Abstract
The basolateral amygdala (BLA) is a site of convergence of negative and positive stimuli, and is critical for emotional behaviors and associations. However, the neural substrate for negative and positive behaviors and relationship between negative and positive representations in the basolateral amygdala is unknown. Here, we identified two genetically distinct, spatially segregated populations of excitatory neurons in the basolateral amygdala (BLA) that participate in valence-specific behaviors and are connected through mutual inhibition. These results identify a genetically-defined neural circuit for the antagonistic control of emotional behaviors and memories.
INTRODUCTION
The basolateral complex of the amygdala consists of two intimately juxtaposed nuclei—the lateral nucleus (LA) and basolateral nucleus (BLA)1, 2. The BLA is a cortical-like brain structure consisting of two-types of nonlaminarly organized excitatory pyramidal, magnocellular and parvocellular neurons segregated in the anterior and posterior BLA (aBLA, pBLA), intermingled with populations of genetically-defined interneurons1, 3–8. The BLA is activated by negative and positive emotional stimuli, and participates in emotional behaviors and associations9–16. Recent studies showed that BLA neurons drive opposing behaviors; therefore, the BLA may be a key site for the regulation of negative and positive behaviors9, 15. Despite the critical role of the BLA in emotional behaviors, it is not established whether the BLA pyramidal neurons that contributes to negative and positive behaviors (negative neurons and positive neurons) are structurally distinct, let alone, genetically distinguishable17, 18. Furthermore, a neural circuit subserving the antagonistic nature of emotional behaviors has yet to be identified.
Recent studies demonstrated that BLA neurons, which express the activity-dependent gene, c-Fos, during a negative or positive stimulus, were capable of driving a behavioral response consistent with the valence of the experience9, 15. Therefore, we reasoned that by utilizing a c-Fos-based genetic expression system, molecular profiles of the putative negative and positive neurons within the BLA can be obtained. Activity-dependent molecular profiles of BLA neurons may reveal genetic markers unique to negative and positive neurons. In turn, the identification of distinguishing genetic markers for negative and positive BLA neurons will provide a foundation for identifying the neural circuits underlying antagonistic behaviors elicited by negative and positive stimuli.
RESULTS
Identification of BLA Genetic Markers
Genetics-based RNA profiling strategies in mammalian models have involved ectopically expressing epitope-tagged RNA associated proteins or exploiting molecular modifications of RNA-associated substrates19–22. In order to obtain transcriptional profiles, we implemented a strategy involving ectopically expressing an epitope-tagged RNA binding protein, poly(A) binding protein with a c-terminus FLAG tag (PABP-FLAG)23. Two AAV9 constructs were used, one containing the tetracyclin-based transcription factor, tTA, under the control of the activity-dependent promoter of c-Fos (AAV9-c-Fos-tTA), and the other containing Pabp-flag under the control of the tetracycline response element TRE (AAV9-TRE-Pabp-flag). Activation of the c-Fos promoter drives the expression of tTA. In the absence of doxycycline (Dox), tTA binds TRE to induce the expression of PABP-FLAG. PABP-FLAG competes with endogenous PABP and bind the polyA tails of mRNA, which then can be isolated via immunoprecipiation using an anti-flag antibody and A/G coated magnetic beads (Fig. 1a).
Figure 1. Activity-dependent transcriptional profiling of BLA neurons.
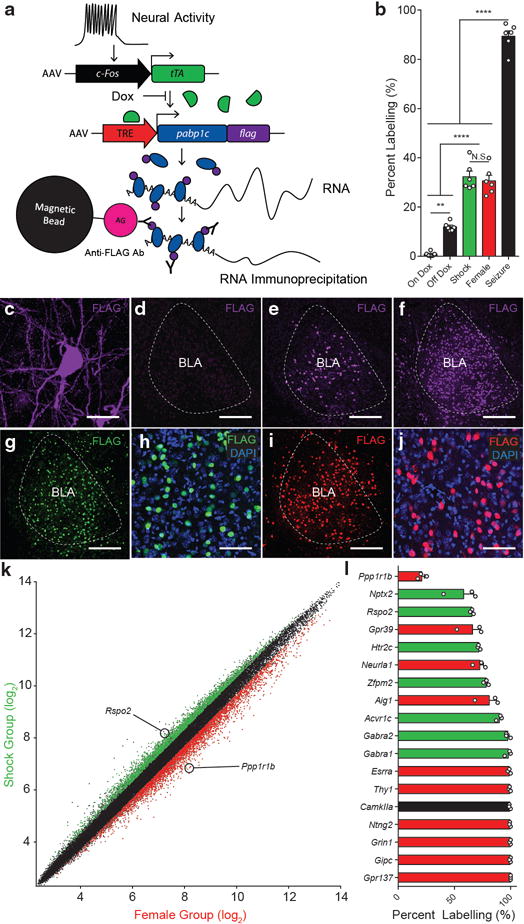
a, Viral-based genetic scheme for activity-dependent transcriptional profiling. c-Fos promoter activity drives the expression of tTA, which in turn, binds TRE and drives the expression of PABP-FLAG in the absence of doxycycline (Dox). b, PABP-FLAG expression in the BLA in mice kept on a Dox diet (On Dox), taken off a Dox diet and exposed to home cage (Off Dox), Shock, Female, Seizure, (one-way ANOVA, F4,25=131.0, P<0.0001, n = 6 per group). Significance for multiple comparisons, **P < 0.01, ****P < 0.0001, not significant (N.S.). c, PABP-FLAG expression in soma and varicosities of a BLA neuron. FLAG expression in the BLA of On Dox (d), Off Dox (e), Seizure (f), Shock (g,h), and Female (i,j) group. FLAG expression and nuclear marker, DAPI, in Shock (h), and Female (j) group. Scale bar 25μm (c), 250μm (d,e,f,g,i), 80 μm (h,j). k, RMA normalizedn RNA expression values from microarray from RNA purified from Shock (n = 3) and Female (n = 3) groups. Red and green points represent enriched genes (>1.25 fold, ANOVA p<.05, log2 scale). l, Quantification of in situ hybridization of BLA expression of candidate genetic markers enriched in shock group (green) and female group (red) (n = 3 mice per group). Positive control genes (black). Results show mean ± s.e.m (b,l).
The putative negative and positive neurons were targeted by exposing male mice to footshocks and a female mouse, respectively. AAV9-c-Fos-tTA and AAV9-TRE-Pabp-flag were introduced into the BLA in mice kept on a Dox diet. Once placed off a Dox diet for 2 days, mice were exposed to footshocks or female mouse, then immediately placed back on a Dox diet for 2 days prior to sacrifice. A similar number of BLA neurons were FLAG+ in the shock and female groups, but were greater than mice that were kept in their home cages or kept on a Dox diet (Fig. 1b,c,d,e,g–j). In contrast, a greater number of BLA neurons were FLAG+ in the mice that underwent kainic acid-induced seizures compare to the shock or female group (Fig. 1b,f–j). This affirms the activity-dependency of the genetic system. Therefore, RNA immunoprecipitation using antibodies against FLAG was performed from the shock and female group. Isolated RNA was reverse-transcribed to cDNA and underwent microarray analysis using Affymetric Mouse 430A chip. After RMA or MAS5 normalization (see Methods), differential gene expression profiles were compared between the shock and female group and were used as the basis of the screen for identifying genetic markers for the putative negative and positive neurons of the BLA (Fig. 1k, Supplemental Fig. 1).
Based on previous observations9, 15, we hypothesized that the putative negative and positive BLA neurons would be non-overlapping; therefore, we sought to select from our potential list of candidate genetic markers a single gene candidate, one for each of the putative negative and positive neuron populations. As a corollary, this posits that each of the gene markers would label a subpopulation (<100%) of BLA principle neurons. First, independent of statistical significance, hundreds of genes whose expression was enriched in the shock and female groups were individually screened on Allen Mouse Brain Atlas24. 37 genes were selected for single label fluorescent in situ hybridization, of which, 16 probes yielded a quantifiable signals in the −1.0 to −1.6 anterior-posterior (AP) plane of the BLA (Supplemental Fig 2). Quantification of gene expression in the BLA revealed that the majority of these candidate genes were expressed in a virtually all BLA neurons (Fig. 1l, Supplemental Fig. 2). Rspo2 (R-spondin 2) was enriched in the shock group (Fig. 1k) and was expressed in less than 100% of BLA neurons (Fig. 1l). Furthermore, Rspo2 expression was specific to the BLA, with little expression in other brain areas. In fact, because of this BLA-specific expression, we had previously generated a BLA-restricted Cre transgenic mouse line using the Rspo2 gene promotor, although we had not further characterized this transgenic mouse with respect to more detailed expression pattern of the Cre expression within the BLA, nor examined the role of Rspo2-expressing BLA neurons in valence-related behavior. Therefore, Rspo2 was selected as a candidate for a negative BLA neuron marker for further anatomical and functional studies of negative BLA neurons. Rspo2+ BLA neurons labeled less than 100%, but greater than 50%, of BLA neurons. Among the candidate genes belonging to the female group, Ppp1r1b (protein phosphatase 1 regulatory inhibitor subunit 1B, which encoded for DARPP-3225) was the only gene that labeled less than 50% of BLA neurons. Furthermore, based on the distribution pattern of Rpso2 and Ppp1r1b, it appeared that these two markers may be expressed in neurons that are non-overlapping. Therefore, we selected Ppp1r1b as a potential marker for positive BLA neurons for further characterization.
Double label single molecule fluorescent in situ hybridization (smFISH) and quantification across the anterior-posterior (AP) axis of the BLA (−0.8 to −2.8mm from bregma) revealed that Rspo2 and Ppp1r1b labeled spatially segregated population of neurons (Fig. 2a–c). Less than 1% of BLA neurons were Rspo2+Ppp1r1b+ (Table 1). Rspo2+ and Ppp1r1b+ BLA neurons are co-labelled with the pyramidal neurons marker, Camk2a, and non-overlapping with the inhibitory neuron marker, Gad1 (Fig. 2d–g, Table 1). Rspo2+ neurons correspond to magnocellular pyramidal neurons in the anterior BLA (aBLA). In contrast, Ppp1r1b+ neurons correspond to the parvocellular pyramidal neurons or posterior BLA (pBLA)1, 26. Double smFISH with a Camk2a probe and the combined probes of both the Rspo2 and Ppp1r1b showed that virtually all Camk2a+ BLA neurons express either Rspo2 or Ppp1r1b (Table 1, Supplemental Fig. 3). Therefore, Rpso2+ and Ppp1r1b+ neurons collectively define the entirety of BLA pyramidal neurons.
Figure 2. Rspo2+ and Ppp1r1b+ BLA neurons define spatially segregated populations of BLA pyramidal neurons.
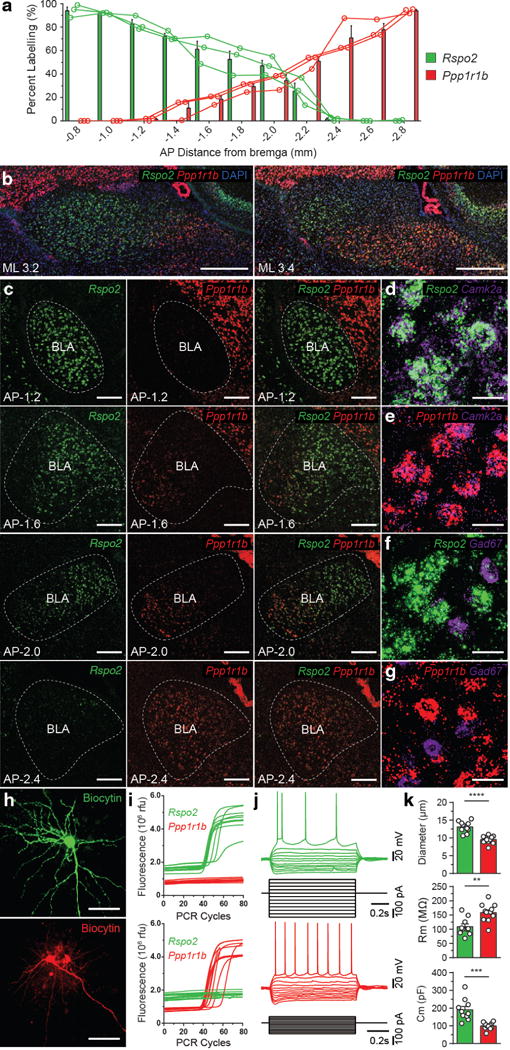
a, Quantification of smFISH of Rspo2 (green) and Ppp1r1b (red) expression across the AP axis (coronal distance from bregma −0.8mm to −2.8mm) of the BLA, bars represent means, line represents individual mice (n = 3). b, Two sagittal views (ML distance from midline, 3.2mm, 3.4mm) of double smFISH of Rspo2 and Ppp1r1b with nuclear marker, DAPI, in the BLA c, Coronal view of double smFISH of Rspo2 and Ppp1r1b across the AP axis of the BLA. Double smFISH of Camk2a and Rspo2 (d), Camk2a and Ppp1r1b (e), Gad1 and Rspo2 (f) Gad1 and Ppp1r1b (g), in the BLA (Larger micrograph in Supplemental Fig. 3). Scale bar 500μm (b), 200μm (c), 25μm (d–g). h, Biocytin-filled magnocellular (top) and parvocellular (bottom) BLA neuron, scale bar 50μm. i, Single-cell qPCR traces of Rpso2 (green) and Ppp1r1b (red), of magnocellular (top) and parvocellular (bottom) BLA neurons, qPCR traces in which amplification occurred for either gene is shown. j, Electrophysiological response to current steps in a Rspo2+ (top) and Ppp1r1b+ (bottom) BLA neuron. k, Comparison of mean soma diameter, membrane resistance (Rm), and membrane capacitance (Cm) of qPCR-confirmed Rpso2+ (green, n = 11) and Ppp1r1b+ (red, n = 12) neurons. Significance for unpaired t-test, **P< 0.01, ***P<0 .001, ****P<0.0001. Results show mean ± s.e.m (a,k).
Table 1. Genetic, anatomical, morphological, and electrophysiological characterization of BLA neurons.
(a) smFISH quantification of Rpso2 and Ppp1r1b expression in the BLA, Rpso2 and Ppp1r1b colocalization with Gad1 in the BLA, combined probes of Rpso2 and Ppp1r1b and their colocalization with Camk2a in the BLA, Rpso2 and Ppp1r1b colocalization with CTB injected mice (CeC, CeL/M, and NAc), cell counts corresponds to the total number of neurons counted in n animals. (b) Rpso2+ and Ppp1r1b+ BLA neurons correspond to magnocellular and parvocellular BLA neurons. Quantification of morphological (soma diameter) and electrophysiological properties (resting membrane potential (Vm), membrane resistance (Rm), capacitance (Cm), spike threshold, and rheobase) of qPCR-unconfirmed magnocellular, parvocellular, and qPCR-confirmed Rpso2+ and Ppp1r1b+ BLA neurons, p-values for unpaired t-test for comparisons, n corresponds to the number of neurons from at least 3 mice per group.
| (a)
| |||
|---|---|---|---|
| Rspo2+ | Ppp1r1b+ | Rspo2+Ppp1r1b+ | |
| Total Neurons (n = 3) | 3611 | 2311 | 54 |
| Mean Proportion (%) | 59.9 ± 1.28 | 39.1 ± 1.14 | 0.970 ± 0.191 |
|
| |||
| Rspo2+ | Gad1+ | Rspo2+Gad1+ | |
|
| |||
| Total Neurons (n = 1) | 303 | 112 | 0 |
|
| |||
| Ppp1r1b+ | Gad1+ | Ppp1r1b +Gad1+ | |
|
| |||
| Total Neurons (n = 1) | 190 | 116 | 0 |
|
| |||
| (Rspo2+Ppp1r1b)+Camk2+ | (Rspo2+Ppp1r1b) −Camk2+ | (Rspo2+Ppp1r1b)+Camk2− | |
|
| |||
| Total Neurons (n = 4) | 2361 | 0 | 0 |
|
| |||
| CTB-CeC+Rspo2+ | CTB-CeC Ppp1r1b+ | ||
|
| |||
| Total Neurons (n = 3) | 423 | 16 | |
| Mean Proportion (%) | 96.2 ± 0.945 | 3.78 ± 0.945 | |
|
| |||
| CTB-CeL/M+Rspo2+ | CTB-CeL/M+Ppp1r1b+ | ||
|
| |||
| Total Neurons (n = 3) | 64 | 1012 | |
| Mean Proportion (%) | 5.58 ± 1.74 | 94.4 ± 1.74 | |
|
| |||
| CTB-NAc+Rspo2+ | CTB-NAc+Ppp1r1b+ | ||
|
| |||
| Total Neurons (n = 3) | 344 | 775 | |
| Mean Proportion (%) | 30.7 ± 3.53 | 69.2 ± 3.53 | |
| (b)
| ||||||
|---|---|---|---|---|---|---|
| Cell type | Soma diameter (μm) | Vm (mV) | Rm (MΩ) | Cm (pF) | Spike threshold (mV) | Rheobase (pA) |
| Magnocellular (n = 37) | 12.8 ± 0.2 | −60.9 ± 0.9 | 103.1 ± 4.7 | 197.6 ± 10 | −37.7 ± 0.4 | 213.7 ± 10.5 |
| Parvocellular (n = 38) | 9.4 ± 0.2 | −55.6 ± 0.8 | 165.5 ± 6.5 | 102.2 ± 4.1 | −35.5 ± 0.5 | 137.7 ± 6.7 |
| Rspo2+ (n = 10) | 13.1 ± 0.5 | −62.5 ± 2.1 | 108.9 ± 9.6 | 190.3 ± 19.1 | −37.6 ± 0.9 | 198.7 ± 18 |
| Ppp1r1b+ (n = 11) | 9.5 ± 0.3 | −57.1 ± 1.5 | 158.7 ± 9.5 | 99.5 ± 5.9 | −34.8 ± 1.1 | 152.3 ± 15.2 |
|
| ||||||
| p-values | Soma diameter (μm) | Vm (mV) | Rm (MΩ) | Cm (pF) | Spike threshold (mV) | Rheobase (pA) |
|
| ||||||
| Magno. (n = 37) vs Parvo. (n= 38) | 7.00E-17 | 0.00003 | 6.00E-11 | 1.00E-11 | 0.0008 | 8.00E-08 |
| Rspo2+ (n = 10) vs Ppp1r1b+(n = 11) | 0.00002 | 0.051 | 0.0016 | 0.0009 | 0.06 | 0.06 |
| Magno. (n = 27) vs Rspo2+(n = 10) | 0.5 | 0.4 | 0.5 | 0.7 | 0.9 | 0.4 |
| Parvo. (n = 27) vs Ppp1r1b+ (n = 11) | 0.6 | 0.3 | 0.5 | 0.6 | 0.5 | 0.2 |
Genetic and Anatomical (a), Morphalogical and Electrophysiological (b) Characterization of Rpso2+ and Ppp1r1b+ BLA neurons
The electrophysiological and morphological properties of Rspo2+ and Ppp1r1b+ neurons were examined using patch clamp recordings. Rspo2+ and Ppp1r1b+ were targeted by patching magnocellular and parvocellular BLA neurons (Fig. 2h). To ascertain genetic identity, Rspo2+ and Ppp1r1b+ neurons were identified by the use of single-cell quantitative polymerase chain reaction (qPCR) from cytoplasmic harvest of patch clamped recorded BLA neurons. Of 37 magnocellular neurons, single cell qPCR yielded 10 Rpso2+ and 0 Ppp1r1b+ neurons; of 38 parvocellular neurons, single cell qPCR yielded 0 Rpso2+ and 11 Ppp1r1b+ neurons (Fig. 2i). Soma diameter was larger in Rspo2+ neurons than Ppp1r1b+ neurons; membrane resistance was smaller in Rspo2+ neurons than Ppp1r1b+ neurons; membrane capacitance was larger in Rspo2+ neurons than Ppp1r1b+ neurons (Fig. 2j,k). qPCR-confirmed Rspo2+ and Ppp1r1b+ neurons were not significantly different from unconfirmed magnocellular and parvocellular neurons, respectively (Table1). Taken together, Rpso2+ and Ppp1r1b+ BLA neurons defined spatially segregated, genetically, morphologically, and electrophysiological distinct cell-types.
BLA Activation by Valence-Specific Stimuli
If Rspo2+ and Ppp1r1b+ neurons represent negative and positive neurons of the BLA, respectively, then stimuli that elicit valence-specific behaviors may differentially activate the aBLA and pBLA. Mice were exposed to the stimuli used to identify BLA gene markers–shocks or female mice—and were sacrificed 90 minutes later. c-Fos+ neurons were quantified sepearately in the aBLA and pBLA (defined by cytoarchitectural boundaries) by measuring the total number of c-Fos + neurons per section at intervals across the AP axis (Fig. 3a–c, Supplemental Fig. 4). The relative c-Fos expression, measured by the number of c-Fos + neurons in the aBLA or pBLA as a percentage of total c-Fos + BLA neurons, was significantly greater in the aBLA in response to footshocks compared to exposure to a female mice or control mice, which received no stimulus in a context (Fig. 3d). Conversely, relative c-Fos expression was significantly greater in the pBLA in response to female mice compared to exposure shock or control, which were exposed to a neutral context (Fig. 3d). In response to valence-specific olfactory stimuli—2,3,5-Trimethyl-3-thiazoline (TMT), or peanut oil—relative c-Fos expression was significantly greater in the aBLA in response to TMT compared to exposure to a neutral odor benzaldehyde (BA) or peanut oil, while relative c-Fos expression was significantly greater in the pBLA in response to peanut oil compared to exposure to a BA or TMT (Fig. 3e). In response to valence-specific gustatory stimuli —quinine (bitter), water, sucrose (sweet)—relative c-Fos expression was significantly greater in the pBLA in response to water and sucrose water compared to mice that received no water or quinine water (Fig. 3f). In contrast, no significant difference was observed in relative c-Fos expression between exposure to quinine water (which did not elicit much water drinking) compared to no water, as well as between sucrose water and water (Fig. 3f). Overall, the aBLA is recruited by stimuli that elicits negative behaviors (shocks, TMT), while the pBLA is recruited by stimuli that elicits positive behaviors (female, water, sucrose, peanut oil).
Figure 3. Rspo2+ and Ppp1r1b+ BLA neurons are activated by valence-specific stimuli.
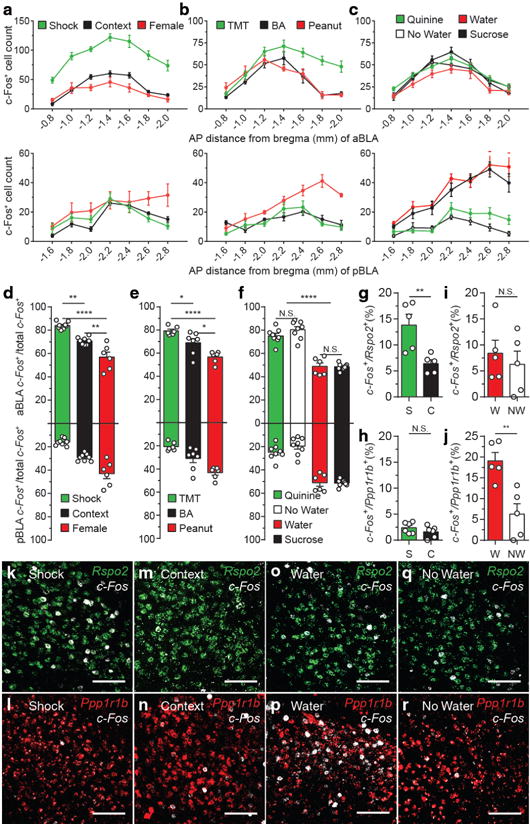
c-Fos expression across the AP axis (coronal distance from bregma −0.8mm to −2.8mm) of the aBLA (top) and pBLA (bottom) in response to shock (n = 8), context (n = 8), female (n = 6) (a); TMT (n = 6), BA (n = 7), peanut oil (n = 6) (b); quinine water (n = 8), no water (n = 8), water (n = 6), sucrose water (n = 8) (c). The total number of c-Fos+ cells is represented for each coronal section of a unilateral BLA (a–c), micrographs found in Supplemental Fig. 4. d, Relative c-Fos expression in the aBLA and pBLA in response to shock, context, female (one-way ANOVA, F2,19=33.91, P<0.0001). e, Relative c-Fos expression in response to TMT, BA, peanut oil (one-way ANOVA, F2,16=16.61, P=0.0001,). f, Relative c-Fos expression in response to quinine water, no water, water, sucrose water (one-way ANOVA, F2,19=33.91, P<.0001). Significance for multiple comparisons (d–f), *P < 0.05, **P < 0.01, ****P < 0.0001, not significant (N.S.). Double-label smFISH (n = 5 in each group) of c-Fos/Rspo2+ (g,k,m) or c-Fos/Ppp1r1b+ (h,l,n) in response to shock (S) or context (C). Double-label smFISH of c-Fos/Rspo2+ (i,o,q) or c-Fos/Ppp1r1b+ (j,p,r) in response to water (W) or no water (NW) (AP axis analysis in Supplemental Fig. 5), y-axis values denote percentage. Significance for unpaired t-test (g–j), **P<0.01, not significant (N.S). Scale bar 125μm (k–r). Results show mean ± s.e.m (a–j). Color scheme corresponds to the valence of the stimuli negative (green) and positive (red) (a–j), Rpso2(green) and Ppp1r1b(red) (k–r).
Double smFISH was performed to directly assess the expression of c-Fos in Rspo2+ or Ppp1r1b+ BLA neurons in response to valence-specific stimuli (stimuli that will be used in subsequent behavioral experiments). Shocks significantly increased c-Fos expression in Rspo2+ (Fig. 3g, k), but not in Ppp1r1b+ neurons (Fig. 3h, l), compared to context (Fig. 3g, h, m, n). In contrast, administration of water significantly increased c-Fos expression in Ppp1r1b+ (Fig. 3j,p), but not in Rspo2+ neurons (Fig. 3i,o), compared no water (Fig. 3i,j,q,r). This shock-specific activation of Rspo2+ aBLA cells and water-specific activation of Ppp1r1b+ pBLA cells were confirmed by further analyses of A/P axis planes within aBLA and pBLA, respectively (Supplemental Fig. 5). However, these analyses also revealed some heterogeneity of Rspo2+ neurons activated by shock within aBLA and Ppp1r1b+ neurons activated by water within pBLA, relative to the neutral stimuli (Supplemental Fig. 5): Rspo2+ cells activated specifically by shock and Ppp1r1b+ neurons activated specifically by water were more distributed posteriorly within aBLA and pBLA, respectively (Supplemental Fig. 5). Nevertheless, these data suggest that negative and positive information is represented by genetically-defined populations of neurons in the BLA that are spatially segregated; Rspo2+ neurons, which predominates in the aBLA, represent negative valence, while Ppp1r1b+ neurons, which predominates in the pBLA, represent positive valence.
BLA in Valence-Specific Behaviors
Valence-specific activation of Rpso2+ and Ppp1r1b+ neurons posits that these populations may be necessary for valence-specific behaviors; therefore, the effects of inhibiting these BLA populations were performed in a fear and reward conditioning paradigm. Rspo2+ and Ppp1r1b+ neurons were genetically targeted using Rspo2-Cre and Cartpt-Cre mice, respectively. Ppp1r1b+ BLA neurons are accessible by Carpt-Cre mice (Supplemental Fig. 6), and hereafter, virus-injected Cartpt-Cre mice will be referred to using “Ppp1r1b”. Light-activated inhibitory ion channel, eArch3.0, was expressed in Rspo2+ (Rspo2-Arch) and Ppp1r1b+ (Ppp1r1b-Arch) BLA neurons using a Cre-dependent viral vector (AAV5-EF1α-DIO-eArch3.0-eYFP) bilaterally targeted to the BLA of Rspo2-Cre and Cartpt-Cre mice, respectively. Control mice (Rspo2-eYFP, Ppp1r1b-eYFP) received a viral vector lacking eArch3.0, (AAV5-EF1α-DIO-eYFP) (Fig. 4a,1,m, Supplemental Fig. 7).
Figure 4. Rspo2+ and Ppp1r1b+ BLA neurons participate in valence-specific behaviors.
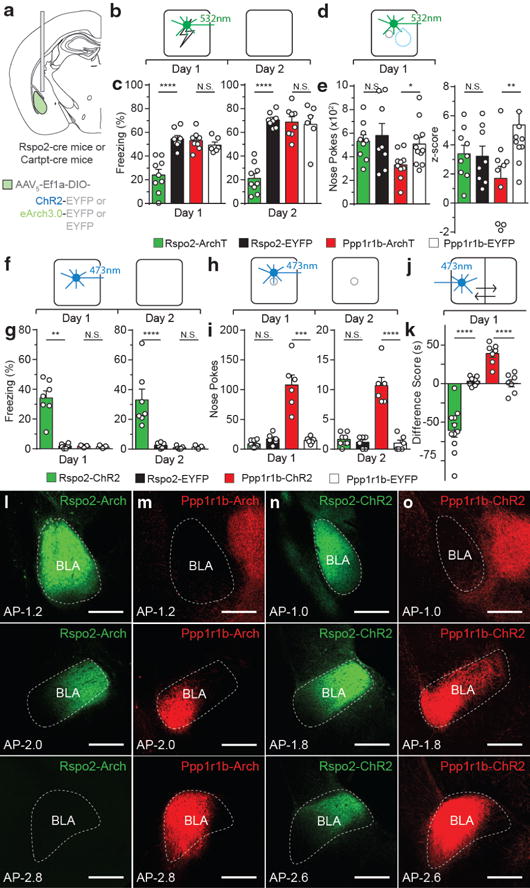
a, Optogenetically targeting Rspo2+ and Ppp1r1b+ BLA neurons. Scheme and results for Rspo2-Arch and Ppp1r1b-Arch mice in a fear (b,c) and reward (d,e) conditioning. c, Rspo2-Arch mice (n = 9) displayed lower freezing on Day 1 and 2 compared to eYFP controls (n = 8), no difference between Ppp1r1b-Arch (n = 8) and Ppp1r1b-eYFP (n = 6) mice. e, Ppp1r1b-Arch mice (n = 10) displayed lower total nose pokes and cue-reward association in nose port (z-score) compared to eYFP controls (n = 11), no difference between Rspo2-Arch (n = 9) and Rspo2-eYFP (n = 8). Scheme and results for Rspo2-ChR2 and Ppp1r1b-ChR2 mice in an optogenetic freezing test (f,g), optogenetic self-stimulation test (h,i), and optogenetic place preference test (j,k). g, Rspo2-ChR2 mice (n = 7) displayed greater freezing levels on Day 1 and 2 compared to eYFP controls (n = 6), no difference between Ppp1r1b-ChR2 (n = 5) and Ppp1r1b-eYFP (n =5) mice. i, Ppp1r1b-ChR2 mice (n = 6) displayed greater levels of nose pokes on day 1 and 2 compared to EYFP controls (n = 6), no difference between Rspo2-ChR2 (n = 8) and Rspo2-eYFP (n = 6) mice. k, Rspo2-ChR2 mice (n = 11) displayed greater preference to light stimulation compared to eYFP controls (n = 8), while Ppp1r1b-ChR2 (n = 7) mice displayed greater preference to light stimulation compared to eYFP controls (n = 7). Significance for unpaired t-test between experimental groups compared to corresponding eYFP controls, *P< 0.05, **P< 0.01, ***P<0 .001, ****P<00001, not significant (N.S), results show mean ± s.e.m (c,e,g,i,k). Expression of eArch-EYFP in Rspo2-Arch mice (l) and Ppp1r1b-Arch mice (m). Expression of ChR2-EYFPin Rspo2-ChR2 mice (n) and Ppp1r1b-ChR2 mice (o). Strong Ppp1r1b+ fibers are found in the central amygdala (m,o). Scale bar, 300μm (l,m,n,o). Color scheme corresponds to the virus-infected transgenic mouse, Rpso2-Cre (green) and Cartpt-Cre (red) (c,e,g,i,k,l–o).
On day 1 of contextual fear conditioning, mice received green light, bilaterally targeted to the BLA, during footshocks (Fig. 4b). Rspo2-Arch mice displayed reduced levels of freezing in response to footshocks compared with Rspo2-eYFP mice. Ppp1r1b-Arch mice displayed similar levels of freezing compared to Ppp1r1b-eYFP mice. On day 2, mice were tested in the context without shock or light stimulation. Reduction of freezing was observed in Rspo2-Arch mice compared to Rspo2-GFP mice, while, similar levels of freezing was observed in Ppp1r1b-Arch mice compared to Ppp1r1b-eYFP mice (Fig. 4c). Thus, Rspo2+, but not Ppp1r1bp+, BLA neuronal activity is critical for freezing to shock stimuli and for the association of a context to freezing behavior.
Reward conditioning took place in an operant conditioning chamber, where water was dispensed contingent on a nose poke following an external light cue (Fig. 4d). Green light was bilaterally delivered into the BLA simultaneously with the presentation of water. Rspo2-Arch and Rspo2-eYFP mice displayed similar levels of nose pokes and cue-reward association (z-score of time spent in the reward port during cue period, see Methods). In contrast, Ppp1r1b-Arch mice displayed reduced levels of nose pokes and cue-reward association compared to Ppp1r1b-eYFP mice (Fig. 4e). Thus, Ppp1r1b +, but not Rpso2+, BLA neuronal activity is critical for reward-seeking behavior and for the association of a conditioned stimulus to appetitive behavior.
Next, the effects of activating these BLA neurons were assessed. Light-activated excitatory ion channel, ChR2, was expressed in Rspo2+ (Rspo2-ChR2) and Ppp1r1b+ (Ppp1r1b-ChR2) BLA neurons using a Cre-dependent viral vector (AAV5-EF1α-DIO-ChR2-eYFP) unilaterally targeted to the BLA of Rspo2-Cre and Cartpt-Cre mice, respectively. Control mice (Rspo2-eYFP, Ppp1r1b-eYFP) received a viral vector lacking ChR2, (AAV5-EF1α-DIO-eYFP) (Fig. 4a,n,o).
On day 1 of the optogenetic freezing test, mice were placed in a neutral context while receiving blue light stimulation (Fig. 4f, see methods). Rspo2-ChR2 mice displayed greater levels of freezing compared to Rspo2-eYFP mice, while Ppp1r1rb-ChR2 and Ppp1r1b-eYFP mice displayed similar levels of freezing (Fig. 4g). On day 2, mice were returned to the context and freezing was measured without shock. Rspo2-ChR2 mice displayed greater levels of freezing compared to Rspo2-eYFP mice, while Ppp1r1rb-ChR2 and Ppp1r1b-eYFP mice displayed similar levels of freezing (Fig. 4g). Thus, Rspo2+, but not Ppp1r1b+, BLA neurons are capable of eliciting freezing, which can be conditioned to a neutral context.
On day 1 of the optogenetic self-stimulation test, mice were placed in an operant conditioning chamber in which blue light stimulation was administered when poking into a nose port (Fig. 4h). Ppp1r1b-ChR2 mice displayed greater number of pokes compared to Ppp1r1b-eYFP mice, while Rspo2-ChR2 and Rpso2-eYFP mice displayed similar number of pokes. On day 2, mice were returned to the operant condition chamber in which no light stimulation was delivered. Ppp1r1b-ChR2 mice displayed greater number of pokes compared to Ppp1r1b-eYFP mice, while Rspo2-ChR2 and Rspo2-eYFP mice displayed similar number of pokes (Fig. 4i). Thus, Ppp1r1b+, but not Rspo2+, BLA neurons are capable of eliciting self-stimulation and support reward conditioning.
In real-time optogenetic place preference test (Fig. 4j), Rspo2-ChR2 mice spent less time in the light-stimulated side compared to corresponding controls, while Ppp1r1b-ChR2 mice spent more time in the light-stimulated side compared to corresponding controls (Fig. 4k). Therefore, Rspo2+ BLA neurons are sufficient to elicit place aversion while Ppp1r1b+ BLA neurons are capable of eliciting place preference.
Antagonism between negative and positive BLA neurons
Rpso2+ and Ppp1r1b+ neurons drive opposing behaviors; therefore, we examined whether these two types of neurons contribute to the antagonistic control of emotional behaviors and memories. For this purpose, we examined the behavioral effects of optogenetically activating Rpso2+ or Ppp1r1b+ neurons during the presence of valence-specific stimuli. On day 1 of contextual fear conditioning, ChR2-expressing mice received bilateral blue light stimulation in the BLA during footshocks (Fig. 5a). Rspo2-ChR2 and Rspo2-eYFP mice displayed similar levels of freezing in response to footshocks, while Ppp1r1b-ChR2 mice displayed lower levels of freezing than Ppp1r1b-eYFP mice (Fig. 5b,c). On day 2, conditioned freezing was assessed by returning mice to the context without footshock or light stimulation. Similar to day 1, no difference in freezing was observed between Rspo2-ChR2 and Rspo2-eYFP mice, while less freezing was observed in Ppp1r1b-ChR2 mice compared Ppp1r1b-eYFP mice (Fig.5b,c). Thus, activation of Ppp1r1b+ BLA neurons is capable of disrupting freezing to footshocks and the association of a conditioned contextual stimulus with footshocks.
Figure 5. Rpso2+ and Ppp1r1b+ BLA neurons antagonize valence-specific behaviors.
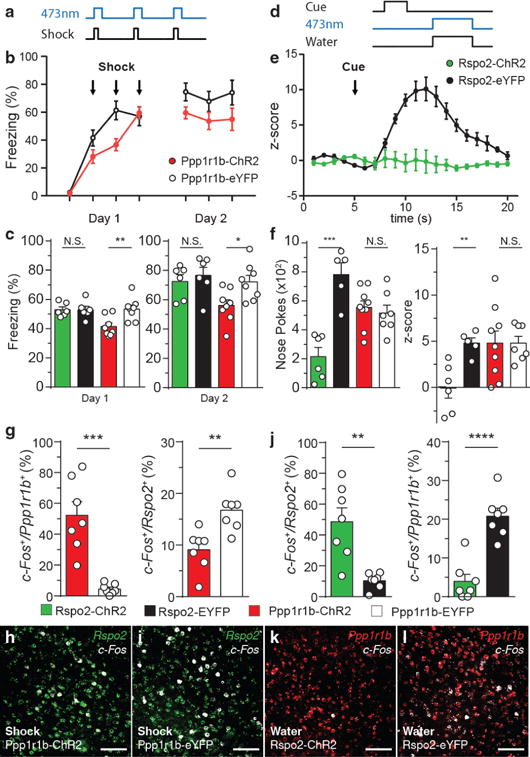
a, Scheme of activation of BLA neurons in Rspo2-ChR2 and Ppp1r1b-ChR2 mice during shocks (Day 1). b, Time course of freezing during day 1 and day 2 in Ppp1r1b-ChR2 (n = 8) and Ppp1r1b-eYFP (n = 8) mice. c, On day 1 and day 2, Ppp1r1b-ChR2 (n = 8) displayed lower freezing levels compared to eYFP controls (n =8), no difference between Rspo2-ChR2 (n = 6) and Rspo2-eYFP (n = 6) mice. d, Scheme of activation of BLA neurons in Rspo2-ChR2 and Ppp1r1b-ChR2 mice during reward conditioning. e, Time course of z-score of poking in Rspo2-ChR2 (n = 6) and Rspo2-eYFP (n = 5) mice. f, Rspo2-ChR2 mice (n = 6) displayed lower total nose pokes and cue-reward association compared to eYFP controls (n = 5), no difference between Ppp1r1b-ChR2 (n = 9) and Ppp1r1b-eYFP (n = 7) mice. Significance for unpaired t-test between experimental groups compared to corresponding eYFP controls, *P< 0.05, **P< 0.01, ***P<0 .001, ****P<0.0001, not significant (N.S), results show mean ± s.e.m. g,h,i Quantification of smFISH of c-Fos in Ppp1r1b+and Rpso2+ neurons in Ppp1r1b-ChR2 and Ppp1r1b-eYFP mice that received shock simultaneously with blue light stimulation. j,k,l, Quantification of smFISH of c-Fos in Rspo2+and Ppp1r1b+ neurons in Rspo2-ChR2 and Rspo2-eYFP mice that received water simultaneously with blue light stimulation. Significance for unpaired t-test (g,j), **P< 0.01, ***P<0 .001, ****P<0.0001, not significant (N.S). Results show mean ± s.e.m. Scale bar, 300μm (h,i,k,l). Color scheme corresponds to the virus-infected transgenic mouse, Rpso2-Cre (green) and Cartpt-Cre (red) (b,c,e,f,g,j) or valence of stimuli negative (green) and positive (red or white) (h,i,k,l).
In reward conditioning, ChR2-expressing mice received blue light stimulation during reward delivery (Fig. 5d). Rspo2-ChR2 displayed reduced levels of nose pokes and cue-reward association compared to Rspo2-eYFP mice, while Ppp1r1b-ChR2 and Ppp1r1bp-eYFP mice displayed similar levels of nose pokes and cue-reward association (Fig. 5e,f). Thus, activation of Rspo2 + BLA neurons is capable of disrupting reward-seeking behaviors and the association of a conditioned stimulus with a reward.
Although Rpso2+ and Ppp1r1b+ neurons antagonize behaviors elicited by stimuli of the opposing valence, behavioral antagonism may be result of interference by downstream circuits rather than by direct interactions between these two neuronal populations. Therefore, the effect of optogenetic activation of one of the two neuronal populations on the activation of the other in response to valence-specific stimuli was measured using c-Fos. In Ppp1r1b-ChR2 mice, which received blue light stimulation in the presence of footshocks, c-Fos was increased in Ppp1r1b+ neurons and decreased in Rpso2+ neurons compared to Ppp1r1b-eYFP mice (Fig. 5g,h,i). In water-deprived Rspo2-ChR2 mice, which received blue light stimulation during the consumption of water, c-Fos was increased in Rspo2+ neurons and decreased in Ppp1r1b+ neurons compared to Rspo2-eYFP mice (Fig 5j,k,l). Thus, Ppp1r1b+ and Rpso2+ neurons are capable of reducing the activity elicited by valence-specific stimuli in the opposite neuronal population.
Antagonism observed at the behavioral and c-Fos activation level of valence-specific BLA neurons was further examined at the microcircuit level by combining patch clamp recording with optogenetic stimulation of valence-specific neurons. The functional relationship between Rspo2+ and Ppp1r1b+ neurons were examined by combining patch clamp recordings with optogenetic stimulation of cell type-specific axons (Fig. 6a–d). Patch clamp recordings of Rspo2+ and Ppp1r1b+ neurons revealed distinct intrinsic physiological properties (Table 1). Therefore, the postsynaptic cell target was recognized based on a combination of anatomical position, soma size, and intrinsic electrophysiological properties (Fig. 6m,n). Electrophysiological recordings of Rspo2+ neurons in response to optogenetic stimulation of Ppp1r1b-ChR2+ fibers and recordings of Ppp1r1b+ neurons in response to stimulation of Rspo2-ChR2+ fibers resulted in inhibitory post-synaptic potentials (IPSPs) (Fig. 6e–h,k,l). The probability of connections of Rpso2+ to Ppp1r1b+and vice versa were 100% and 100% inhibitory, respectively (Fig. 6i,j), of which 25% of connections of Ppp1r1b+ to Rpso2+BLA neurons and 17% of connections of Rpso2+to Ppp1r1b+were both inhibitory and excitatory (Fig. 6i,j). These data suggest that these two populations interact predominantly through mutual inhibition.
Figure 6. Rspo2+ and Ppp1r1b+ BLA neurons establish reciprocal inhibitory connections.
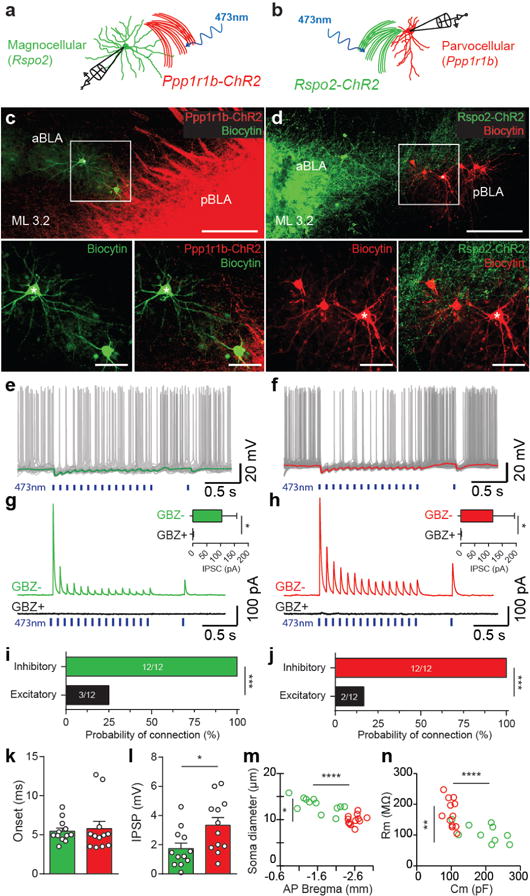
a,b, Scheme for the experimental setup for recording in magnocellular (Rspo2+) (a) and parvocellular (Ppp1r1b+) (b) neurons, while stimulating Ppp1r1b+ (Ppp1r1b-ChR2 mice) and Rpso2+ (Rspo2-ChR2 mice) neurons, respectively. c,d, Sagittal view of biocytin-filled Rspo2+ BLA neurons in Ppp1r1b-ChR2 mice (c) and Ppp1r1b+ BLA neurons in Rspo2-ChR2 mice (d). Scale bar 200μm, inset: 50 μm (c,d). AsterisksA denotes the electrophysiological traces in e and f.f Inhibitory postsynaptic potentials (IPSPsP) recorded in Rspo2+ (e) and Ppp1r1b+ (f) BLA neurons by 10Hz optogenetic stimulation of Ppp1r1b-ChR2 (e) and Rspo2-ChR2 (f) fibers. Green.G (e) and red (f) traces represent average trace of 20 sweeps recorded during periods without spikes. Inhibitory postsynaptic currents (IPSCs) recorded in Rpso2+ (magnocellular) (g) and Ppp1r1b+ (parvocellular) (h) BLA neurons (clamped at 0 mV) in response to optogenetic stimulation (10Hz train) of Ppp1r1b-ChR2 (g) and Rspo2-ChR2 (h) fibers. Currents are blocked by bath application of gabazine (GBZ, 10 μM), insets: IPSCs amplitude before (GBZ-) and after GBZ (GBZ+) application (for both magnocellular (n = 6) (g) and parvocellular (n = 6) (h), Wilcoxon signed-rank test, *P < 0.05. Probability of connection, Ppp1r1b+ to Rspo2+ connection (i) and Rspo2+ to Ppp1r1b+ connection (j). The two groups interact predominately by mutual inhibition rather than excitation, Fisher exact test, ***P <0.001 (i,j). IPSC onset in Rspo2+ (green) and Ppp1r1b+ (red) neurons were similar (k). IPSC amplitude was greater in Ppp1r1b+ (red) than in Rspo2+ (green) neurons (l), unpaired t-test *P<0.05. Recorded magnocellular (green) and parvocellular (red) neurons were confirmed using soma diameter and anatomical position (m); membrane resistance (Rm) and membrane capacitance (Cm) (n). Rspo2+ and Ppp1r1b+ cells were statistically distinct in all four parameters and consistent with values characterized in Fig. 2, significance for unpaired t-test *P<0.05, **P<0.01, ****P<0.0001 (n, m). Results show mean ± s.e.m (g,h,,k,l). Color scheme corresponds to the neurons recorded or virus-infected transgenic mouse, Rpso2+ neurons or Rpso2-Cre (green) and Ppp1r1b+ neurons or Cartpt-Cre (red) (a–n).
Projections from negative and positive BLA neurons
The distinct projection targets of the Rspo2+ and Ppp1r1b+ neurons may reveal divergent brain structures that mediate negative and positive behaviors. Therefore, retrograde tracing from putative projection targets was examined using cholera toxin subunit b (CTB). CTB targeted to the capsular nucleus of the central amygdala (CeC), revealed CTB+ neurons primarily in the aBLA (Fig. 7a,c,d). CTB targeted to the lateral/medial nucleus of the central amygdala (CeL/CeM), resulted in CTB+ neurons distributed along the lateral side of the pBLA (Fig. 7a,e,f). CTB targeted to the nucleus accumbens (NAc), resulted in CTB+ neurons distributed along the medial side of the BLA, spanning the posterior end of the aBLA to the posterior end of the pBLA (Fig. 7a,g,h). Dual-labelled CTB targeted to the prelimbic (PL) and inframlimbic (IL) cortex resulted in spatially segregated distribution of CTB+ neurons in the BLA—PL-CTB+ neurons primarily in the aBLA, IL-CTB+ neurons primarily in the pBLA (Fig. 7b,i,j). smFISH of Rspo2 or Ppp1r1b probe in CTB injected mice, revealed that CeC-CTB+ BLA neurons are 96% Rpso2+ and 4% Ppp1r1b+; CeL/CeM-CTB+ neurons are 6% Rspo2+ and 94% Ppp1r1b+; NAc CTB+ neurons are 30% Rpso2+ and 70% Ppp1r1b+ (Fig. 7k–n, Supplemental Fig. 8, Table 1).
Figure 7. Rspo2+ and Ppp1r1b+ BLA neurons project to distinct amygdaloid nuclei and prefrontal areas.
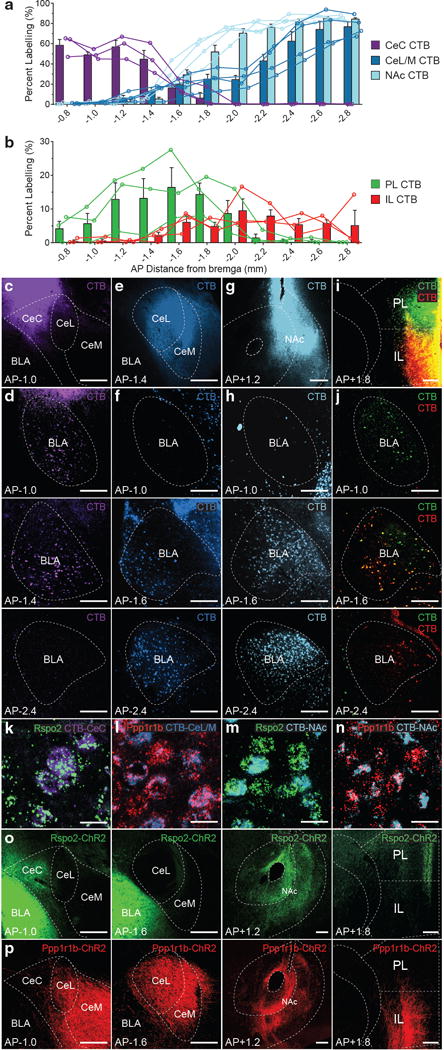
Quantification of CTB+ neurons across the AP axis (coronal distance from bregma −0.8mm to −2.8mm) of the BLA from CTB targeted to the amygdala and supplemental amygdala areas (a)—CeC (c,d), CeL/CeM (e,f), NAc (g,h), or dual CTB targeted to prefrontal cortex (b)—PL and IL (i,j) (bar represent mean, lines represent individual mice, n = 3 mice per group). Injections site of CTB (c,e,g,i) and CTB+ BLA neurons (d,f,h,j). Co-labelling of Rspo2 mRNA in the BLA with CTB targeted to the CeC (k) and NAc (m). Co-labelling of Ppp1r1b mRNA in the BLA with CTB injected into the CeL/CeM (l) and NAc (n), quantification in Table 1, micrographs in Supplemental Fig.7. Rspo2-ChR2+ fibers are found in the CeC, NAc, and PL (o). Ppp1r1b-ChR2+ fibers are found in the CeL, CeM, NAc, and IL (p). Scale bar 250 μm (c-j,o,p), 25 μm(k–n). Results show mean ± s.e.m. Color scheme corresponds to the virus-infected transgenic mouse, Rpso2-Cre (green) and Cartpt-Cre (red) (o,p).
For anterograde characterization, ChR2-eYFP+ fibers in Rspo2-ChR2 and Ppp1r1b-ChR2 mice was examined. In Rspo2-ChR2 mice, dense fibers were found in the CeC, NAc, PL, but not in the CeL, CeM, or IL (Fig. 7o). In Ppp1r1b-ChR2 mice, dense fibers were found in the CeL, CeM, NAc, and IL but not in the CeC or PL (Fig. 7p). Together, CTB retrograde tracing and anterograde characterization of projection fibers suggest that Rspo2+ distinctly project to the CeC and PL, Ppp1r1b+ neurons distinctly project to the CeL, CeM, and IL, while both Rspo2+ and Ppp1r1b+ BLA neurons both project to the NAc.
DISCUSSION
Here, we employed a forward genetic strategy in order to transcriptionally profile active neurons in BLA. This approach revealed genetic markers for distinct populations of BLA neurons and was predictive of neuronal function. Rspo2+ BLA neurons are activated by stimuli that elicit negative behaviors, while Ppp1r1b+ BLA neurons are activated by stimuli that elicit positive behaviors. Rspo2+ BLA neurons are crucial for negative behaviors and associations, while Ppp1r1b+ BLA neurons are crucial for positive behaviors and associations. Rspo2+ and Ppp1r1b+ neurons are antagonistic at the behavioral, neuronal population, and electrophysiological levels. They not only drive opposing behaviors, but also antagonize valence-specific behaviors, antagonize the overall activation of the opposing neurons and interact through reciprocal feedforward inhibition. Collectively, these results support a model in which mutually inhibitory Rspo2+ and Ppplr1b+ neurons are the principle neurons that represent and elicit negative and positive behaviors, respectively.
Previous inactivation studies have implicated a greater contribution of the aBLA in contextual fear conditioning12, and the pBLA in reward conditioning27. Here, we dissociated, using specific genetic markers for cell-type specific manipulations, the aBLA and pBLA in negative and positive behaviors, respectively. Although Rpso2+ and Ppp1r1b+ neurons constitute virtually all BLA pyramidal neurons, there may be further functional, genetic, and or structural diversity within each of these two cell-types. Other genetic markers found as candidates on our screen were not further pursued in this study, but further studies could be performed to examine the role of other genetically distinct BLA neurons, and these studies may reveal functionally distinct subsets of neurons within Rspo2+ negative neurons or within Ppp1r1b+ positive neurons. However, from the examination of Rpso2+ and Ppp1r1b+ neurons in a set of behavioral assays (Fig. 4), we found no evidence suggesting that Rspo2+ and Ppp1r1b+ neurons participate in behaviors or associations across valence. Together, these findings add to the growing evidence of the spatially segregated representation of negative and positive information in the brain, as demonstrated in the medial amygdala28, cortical amygdala29, gustatory cortex in mice30, and dopaminergic neurons in Drosophila31, 32. Thus, spatially segregated representation of negative and positive information may be a common motif throughout the central nervous system and across invertebrate and vertebrate species.
Previous in vivo electrophysiology and stimulus-dependent studies suggested that negative and positive BLA neurons may be intermingled15, 16. However, our results suggest that negative and positive neurons are spatially segregated into the aBLA and pBLA, respectively. At the transition between the aBLA and pBLA, these two types of neurons can be considered intermingled; however, examination of the entire BLA showed that Rspo2 and Ppp1r1b are spatially segregated in the BLA and define what has previously been defined as the anterior and posterior subfields of the BLA and correspond to magnocellular and parvocellular neurons, respectively4.
The identification of a mutually inhibitory microcircuit between negative and positive neurons suggests that the BLA is a key site for the antagonistic control of affective emotional states and emotional memories. The antagonistic BLA circuit provides a circuit mechanism of representing and associating a continuous range of negative and positive information based on the balance of excitation between these two populations. Studies have correlated negative affective states with elevated excitability in the BLA33, 34, which suggest that the imbalance of excitation between negative and positive BLA neurons may be an underlying mechanism in emotional disorders. Therefore, the identification of distinct molecular markers for neurons that participate in an antagonistic circuit provides an avenue for more precise cellular and pharmacological targeting for the treatment of disease in addition to genetic models for further elucidation of the circuitry and mechanisms underlying emotional behavior and memories.
Previous studies have targeted BLA neurons for the study of negative and positive behaviors using projection target-based criteria18. These studies suggested that nucleus accumbens (NAc) projections may be a defining feature of positive BLA neurons18. However, retrograde and anterograde projection experiments showed that ~30% of BLA neurons that project to NAc, are Rspo2+ BLA neurons. Furthermore, stimulation of Rpso2+ somas or their NAc projections resulted in negative behaviors (Supplemental Fig.9). These findings demonstrate that such a projection-based definition is insufficient for distinguishing negative and positive BLA neurons34. Previous observations that BLA to NAc projections mediate positive behaviors8,16 are likely due to the observation that a larger proportion of NAc-projecting BLA neurons are Ppp1r1b+ (Table 1).
It is widely thought that the amygdala fear circuit involves direct transmission of negative information from BLA principle neurons to CeL neurons and/or effector neurons in the CeM18, 35–38. Contrary to these previous hypotheses, our data suggest that positive, but not negative BLA neurons project to the CeM and CeL, while negative, but not positive BLA neurons, project to the CeC. Here, previous projection-based definition of BLA neurons—namely that the neurons projecting directly to the CeM drive negative behavior18, 35–38—is not supported by our findings. In regards to CeM and CeL projections, our findings are consistent with anatomical studies demonstrating that parvocellular BLA neurons (which are Ppp1r1b+) send strong projections to the CeL and CeM and provide further support for the role of the central amygdala in appetitive behaviors26, 39–43. In regards to connections from negative BLA neurons to effector neurons in the CeM, our findings suggest that this may be an indirect route through the CeC. A recent study identified a population of Calcrl+ neurons in the CeC/CeL, which supports similar negative behaviors as Rpso2+ BLA neurons, and, thus, may be an intermediate between negative BLA neurons and the putative CeM effector neurons44.
Overall, the identification of genetic markers for distinct populations of BLA neurons has permitted the functional and anatomical dissociation of the circuit underlying negative and positive behaviors, in turn, providing a revised functional and structural model of the BLA (Supplemental Fig. 10).
Supplementary Material
Acknowledgments
A. Wagatsuma, R.L. Redondo for help with the designing of behavioral apparatuses, MIT BioMicroCenter for support on collecting the RNA array data, X. Liu on the cloning of the c-Fos-tTA plasmid, T.J. Ryan, D.S. Roy for comments on the manuscript, and all the members of the Tonegawa lab for their support. This work is supported in part by the NIH Pre-Doctoral Training Grant T32GM007287 (to J.K.) and by the RIKEN Brain Science Institute, the Howard Hughes Medical Institute and the JPB Foundation (to S.T.).
Footnotes
Author Contributions J.K., and S.T. conceived the study. J.K. identified gene markers. S.I. generated the transgenic Rspo2-Cre mouse. J.K. collected and analyzed histological data. J.K., S.X., collected and analyzed behavioral data. M.P. collected and analyzed electrophysiological data. J.K., S.T., wrote the manuscript. J.K., M.P., S.X., S.I., S.T., discussed and commented on the manuscript. Authors declare no conflict of interest.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Accession codes. RNA array data can be obtained on NCBI Gene Expression Omnibus (GEO), accession number GSE78137.
References
- 1.Pitkanen A, Savander V, LeDoux JE. Organization of intra-amygdaloid circuitries in the rat: an emerging framework for understanding functions of the amygdala. Trends in neurosciences. 1997;20:517–523. doi: 10.1016/s0166-2236(97)01125-9. [DOI] [PubMed] [Google Scholar]
- 2.McDonald AJ. Neuronal organization of the lateral and basolateral amygdaloid nuclei in the rat. The Journal of comparative neurology. 1984;222:589–606. doi: 10.1002/cne.902220410. [DOI] [PubMed] [Google Scholar]
- 3.Swanson LW, Petrovich GD. What is the amygdala? Trends in neurosciences. 1998;21:323–331. doi: 10.1016/s0166-2236(98)01265-x. [DOI] [PubMed] [Google Scholar]
- 4.Hall E. The amygdala of the cat: a Golgi study. Zeitschrift fur Zellforschung und mikroskopische Anatomie. 1972;134:439–458. doi: 10.1007/BF00307668. [DOI] [PubMed] [Google Scholar]
- 5.McDonald AJ. Neurons of the lateral and basolateral amygdaloid nuclei: a Golgi study in the rat. The Journal of comparative neurology. 1982;212:293–312. doi: 10.1002/cne.902120307. [DOI] [PubMed] [Google Scholar]
- 6.Carlsen J, Heimer L. The Basolateral Amygdaloid Complex as a Cortical-Like Structure. Brain Res. 1988;441:377–380. doi: 10.1016/0006-8993(88)91418-7. [DOI] [PubMed] [Google Scholar]
- 7.Mascagni F, McDonald AJ. A novel subpopulation of 5-HT type 3A receptor subunit immunoreactive interneurons in the rat basolateral amygdala. Neuroscience. 2007;144:1015–1024. doi: 10.1016/j.neuroscience.2006.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald AJ, Muller JF, Mascagni F. GABAergic innervation of alpha type II calcium/calmodulin-dependent protein kinase immunoreactive pyramidal neurons in the rat basolateral amygdala. The Journal of comparative neurology. 2002;446:199–218. doi: 10.1002/cne.10204. [DOI] [PubMed] [Google Scholar]
- 9.Redondo RL, et al. Bidirectional switch of the valence associated with a hippocampal contextual memory engram. Nature. 2014;513:426–430. doi: 10.1038/nature13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuber GD, et al. Excitatory transmission from the amygdala to nucleus accumbens facilitates reward seeking. Nature. 2011;475:377–380. doi: 10.1038/nature10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambroggi F, Ishikawa A, Fields HL, Nicola SM. Basolateral amygdala neurons facilitate reward-seeking behavior by exciting nucleus accumbens neurons. Neuron. 2008;59:648–661. doi: 10.1016/j.neuron.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goosens KA, Maren S. Contextual and auditory fear conditioning are mediated by the lateral, basal, and central amygdaloid nuclei in rats. Learning & memory. 2001;8:148–155. doi: 10.1101/lm.37601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cador M, Robbins TW, Everitt BJ. Involvement of the Amygdala in Stimulus Reward Associations - Interaction with the Ventral Striatum. Neuroscience. 1989;30:77–86. doi: 10.1016/0306-4522(89)90354-0. [DOI] [PubMed] [Google Scholar]
- 14.Killcross S, Robbins TW, Everitt BJ. Different types of fear-conditioned behaviour mediated by separate nuclei within amygdala. Nature. 1997;388:377–380. doi: 10.1038/41097. [DOI] [PubMed] [Google Scholar]
- 15.Gore F, et al. Neural Representations of Unconditioned Stimuli in Basolateral Amygdala Mediate Innate and Learned Responses. Cell. 2015;162:134–145. doi: 10.1016/j.cell.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paton JJ, Belova MA, Morrison SE, Salzman CD. The primate amygdala represents the positive and negative value of visual stimuli during learning. Nature. 2006;439:865–870. doi: 10.1038/nature04490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zirlinger M, Kreiman G, Anderson DJ. Amygdala-enriched genes identified by microarray technology are restricted to specific amygdaloid subnuclei. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5270–5275. doi: 10.1073/pnas.091094698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Namburi P, et al. A circuit mechanism for differentiating positive and negative associations. Nature. 2015;520:675–678. doi: 10.1038/nature14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knight ZA, et al. Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell. 2012;151:1126–1137. doi: 10.1016/j.cell.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gay L, et al. Mouse TU tagging: a chemical/genetic intersectional method for purifying cell type-specific nascent RNA. Genes & development. 2013;27:98–115. doi: 10.1101/gad.205278.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z, Edenberg HJ, Davis RL. Isolation of mRNA from specific tissues of Drosophila by mRNA tagging. Nucleic acids research. 2005;33:e148. doi: 10.1093/nar/gni149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 25.Hemmings HC, Jr, Greengard P, Tung HY, Cohen P. DARPP-32, a dopamine-regulated neuronal phosphoprotein, is a potent inhibitor of protein phosphatase-1. Nature. 1984;310:503–505. doi: 10.1038/310503a0. [DOI] [PubMed] [Google Scholar]
- 26.Savander V, Go CG, Ledoux JE, Pitkanen A. Intrinsic Connections of the Rat Amygdaloid Complex - Projections Originating in the Basal Nucleus. Journal of Comparative Neurology. 1995;361:345–368. doi: 10.1002/cne.903610211. [DOI] [PubMed] [Google Scholar]
- 27.Kantak KM, Black Y, Valencia E, Green-Jordan K, Eichenbaum HB. Dissociable effects of lidocaine inactivation of the rostral and caudal basolateral amygdala on the maintenance and reinstatement of cocaine-seeking behavior in rats. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2002;22:1126–1136. doi: 10.1523/JNEUROSCI.22-03-01126.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi GB, et al. Lhx6 delineates a pathway mediating innate reproductive behaviors from the amygdala to the hypothalamus. Neuron. 2005;46:647–660. doi: 10.1016/j.neuron.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Root CM, Denny CA, Hen R, Axel R. The participation of cortical amygdala in innate, odour-driven behaviour. Nature. 2014;515:269–273. doi: 10.1038/nature13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng Y, et al. Sweet and bitter taste in the brain of awake behaving animals. Nature. 2015;527:512–515. doi: 10.1038/nature15763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aso Y, et al. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife. 2014;3:e04580. doi: 10.7554/eLife.04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohn R, Morantte I, Ruta V. Coordinated and Compartmentalized Neuromodulation Shapes Sensory Processing in Drosophila. Cell. 2015;163:1742–1755. doi: 10.1016/j.cell.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duvarci S, Pare D. Glucocorticoids enhance the excitability of principal basolateral amygdala neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:4482–4491. doi: 10.1523/JNEUROSCI.0680-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis M, Rainnie D, Cassell M. Neurotransmission in the rat amygdala related to fear and anxiety. Trends in neurosciences. 1994;17:208–214. doi: 10.1016/0166-2236(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 35.Herry C, Johansen JP. Encoding of fear learning and memory in distributed neuronal circuits. Nature neuroscience. 2014;17:1644–1654. doi: 10.1038/nn.3869. [DOI] [PubMed] [Google Scholar]
- 36.Duvarci S, Pare D. Amygdala microcircuits controlling learned fear. Neuron. 2014;82:966–980. doi: 10.1016/j.neuron.2014.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tovote P, Fadok JP, Luthi A. Neuronal circuits for fear and anxiety. Nature reviews Neuroscience. 2015;16:317–331. doi: 10.1038/nrn3945. [DOI] [PubMed] [Google Scholar]
- 38.Janak PH, Tye KM. From circuits to behaviour in the amygdala. Nature. 2015;517:284–292. doi: 10.1038/nature14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkinson JA, Robbins TW, Everitt BJ. Dissociable roles of the central and basolateral amygdala in appetitive emotional learning. The European journal of neuroscience. 2000;12:405–413. doi: 10.1046/j.1460-9568.2000.00960.x. [DOI] [PubMed] [Google Scholar]
- 40.Cai H, Haubensak W, Anthony TE, Anderson DJ. Central amygdala PKC-delta(+) neurons mediate the influence of multiple anorexigenic signals. Nature neuroscience. 2014;17:1240–1248. doi: 10.1038/nn.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallagher M, Graham PW, Holland PC. The amygdala central nucleus and appetitive Pavlovian conditioning: lesions impair one class of conditioned behavior. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1990;10:1906–1911. doi: 10.1523/JNEUROSCI.10-06-01906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knapska E, et al. Differential involvement of the central amygdala in appetitive versus aversive learning. Learning & memory. 2006;13:192–200. doi: 10.1101/lm.54706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knapska E, Radwanska K, Werka T, Kaczmarek L. Functional internal complexity of amygdala: focus on gene activity mapping after behavioral training and drugs of abuse. Physiological reviews. 2007;87:1113–1173. doi: 10.1152/physrev.00037.2006. [DOI] [PubMed] [Google Scholar]
- 44.Han S, Soleiman MT, Soden ME, Zweifel LS, Palmiter RD. Elucidating an Affective Pain Circuit that Creates a Threat Memory. Cell. 2015;162:363–374. doi: 10.1016/j.cell.2015.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


