Abstract
The oligomeric state of active human immunodeficiency virus type 1 (HIV-1) integrase (IN) has not been clearly elucidated. We analyzed the activity of the different purified oligomeric forms of recombinant IN obtained after stabilization by platinum crosslinking. The crosslinked tetramer isolated by gel chromatography was able to catalyze the full-site integration of the two viral LTR ends into a target DNA in vitro, whereas the isolated dimeric form of the enzyme was involved in the processing and integration of only one viral end. Accurate concerted integration by IN tetramers was confirmed by cloning and sequencing. Kinetic studies of DNA-integrase complexes led us to propose a model explaining the formation of an active complex. Our data suggest that the tetrameric IN bound to the viral DNA ends is the minimal complex involved in the concerted integration of both LTRs and should be the oligomeric form targeted by future inhibitors.
INTRODUCTION
Integration of viral DNA into the host genome is a critical step in HIV infection. Integrase (IN) is essential for viral replication and necessary and sufficient for the integration reaction in vitro. The in vivo integration of linear retroviral DNA catalyzed by integrase occurs by a concerted mechanism known as full-site or two-ended integration. The insertion of viral DNA is a process in which two catalytic reactions are performed at each end of the viral genome: 3′-processing and strand transfer. In vitro IN can efficiently perform both the 3′-processing and the insertion of one viral extremity. The purified recombinant enzyme also accurately reproduces concerted integration, but the in vitro efficiency is still far below that expected for integration in vivo.
Early reconstitution experiments using purified IN, a linear retrovirus-like DNA donor and a target DNA were successful but yielded low levels of concerted integration (1,2). The efficiency of full-site integration was significantly increased later on by employing non-ionic detergent IN-containing lysates of HIV-1 virions (3), but the proficiency and reliability of the reaction vary with the various reconstitution systems (4). Although purified HIV-1 IN can perform full-site integration in vitro, retroviral proteins, such as the nucleocapsid protein (NCp) (5), or factors, such as HMGA1, belonging to cellular high mobility group proteins have been shown to mediate efficient full-site integration (6). The lack of these factors in in vitro integration assays may account for the differences observed. Moreover, deficient protein-folding or incorrect oligomerization of purified IN may also prevent native assembly of complexes with viral DNA ends, which is essential for full-site integration (7,8).
The aim of our study was to determine whether efficiency of the full-site integration activity is correlated with the oligomerization state of IN. For that purpose we stabilized the multimeric structure of the enzyme by chemical crosslinking. We determined the activity of the isolated different IN monomers and multimers and observed that the cross-linked tetrameric form of IN is the minimal oligomer that is able to perform full-site integration of a substrate carrying the two LTRs.
MATERIALS AND METHODS
Bacteria and DNA
The Escherichia coli strain DH5α was used for plasmid amplification. MC1060/P3 E.coli strain (Invitrogen) was used for cloning the integration products. DNA was extracted and purified as previously described (9). The HIV-1 IN gene was obtained from a cloned genomic provirus from a San Francisco isolate (SF2) (10). The expression vector pHIV1SF2IN was derived from the yeast/E.coli shuttle plasmid pBS24.1 (11).
Purification of IN
HIV-1 IN was expressed in yeast and purified as previously described (12).
Gel filtration chromatography
Purified IN was diluted in 1 ml loading solution (50 mM HEPES pH 7.5; 7 mM CHAPS; 1 mM DTT; 150 mM NaCl; 0.1 mM EDTA) at a final concentration of 150 nM and chromatographed through a Smart Superose 12 (Pharmacia-LKB) on the Ettan LC system. The void volume was determined with blue dextran (>2000 kDa) and the column was calibrated with catalase (232 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa) and chymotrypsinogen A (25 kDa) (Pharmacia). Proteins were eluted with a flow rate of 0.04 ml/min and recorded by continuously monitoring the absorbance at 280 nm. Prior to chromatography, samples were centrifuged for 10 min at 10 000 rpm to remove large protein aggregates. The protein composition of the pooled fractions was confirmed by mass spectrometry.
Concerted integration DNA substrates
Both target and donor plasmids were kind gifts from Dr Karen Moreau (Université Claude Bernard-Lyon I, France). The target corresponds to the plasmid pBSK+ (Stratagene, La Jolla, California) carrying the zeocin resistance encoding gene. The donor substrate was obtained by cleavage of the pUC19supF plasmid by NdeI giving a 294 bp DNA substrate that contained the supF tRNA gene flanked by two pre-cleaved extremities mimicking the 3′-processed U3 and U5 LTR sequences. The DNA substrate without the LTR sequences was generated by PCR using the pUC19-SupF plasmid as template and primers A (5′-TTGAGCGTCGATTTTTGTGAT-3′) and B (5′-TACGTTGCCCGGATCCGGTCG-3′). The DNA substrate carrying one LTR was obtained similarly, but primer A was replaced by primer C (5′- TATGCTAGAGATTTTCCACATTGAGCGTCGATTTTTGTGAT-3′).
Integration reactions
The concerted integration reaction conditions were similar to those described in reference 13, except that no cellular proteins were added and the HIV-1 system was used. Briefly, purified HIV-1 IN (250 nM) was preincubated with both the 5′-end-labeled donor DNA (10 ng) containing the 3′-processed U3 and U5 LTR sequences and the target DNA plasmid pBSK+ (100 ng) at 0°C during 20 min in a total volume of 5 μl. Then the reaction mixture (20 mM HEPES, pH 7.5; 10 mM DTT; 7.5 mM MgCl2; 10% DMSO; 8% PEG) was added and the reaction continued for 90 min. Incubation was stopped by adding a phenol/isoamyl alcohol/chloroform mix (24/1/25 v/v/v). The aqueous phase was loaded on a vertical 1% agarose gel in the presence of 1% bromophenol blue and 1 mM EDTA. After separation of the products the gel was treated with 5% TCA for 20 min, dried and autoradiographied. IN activity was quantified by scanning the bands using the NIH software.
The 3′ processing and strand transfer reactions were performed as described (8). All assays were performed in 20 mM HEPES pH 8, 10 mM DTT, 7.5 mM MnCl2, 0.05% NP40 in a total volume of 20 μl. The reaction mixture was incubated at 37°C for 1 h in the presence of IN (1–5 pmol) and radiolabeled oligonucleotides (1 pmol) and the incubation was stopped by adding 10 μl of loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue) and heating at 90°C for 5 min. The reaction products were analyzed by electrophoresis on 15% polyacrylamide gels with 7 M urea in Tris-borate-EDTA (TBE) pH 7.6 and autoradiographied. The sequence of the ODNs used to perform the processing and strand transfer assays were the following: ODN 70: 5′GTGTGGAAAATCTCTAGCAGT3′, ODN 71: 5′GTGTGGAAAATCTCTAGCA3′, ODN 72: 5′ACTGCTAGAGATTTTCCACAC3′. To perform the 3′ processing assay, the 5′ radiolabeled ODN 70 hybridized to ODN 72 was used as a substrate while the 5′ radiolabeled ODN 71 hybridized to ODN 72 was used as a substrate in the strand transfer reaction. All IN activities were quantified by scanning of the bands after gel electrophoresis and autoradiography using the NIH software.
Cloning and sequencing of integration products
After concerted integration the products were purified on a DNA purification system column (Promega) as described by the supplier and then introduced into a MC1060/P3 E.coli strain, which contained ampicillin, tetracycline and kanamycin resistance genes. Both ampicillin and tetracycline resistance genes carry an amb mutation. These proteins are thus expressed only in the presence of the supF gene products. Integration clones carrying both zeocin-resistant and supF genes were therefore selected in the presence of 40 μg/ml ampicillin, 10 μg/ml tetracycline, 15 μg/ml kanamycin and 25 μg/ml zeocin. Plasmids were isolated from quadruple-resistant colonies and donor–acceptor DNA junctions were checked by polymerase chain reaction-based sequencing (ABI Prism big dye terminator cycle sequencing ready reaction kit, Applied Biosystems) using the U3 primer (5′-TATGGAAGGGCTAATTCACT-3′) and the U5 primer (5′-TATGCTAGAGATTTTCCACA-3′).
Chemical crosslinking
Highly purified IN (5 pmol) was incubated alone or with radiolabeled ODNs (1 pmol) in the presence of 300 μM cis-aquahydroxydiamino-platinum (AHDAP), 0.05% NP40, 0.5 M Mg acetate, 10 mM DTT, 20 mM HEPES pH 7.5 at 37°C in the dark in a final volume of 10 μl. The platinum derivative was prepared as described before (14). The reaction was stopped by eliminating the excess of AHDAP after crosslinking by elution through a G25 MicroSpin column (Amersham).
RESULTS
Recombinant HIV-1 IN purified from yeast can perform in vitro full-site integration
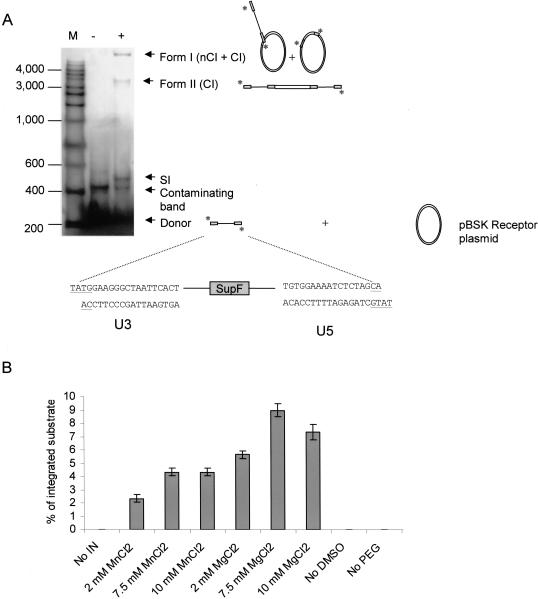
To determine whether the yeast recombinant IN was able to catalyze the in vitro concerted integration reaction, the purified enzyme was assayed as described in the Material and Methods section. The donor–target products were visualized by agarose gel electrophoresis using radioactively labeled donors. As our recombinant HIV-1 IN was not able to catalyze concerted integration from a blunt ended DNA substrate, we used a two LTRs pre-cleaved substrate mimicking the 3′ processed viral DNA and carrying 18 bp from U3 and U5 along with a two bases overgang TA at each 5′end (Figure 1A). This shows that this enzyme was less efficient for performing the whole integration reaction than the purified preintegration complex (PIC) or INs from other retroviral sources (3,13).
Figure 1.
Concerted integration reaction catalyzed by yeast recombinant HIV-1 IN. (A) Gel analysis of the integration products. Reactions were performed in the presence (+) or in absence (−) of 250 nM of IN. The target DNA is indicated as pBSK receptor plasmid. The donor DNA was 5′-end labeled. Products were submitted to 1% agarose gel electrophoresis. nCI, CI: respectively non-concerted and concerted full-site integration products. SI: Self-integration products. M: Markers (base pairs). The structure of the expected products are also described in this figure. (B) Effect of reaction conditions on the integration reaction catalyzed by recombinant HIV-1 IN. For each condition one parameter was changed independently (MgCl2 or MnCl2 concentration, presence of DMSO or PEG). The percentage of the total integrated substrate was reported on the graph. Results are the mean +/− SD of three separate experiments.
Various integration products were obtained in the presence of the recombinant enzyme [Figure 1A, lane (+)]. Restriction analyses were performed to assess the nature of the products (data not shown). Form I comprises circular molecules resulting from the integration of one LTR (one-site or non-concerted integration, nCI) and of two LTRs (concerted integration, CI) from the same donor into the receptor plasmid. Form II corresponds to linear full-site concerted integration products of two LTRs from two donor molecules into the receptor plasmid. The band termed SI (self-integration products) corresponds to integration of one-LTR DNA into another donor molecule. Since form I includes the sum of different integration products (concerted and non-concerted) and all the linear products found in form II may be considered as originating from concerted integration, the latter form was considered as representative of integration of two LTR into the target DNA.
Efficient integration was observed only in the presence of high concentrations of PEG and DMSO, respectively 8% and 15% (w/v) (Figure 1B). The preferred cation was Mg++ (optimum at 7.5 mM). Gel analysis permits quantification of global integration efficiency but does not make it possible to distinguish one-site insertion from the concerted integration products. To address this issue, integration products can be cloned into MC1061/P3 E.coli that contains drug resistance markers with amber mutations. Only DNA products carrying the amber mutation suppressor gene (supF) should be able to replicate and form colonies under drug selection. Thus, only the circular concerted integrated products corresponding to form I should replicate in bacteria. Total products from the integration reaction described in Figure 1 were cloned in MC1061/P3 bacteria and 20 resistant clones were selected. In contrast, no colonies were selected when the integration reaction was performed using an inactive IN that exhibits the D116A mutation. We sequenced the donor DNA-acceptor plasmid junctions of the isolated integration products to check the accuracy of the integration reaction (duplication of short acceptor DNA sequence). As reported in Table 1, of 20 clones, 12 exhibited a target DNA duplication of 5 bp, the hallmark of HIV-1 IN in vivo integration activity, 6 clones showed a duplication of different sizes and 2 clones carried a short deletion of the target DNA. These results indicate that INs purified from yeast shares the same in vitro characteristics as other recombinant INs obtained from bacteria (15,16).
Table 1.
Sequence analysis of donor–target junctions from clones produced by wild type IN
| Total number of clones | 20 |
|---|---|
| Number of sequenced clones | 20 |
| Duplications with 5 bp | 12 |
| Other duplications | |
| 3 bp | 3 |
| 4 bp | 3 |
| Deletions | 2 |
Total integration products obtained in Figure 1 by wild type IN were introduced in MC1061/P3 E.coli strain and the plasmid DNA from the isolated clones was purified and sequenced as described in the Materials and Methods section.
Although the level of concerted integration obtained with recombinant IN was similar to that described before, maximum activity corresponded to <10% of the total substrate integrated which is a relatively low figure compared to the activity obtained with lyzed virions (3). This could be due to: (i) a need for auxiliary, viral or cellular proteins that are absent from the in vitro preparation; (ii) differences between the specific activities of virion and recombinant integrases and (iii) inefficient folding or oligomerization of the enzyme. We investigated this by studying the possible link between IN multimerization and its in vitro integration activity and the in vitro activity of pre-stabilized IN oligomers.
Purification of the integrase oligomeric forms
To address the question of the enzymatic activity versus the oligomerization state of HIV-1 IN, the enzyme was crosslinked to form stable multimers. The chemical crosslinking agent used was cis-aquahydroxydiamino platinum. This technique allows the formation of covalent bonds between platinum and the potential acceptors on proteins and nucleic acids, mainly the sulfur-containing groups of cysteine or methionine residues, the imidazole rings in histidine residues, and the N group of guanine, adenine and cytosine (17).
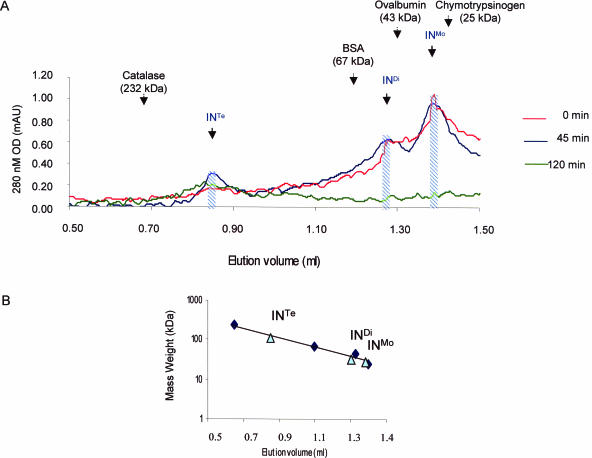
IN was treated with AHDAP for different lengths of time and chromatographed through a Superose 12 gel filtration column. The elution profile of the different forms of IN was checked with known molecular weight protein markers (Figure 2A). Before the crosslinking treatment IN appeared in solution mainly as a monomer–dimer equilibrium while a lower but significant amount of tetramer was detected (Figure 2A, 0 min) indicating that our yeast recombinant IN has the same oligomeric structure in solution as recombinant bacterial IN (18,19). As expected, the time duration of the crosslinking treatment increased the proportion of oligomeric forms of the enzyme including tetramers of IN (Figure 2A, 45 and 120 min). High molecular weight aggregates were eliminated through centrifugation before loading onto the column.
Figure 2.
(A) Gel filtration chromatography of crosslinked HIV-1 IN. IN (150 pmol) was crosslinked for different lengths of time and products were loaded on a Superose 12 column. Migration of the molecular weight markers is indicated. Monomers (Mo), dimers (Di) and tetramers (Te) of IN are indicated by comparison with markers. The fractions containing monomers (INMo), dimers (INDi) and tetramers (INTe) of IN were pooled and used to assay the IN activity. (B) Size of the IN oligomers determined by gel filtration chromatography. 25 μl aliquots of the fractions INMo, INDi and INTe obtained in A were chromatographed on a Superose 12 column under similar conditions. The elution position of each fraction was reported (shown as triangles) in this figure and compared to elution profile of known molecular weight markers (shown as clubs in the figure): catalase (232 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa) and chymotrypsinogen A (25 kDa).
To obtain stable oligomers we performed a 45-min incubation of IN in the presence of AHDAP and chromatographed through the Superose column allowing sufficient separation in order to isolate tetramers, dimers and monomers. Fractions containing the different oligomeric forms of IN were collected and their nature was checked by loading an aliquot of each fraction onto a new Superose column. As reported in Figure 2B the fractions migrated as expected from their corresponding oligomeric size. The multimeric fractions were called INMo, INDi and INTe respectively. Mass spectrometry analysis further confirmed the presence of 90–95% of monomers, crosslinked dimers and tetramers of IN in the corresponding pooled fractions. The remaining 5–10% showed a mass weight lower than the IN monomer and thus correspond probably to minor degradation products.
Purified fractions of crosslinked IN oligomers were obtained and stocked at sub-micromolar protein concentrations (from 400 to 500 nM). Those concentrations were maintained during all subsequent studies. Since under these conditions wild type IN appears mainly as a monomer, as described previously by time-resolved fluorescence anisotropy (18), no re-association of the purified crosslinked oligomers was expected to take place during further steps of this study.
In vitro activities of oligomeric forms of integrase
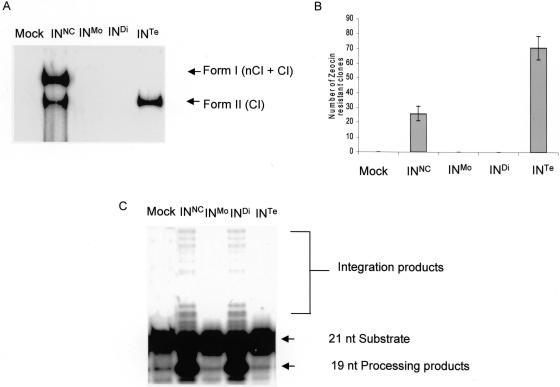
The fractions were tested for concerted integration (Figure 3A). We focused our interest on integration products I and II since they are the most representative of the physiological integration reaction. Interestingly, amongst the crosslinked forms of IN, only the fraction containing tetramers (lane INTe) was able to catalyze the formation of the full-site concerted integration form II product. A control with IN that was not crosslinked (lane INNC) showed that this fraction was also able to perform the concerted integration reaction in addition to non-concerted integration.
Figure 3.
In vitro activities of the HIV-1 IN oligomers. Reactions were performed under standard conditions using no protein (Mock), 250 nM of non-crosslinked IN (INNC), monomers (INMo), or crosslinked dimers (INDi) and tetramers (INTe) purified by gel filtration. (A) In vitro concerted integration assay; (B) number of zeocin-resistant clones obtained after cloning of the total reaction products in MC1061/P3 E.coli strain (mean +/− SD of three experiments; (C) processing and strand transfer assays.
Integration products were further analyzed by cloning them into bacteria. As reported in Figure 3B, a mean value of 25 clones were recovered from the cloning of the integration products obtained with non-crosslinked IN and 70 with crosslinked tetramers indicating a higher level of concerted integration catalyzed by the tetramer of IN as compared with the non-crosslinked protein. No clones were recovered from the INMo and the INDi enzymes confirming that they were not active. Although no form I products corresponding to INTe were observed in the gel, bacterial clones of the tetrameric form were isolated indicating that even when form I products were undetected on agarose gel, they were actually present confirming that this IN multimer performed a ‘true’ concerted integration of a complete viral substrate.
In order to identify the structure of the integration products, 20 clones were isolated and sequenced for each condition (Table 2). From the 20 clones obtained with non-crosslinked IN, 11 exhibited a target DNA duplication of 5 bp as expected from HIV-1 IN activity, 6 exhibited a duplication comprising sizes from 3 to 6 bp and 3 exhibited a deletion in the acceptor DNA. In sharp contrast, all the clones obtained with products of the reaction catalyzed by the IN tetramers exhibited the HIV-1 IN 5 bp duplication hallmark. This striking result strongly supports the idea that crosslinked IN tetramers display a higher concerted integration activity than non-crosslinked IN tetramers thereby suggesting the involvement of the tetrameric forms in the physiological integration of both LTR viral ends in the infected cell.
Table 2.
Sequence analysis of donor–target junctions from clones produced by non-crosslinked and crosslinked tetrameric IN
| INNC | INTe | |
|---|---|---|
| Total number of clones | 20 | 55 |
| Number of sequenced clones | 20 | 20 |
| Duplications with 5 bp | 11 | 20 |
| Other duplications | ||
| 3 bp | 3 | |
| 6 bp | 3 | |
| Deletions | 3 | - |
Total integration products obtained in Figure 3A were introduced in MC1061/P3 E. coli strain. Plasmid DNA from the selected clones was purified and sequenced as described in Materials and Methods.
The purified crosslinked oligomers of IN were also tested using the one-LTR 3′-processing and one viral end strand transfer substrates in the in vitro reactions (Figure 3C). Both processing and strand transfer activities were detected only with the dimeric form of IN. These results were similar to those obtained with the non-crosslinked IN (compare lanes INDi and INNC). Monomers and tetramers did not show any 3′-processing cleavage or strand transfer activities (lanes INMo and INTe).
Taken together these results show that the different oligomeric forms of IN do not share the same in vitro catalytic properties. Monomers were inactive for all specific IN activities. Dimers were able to catalyze the 3′-processing and the insertion of one LTR into a short target DNA, but could not integrate viral extremities in the longer substrates used for the concerted integration reaction. In contrast, the full-site integration of both LTR extremities required at least one tetramer.
The in vitro requirements of the tetrameric form of HIV-1 IN are less stringent concerning non-physiological molecules
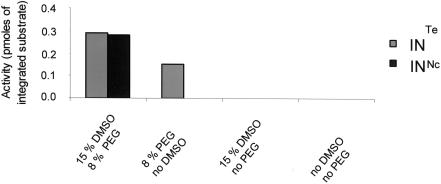
The requirement of high concentrations of non-physiological molecules like PEG and DMSO to obtain efficient in vitro integrations catalyzed by wild type INNC suggests that this recombinant protein might need the presence of these agents to attain a significant degree of activity. Thus, the role of PEG and DMSO may be to overcome the lack of cellular factors or the misfolding of the enzyme in vitro. To investigate this hypothesis, we tested the effect of these compounds on the full-site integration reaction catalyzed with the purified INTe. As shown in Figure 4, concerted integration was observed with INTe, even in the absence of DMSO, under conditions where INNC showed no concerted integration activity, while the presence of PEG was still needed by the tetrameric form to perform this reaction.
Figure 4.
Effect of reaction conditions on concerted integration activity. Standard conditions described in Materials and Methods section were used, but either DMSO or PEG or both were omitted. The total integrated donor DNA into the receptor plasmid was determined by quantifying the bands corresponding to form I (nCI + CI) and form II (CI) using the NIH software.
These results indicate that: (i) the tetrameric form of IN possesses biochemical properties closer to the enzyme found in the infected cell than to the native non-crosslinked IN preparation and (ii) DMSO could compensate for deficient oligomerization of the non-crosslinked IN required for efficient in vitro integration. In contrast, PEG may act directly on the integration process most probably via a molecular crowding effect.
Oligomeric state of the integrase bound to DNA
The oligomeric state of HIV-1 can be affected by the presence of DNA (19). Thus, we used the crosslinking approach to analyze the oligomeric forms of the enzyme bound to different DNA substrates.
Three 294 bp labeled DNA oligonucleotides carrying one-, two- or no LTR ends were crosslinked to IN for different times. The products were subjected to DNase I treatment, in order to eliminate non-IN-protected DNA tails and to distinguish between monomers and higher oligomeric forms of IN bound to the oligonucleotide. The labeled crosslinked complexes were visualized by SDS-PAGE and autoradiography (Figure 5). Different IN–DNA complexes were observed depending on the substrate DNA. High molecular weight multimeric products unable to enter the gel were obtained when IN was crosslinked to the DNA oligonucleotide without a viral sequence (Figure 5A). In contrast, the crosslink of IN to DNAs carrying one or two LTRs led to the formation of intermediary products corresponding in size to oligomers of IN bound to DNA (as determined by comparison with protein markers). With the ODN carrying one-LTR, two crosslinked products were detected prior to the formation of high-order complexes (Figure 5B). The first band, termed (a), which presents a size that corresponds to a monomer of IN, was observed before the formation of the second IN–DNA complex, termed (b). The results of the migration taken together with the DNase I treatment suggest that band (b) migrates as a complex formed by a dimer of IN bound to one-LTR viral extremity. When the two-LTR oligonucleotide was used, the binding to IN was stronger, as observed by the intensity of the bands (Figure 5C). In addition to bands (a) and (b), band (c) corresponding to the size of a tetramer of IN was detected. In a control performed only with DNA and the crosslinking agent, only aggregates of crosslinked material were obtained. No discrete bands such as (a), (b) and (c) were present (data not shown). The activities of isolated INMo, INDi and INTe were also analyzed by protein–DNA crosslink. While INMo formed exclusively band (a) with the single LTR substrate, INDi and INTe yielded high molecular weight aggregates (data not shown).
Figure 5.
SDS-PAGE analysis of crosslinked IN–DNA complexes. IN (5 pmol) was preincubated with the 5 ′-end radiolabeled 294 bp DNA substrate (1 pmol) carrying either no viral LTR (A), one viral LTR (B) or two viral LTRs sequences (C) for 0 to 60 min (lanes 0 to 60) in the presence of AHDAP at 37°C (final volume 20 μl). After 1 h of DNase 1 treatment, products were separated by electrophoresis on 12% SDS-PAGE gel. The gel was then dried and autoradiographed. The positions of bands a, b and c were compared with the migration of protein weight markers (BIO-RAD) submitted to electrophoresis under the same conditions and reported in the right part of each gel. The migration front of monomers, dimers and tetramers of IN obtained by crosslink of the enzyme by AHDAP is also reported (stars in the figure).
The kinetic analysis of the formation of different IN–DNA complexes suggested that the presence of one LTR induces the binding of monomers prior to the formation of IN dimers bound to the DNA. In the presence of two LTRs, monomer and dimer binding is increased, most probably due to the possibility of binding to both LTR ends leading to the formation of a tetrameric IN–DNA complex. In the absence of LTR, only a non-specific binding of DNA to IN leading to aggregates is observed. These results indicate that binding of DNA induces IN oligomerization whose level depends on the presence of the specific LTR sequences.
DISCUSSION
Determination of the oligomeric state of physiologically active IN is crucial to understand the retroviral integration reaction mechanism and to modelize efficient inhibitors of the enzyme. The activity of recombinant HIV-1 IN in solution is related to an equilibrium between different oligomeric forms and depends on the concentration of proteins, cations, detergent and the presence of DNA substrate(s) (18–21). Previous studies have provided important data concerning the IN multimerization process using dynamic approaches (18–20). The crosslinking method used here allows the chemical stabilization, further isolation and assay of specific oligomeric forms of the protein. We isolated stable crosslinked oligomers and tested their in vitro activities. Dimers of HIV-1 IN catalyzed 3′-processing and integration of only one LTR extremity, while full integration of both viral extremities required at least IN tetramers, suggesting that the reactions catalyzed by IN could involve different enzyme quaternary structures. A lower activity of the crosslinked enzyme when compared with the native one was expected since a fraction of inactive enzyme should result from the crosslinking reaction. We also showed that crosslinked tetramers of IN lead to a higher percentage of integrated products bearing the HIV-1 IN hallmark of 5 bp duplication when compared with non-crosslinked IN. These data strongly suggest that this multimeric form of HIV-1 IN is the minimal active form involved in nuclear proviral integration.
However, a surprising result was obtained with the tetrameric form: the enzyme was unable to perform the one-LTR integration, in contrast to the ability of the dimeric form to perform the reaction (Figure 3). One hypothesis to explain this observation is that dimers bind only to one LTR, leading to processing and strand transfer whereas the conformation of the tetramers allow only the binding to two viral ends close to each other. It could thus suggest that the structure of the isolated dimer and the dimer involved in a tetrameric complex are different. Alternatively, we cannot completely rule out the possibility that the isolated tetramers were defective in the single LTR processing assay due to an artifact of crosslink.
On the basis of complementation studies, the active form of IN has been shown to be at least one dimer (22–24). Moreover, the DNA binding ability of IN has been linked to oligomerization (25). Stable complexes have been obtained between DNA substrate carrying one-LTR extremity and IN monomers and dimers (19). These studies did not rule out the possibility that a higher order organization might be responsible for the in vivo activity.
Structural modeling makes it unlikely that a dimer may mediate the concerted integration of both viral DNA ends (26). The site of insertion on each strand of target DNA is separated by 5 bp, corresponding to a distance of 15 Å in the helical B-form of DNA. The functional unit of IN should therefore have two active sites separated by a spacing around 15 Å. However, in the crystal structure of HIV-1 IN, the active sites in the dimer are separated by more than 30 Å. Assuming that the dimer interface in crystals is maintained in the functional IN multimer, at least one tetramer of the enzyme must be required to fulfill the complete integration mechanism. Modeling and biochemical experiments support this idea. Mutations of amino acids involved in the interface of two HIV-1 IN dimers leading to tetramers have a deleterious effect on in vitro integration activity, without any detectable effect on partial 3′-processing (27).
HIV-1 IN is a member of a family of polynucleotidyl transferase enzymes sharing certain functional aspects and similar topology. This family also includes other retroviral integrases, bacteriophage Mu transposase, RuvC, E.coli RNase H and the closely related RNase H domain of HIV-1 RT. Catalytically active recombinant HIV-1 IN can exist in a dynamic equilibrium of monomers, dimers, tetramers and higher order species. Numerous studies indicate that the enzyme functions as a multimer, minimally a dimer. In the case of the avian retroviral IN, it has been suggested that tetramer formation may be a pre-requisite step during catalysis (28). By using presteady-state active site titrations, it was shown that four IN protomers were required for a single catalytic turnover. The volumetric determination of IN–DNA complexes as determined by atomic force microscopy revealed substrate-induced assembly of a tetramer (28). Moreover, avian IN mutants, in which tetramer formation is altered, are less efficient at performing two-ended concerted DNA integration (13). Tetramers and dimers have been reported in Mu transposase. Analysis of the structure of the stable intermediates in Mu transposition revealed that recombination is catalyzed by a tetramer of Mu transposase bound to the ends of the Mu DNA (29,30). Other transposases, however, are dimeric (31). Structural studies have shown that the Tn5 transposase complexed to DNA adopts a dimeric form. Interestingly, comparison of the bacterial Tn5 transposase dimer with the tetrameric structure of HIV-1 IN shows a close resemblance (32).
Even if the tetramer of HIV-1 IN is required for the integration of both viral ends into cellular DNA, our data showed that it may not be involved in all the integration steps. The crosslinked tetramer is not active for 3′-viral end processing in contrast to the IN dimer. This strongly suggests that a sequential formation of different complexes, accompanied by conformational changes as described previously (33–36), are crucial for performing the whole integration reaction. Following terminal cleavage of viral DNA, complexes are more stable than those present at earlier reaction steps and probably help to promote correct assembly and avoid reversal of this reaction (37).
The chemical crosslink data illustrates the formation of a stable complex between the two DNA LTRs and IN tetramers. On the basis of data from the literature and our own results, we propose a model of the formation of the DNA–IN complex (Figure 6). IN monomers bind each LTR allowing the dimerization of IN. The two dimers can perform 3′-processing of the viral ends and can then interact together to form a tetramer capable of integrating the two LTRs in the target DNA. In the case of MLV virus it has been reported that mutations of one DNA end inhibited processing of the other intact LTR wild type end (38–39). This result suggests that the two viral ends may be synapsed together before the processing reaction. IN could thus engage both LTR ends before being active to cleave either end. Taken together our results and these data suggest that processing could also occur in an additional step of our model between E and F and before formation of tetramers. Each dimer engaged with one LTR could interact with the second viral end engaged with the second dimer forming a complex able to catalyze the concerted 3′processing of each viral LTR. Then remodelization of this complex could lead to the formation of an active tetramer for integration of both viral ends.
Figure 6.
Model for the in vitro formation of an active IN–DNA complex. HIV-1 IN exists in solution as an equilibrium between monomers and dimers (A). Each LTR extremity might bind to a correctly folded monomer (B), allowing the correct dimerization of IN (C). The 3′-processing of each extremity might be performed by dimers (D). Both dimers might interact together (E), forming a tetramer ready to integrate the two LTRs in the target DNA (F). Black circles indicate the 3′-processed viral ends.
This model is in agreement with others obtained with different approaches (19,27). Mutagenesis studies of HIV-1 IN performed recently in our laboratory confirmed the involvement of different IN structures in the different integration steps, leading to similar conclusions and to the proposal of a complementary model (40). Crosslinked IN tetramers were able to catalyze the two-LTR integration in vitro suggesting that the two viral extremities could be joined in the absence of a dimer intermediary.
The oligomeric association of IN in vivo is likely to be governed by cellular proteins that might stabilize a tetrameric organization of the enzyme. We have previously described the involvement of the cellular chaperone HSP60 in the stabilization of the active structure of IN (8). Such proteins, absent in the in vitro integration assays used in this work, should play an important role in the cellular integration process. The absence of cellular proteins can explain the differences observed between the in vitro IN biochemical characteristics and the in vivo proviral integration conditions (cation preference, DMSO and PEG requirements, etc.). In our crosslinking experiments the lack of cellular factors could be compensated by the increased stability of the IN crosslinked oligomers. While this work was under progress, the importance of cellular proteins was elegantly demonstrated by results showing that LEDGF/p75, a cellular protein able to interact strongly with IN and found associated with HIV-1 IN in human cells, may be involved in the formation of stable IN multimers (41).
In conclusion, we have isolated a minimal crosslinked active form of HIV-1 IN capable of catalyzing the in vitro concerted full-site integration and have identified it as a tetramer. A significant fraction of the crosslinked tetramer is enzymatically active in the concerted integration reaction and, thus, could be used as a tool for determining the structure of the active multimer of HIV-1 integrase and for identifying the region of interaction between monomers in the active complex. Identification of the monomer–monomer interaction domains inside the crosslinked tetramer is underway in our laboratory using isotopic exchange coupled with mass spectrometry. Data described here and experiments in progress will provide the basis for the future modeling of ligands/inhibitors of IN and should enlighten the reaction mechanism.
Acknowledgments
This work was supported by the French National Research Agency against AIDS (ANRS), the CNRS and the University of Bordeaux 2. A kind donation from “Les Vignobles Internationaux – Bordeaux” is greatly acknowledged. V.P. benefited from a post-doctoral fellowship from ‘Ensemble contre le SIDA. SIDACTION’. A.F. benefited from a doctoral fellowship from ANRS. We thank Dr D. P. Grandgenett (St. Louis University, USA) for useful comments and to Professors R. Cook and J. Pageze (English Department, University of Bordeaux 2) for their editorial assistance. Funding to pay the Open Access publication charges for this article was provided by CNRS.
REFERENCES
- 1.Craigie R., Fujiwara T., Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 2.Katz R.A., Merkel G., Kulkosky J., Leis J., Skalka A.M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 3.Goodarzi G., Im G.J., Brackmann K., Grandgenett D. Concerted integration of retrovirus-like DNA by human immunodeficiency virus type 1 integrase. J. Virol. 1995;69:6090–6097. doi: 10.1128/jvi.69.10.6090-6097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vora A.C., Grandgenett D.P. Assembly and catalytic properties of retrovirus integrase-DNA complexes capable of efficiently performing concerted integration. J. Virol. 1995;69:7483–7488. doi: 10.1128/jvi.69.12.7483-7488.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carteau S., Gorelick R.J., Bushman F.D. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hindmarsh P., Leis J. Retroviral DNA integration. Microbiol. Mol. Biol. Rev. 1999;63:836–843. doi: 10.1128/mmbr.63.4.836-843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craigie R. HIV integrase, a brief overview from chemistry to therapeutics. J. Biol. Chem. 2001;276:23213–23216. doi: 10.1074/jbc.R100027200. [DOI] [PubMed] [Google Scholar]
- 8.Parissi V., Calmels C., De Soultrait V.R., Caumont A., Fournier M., Chaignepain S., Litvak S. Functional interactions of human immunodeficiency virus type 1 integrase with human and yeast HSP60. J. Virol. 2001;75:11344–11353. doi: 10.1128/JVI.75.23.11344-11353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez-Pescador R., Power M.D., Barr P.J., Steimer K.S., Stempien M.M., Brown-Shimer S.L., et al. Nucleotide sequence and expression of an AIDS-associated retrovirus (ARV-2) Science. 1985;227:484–492. doi: 10.1126/science.2578227. [DOI] [PubMed] [Google Scholar]
- 11.Caumont A.B., Jamieson G.A., Pichuantes S., Nguyen A.T., Litvak S., Dupont C. Expression of functional HIV-1 integrase in the yeast Saccharomyces cerevisiae leads to the emergence of a lethal phenotype: potential use for inhibitor screening. Curr. Genet. 1996;29:503–510. doi: 10.1007/BF02426953. [DOI] [PubMed] [Google Scholar]
- 12.Caumont A., Jamieson G., De Soultrait V.R., Parissi V., Fournier M., Zakharova O.D., Bayandin R., Litvak S., Tarrago-Litvak L., Nevinsky G.A. High affinity interaction of HIV-1 integrase with specific and non-specific single-stranded short oligonucleotides. FEBS Lett. 1999;455:154–158. doi: 10.1016/s0014-5793(99)00859-5. [DOI] [PubMed] [Google Scholar]
- 13.Moreau K., Faure C., Violot S., Verdier G., Ronfort C. Mutations in the C-terminal domain of ALSV (Avian Leukemia and Sarcoma Viruses) integrase alter the concerted DNA integration process in vitro. Eur. J. Biochem. 2003;270:4426–4438. doi: 10.1046/j.1432-1033.2003.03833.x. [DOI] [PubMed] [Google Scholar]
- 14.Dufour E., El Dirani-Diab R., Boulme F., Fournier M., Tarrago-Litvak L., Litvak S., Andreola M.L. p66/p51 and p51/p51 recombinant forms of reverse transcriptase from human immunodeficiency virus type 1—interaction with primer tRNA(Lys3), initiation of cDNA synthesis, and effect of inhibitors. Eur. J. Biochem. 1998;251:487–495. doi: 10.1046/j.1432-1327.1998.2510487.x. [DOI] [PubMed] [Google Scholar]
- 15.Aiyar A., Hindmarsh P., Skalka A.M., Leis J. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J. Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinha S., Pursley M.H., Grandgenett D. Efficient concerted integration by recombinant human immunodeficiency virus type 1 integrase without cellular or viral cofactors. J. Virol. 2002;76:3105–3113. doi: 10.1128/JVI.76.7.3105-3113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petsko G.A. Preparation of isomorphous heavy-atom derivatives. Meth. Enzymol. 1985;114:147–156. doi: 10.1016/0076-6879(85)14015-2. [DOI] [PubMed] [Google Scholar]
- 18.Deprez E., Tauc P., Leh H., Mouscadet J.F., Auclair C., Brochon J.C. Oligomeric states of the HIV-1 integrase as measured by time-resolved fluorescence anisotropy. Biochemistry. 2000;39:9275–9284. doi: 10.1021/bi000397j. [DOI] [PubMed] [Google Scholar]
- 19.Deprez E., Tauc P., Leh H., Mouscadet J.F., Auclair C., Hawkins M.E., Brochon J.C. DNA binding induces dissociation of the multimeric form of HIV-1 integrase: a time-resolved fluorescence anisotropy study. Proc. Natl Acad. Sci. USA. 2001;98:10090–10095. doi: 10.1073/pnas.181024498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leh H., Brodin P., Bischerour J., Deprez E., Tauc P., Brochon J.C., LeCam E., Coulaud D., Auclair C., Mouscadet J.F. Determinants of Mg2+-dependent activities of recombinant human immunodeficiency virus type 1 integrase. Biochemistry. 2000;39:9285–9294. doi: 10.1021/bi000398b. [DOI] [PubMed] [Google Scholar]
- 21.Lee S.P., Xiao J., Knutson J.R., Lewis M.S., Han M.K. Zn2+ promotes the self-association of human immunodeficiency virus type- 1 integrase in vitro. Biochemistry. 1997;36:173–180. doi: 10.1021/bi961849o. [DOI] [PubMed] [Google Scholar]
- 22.van Gent D.C., Vink C., Groeneger A.A., Plasterk R.H. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12:3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelman A., Bushman F.D., Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Ent F.M., Vos A., Plasterk R.H. Dissecting the role of the N-terminal domain of human immunodeficiency virus integrase by trans-complementation analysis. J. Virol. 1999;73:3176–3183. doi: 10.1128/jvi.73.4.3176-3183.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vercammen J., Maertens G., Gerard M., De Clercq E., Debyser Z., Engelborghs Y. DNA-induced polymerization of HIV-1 integrase analyzed with fluorescence fluctuation spectroscopy. J. Biol. Chem. 2002;277:38045–38052. doi: 10.1074/jbc.M205842200. [DOI] [PubMed] [Google Scholar]
- 26.Heuer T.S., Brown P.O. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry. 1998;37:6667–6678. doi: 10.1021/bi972949c. [DOI] [PubMed] [Google Scholar]
- 27.Podtelezhnikov A.A., Gao K., Bushman F.D., McCammon J.A. Modeling HIV-1 integrase complexes based on their hydrodynamic properties. Biopolymers. 2003;68:110–120. doi: 10.1002/bip.10217. [DOI] [PubMed] [Google Scholar]
- 28.Bao K.K., Wang H., Miller J.K., Erie D.A., Skalka A.M., Wong I. Functional oligomeric state of avian sarcoma virus integrase. J. Biol. Chem. 2003;278:1323–1327. doi: 10.1074/jbc.C200550200. [DOI] [PubMed] [Google Scholar]
- 29.Lavoie B.D., Chan B.S., Allison R.G., Chaconas G. Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. EMBO J. 1991;10:3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizuuchi M., Baker T.A., Mizuuchi K. Assembly of the active form of the transposase-Mu DNA complex: a critical control point in Mu transposition. Cell. 1992;70:303–311. doi: 10.1016/0092-8674(92)90104-k. [DOI] [PubMed] [Google Scholar]
- 31.Davies D.R., Goryshin I.Y., Reznikoff W.S., Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediates. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 32.Wang J.-Y., Ling H., Yang W., Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellison V., Brown P.O. A stable complex between integrase and viral DNA ends mediates human immunodeficiency virus integration in vitro. Proc. Natl Acad. Sci. USA. 1994;91:7316–7320. doi: 10.1073/pnas.91.15.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asante-Appiah E., Seeholzer S.H., Skalka A.M. Structural determinants of metal-induced conformational changes in HIV-1 integrase. J. Biol. Chem. 1998;273:35078–35087. doi: 10.1074/jbc.273.52.35078. [DOI] [PubMed] [Google Scholar]
- 35.Espeseth A.S., Felock P., Wolfe A., Witmer M., Grobler J., Anthony N., Egbertson M., Melamed J.Y., Young S., Hamill T., Cole J.L., Hazuda D.J. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc. Natl Acad. Sci. USA. 2000;97:11244–11249. doi: 10.1073/pnas.200139397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hazuda D.J., Felock P., Witmer M., Wolfe A., Stillmock K., Grobler J.A., Espeseth A., Gabryelski L., Schleif W., Blau C., Miller M.D. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 37.Gao K., Butler S.L., Bushman F. Human immunodeficiency virus type 1 integrase: arrangement of protein domains in active cDNA complexes. EMBO J. 2001;20:3565–3576. doi: 10.1093/emboj/20.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy J.E., Goff S.P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J. Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei S.Q., Mizuuchi K., Craigie R. Footprints on the viral DNA ends in moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc. Natl Acad. Sci. USA. 1998;94:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calmels C., De Soultrait V.R., Caumont A., Desjobert C., Faure A., Fournier M., Tarrago-Litvak L., Parissi V. Biochemical and random mutagenesis analysis of the region carrying the catalytic E152 amino acid of HIV-1 integrase. Nucleic Acid Res. 2004;32:1527–1538. doi: 10.1093/nar/gkh298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]