Abstract
Thioredoxins are small, highly conserved oxidoreductases which are required to maintain the redox homeostasis of the cell. Saccharomyces cerevisiae contains a cytoplasmic thioredoxin system (TRX1, TRX2, and TRR1) as well as a complete mitochondrial thioredoxin system, comprising a thioredoxin (TRX3) and a thioredoxin reductase (TRR2). In the present study we have analyzed the functional overlap between the two systems. By constructing mutant strains with deletions of both the mitochondrial and cytoplasmic systems (trr1 trr2 and trx1 trx2 trx3), we show that cells can survive in the absence of both systems. Analysis of the redox state of the cytoplasmic thioredoxins reveals that they are maintained independently of the mitochondrial system. Similarly, analysis of the redox state of Trx3 reveals that it is maintained in the reduced form in wild-type cells and in mutants lacking components of the cytoplasmic thioredoxin system (trx1 trx2 or trr1). Surprisingly, the redox state of Trx3 is also unaffected by the loss of the mitochondrial thioredoxin reductase (trr2) and is largely maintained in the reduced form unless cells are exposed to an oxidative stress. Since glutathione reductase (Glr1) has been shown to colocalize to the cytoplasm and mitochondria, we examined whether loss of GLR1 influences the redox state of Trx3. During normal growth conditions, deletion of TRR2 and GLR1 was found to result in partial oxidation of Trx3, indicating that both Trr2 and Glr1 are required to maintain the redox state of Trx3. The oxidation of Trx3 in this double mutant is even more pronounced during oxidative stress or respiratory growth conditions. Taken together, these data indicate that Glr1 and Trr2 have an overlapping function in the mitochondria.
The process of respiration requires the uptake of molecular oxygen. While this energy-efficient process is necessary for aerobic organisms, it has also meant that they are exposed to the damaging effects of oxygen. Relatively unreactive and harmless in its ground state, oxygen (O2) is capable of undergoing excitation or partial reduction to form a number of highly reactive species, including the superoxide anion (O2−), singlet oxygen (O21), ozone (O3), and the hydroxyl radical (·OH). As a consequence, organisms have evolved a broad range of responses which can detoxify reactive oxygen species (ROS), reduce the rate of their production, and repair the damage caused by them (16, 21, 45). An oxidative stress is said to occur when a proportion of the ROS evades these host defenses, resulting in damage to various cellular macromolecules including lipids, proteins, and nucleic acids. A number of reports have highlighted the key role played by sulfydryl groups (-SH) in the response to oxidative stress and, in particular, the roles of the glutathione (GSH)-glutaredoxin and thioredoxin systems in maintaining the redox homeostasis of the cell (3, 11).
Glutaredoxins and thioredoxins are small, heat-stable oxidoreductases containing two conserved cysteine residues in their active sites (18). They were originally identified as hydrogen donors for ribonucleotide reductase but also act upon a number of metabolic enzymes that form a disulfide as part of their catalytic cycle (38). They have proposed roles in many cellular processes including protein folding and regulation, reduction of dehydroascorbate, repair of oxidatively damaged proteins, and sulfur metabolism (18, 38). Glutaredoxins and thioredoxins are structurally similar and have been conserved throughout evolution (19). However, despite considerable functional overlap, they are regulated differentially. The oxidized disulfide form of thioredoxin is reduced directly by NADPH and thioredoxin reductase, whereas glutaredoxin is reduced by GSH with electrons donated by NADPH.
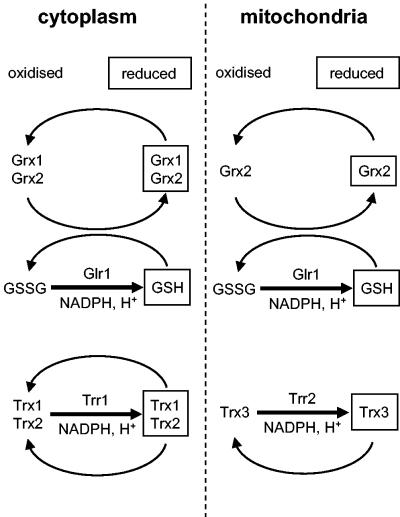
The main cytoplasmic and mitochondrial redox regulatory systems that have been described in Saccharomyces cerevisiae are shown in Fig. 1. Yeast contains two genes encoding glutaredoxins (GRX1 and GRX2) sharing extensive homology with the glutaredoxins from bacterial and mammalian species (25). Strains with GRX1 and GRX2 deleted are viable but lack heat-stable oxidoreductase activity measured using a model disulfide substrate. Grx1 and Grx2 act as antioxidants and have activity as general hydroperoxidases and GSH S-transferases (5, 6, 25). Yeast also contains two genes encoding cytoplasmic thioredoxins (TRX1 and TRX2), which are dispensable during normal growth conditions (8, 31). However, deletion of TRX1 and TRX2 affects the cell cycle, prolonging S phase and shortening the G1 interval (31). Thioredoxins are also required for protection against ROS and provide reducing power for various thioredoxin peroxidase isoenzymes (33). Genetic screens have identified the yeast cytoplasmic thioredoxin reductase (TRR1) (27, 34), but little is known about its role in maintaining the cellular redox environment. Yeast, like most eukaryotes, contains a complete mitochondrial thioredoxin system including a thioredoxin (TRX3) and thioredoxin reductase (TRR2), which is thought to function in protection against oxidative stress generated during respiratory metabolism (35).
FIG. 1.
GSH-glutaredoxin and thioredoxin systems. Yeast contains two gene pairs encoding cytoplasmic glutaredoxins (GRX1 and GRX2) and thioredoxins (TRX1 TRX2). The oxidized disulfide form of thioredoxin is reduced directly by NADPH and thioredoxin reductase (Trr1). In contrast, oxidized glutaredoxin is reduced by GSH, and oxidized GSSG is in turn reduced in an NADPH-dependent reaction catalyzed by GSH reductase (Glr1). Yeast also contains a complete mitochondrial thioredoxin system including a thioredoxin (TRX3) and a thioredoxin reductase (TRR2). Grx2 and Glr1 have been colocalized to the cytosol and mitochondria (32, 35). The redox status of GSH may provide a functional link between the GSH-glutaredoxin and thioredoxin systems, since cytoplasmic thioredoxins function along with Glr1 to maintain the high intracellular GSH/GSSG ratio (9, 29). In addition, the redox state of cytoplasmic Trx1 and Trx2 is maintained independently of the GSH-glutaredoxin system, whereas there is a strong correlation between the redox state of glutaredoxins and the oxidation state of the GSSG-2GSH redox couple (43).
The first evidence that the thioredoxin and GSH-glutaredoxin systems have an overlapping function in redox regulation came from the identification of GLR1, encoding GSH reductase, in a genetic screen for mutations which confer a requirement for thioredoxins (29). Additionally, loss of TRX1 and TRX2 has been shown to result in elevated GSH levels, indicating a link between the thioredoxin system and GSH metabolism in the cell (9, 29). Furthermore, deletion analysis has shown that a quadruple trx1 trx2 grx1 grx2 mutant is inviable and a single functional disulfide reductase system is necessary for viability (7). To further test the requirement for components of the thioredoxin and GSH-glutaredoxin systems, we have attempted to construct mutants lacking components of each system. Strains completely lacking the cytoplasmic thioredoxin (trx1 trx2 trr1) or GSH (lacking GLR1 and GSH1 encoding the enzyme for the first step in GSH biosynthesis) system are viable, but strains having components of both systems simultaneously deleted (gsh1 trx1 trx2 and glr1 trr1) are inviable (43). The overlapping nature of the two systems raises important questions regarding the reasons for this apparent redundancy. Analysis of the redox state of cytoplasmic thioredoxins and glutaredoxins has revealed that cytoplasmic thioredoxins are maintained independently of the GSH-glutaredoxin system. In contrast, there is a strong correlation between the redox state of glutaredoxins and the oxidation state of the GSSG-2GSH redox couple (43). We have proposed a model in which independent redox regulation of thioredoxins enables cells to survive under conditions where the GSH-glutaredoxin system is oxidized.
In this study, we have analyzed the overlapping roles of the cytoplasmic and mitochondrial redox regulatory systems. Surprisingly, these studies have shown that the redox state of mitochondrial Trx3 is unaffected in strains having the cytoplasmic (Trr1) or mitochondrial (Trr2) thioredoxin reductases deleted. However, loss of both TRR2 and GLR1 shifts the redox state of Trx3 to a more oxidized form. These data indicate that Glr1 and Trr2 have an overlapping function in the mitochondria and provide the first in vivo evidence that the GSH-glutaredoxin system can influence the redox state of a thioredoxin.
MATERIALS AND METHODS
Yeast strains.
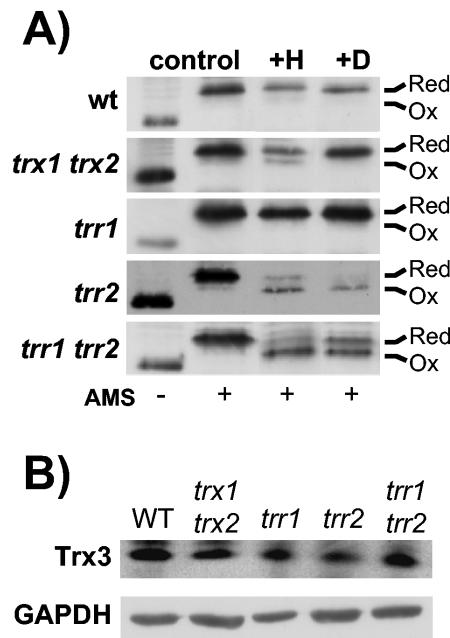
S. cerevisiae strains used in this study were all isogenic derivatives of CY4 (MATa ura3-52 leu2-3 leu2-112 trp1-1 ade2-1 his3-11 can1-100) (13). Strains with deletions of thioredoxins (trx1::TRP1 trx2::URA3), thioredoxin reductase (trr1::HIS3), and GSH reductase (glr1::TRP1) have been described before (44). Strain CY760, which has TRR2 deleted (trr2::HIS3), was constructed using a one-step PCR amplification protocol that replaced the entire TRR2 open reading frame with the yeast HIS3 gene (1). Strain CY891, which has TRX3 deleted, was made by backcrossing CY4 with a EUROSCARF strain (MATα his3D1 leu2D0 lys2D0 ura3D0 trx3::kanMX4). Standard yeast genetic techniques were used to construct mutants lacking multiple genes encoding the GSH and thioredoxin systems. Trx3 was epitope tagged with a three-Myc tag, and Trx1 and Trx2 were epitope tagged with a three-His tag by a PCR-based strategy (22). In Fig. 6 we show that deleting TRX3 in a glr1 trr2 mutant partially restores the slow growth observed under respiratory conditions. In contrast, epitope tagging Trx3 with the three-Myc tag does not affect the slow growth of the glr1 trr2 mutant, confirming that tagged Trx3 is biologically active (data not shown).
FIG. 6.
Redox state of Trx3 in thioredoxin mutants. (A) The indicated strains were grown to exponential phase in SD medium (control) and treated with 2 mM H2O2 for 1 h (+H) or 2 mM diamide for 1 h (+D). Proteins were precipitated with TCA, and free thiols were modified by reaction with AMS. Samples were separated using SDS-18% PAGE, and Trx3 was detected by Western blot analysis. Oxidized and reduced proteins are indicated. Trx3 contains two redox active Cys residues, as well as two additional Cys residues, and fully oxidized Trx3 was never detected. (B) Western blot analysis of Trx3 protein levels. Trx3 protein concentrations were measured in wild-type and trx1 trx2, trr1, trr2, and trr1 trr2 mutant strains by Western blot analysis with antibodies specific for c-Myc (Trx3) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
Growth conditions.
Strains were grown in rich YEPD medium (2% [wt/vol] glucose, 2% [wt/vol] Bacto peptone, 1% [wt/vol] yeast extract) or minimal SD medium (0.17% [wt/vol] yeast nitrogen base without amino acids, 5% [wt/vol] ammonium sulfate, 2% [wt/vol] glucose supplemented with appropriate amino acids and bases) (41). For growth on nonfermentable carbon sources, SGE contained 3% (vol/vol) glycerol and 1% (vol/vol) ethanol. Media were solidified by the addition of 2% (wt/vol) agar.
Sensitivity to oxidants was determined by growing cells to exponential phase (A600 = 1) or stationary phase (48-h growth) and spotting them onto agar plates containing various concentrations of H2O2. Growth to an A600 of 1 in minimal SD medium was defined as exponential-phase growth based on our previous growth curve experiments for the various glutaredoxin and thioredoxin mutants (7). Similarly, stationary phase was defined as 48-h growth in minimal SD medium, since at this time point the cells had passed through the diauxic phase of growth and there was no longer any change in cell density (7).
Determination of redox states.
Reduced and oxidized GSH levels were determined as described previously (15). The redox state of thioredoxins was measured by covalent modification with the thiol-reactive probe 4-acetamido-4′maleimidyldystilbene-2,2′-disulfonic acid (AMS; Molecular Probes) as described previously (43). Cytoplasmic thioredoxins were detected using rabbit anti-yeast thioredoxin antibody (43). This antibody was generated against recombinant yeast Trx1 but recognizes both Trx1 and Trx2. Mitochondrial Trx3 was detected using mouse anti-c-Myc antibody (Roche), and cytoplasmic Trx1 and Trx2 were detected using mouse monoclonal antibody HA.11 (Covance).
RESULTS
Overlap between the cytoplasmic and mitochondrial thioredoxin systems.
Previous studies have shown that strains completely lacking the cytoplasmic thioredoxin (trx1 trx2 trr1) or GSH (gsh1 glr1) systems are viable, but strains having components of both systems simultaneously deleted (gsh1 trx1 trx2) and (glr1 trr1) are inviable (43). We therefore tested the overlap between the cytoplasmic and mitochondrial thioredoxin systems by attempting to construct mutant strains that have components of both systems deleted. Strains lacking both thioredoxin reductases (trr1 trr2) or all three thioredoxins (trx1 trx2 trx3) are viable, indicating that cells can survive in the absence of both the cytoplasmic and mitochondrial thioredoxin systems.
Growth phenotypes of thioredoxin mutants.
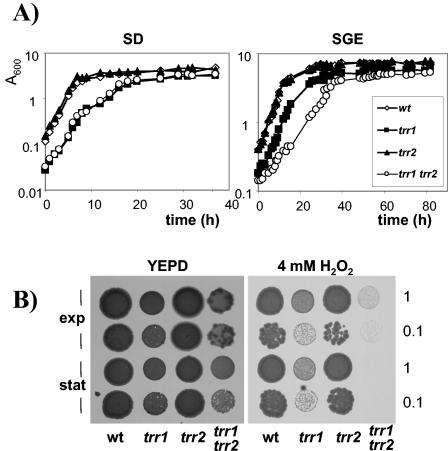
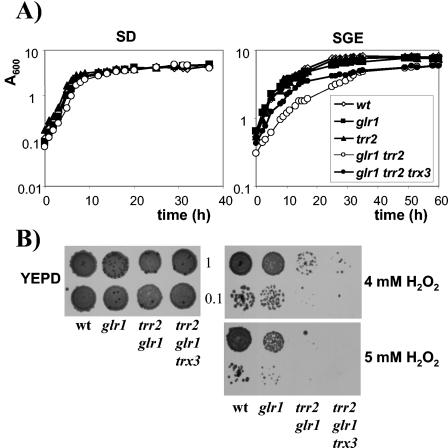
The growth kinetics of the trr1 trr2 mutant strain were analyzed to determine the relative importance of cytoplasmic Trr1 and mitochondrial Trr2 during normal cellular function. Cells were inoculated to the same initial starting density in minimal SD medium and monitored throughout the various phases of growth. The strain with TRR2 deleted grew at the same rate and reached a similar final cell yield as did the wild-type strain (Fig. 2A). In agreement with previous observations, the strain with TRR1 deleted showed a longer lag phase and reached a lower final cell yield (44). The trr1 trr2 double mutant displayed the same growth kinetics as the trr1 mutant strain did. We reasoned that any growth requirement for mitochondrial Trr2 might become apparent during respiratory growth conditions which would be expected to generate intracellular ROS. The growth of strains was therefore compared in minimal media containing glycerol and ethanol as respiratory carbon sources (Fig. 2A). However, similar to fermentative growth, the kinetics of the trr2 mutant (doubling time = 3.6 h) were similar to those of the wild-type strain (doubling time = 3.8 h). Interestingly, the trr1 trr2 mutant showed a lower growth rate (doubling time = 7.7 h) than the trr1 mutant did (doubling time = 4.0 h) and reached a lower final cell yield, indicating that mitochondrial Trr2 is required in the absence of cytoplasmic Trr1 during respiratory growth.
FIG. 2.
Growth and hydrogen peroxide sensitivity of thioredoxin reductase mutants. (A) Wild-type and trr1, trr2, and trr1 trr2 mutant cells were inoculated to the same initial cell density in minimal glucose medium (SD; initial A600 = 0.0025) or minimal medium containing glycerol plus ethanol (SGE; initial A600 = 0.05), and growth was monitored by A600. (B) Sensitivity to hydrogen peroxide was determined by spotting the same strains onto YEPD plates containing 4 mM H2O2. Cultures of strains were grown into exponential phase or stationary phase and adjusted to an A600 of 1.0 or 0.1 before spotting onto appropriate plates. Plates were incubated at 30°C for 3 days before scoring growth.
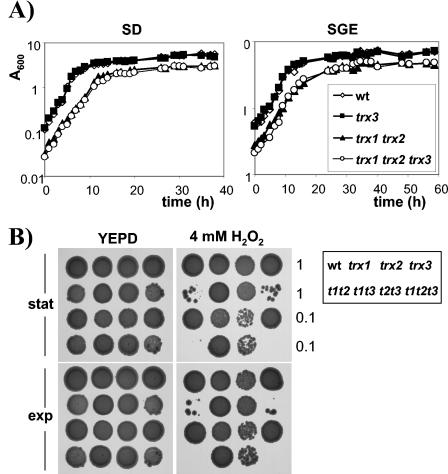
As described previously (7, 31), strains having cytoplasmic TRX1 and TRX2 deleted display a longer lag phase and lower final cell yield than does the wild-type strain when grown on SD medium (Fig. 3A). Loss of mitochondrial TRX3 did not affect the growth kinetics of either the wild-type or trx1 trx2 mutant strain. Similarly, loss of TRX1 and TRX2 resulted in a longer lag phase and lower final cell yield on respiratory medium (SGE), but there was no additional growth defect resulting from the loss of TRX3 in the wild-type or trx1 trx2 mutant strain (Fig. 3A). Thus, there is no requirement for the mitochondrial thioredoxin in the absence of the cytoplasmic thioredoxins during fermentative or respiratory growth.
FIG. 3.
Growth and hydrogen peroxide sensitivity of thioredoxin mutants. (A) Wild-type and trx1, trx2, trx3, trx1 trx2, trx1 trx3, trx2 trx3, and trx1 trx2 trx3 mutant cells were inoculated to the same initial cell density in minimal glucose medium (SD; initial A600 = 0.0025) or minimal medium containing glycerol plus ethanol (SGE; initial A600 = 0.05), and growth was monitored by A600. (B) Sensitivity to hydrogen peroxide was determined by spotting the same strains onto YEPD plates containing 4 mM H2O2. Cultures of strains were grown into exponential phase or stationary phase and adjusted to an A600 of 1.0 or 0.1 before spotting onto appropriate plates. Plates were incubated at 30°C for 3 days before scoring growth.
To characterize the possible functional overlap of the cytoplasmic and mitochondrial thioredoxin systems as antioxidants, we tested the sensitivity of the mutants to oxidative stress. Strains were grown to exponential or stationary phase and spotted onto YEPD plates containing H2O2. On 4 mM H2O2, strains lacking TRR2 showed a wild-type level of resistance, whereas the trr1 mutant was sensitive during both growth phases (Fig. 2B). Interestingly, the trr1 trr2 mutant was hypersensitive to H2O2 and did not grow. Strains lacking the cytoplasmic thioredoxins were also sensitive to 4 mM H2O2 (Fig. 3B). However, loss of TRX3 did not affect the sensitivity of either the wild type or the cytoplasmic thioredoxin mutant strain. These data indicate that, in the absence of the cytoplasmic thioredoxin reductase (Trr1), the mitochondrial thioredoxin reductase (Trr2) is required for respiratory growth and resistance to oxidative stress. In contrast, there is no apparent requirement for the mitochondrial thioredoxin (Trx3).
Loss of the mitochondrial thioredoxin system does not influence the redox state of the cytoplasmic thioredoxin system.
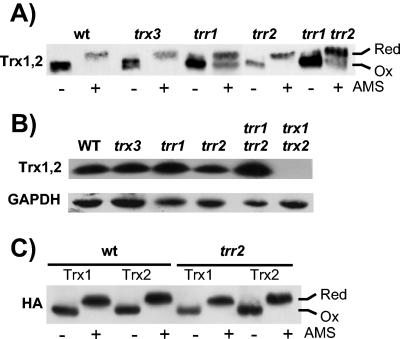
We next examined whether the in vivo oxidation state of the cytoplasmic thioredoxins is affected by loss of the mitochondrial thioredoxin system. The cellular oxidation state was preserved by rapidly treating cells with trichloroacetic acid (TCA), which protonates free thiol groups. Extracts were then reacted with the thiol-specific probe AMS, followed by separation with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis. The antibody used in this experiment was generated against recombinant yeast Trx1 but recognizes both Trx1 and Trx2. In agreement with previous observations (43), the migration of all cytoplasmic Trx protein from the wild-type strain was decreased following treatment with AMS, indicating that the vast majority is present in the reduced form (Fig. 4A). As might be expected, loss of cytoplasmic TRR1 shifted the thioredoxin redox balance to a more oxidized state, with approximately equal amounts of reduced and oxidized forms present. Loss of mitochondrial TRX3 or TRR2 did not affect the redox state of the cytoplasmic thioredoxins, and similarly, there was no additive effect resulting from the loss of TRR2 in the trr1 mutant. The AMS treatment appeared to affect the levels of thioredoxin proteins detected in these experiments. It is unclear what the reason is for this effect, but it may be due to solubility problems with the AMS-modified thioredoxin or, alternatively, due to a difference in recognition of the modified protein by the antibody. Western blot analysis was therefore used to confirm that there were no differences in cytoplasmic thioredoxin protein levels in the mitochondrial thioredoxin mutants (Fig. 5B).
FIG. 4.
Redox state of mitochondrial thioredoxin system mutants. (A) Redox state of cytoplasmic thioredoxins. The indicated strains were grown to exponential phase (A600 = 1.0) in SD medium. Proteins were precipitated with TCA, and free thiols were modified by reaction with AMS. Samples were separated using SDS-18% PAGE, and cytoplasmic thioredoxins (Trx1 and Trx2) were detected by Western blot analysis with rabbit anti-yeast thioredoxin antibody (Trx1,2). Fully oxidized and fully reduced proteins are indicated. (B) Western blot analysis of thioredoxin protein levels. Thioredoxin protein concentrations were measured in wild-type and trx3, trr1, trr2, trr1 trr2, and trx1 trx2 mutant strains by Western blot analysis with antibodies specific for thioredoxins (Trx1,2) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). (C) Trx1 and Trx2 were epitope tagged with a hemagglutinin (HA) tag to determine the redox state of each cytoplasmic thioredoxin isoform. Trx1 and Trx2 are both present in the fully reduced form in a wild-type strain and a strain lacking mitochondrial Trr2.
FIG. 5.
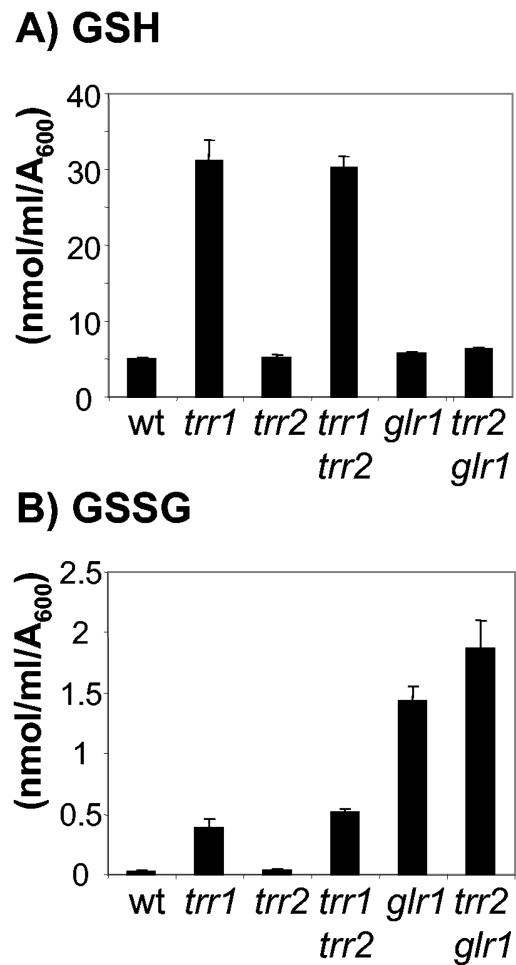
Redox state of the GSH system in mitochondrial thioredoxin mutants. The indicated strains were grown to exponential phase in minimal SD medium. The levels of reduced GSH (A) and oxidized GSH (GSSG) (B) were determined. Values shown are the means of at least three independent determinations and are given in nanomoles per milliliter per A600 unit.
To examine the redox state of each cytoplasmic thioredoxin individually, Trx1 and Trx2 were epitope tagged with a hemagglutinin tag (see Materials and Methods). Trx1 and Trx2 were present in a fully reduced form in both the wild type and mitochondrial trr2 mutant (Fig. 4C). In addition, loss of the mitochondrial thioredoxin system does not affect the cytoplasmic system during respiratory growth conditions (data not shown). These data indicate that the mitochondrial thioredoxin system is not required to maintain the redox state of the cytoplasmic thioredoxins during normal growth conditions.
Loss of the mitochondrial thioredoxin system does not affect the GSH redox couple.
Loss of cytoplasmic thioredoxins (TRX1 TRX2) or thioredoxin reductase (TRR1) has previously been shown to result in increased cellular concentrations of reduced (GSH) and oxidized (GSSG) GSH and to shift the GSH redox ratio (GSH/GSSG) to a more oxidized state (9, 29). We therefore determined whether loss of the mitochondrial thioredoxin system has a similar effect. However, the trr2 mutant was found to contain wild-type levels of GSH and GSSG (Fig. 5). In addition, deletion of TRR2 in the trr1 mutant did not cause any additional effect on the levels of GSH or GSSG. Similarly, loss of TRX3 in a wild-type or trx1 trx2 mutant strain did not alter GSH levels or redox state (data not shown). Loss of the mitochondrial thioredoxin system was also found to have no effect on the GSH system during respiratory growth conditions (data not shown). Given that loss of the mitochondrial thioredoxin system does not influence the redox state of the cytoplasmic thioredoxin system or the GSH redox couple, we next determined whether these systems influence the redox state of the mitochondrial thioredoxin system.
Redox state of the mitochondrial thioredoxin.
Mitochondrial Trx3 contains two redox active cysteine residues within the Cys-Gly-Pro-Cys motif that is conserved among thioredoxins from all species. In addition, Trx3 contains two extra Cys residues which are not present in cytoplasmic Trx1 and Trx2. Analysis of the redox state of Trx3 indicated that it is predominantly present in the reduced state in a wild-type strain during normal growth conditions (Fig. 6A). Loss of the cytoplasmic thioredoxins (trx1 trx2) or thioredoxin reductase (trr1) did not alter the redox state of Trx3. Surprisingly, the redox state of Trx3 is also unaffected by the loss of TRR2. This is in contrast to the cytoplasmic thioredoxins, which are shifted to a more oxidized state in the absence of cytoplasmic Trr1 (Fig. 4). Similarly, the redox state of mitochondrial Trx3 was unaffected by the simultaneous loss of both TRR1 and TRR2. Western blot analysis revealed that the cellular concentrations of mitochondrial Trx3 were unaffected in mutants lacking the cytoplasmic thioredoxins (TRX1 TRX2), thioredoxin reductase (TRR1), or mitochondrial thioredoxin reductase (TRR2) (Fig. 6B).
Given that Trx3 is maintained in a reduced form in the various thioredoxin mutants, we examined whether the redox state is altered in response to oxidative stress conditions. Thioredoxin redox state was determined in exponential-phase cells exposed to H2O2 or the thiol oxidant diamide. The redox state of the cytoplasmic thioredoxins was unaffected by treatment with diamide, whereas treatment with H2O2 shifted the redox balance to a more oxidized state, with approximately equal amounts of reduced and oxidized forms present (data not shown). In contrast, the redox state of mitochondrial Trx3 in the wild-type strain was unaffected by both the H2O2 and diamide treatments (Fig. 6A). However, the oxidized form of Trx3 did accumulate in the trr2 and trr1 trr2 mutants exposed to both H2O2 and diamide. A small amount of oxidized Trx3 was also observed in the trx1 trx2 mutant exposed to H2O2. These data indicate that Trx3 is largely maintained in the reduced form, even in the absence of TRR2, unless cells are exposed to an oxidative stress. We next determined whether the reduced state of Trx3 is dependent on an intact GSH system.
GSH reductase is required to maintain the redox state of Trx3.
GSH reductase is the key enzyme determining the redox state of the GSH system and has recently been shown to colocalize to the cytosol and mitochondria (32). We therefore examined the effect of deleting GLR1 in the trr2 mutant to determine whether the GSH system is required to maintain the redox state of mitochondrial Trx3. The resulting trr2 glr1 mutant is viable, and the kinetics of growth were determined to compare the mutant strains (Fig. 7A). In agreement with previous observations, the strain having GLR1 deleted is unaffected in growth on fermentative or respiratory carbon sources (14, 44). The trr2 glr1 double mutant also grows with wild-type kinetics on SD medium. However, the trr2 glr1 mutant displays a lower growth rate (doubling time = 5.0 h) and lower final cell yield on SGE medium than does the wild type (doubling time = 3.8 h), glr1 mutant (doubling time = 3.7 h), or trr2 mutant (doubling time = 3.5 h), indicating that Trr2 and Glr1 are both required for growth on respiratory carbon sources. To test for any overlapping requirement as antioxidants, the mutants were tested for sensitivity to oxidative stress conditions. In agreement with previous observations (14), the glr1 mutant is sensitive to 5 mM H2O2 (Fig. 7B). Loss of TRR2 in the glr1 mutant was found to exacerbate this oxidant sensitivity, and the double mutant was unable to grow on 4 mM H2O2.
FIG. 7.
Growth and hydrogen peroxide sensitivity of GSH reductase mutants. The indicated strains were inoculated to the same initial cell density in minimal glucose medium (SD; initial A600 = 0.0025) or minimal medium containing glycerol plus ethanol (SGE; initial A600 = 0.05), and growth was monitored by A600. (B) Sensitivity to hydrogen peroxide was determined by spotting the same strains onto YEPD plates containing 4 mM H2O2. Cultures of strains were grown into stationary phase and adjusted to an A600 of 1.0 or 0.1 before spotting onto appropriate plates. Plates were incubated at 30°C for 3 days before scoring growth.
Oxidized redox state of the glr1 trr2 mutant.
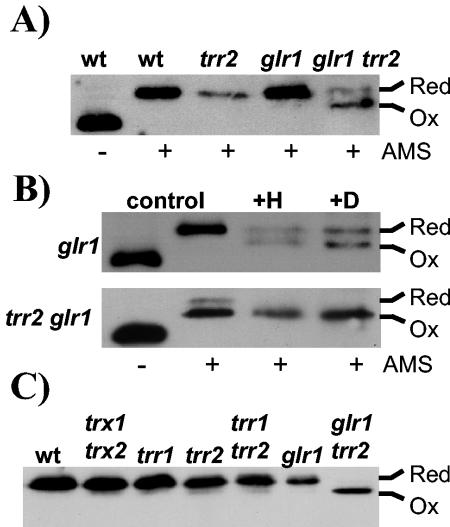
Analysis of the redox state of mitochondrial Trx3 revealed that it is unaffected by the loss of GLR1 (Fig. 8A). However, Trx3 is shifted to a more oxidized state in the trr2 glr1 double mutant, indicating that Trr2 and Glr1 are required to maintain the redox state of Trx3 during normal growth conditions. In addition, exposure of the glr1 mutant to diamide or H2O2 resulted in an accumulation of oxidized Trx3 (Fig. 8B). This effect is even more pronounced in the trr2 glr1 mutant, where Trx3 is predominantly present in an oxidized form following exposure to oxidative stress. Finally, we examined the redox state of mitochondrial Trx3 during respiratory growth conditions (Fig. 8C). Trx3 was found to be maintained in a reduced form in the wild-type strain, cytoplasmic thioredoxin mutant (trx1 trx2), thioredoxin reductase mutants (trr1, trr2, and trr1 trr2), and GSH reductase mutant (glr1) during respiratory growth. Interestingly, the trr2 glr1 mutant is unable to maintain the redox state of Trx3, which is predominantly present in the oxidized form during respiratory growth.
FIG. 8.
Redox state of mitochondrial Trx3 in GSH reductase mutants. The redox state of Trx3 was detected as described for Fig. 5. (A) The indicated strains were grown to exponential phase in SD medium. (B) The glr1 and glr1 trr2 mutants were grown to exponential phase in SD medium (control) and treated with 2 mM H2O2 for 1 h (+H) or 2 mM diamide for 1 h (+D). (C) The indicated strains were grown to exponential phase in SGE medium.
Mutants lacking GLR1 contain elevated levels of GSSG, consistent with the role of Glr1 in recycling oxidized GSSG to reduced GSH (Fig. 5) (12, 29). We therefore examined whether loss of TRR2 in the glr1 mutant has any influence on GSH levels. The high concentrations of GSSG detected in the glr1 mutant are further elevated in the glr1 trr2 double mutant (Fig. 5B). Taken together, these data indicate that the simultaneous loss of GLR1 and TRR2 results in an accumulation of oxidized Trx3 and GSSG.
Given that oxidized Trx3 accumulates in the glr1 trr2 mutant, we examined whether it could account for the slow growth and oxidant sensitivity of the mutant. The triple trr2 glr1 trx3 mutant was found to be viable but shows the same sensitivity to H2O2 as the trr2 glr1 mutant (Fig. 7B). However, loss of TRX3 in the trr2 glr1 mutant increased the growth rate of the mutant during respiratory conditions, indicating that oxidized Trx3 accounts, in part at least, for the slow growth of the trr2 glr1 mutant under respiratory conditions.
DISCUSSION
Complete thioredoxin and GSH-glutaredoxin systems have now been identified in the cytosol and mitochondria of many eukaryotic organisms (10, 26, 35, 42). However, it is as yet unclear whether there is any cross talk between the different organellar systems. We have previously examined the overlap between the cytoplasmic systems by measuring the redox state of thioredoxins and glutaredoxins in mutants lacking GSH reductase (Glr1) and thioredoxin reductase (Trr1) (43). Glr1 and Trr1 are the key regulatory enzymes determining the redox state of the GSH-glutaredoxin and thioredoxin systems, respectively. This analysis revealed that cytoplasmic thioredoxins are maintained independently of the GSH-glutaredoxin system. In contrast, there is a strong correlation between the redox state of glutaredoxins and the oxidation state of the GSSG-2GSH redox couple. A model was proposed in which independent redox regulation of thioredoxins enables cells to survive under conditions where the GSH-glutaredoxin system is oxidized (43). In this present study, we have shown that the redox states of the mitochondrial and cytoplasmic thioredoxin systems are maintained independently, since loss of one system does not affect the redox state of the other. In contrast, the redox state of the mitochondrial thioredoxin system was found to depend on the presence of an intact GSH system.
GSH is ubiquitous in eukaryotic cells and has long been considered the major cellular redox buffer, with the redox state of the GSH system being taken as an indicator of the cellular redox environment (28). Mutants having GLR1, TRR1, or TRX1 TRX2 deleted contain elevated levels of GSSG (9, 29, 44). In contrast, loss of Grx1 or Grx2 does not affect the GSH redox balance (25). Additionally, loss of cytoplasmic thioredoxins or thioredoxin reductase results in elevated levels of reduced GSH, consistent with the known Yap1-mediated increase in GSH1 and GLR1 expression in thioredoxin mutants. Yap1 is a redox-sensitive bZip transcription factor which regulates the expression of many antioxidant genes and is required for Yap1 deactivation (reviewed in reference 3). In the present study we have shown that loss of the mitochondrial thioredoxin system does not alter GSH levels or redox balance during normal aerobic growth. Similarly, loss of mitochondrial TRR2 does not affect Glr1 enzyme activity (data not shown). This result indicates that, unlike the cytoplasmic thioredoxin system, the mitochondrial thioredoxin system does influence the expression of genes like GLR1 which are regulated by Yap1. Relatively few studies have directly examined the redox state of isolated organelles, such as mitochondria, mainly due to the difficulties encountered in maintaining the redox balance during prolonged extraction and purification procedures. Analysis of yeast and mammalian cells has indicated that the ratio of oxidized GSSG to reduced GSH is somewhat higher in mitochondria than in the cytosol, in accord with the idea that mitochondria are the major source of ROS (24, 32). Our studies have measured the total concentrations of oxidized and reduced GSH in cells and hence give an indication of the overall redox environment. The cytosol is the largest compartment in the cell and is therefore likely to be the principal determinant of the redox environment (40). However, mitochondria are the major source of ROS in cells, and during respiration, complex III of the respiratory chain is thought to be responsible for more than 80% of ROS production (2, 4). Thus, any increases in mitochondrial GSSG should be apparent in these measurements.
Deletion of cytoplasmic TRX1 and TRX2 has been shown to result in a prolonged S phase and shortened G1 interval of the cell cycle (31). This cell cycle defect does not result from alterations in the levels of deoxyribonucleotides but requires a redox-dependent function (30). Similarly, loss of TRR1 results in slow growth presumably due to an inability to recycle oxidized thioredoxins to the reduced form (44). In contrast, loss of the mitochondrial thioredoxin system does not affect growth during normal aerobic conditions. Interestingly, mitochondrial Trr2 was found to be required for growth during respiratory conditions in the absence of the cytoplasmic thioredoxin system. This requirement for Trr2 appears to be independent of mitochondrial Trx3, since loss of TRX3 does not exacerbate the slow growth of the trx1 trx2 mutant and oxidized Trx3 cannot be detected in the trr1 trr2 mutant during respiratory growth conditions. As in other species, yeast Trx1 and Trx2 are active as antioxidants and play key roles in protection against oxidative stress induced by various ROS (20, 23). TRX2 appears to play the predominant role, since mutants lacking TRX2 are hypersensitive to hydroperoxides and mutants containing TRX2, in the absence of TRX1, are resistant to these oxidants (9). A previous study has shown that a trr2 mutant, but not a trx3 mutant, is sensitive to an acute oxidative stress treatment induced by exposure to H2O2 (35). In the present study we have extended these observations to show that neither Trx3 nor Trr2 is required for protection against long-term exposure to H2O2. In contrast, the cytoplasmic thioredoxins are required for growth under these conditions. However, a requirement for TRR2 is observed in the trr1 mutant since the trr1 trr2 mutant is hypersensitive to H2O2. Taken together, these data indicate that mitochondrial Trr2 must have some function, in addition to its role in the reduction of Trx3, which is required during respiratory growth conditions and for protection against oxidative stress. Potential functions include direct reduction of hydroperoxides (35) or activity with other redox active molecules, such as Grx2 or Grx5, which localize to mitochondria (36, 39).
The thioredoxin and GSH-glutaredoxin systems are thermodynamically linked to each other since they both use NADPH as a source of reducing equivalents. Thioredoxin reductase uses NADPH to reduce thioredoxins but is unable to reduce glutaredoxins (18). In addition, low-molecular-weight disulfides such as GSSG are poor substrates for the thioredoxin system (17). GSH reductase uses NADPH to reduce GSH, which in turn maintains the redox state of glutaredoxins. Since the GSH system is unable to reduce thioredoxins, the GSH-glutaredoxin and thioredoxin systems have long been considered as two separate disulfide reduction systems (17, 18, 37). We have previously shown that shifting the GSH pool to a more oxidized state in the glr1 mutant does not affect the redox state of cytoplasmic thioredoxins, whereas complete GSH depletion does result in cytoplasmic thioredoxin oxidation (43). In addition, mutation studies indicate that there is an essential cellular requirement for either an active cytoplasmic thioredoxin or GSH-glutaredoxin system. This does not appear to be a requirement to prevent the accumulation of toxic disulfide bonds, since the lethality of a glr1 trr1 mutant cannot be rescued by anaerobic growth or addition of exogenous GSH. The basis of this lethality is therefore thought to be due to the loss of activity of an essential metabolic enzyme such as ribonucleotide reductase (43).
The present study has shown that, in contrast to a trr1 glr1 mutant, the trr2 glr1 mutant is viable. However, the trr2 glr1 mutant is hypersensitive to H2O2 and grows slowly during respiratory growth conditions, indicating that there is an overlapping requirement for Glr1 and Trr2. Part of the growth defect of this mutant appears to result from the toxicity of oxidized mitochondrial Trx3, since deletion of TRX3 restores the growth of a glr1 trr2 mutant during respiratory growth conditions. Oxidized Trx3 may therefore promote disulfide formation in the mitochondria. The overlap between the GSH and mitochondrial thioredoxin systems is further demonstrated by the redox state of the trr2 glr1 mutant, since GSH and Trx3 are both shifted to a more oxidized state in this mutant. An accumulation of oxidized GSH and thioredoxin may account for the growth defects of this mutant. The overlapping function of GSH and mitochondrial Trx3 is unknown at present but may indicate that they have a common mitochondrial substrate. In summary, there is now increasing evidence that, rather than representing separate systems, the various cellular redox regulatory systems have many overlapping features in terms of both regulation and substrate specificity.
REFERENCES
- 1.Baudin, A., O. Ozier-Kalogeropoulos, A. Danouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boveris, A., and E. Cadenas. 1982. Production of superoxide radicals and hydrogen peroxide in mitochondria, p. 15-30. In L. W. Oberley (ed.), Superoxide dismutases, vol. 2. CRC Press, Boca Raton, Fla. [Google Scholar]
- 3.Carmel-Harel, O., and G. Storz. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54:439-461. [DOI] [PubMed] [Google Scholar]
- 4.Chance, B., H. Sies, and A. Boveris. 1979. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 59:527-605. [DOI] [PubMed] [Google Scholar]
- 5.Collinson, E. J., and C. M. Grant. 2003. Role of yeast glutaredoxins as glutathione S-transferases. J. Biol. Chem. 278:22492-22497. [DOI] [PubMed] [Google Scholar]
- 6.Collinson, E. J., G. Wheeler, E. O. Garrido, A. M. Avery, S. V. Avery, and C. M. Grant. 2002. The yeast glutaredoxins are active as glutathione peroxidases. J. Biol. Chem. 277:16712-16717. [DOI] [PubMed] [Google Scholar]
- 7.Draculic, T., I. W. Dawes, and C. M. Grant. 2000. A single glutaredoxin or thioredoxin is essential for viability in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 36:1167-1174. [DOI] [PubMed] [Google Scholar]
- 8.Gan, Z.-R. 1991. Yeast thioredoxin genes. J. Biol. Chem. 266:1692-1696. [PubMed] [Google Scholar]
- 9.Garrido, E. O., and C. M. Grant. 2002. Role of thioredoxins in the response of Saccharomyces cerevisiae to oxidative stress induced by hydroperoxides. Mol. Microbiol. 43:993-1003. [DOI] [PubMed] [Google Scholar]
- 10.Gladyshev, V. N., A. Liu, S. V. Novoselov, K. Krysan, Q. A. Sun, V. M. Kryukov, G. V. Kryukov, and M. F. Lou. 2001. Identification and characterization of a new mammalian glutaredoxin (thioltransferase), Grx2. J. Biol. Chem. 276:30374-30380. [DOI] [PubMed] [Google Scholar]
- 11.Grant, C. M. 2001. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol. Microbiol. 39:533-541. [DOI] [PubMed] [Google Scholar]
- 12.Grant, C. M., L. P. Collinson, J.-H. Roe, and I. W. Dawes. 1996. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol. Microbiol. 21:171-179. [DOI] [PubMed] [Google Scholar]
- 13.Grant, C. M., F. H. MacIver, and I. W. Dawes. 1996. Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr. Genet. 29:511-515. [DOI] [PubMed] [Google Scholar]
- 14.Grant, C. M., F. M. MacIver, and I. W. Dawes. 1996. Stationary phase induction of GLR1 expression is mediated by the yAP-1 transcriptional protein in Saccharomyces cerevisiae. Mol. Microbiol. 22:739-746. [DOI] [PubMed] [Google Scholar]
- 15.Grant, C. M., G. Perrone, and I. W. Dawes. 1998. Glutathione and catalase provide overlapping defenses for protection against hydrogen peroxide in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 253:893-898. [DOI] [PubMed] [Google Scholar]
- 16.Halliwell, B., and J. M. C. Gutteridge. 1989. Free radicals in biology and medicine, 2nd ed. Oxford University Press, Oxford, United Kingdom.
- 17.Holmgren, A. 1979. Reduction of disulfides by thioredoxin. Exceptional reactivity of insulin and suggested functions of thioredoxin in mechanism of hormone action. J. Biol. Chem. 254:9113-9119. [PubMed] [Google Scholar]
- 18.Holmgren, A. 1989. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 264:13963-13966. [PubMed] [Google Scholar]
- 19.Holmgren, A., and F. Aslund. 1995. Glutaredoxin. Methods Enzymol. 252:283-292. [DOI] [PubMed] [Google Scholar]
- 20.Izawa, S., K. Maeda, K.-I. Sugiyama, J. Mano, Y. Inoue, and A. Kimura. 1999. Thioredoxin deficiency causes the constitutive activation of Yap1, an AP-1-like transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 274:28459-28565. [DOI] [PubMed] [Google Scholar]
- 21.Jamieson, D. J. 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14:1511-1527. [DOI] [PubMed] [Google Scholar]
- 22.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 23.Kuge, S., and N. Jones. 1994. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenton, K. J., H. Therriault, and J. R. Wagner. 1999. Analysis of glutathione and glutathione disulfide in whole cells and mitochondria by postcolumn derivatization high-performance liquid chromatography with ortho-phthalaldehyde. Anal. Biochem. 274:125-130. [DOI] [PubMed] [Google Scholar]
- 25.Luikenhuis, S., I. W. Dawes, and C. M. Grant. 1997. The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol. Biol. Cell 9:1081-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundberg, M., C. Johansson, J. Chandra, M. Enoksson, G. Jacobsson, J. Ljung, M. Johansson, and A. Holmgren. 2001. Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. J. Biol. Chem. 276:26269-26275. [DOI] [PubMed] [Google Scholar]
- 27.Machado, A. K., B. A. Morgan, and G. F. Merril. 1997. Thioredoxin reductase-dependent inhibition of MCB cell cycle box activity in Saccharomyces cerevisiae. J. Biol. Chem. 272:17045-17054. [DOI] [PubMed] [Google Scholar]
- 28.Meister, A., and M. E. Anderson. 1983. Glutathione. Annu. Rev. Biochem. 52:711-760. [DOI] [PubMed] [Google Scholar]
- 29.Muller, E. G. D. 1996. A glutathione reductase mutant of yeast accumulates high levels of oxidized glutathione and requires thioredoxin for growth. Mol. Biol. Cell 7:1805-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller, E. G. D. 1995. A redox-dependent function of thioredoxin is necessary to sustain a rapid rate of DNA synthesis in yeast. Arch. Biochem. Biophys. 318:356-361. [DOI] [PubMed] [Google Scholar]
- 31.Muller, E. G. D. 1991. Thioredoxin deficiency in yeast prolongs S phase and shortens the G1 interval of the cell cycle. J. Biol. Chem. 266:9194-9202. [PubMed] [Google Scholar]
- 32.Outten, C. E., and V. C. Culotta. 2004. Alternative start sites in the S. cerevisiae GLR1 gene are responsible for mitochondrial and cytosolic isoforms of glutathione reductase. J. Biol. Chem. 279:7785-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, S. G., M.-K. Cha, W. Jeong, and I.-H. Kim. 2000. Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J. Biol. Chem. 275:5723-5732. [DOI] [PubMed] [Google Scholar]
- 34.Pearson, G. D., and G. F. Merril. 1998. Deletion of the Saccharomyces cerevisiae TRR1 gene encoding thioredoxin reductase inhibits p53-dependent reporter gene expression. J. Biol. Chem. 273:5431-5434. [DOI] [PubMed] [Google Scholar]
- 35.Pedrajas, J. R., E. Kosmidou, A. Miranda-Vizuete, J.-A. Gustafsson, A. P. H. Wright, and G. Spyrou. 1999. Identification and functional characterization of a novel mitochondrial thioredoxin system in Saccharomyces cerevisiae. J. Biol. Chem. 274:6366-6373. [DOI] [PubMed] [Google Scholar]
- 36.Pedrajas, J. R., P. Porras, E. Martinez-Galisteo, C. A. Padilla, A. Miranda-Vizuete, and J. A. Barcena. 2002. Two isoforms of Saccharomyces cerevisiae glutaredoxin 2 are expressed in vivo and localize to different subcellular compartments. Biochem. J. 364:617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prinz, W. A., F. Aslund, A. Holmgren, and J. Beckwith. 1997. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulphide bonds in Escherichia coli. J. Biol. Chem. 272:15661-15667. [DOI] [PubMed] [Google Scholar]
- 38.Rietsch, A., and J. Beckwith. 1998. The genetics of disulfide bond metabolism. Annu. Rev. Genet. 32:163-184. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-Manzaneque, M. T., J. Tamarit, G. Belli, J. Ros, and E. Herrero. 2002. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell 13:1109-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schafer, F. Q., and G. R. Buettner. 2001. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 30:1191-1212. [DOI] [PubMed] [Google Scholar]
- 41.Sherman, F., G. R. Fink, and C. W. Lawrence. 1974. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Tanaka, T., F. Hosoi, Y. Yamaguchi-Iwai, H. Nakamura, H. Masutani, S. Ueda, A. Nishiyama, S. Takeda, H. Wada, G. Spyrou, and J. Yodoi. 2002. Thioredoxin-2 (TRX-2) is an essential gene regulating mitochondria-dependent apoptosis. EMBO J. 21:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trotter, E. W., and C. M. Grant. 2003. Non-reciprocal regulation of the redox state of the glutathione/glutaredoxin and thioredoxin systems. EMBO Rep. 4:184-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trotter, E. W., and C. M. Grant. 2002. Thioredoxins are required for protection against a reductive stress in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 46:869-878. [DOI] [PubMed] [Google Scholar]
- 45.Yu, B. P. 1994. Cellular defenses against damage from reactive oxygen species. Physiol. Rev. 74:139-162. [DOI] [PubMed] [Google Scholar]