.
To analyze the role of rop1 during pathogenic development, we performed plant infections with crosses of compatible rop1 deletion strains. As prf1 deletion strains are nonpathogenic and rop1 is essential for prf1 gene expression, we were surprised to observe disease symptoms with compatible Δrop1 strains which were as severe as symptoms induced by wild-type strains (Table 3). This result suggests that rop1 is dispensable for prf1 gene expression during infection.
TABLE 3.
Pathogenicity of rop1 deletion strains
| Strain or cross | No. of plants infected | No. of plants with tumors | % with tumors |
|---|---|---|---|
| FB1 × FB2 | 31 | 29 | 94 |
| FB1Δrop1 × FB2Δrop1 | 65 | 61 | 94 |
| SG200 | 24 | 23 | 96 |
| SG200Δrop1 | 44 | 41 | 93 |
To monitor prf1 gene expression during axenic and pathogenic growth, compatible wild-type and rop1 deletion strains encoding a gfp-reporter gene under the control of the prf1 promoter were generated. The resulting haploid strains were first analyzed for gfp expression after treatment with pheromone or the solvent dimethyl sulfoxide as a control (Fig. 7A). The wild-type strains showed weak basal fluorescence and formed brightly fluorescent conjugation hyphae upon pheromone stimulation. The rop1 deletion strains did not form conjugation hyphae, and no GFP fluorescence was detectable (Fig. 7A). Moreover, the expression levels of the Pprf1::gfp reporter were quantified by fluorimetric measurement. This revealed a 13-fold induction in FB2 treated with a1 pheromone and a 36-fold induction in FB1 treated with a2 pheromone (Table 2).
FIG. 7.
rop1 is dispensable for prf1 gene expression and fungal development on the plant surface. All strains carried one copy of a transcriptional prf1 promoter-gfp fusion (Pprf1::gfp) integrated into the ip locus. (A) The FB2 (a2 b2)-derived strains are indicated on top; they were stimulated with compatible a1 pheromone for 5 h (lower panel) or treated with dimethyl sulfoxide for the same period of time (upper panel). The right panels show GFP fluorescence of the same cells depicted on the left. All pictures were taken with the same magnification. Bar, 4 μm. (B) Mixtures of compatible wild-type (wt) or compatible Δrop1 strains carrying the reporter (Pprf1::gfp) were spotted on charcoal-containing PD plates. The cell mixtures indicated on top were microscoped after 1 day of incubation. Lower panels show GFP fluorescence of the differential interference contrast-visualized cells depicted in the upper panels. All pictures were taken at the same magnification. Bar, 20 μm. (C and D) Mixtures of compatible strains at various stages of development (sporidia, conjugation hyphae, matings, and appressoria) on the plant surface. (C) Wild-type strains. (D) Δrop1 mutants. Pictures were taken as indicated on the right with a DAPI filter after Calcofluor staining (upper panels) or a GFP filter to visualize reporter gene expression (lower panels). All pictures were taken at the same magnification. Bar, 4 μm.
In Δrop1 strains, basal expression of the reporter gene was reduced twofold and fivefold in the FB2- and FB1-derived strains, respectively, and pheromone treatment did not elicit an increase in Pprf1::gfp expression (Table 2). These results confirmed the Northern data on prf1 gene expression (Fig. 4A, lanes 3 to 6). To monitor gfp expression during mating conditions in axenic culture, mixtures of FB1Pprf1::gfp and FB2Pprf1::gfp as well as FB1Δrop1Pprf1::gfp and FB2Δrop1 Pprf1::gfp were cospotted on PD-charcoal plates for 24 h and 48 h at 28°C and then subjected to microscopic analysis. While the wild-type strains efficiently formed dikaryotic hyphae which showed strong GFP fluorescence (Fig. 7B), not a single mating event or dikaryon could be observed in mixtures of Δrop1 strains, and the budding cells displayed little or no GFP fluorescence (Fig. 7B). These findings underscore that rop1 is essential for prf1 gene expression during axenic growth. However, when the same strain combinations were used to infect plants, conjugation hyphae, dikaryotic filaments, and appressorium-like structures could be detected in wild-type (Fig. 7C) as well as Δrop1 derivatives (Fig. 7D). In addition, GFP fluorescence was visible in all these structures (Fig. 7C and D). This demonstrates that the prf1 gene is expressed in the plant environment even in the absence of rop1.
DISCUSSION
In this study we have identified two additional sequence-specific HMG box proteins, Rop1 and Hmg3, of which Rop1 was further analyzed. We provide evidence that rop1 is an essential regulator for prf1 gene expression during mating in axenic culture. Epistasis analyses positioned rop1 downstream of the Kpp2 MAP kinase cascade, and in vitro data showed that Rop1 binds specifically to the prf1 promoter. Despite the crucial role of Rop1 for prf1 transcription during growth in culture, rop1 proved dispensable for prf1 gene expression on the plant surface.
HMG domain proteins as developmental regulators in fungi.
The two additional transcription factors, Rop1 and Hmg3, identified in this study belong to the sequence-specific DNA binding class of HMG domain proteins. We demonstrate that rop1 is essential for basal and induced expression of prf1, and this explains all phenotypes of the rop1 deletion strains associated with mating, pheromone-responsive gene expression, and dikaryon formation during axenic growth. The observation that constitutive expression of prf1 complements the mating defect of rop1 deletion strains reinforces the notion that Rop1 is a transcriptional activator for prf1, and this was further substantiated by demonstrating direct binding to the prf1 promoter (see below). Therefore, we conclude that Rop1 directly activates the transcription of prf1.
A similar situation exists in Schizosaccharomyces pombe, where the HMG domain protein Ste11 (45, 46) directly activates the expression of mat1-mc. mat1-mc also encodes a sequence-specific HMG domain protein which acts as a transcriptional activator of M-cell-specific genes required for mating, sporulation, and meiosis (25, 27). In contrast to rop1 deletion strains, Δhmg3 strains were only slightly impaired in cell fusion, and it is presently unclear which component of the fusion machinery is affected in these strains. The finding that neither rop1 nor hmg3 nor prf1 (37) is required for conjugation tube formation when this pathway is genetically activated by expressing the constitutively active MAPKK Fuz7DD argues against an involvement of sequence-specific HMG domain proteins as mediators of this morphological transition.
Rop1 target sites.
The prf1 promoter has previously been shown to underlie a complex regulation. While the PREs in the proximal part of the prf1 promoter most likely confer autoregulation through the prf1 gene product itself, the distal UAS has been shown to be essential for the transcription of prf1 and is subject to nutritional regulation (19). As rop1 deletion strains show the same level of UAS::gfp reporter gene expression as the wild-type strain HA232, we conclude that Rop1 does not regulate prf1 gene expression via the UAS. Instead, electrophoretic mobility shift assays combined with quantitative competition experiments applying specific and unspecific oligonucleotides demonstrate that the Rop1 HMG domain specifically binds to RRS elements in the prf1 promoter. The RRS element (consensus: ATTGT[C/T][C/T]T[A/T]TC) is distinct from the PRE boxes (consensus: TCCCTTTGT) and is not recognized by the HMG domain of Prf1. For the two HMG domain proteins Ste11 and Mat1-Mc of S. pombe, it has been demonstrated that they both bind the TR element (consensus: ttTCTTTGTT) with approximately the same affinity (27).
When compared with the RRS motif sequence, similarity is most prominent in the 3′ portion. The 5′ portion of the RRS shows striking similarity to the motif YYYATTGTTCTC, which is recognized by Rox1p, the sole sequence-specific HMG domain protein in S. cerevisiae (2). In contrast to Rop1, which acts as an activator of prf1 expression, Rox1p functions as a repressor for genes required during oxygen limitation (57). Interestingly, in the human pathogen C. albicans the HMG domain protein Rfg1, which recognizes the same sequence element as Rox1p in S. cerevisiae, is not involved in the response to low oxygen but controls hyphal growth (26). The Rop1 binding RRS motif identified in this study is also found in a number of other promoter regions in U. maydis. This implicates Rop1 in the transcriptional regulation of a variety of other genes besides prf1. The fact that we were unable to detect additional phenotypes except the effects on prf1 may indicate that the expression of this set of genes is not relevant under the culture conditions applied.
Regulation of rop1 gene expression and protein activity.
The observation that rop1 is essential for prf1 and pheromone-responsive gene expression in strains with a genetically activated MAP kinase cascade makes it likely that rop1 functions downstream of the MAP kinase Kpp2. This notion is supported by the observation that rop1 gene expression is regulated by this pathway. Of the three rop1 transcripts detected by Northern blot, only the smallest appears to be polyadenylated and completely spliced and is therefore likely to represent mature rop1 mRNA. In kpp2 deletion strains, the induction of fuz7DD by the shift from glucose to arabinose medium does not allow us to detect spliced rop1 transcripts, while these are readily detectable when the same experiment is done in the presence of Kpp2. This rules out that the shift to arabinose-containing medium is promoting the processing of rop1 message and implicates Kpp2 in the generation of these spliced transcripts. How this is achieved is presently unknown. However, precedence for a role of a MAP kinase module in splicing exists. In human cell lines, alternative splicing of CD44 pre-mRNA is regulated via phosphorylation of the RNA-binding protein Sam68 by the ERK kinase (34, 54).
The presence of potential MAP kinase phosphorylation sites in Rop1 may indicate that Kpp2 is not only involved in generating the mature rop1 transcript but may also affect the activity of Rop1 protein.
Rop1 is a critical factor for the regulation of prf1.
While Rop1 is a critical factor for the regulation of prf1 during mating and dikaryon formation in axenic culture, rop1 is dispensable for pathogenic development. This came as a surprise because prf1 is not only essential during mating in axenic culture but also crucial for mating on the leaf surface and subsequent pathogenic development (18). This implies that on the leaf surface, prf1 gene expression must be induced by the perception of an as yet unidentified environmental signal not present during axenic growth. In the phytopathogenic fungi M. grisea, Colletotrichum gloeosporioides, and Uromyces appendiculatus, surface hydrophobicity and leaf topography, plant hormones, and wax components have been reported to be perceived and to induce appressorial differentiation (28, 50).
In U. maydis there is no evidence that hydrophobicity alone is sufficient to stimulate prf1 gene expression in the absence of rop1 (T. Brefort, unpublished data), which could indicate that chemical rather than physical signals serve as major stimuli. In any case, the signal must bypass the need for rop1. This points to the existence of an as yet unidentified transcription factor with an essential role in prf1 expression during fungal growth on the plant surface. The transmission of the postulated environmental signal might occur via the described cAMP or MAP kinase pathways (37) or an additional pathway acting in parallel. In this scenario, the UAS-mediated regulation of prf1 gene expression could be important. Interestingly, the IME2-like MAP kinase Crk1 was recently described to regulate prf1 expression via the UAS (14, 15). It is thus conceivable that Crk1 might be responsible for activation of the unknown transcription factor acting via the UAS. We are currently trying to isolate the regulator that promotes prf1 transcription through the UAS to sort out the complex mode of regulation affecting the prf1 promoter.
Acknowledgments
Our work was supported by the German Science Foundation (DFG) through grant SFB369 and by BayerCropScience AG.
REFERENCES
- 1.Andrews, D. L., J. D. Egan, M. E. Mayorga, and S. E. Gold. 2000. The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol. Plant-Microbe Interact. 1 3:781-786. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanian, B., C. V. Lowry, and R. S. Zitomer. 1993. The Rox1 repressor of the Saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility group motif. Mol. Cell. Biol. 1 3:6071-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banuett, F., and I. Herskowitz. 1989. Different a-Alleles are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86:5878-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banuett, F., and I. Herskowitz. 1994. Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 8:1367-1378. [DOI] [PubMed] [Google Scholar]
- 5.Bolker, M., H. U. Bohnert, K. H. Braun, J. Gorl, and R. Kahmann. 1995. Tagging pathogenicity genes in Ustilago maydis by restriction enzyme-mediated integration (REMI). Mol. Gen. Genet. 2 48:547-552. [DOI] [PubMed] [Google Scholar]
- 6.Bolker, M., M. Urban, and R. Kahmann. 1992. The a mating type locus of U. maydis specifies cell signaling components. Cell 68:441-450. [DOI] [PubMed] [Google Scholar]
- 7.Bottin, A., J. Kamper, and R. Kahmann. 1996. Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet. 2 53:342-352. [DOI] [PubMed] [Google Scholar]
- 8.Brachmann, A., J. Konig, C. Julius, and M. Feldbrugge. 2004. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 272:216-226. [DOI] [PubMed] [Google Scholar]
- 9.Brachmann, A., J. Schirawski, P. Muller, and R. Kahmann. 2003. An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 22:2199-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brachmann, A., G. Weinzierl, J. Kamper, and R. Kahmann. 2001. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42:1047-1063. [DOI] [PubMed] [Google Scholar]
- 11.Bustin, M., and R. Reeves. 1996. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 54:35-100. [DOI] [PubMed] [Google Scholar]
- 12.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 2001. The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence. Proc. Natl. Acad. Sci. USA 98:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark-Lewis, I., J. S. Sanghera, and S. L. Pelech. 1991. Definition of a consensus sequence for peptide substrate recognition by p44mpk, the meiosis-activated myelin basic protein kinase. J. Biol. Chem. 2 66:15180-15184. [PubMed] [Google Scholar]
- 14.Garrido, E., and J. Perez-Martin. 2003. The crk1 gene encodes an Ime2-related protein that is required for morphogenesis in the plant pathogen Ustilago maydis. Mol. Microbiol. 47:729-743. [DOI] [PubMed] [Google Scholar]
- 15.Garrido, E., U. Voss, P. Muller, R. Kahmann, and J. Pérez-Martín. 2004. The induction of sexual development and virulence in the smut fungus Ustilago maydis depends on Crk1, a novel MAP kinase protein. Genes Dev. 18:3117-3130. [DOI] [PMC free article] [PubMed]
- 16.Gillissen, B., J. Bergemann, C. Sandmann, B. Schroeer, M. Bolker, and R. Kahmann. 1992. A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68:647-657. [DOI] [PubMed] [Google Scholar]
- 17.Grosschedl, R., K. Giese, and J. Pagel. 1994. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1 0:94-100. [DOI] [PubMed] [Google Scholar]
- 18.Hartmann, H. A., R. Kahmann, and M. Bolker. 1996. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 1 5:1632-1641. [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmann, H. A., J. Kruger, F. Lottspeich, and R. Kahmann. 1999. Environmental signals controlling sexual development of the corn Smut fungus Ustilago maydis through the transcriptional regulator Prf1. Plant Cell 11:1293-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 21.Holliday, R. 1974. Ustilago maydis, p. 575-595. In R. C. King (ed.), Handbook of genetics, vol. 1. Plenum Press, New York, N.Y. [Google Scholar]
- 22.Kaffarnik, F., P. Muller, M. Leibundgut, R. Kahmann, and M. Feldbrugge. 2003. PKA and MAPK phosphorylation of Prf1 allows promoter discrimination in Ustilago maydis. EMBO J. 22:5817-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamper, J. 2004. A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis Mol. Genet. Genomics 271:103-110. [DOI] [PubMed] [Google Scholar]
- 24.Kamper, J., M. Reichmann, T. Romeis, M. Bolker, and R. Kahmann. 1995. Multiallelic recognition: non-self-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81:73-83. [DOI] [PubMed] [Google Scholar]
- 25.Kelly, M., J. Burke, M. Smith, A. Klar, and D. Beach. 1988. Four mating type genes control sexual differentiation in the fission yeast. EMBO J. 7:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khalaf, R. A., and R. S. Zitomer. 2001. The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics 1 57:1503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kjaerulff, S., D. Dooijes, H. Clevers, and O. Nielsen. 1997. Cell differentiation by interaction of two HMG-box proteins: Mat1-Mc activates M cell-specific genes in S. pombe by recruiting the ubiquitous transcription factor Ste11 to weak binding sites. EMBO J. 1 6:4021-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolattukudy, P. E., L. M. Rogers, D. Li, C. S. Hwang, and M. A. Flaishman. 1995. Surface signaling in pathogenesis. Proc. Natl. Acad. Sci. USA 92:4080-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kruger, J., G. Loubradou, E. Regenfelder, A. Hartmann, and R. Kahmann. 1998. Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol. Gen. Genet. 2 60:193-198. [DOI] [PubMed] [Google Scholar]
- 30.Laudet, V., D. Stehelin, and H. Clevers. 1993. Ancestry and diversity of the HMG box superfamily. Nucleic Acids Res. 21:2493-2501. [DOI] [PMC free article] [PubMed]
- 31.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 2 66:1723-1726. [DOI] [PubMed] [Google Scholar]
- 32.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 33.Loubradou, G., A. Brachmann, M. Feldbrugge, and R. Kahmann. 2001. A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis. Mol. Microbiol. 40:719-730. [DOI] [PubMed] [Google Scholar]
- 34.Matter, N., P. Herrlich, and H. Konig. 2002. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420:691-695. [DOI] [PubMed] [Google Scholar]
- 35.Mayorga, M. E., and S. E. Gold. 1999. A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol. 34:485-497. [DOI] [PubMed] [Google Scholar]
- 36.Muller, P., C. Aichinger, M. Feldbrugge, and R. Kahmann. 1999. The MAP kinase Kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34:1007-1017. [DOI] [PubMed] [Google Scholar]
- 37.Muller, P., G. Weinzierl, A. Brachmann, M. Feldbrugge, and R. Kahmann. 2003. Mating and pathogenic development of the smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot. Cell 2:1187-1199. [DOI] [PMC free article] [PubMed]
- 38.Murata, Y., M. Fujii, M. E. Zolan, and T. Kamada. 1998. Molecular analysis of pcc1, a gene that leads to A-regulated sexual morphogenesis in Coprinus cinereus. Genetics 1 49:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park, G., C. Xue, L. Zheng, S. Lam, and J. R. Xu. 2002. MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 1 5:183-192. [DOI] [PubMed] [Google Scholar]
- 40.Pevny, L. H., and R. LovellBadge. 1997. Sox genes find their feet. Curr. Opin. Genet. Dev. 7:338-344. [DOI] [PubMed] [Google Scholar]
- 41.Regenfelder, E., T. Spellig, A. Hartmann, S. Lauenstein, M. Bolker, and R. Kahmann. 1997. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 1 6:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Schulz, B., F. Banuett, M. Dahl, R. Schlesinger, W. Schafer, T. Martin, I. Herskowitz, and R. Kahmann. 1990. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60:295-306. [DOI] [PubMed] [Google Scholar]
- 44.Sinclair, A. H., P. Berta, M. S. Palmer, J. R. Hawkins, B. L. Griffiths, M. J. Smith, J. W. Foster, A. M. Frischauf, R. Lovell-Badge, and P. N. Goodfellow. 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346:240-244. [DOI] [PubMed] [Google Scholar]
- 45.Sipiczki, M. 1988. The role of sterility genes (ste and aff) in the initiation of sexual development in Schizosaccharomyces pombe. Mol. Gen. Genet. 21 3:529-534. [DOI] [PubMed] [Google Scholar]
- 46.Sugimoto, A., Y. Iino, T. Maeda, Y. Watanabe, and M. Yamamoto. 1991. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 5:1990-1999. [DOI] [PubMed] [Google Scholar]
- 47.Szabo, Z., M. Tonnis, H. Kessler, and M. Feldbrugge. 2002. Structure-function analysis of lipopeptide pheromones from the plant pathogen Ustilago maydis. Mol. Genet. Genomics 2 68:362-370. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X-Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 2 5:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Travis, A., A. Amsterdam, C. Belanger, and R. Grosschedl. 1991. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function. Genes Dev. 5:880-894. [DOI] [PubMed] [Google Scholar]
- 50.Tucker, S. L., and N. J. Talbot. 2001. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39:385-417. [DOI] [PubMed] [Google Scholar]
- 51.Urban, M., R. Kahmann, and M. Bolker. 1996. Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet. 251:31-37. [DOI] [PubMed] [Google Scholar]
- 52.van de Wetering, M., M. Oosterwegel, D. Dooijes, and H. Clevers. 1991. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1 0:123-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wedlich-Soldner, R., M. Bolker, R. Kahmann, and G. Steinberg. 2000. A putative endosomal t-SNARE links exo- and endocytosis in the phytopathogenic fungus Ustilago maydis. EMBO J. 1 9:1974-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weg-Remers, S., H. Ponta, P. Herrlich, and H. Konig. 2001. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J. 2 0:4194-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wickes, B. L., U. Edman, and J. C. Edman. 1997. The Cryptococcus neoformans STE12alpha gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol. Microbiol. 2 6:951-960. [DOI] [PubMed] [Google Scholar]
- 56.Yue, C., L. M. Cavallo, J. A. Alspaugh, P. Wang, G. M. Cox, J. R. Perfect, and J. Heitman. 1999. The STE12alpha homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics 1 53:1601-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zitomer, R. S., P. Carrico, and J. Deckert. 1997. Regulation of hypoxic gene expression in yeast. Kidney Int. 51:507-513. [DOI] [PubMed] [Google Scholar]