Abstract
The yeast DEAD-box protein Has1p is required for the maturation of 18S rRNA, the biogenesis of 40S r-subunits and for the processing of 27S pre-rRNAs during 60S r-subunit biogenesis. We purified recombinant Has1p and characterized its biochemical activities. We show that Has1p is an RNA-dependent ATPase in vitro and that it is able to unwind RNA/DNA duplexes in an ATP-dependent manner. We also report a mutational analysis of the conserved residues in motif I (86AKTGSGKT93), motif III (228SAT230) and motif VI (375HRVGRTARG383). The in vivo lethal K92A substitution in motif I abolishes ATPase activity in vitro. The mutations S228A and T230A partially dissociate ATPase and helicase activities, and they have cold-sensitive and lethal growth phenotypes, respectively. The H375E substitution in motif VI significantly decreased helicase but not ATPase activity and was lethal in vivo. These results suggest that both ATPase and unwinding activities are required in vivo. Has1p possesses a Walker A-like motif downstream of motif VI (383GTKGKGKS390). K389A substitution in this motif significantly increases the Has1p activity in vitro, which indicates it potentially plays a role as a negative regulator. Finally, rRNAs and poly(A) RNA serve as the best stimulators of the ATPase activity of Has1p among the tested RNAs.
INTRODUCTION
Members of the DExD/H families of helicases are found in almost all organisms, and they are involved in many different biological processes including transcription, DNA repair, pre-rRNA processing and splicing. While some DExH proteins are promiscuous in NTP usage, DEAD-box proteins use ATP hydrolysis to remodel macromolecular interactions involving RNA and proteins (1–3). Nevertheless, specific RNA substrates and biological roles are not known for the vast majority of these proteins (4).
The DEAD-box family of RNA helicases is defined by the presence of nine conserved motifs (1,4) that are important for ATP binding and hydrolysis, for RNA binding and RNA unwinding. Genetic and biochemical data, as well as information from solved crystal structures of DExD/H proteins (5–8) indicate various roles for the motifs. The Q-motif (9) and motifs I and II (Walker motif A and B) are involved in ATP binding and hydrolysis (10). Motif III is thought to link ATP hydrolysis with helicase activity by linking conformational changes of the protein with ATP hydrolysis, which ultimately leads to RNA unwinding or RNA–protein dissociation. Motif VI is believed to be involved in binding the phosphates of ATP, and mutations in this motif impair ATP hydrolysis in some DEAD-box proteins (11,12). The solved crystal structures of a substrate-bound helicase (13) indicate that the remaining motifs (Ia, Ib, IV and V) are probably involved in RNA binding and in stimulation of ATP hydrolysis by RNA binding, and this is consistent with biochemical studies (14–17).
In vitro RNA-dependent ATPase activity has been demonstrated for several RNA helicases; however, in vitro ATP-dependent unwinding has only been demonstrated for a limited number of them [discussed in (4,18)]. To date, 18 putative RNA helicases have been identified that are involved in yeast ribosome biogenesis, where 8 of them are required for the synthesis of 40S r-subunits (19–24) and 10 of them are involved in the synthesis of 60S r-subunits (25–34). Of these proteins, only Rok1p, which is a DEAD-box protein involved in 40S r-subunit biogenesis, has been shown to have ATPase activity (35).
Has1p is a yeast DEAD-box protein involved in the early events of processing of the 35S pre-rRNA transcript, which leads to the proper maturation of 18S rRNA and the formation of the 40S ribosomal subunit (36). The analysis of pre-rRNA processing indicated that the formation of 18S RNA is severely altered due to the inhibition of processing of 35S pre-rRNA at early cleavage sites A0, A1 and A2. In the early pre-ribosome complexes, dynamic rearrangements of RNA–RNA contacts are required for proper processing of the pre-rRNA transcript. The role of RNA helicases may be to facilitate the rearrangements between the different 35S pre-rRNA regions, to alter pre-rRNA/snoRNA interactions, as well as to release trapped RNA–protein complexes. To date, neither the RNA substrate for Has1p nor its precise role in this large RNP complex is known. A particularly interesting feature of this essential trans-acting factor is its physical association not only with the pre-40S maturation pathway (36,37) but also with the pre-60S maturation pathway, as shown by the different proteomic approaches and by co-sedimentation experiments (36–40). In accordance with these observations, processing of 27SA3 and 27SB pre-rRNAs is also delayed upon Has1p depletion, but with no obvious phenotype on the production of the mature 60S subunits. This dual appearance of a DEAD-box protein in ribosome biogenesis is an unusual feature, and it offers the possibility of a regulatory role for Has1p. Nevertheless, the significance of this dual role still remains to be elucidated.
It was previously suggested that the enzymatic activity of Has1p may be required for its in vivo function (36). A mutant with a substitution in motif I that is known to abolish ATPase activity in other related helicases does not support growth of a ΔHAS1 strain. We have purified recombinant Has1p and characterized its biochemical activities to address the following questions: (i) Is Has1p an RNA-dependent ATPase in vitro? (ii) Is Has1p able to unwind RNA/DNA duplexes in an ATP-dependent manner? (iii) What are the kinetic properties of the enzyme in the context of previously reported biochemical studies of other DExD/H proteins? and (iv) How do mutations in conserved residues affect the ATPase and unwinding activities of Has1p? Moreover, we have also identified a Walker A-like motif downstream of motif VI and addressed its importance. Finally, we used the RNA-dependent ATPase activity of Has1p to search for specifically stimulating RNA species.
MATERIALS AND METHODS
Targeted mutagenesis of HAS1
Mutations were introduced into the HAS1 gene by fusion PCR using YCplac111-HA-HAS1 as a template. Pairs of primers used were as follows: (i) 5′ primer at position −300 from HAS1 open reading frame (ORF) and 3′ primer bearing the mutation; (ii) 5′ primer bearing the mutation and 3′ primer at position +200 respective to HAS1 ORF, as described previously (36). Residues targeted for mutagenesis were K92A (AAA→GCT), S228A (TCA→GCA), T230A (ACA→GCA), H375E (CAT→GAA), R376A (AGA→GCA) and K389A (AAA→GCA).
The mutated DNA products from the second stage amplification were co-transformed with linearized YCplac111 plasmid carrying the upstream and downstream regions but not the HAS1 ORF (41), resulting in YCplac111-HA-has1 (YCplac111-HA-K92A, YCplac111-HA-S228A, etc.). The presence of the desired mutation was confirmed by DNA sequencing of the complete ORF in order to avoid acquisition of unwanted mutations during amplification or cloning.
Test of mutational effects in vivo by plasmid shuffling
Viability of the ΔHAS1 strain (JDY10-3A: MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 has1::HIS3) was ensured by the presence of the plasmid pHT4467-HAS1 (HAS1 URA3 CEN) (36). The ΔHAS1 strain was transformed with YCplac111 (LEU2, CEN) carrying the various has1 mutants expressed under their own promoters. Leu+ transformants were selected and streaked on plates containing 5-fluoroorotic acid (5-FOA) to select against the URA3 HAS1 plasmid (42). The ability of mutant has1 alleles to support growth on 5-FOA was tested at 18, 30 and 37°C. Survivors on 5-FOA were streaked on YPD medium and incubated at different temperatures.
Cloning, expression and purification of recombinant wild-type and mutant Has1p
HAS1 wild-type and mutant genes were amplified by PCR using YCplac111-HA-HAS1 and YCplac111-HA-has1 as templates, respectively. The upstream primer contained the first 18 nt of HAS1 and an NdeI site (5′-GTAATAAACATATGGCTACCCCGTCAAAT-3′), and the downstream primer contained the last 18 nt from HAS1 and BamHI site (5′-GCTGGATCCTTACTTATGAGTTTTACG-3′). The resulting DNA fragments were cut with NdeI and BamHI and cloned into pET15b to produce pET15b-HAS1 as well as the various pET15b-has1 plasmids. These plasmids expressed the N-terminal His-tagged version of the wild-type and mutant Has1p in bacteria under the control of a T7 promoter (43). The expression plasmids were transformed into the Escherichia coli strain Rosetta (DE3) [F− ompT hsdSB(rB− mB−) gal dcm lacY1 (DE3) pRARE (CmR); Novagen]. Cultures were inoculated from single colonies and grown overnight in Luria–Bertani medium containing ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml). An aliquot of 500 ml of fresh medium was then inoculated with 5 ml of overnight culture and incubated at 37°C. At OD600 ∼0.6, isopropyl β-d-thiogalactopyranoside (IPTG) was added to the final concentration of 0.6 mM and cultures were incubated for a further 3 h at 30°C. The cells were harvested by centrifugation and pellets were stored until needed at −20°C. The pellets were resuspended in 5 ml of lysis buffer containing 20 mM Tris–HCl, pH 8, 500 mM NaCl, 10 mM imidazole, 0.1% NP40, 2 mM DTT and 0.2 mM phenylmethlysulfonyl fluoride (PMSF). Lysozyme was added to a final concentration of 10 μg/ml and the suspensions were kept on ice for 30 min with occasional mixing. The lysates were sonicated to reduce the viscosity. After centrifugation for 30 min at 15 000 r.p.m. in a Sorvall centrifuge, the supernatant was loaded onto 1 ml Ni-NTA agarose column (Qiagen) equilibrated in the lysis buffer. The column was washed two times with 10 ml of washing buffer (20 mM Tris–HCl, pH 8, 500 mM NaCl, 30 mM imidazole, 0.1% NP40, 2 mM DTT and 0.2 mM PMSF) and the protein was eluted with 3 ml of elution buffer (20 mM Tris–HCl, pH 8, 500 mM NaCl, 100 mM imidazole, 0.1% NP40, 2 mM DTT and 0.2 mM PMSF). Six fractions of 0.5 ml were collected. Usually, Has1p was found in the second and third fractions. Glycerol was added to the fractions with Has1p to a final concentration of 40% and proteins were stored at −80°C. Concentrations of the proteins were determined using the Bio-Rad Protein Assay with BSA as the standard. Purity and the concentrations were verified on 10% SDS–PAGE gels.
ATPase assay
For the determination of the ATPase activity, a colorimetric assay based on the Molybdate–Malachite green reaction system was used as described previously (9,44). Buffer conditions were optimized for Has1p (50 mM potassium acetate, 20 mM HEPES, pH 6.5, 2 mM MgCl2, 2 mM DTT and 0.1 mg/ml BSA). The enzyme concentrations were from 200 to 400 nM, ATP was 1 mM and total yeast RNA Type III (Sigma) was 400 ng/μl. To test whether DNA can stimulate the Has1p ATPase activity, 1 μM 61mer DNA oligonucleotide was added to the reaction instead of RNA. The reactions were performed at 30°C in a final volume of 30 μl and stopped by pipetting 25 μl aliquots into the well of the microtiter plate preloaded with 5 μl of 0.5 mM EDTA, pH 8. To each well, 150 μl of Malachite green was added and absorbance was measured at 630 nm.
For the determination of KM, ATP concentrations were varied from 0.05 to 3 mM. In the competition assay with ADP, concentrations of ADP and ATP were 1 mM. For the determination of the concentration of RNA required for half of the maximal activity (K1/2), total yeast RNA (Sigma) was used at concentrations from 0.02 to 2 μg/ml. For the determination of K1/2 for the RNA transcript (RNA strand from the 3′ duplex used in the unwinding assay), the 44mer concentration was varied from 10 to 500 nM. All the kinetic results were obtained from Hanes–Woolf linearization and also fitted to Michaelis–Menten nonlinear fit using GraphPad Prism 4 (GraphPad Software). Values were obtained from at least three independent measurements.
Unwinding assay
Preparation of substrates for unwinding was as described previously (45). Briefly, to prepare RNA/DNA heteroduplexes, a T7 run-off transcript (5′-GGGCGAAUUCAAAACAAAACAAAACUAGCACCGUAAAGCAAGCU-3′) was annealed to a 32P-labeled DNA oligonucleotide (16mer), complementary to the RNA (underlined). Hereafter, we call this heteroduplex ‘3′ duplex’ to emphasize that the double-stranded region is at the 3′ end of RNA strand. Using the same plasmid and the same procedure, an SP6 run-off transcript was produced (5′-GAAUACUCAAGCUUGCUUUACGGUGCUAGUUUUGUUUUGUUUUGAAUU-3′) and aligned to a 32P-labeled DNA oligonucleotide (17mer), complementary to the RNA (underlined). This substrate is called ‘5′ duplex’, having the double-stranded region at the 5′ end of the RNA transcript. In both the cases, the DNA oligonucleotide was radioactively labeled at the 5′ end to follow the course of the unwinding. The reaction mixture contained 50 nM DNA/RNA duplex, 1 μM cold DNA oligonucleotide (trap DNA oligonucleotide), 50 mM KOAc, 20 mM HEPES, pH 6.5, 2 mM MgCl2, 0.1 mg/ml BSA and 2 mM DTT. ATP was either omitted or added to a final concentration of 1 mM. Time-course reactions were performed at 30°C with a constant concentration of the protein (10, 50 or 200 nM). Reactions were quenched by the addition of 5 μl of stop solution (40% glycerol, 10 mM EDTA, pH 8, 0.025% bromophenol blue and 0.025% xylenecyanol blue) and loaded onto a nondenaturing 15% acrylamide-bisacrylamide gel (29:1) in 100 mM Tris-base, 90 mM boric acid and 1 mM EDTA (TBE). Gels were run at 4°C for 1 h at 16 W, dried and exposed to a phosphoimager screen or alternatively to film for autoradiography. Quantification was carried out using OptiQuant software (Packard).
Polysome analysis, rRNA fractionation and RNA specificity
Polysome analysis was performed as described previously (30). For the isolation of ribosomal RNA, 0.5 ml fractions were collected after ultracentrifugation. RNA was phenol extracted, precipitated with the addition of sodium acetate and ethanol, and about one-tenth of each fraction was analyzed on an agarose gel. After measuring the A260, equal concentrations of selected fractions were used in the ATPase assay.
To determine the extent of activation of the Has1p ATPase activity by the different RNAs, poly(A) RNA was prepared from total yeast RNA by using an oligo-dT cellulose column (18). Yeast tRNA was purchased from Roche-Diagnostics. The ATPase assay was carried out as described above, except the final concentrations of different RNAs were either 10 or 20 μg/ml.
RESULTS
Conserved residues in motifs III and VI are important for Has1p function in vivo
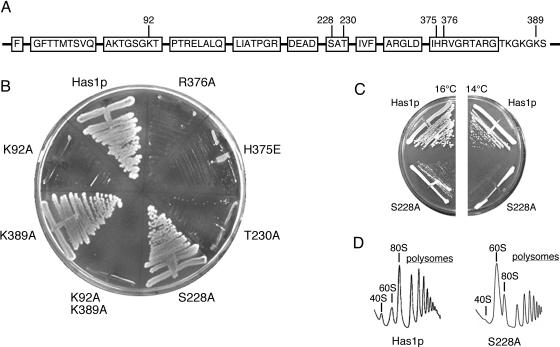
In our previous report (36), we showed that the substitution of the highly conserved K92 by alanine in motif I (GKT→GAT) abolished Has1p function in vivo, which indicated that this residue was important for the enzymatic activity of Has1p. To expand upon this analysis, we constructed two point mutants in motif III (S228A and T230A) and two point mutants in motif VI (H375E and R376A) (Figure 1A). The histidine in motif VI is the least studied residue of those selected; therefore, to assure that mutational effect will be obvious, the histidine was changed to glutamate to provide a change in the residue charge. The in vivo function of the mutant alleles was tested by plasmid shuffling (Figure 1B). Cells containing wild-type Has1p on a LEU CEN plasmid grew on 5-FOA plates. The K92A mutant, previously shown to be lethal in vivo, was used as a negative control. The mutant S228A (motif III) was able to form colonies on 5-FOA at 30°C, but it displayed slow growth at 18 and 16°C, and no growth at 14°C (Figure 1C). The other mutation (T230A) in motif III was lethal, as were both mutations in motif VI (H375E and R376A), i.e. Leu+ cells were not able to grow on 5-FOA at temperatures between 16 and 37°C.
Figure 1.
Mutational analysis of Has1p. (A) Boxes represent the conserved helicase motifs. The putative second Walker A motif is also shown. Positions of the mutations are indicated. (B) Cells containing wild-type HAS1 and either of the indicated has1 alleles were streaked on 5-FOA plates and incubated at 30°C. Only S228A and K389A mutants were able to support growth. (C) Strain with the S228A mutant displays a conditional phenotype, with slow growth at 16°C and no growth at 14°C. (D) Polysome analysis of the S228A mutant grown at 30°C shows a severe effect on the 40S r-subunit production and on the total level of translation. Peaks representing the ribosome subunits, mature ribosomes and polysomes are as indicated.
A polysome analysis was performed at 30°C to further test the phenotype of the only complementing mutant S228A. Although this is a permissive temperature for growth, severe effects were observed on the production of 40S r-subunits, with a proportional increase of free 60S r-subunits and an overall decrease in the 80S peak and in the polysome peaks (Figure 1D). This is similar to the phenotype observed upon Has1p depletion in the cells (36). This drastic decrease in the total amount of the 40S r-subunits indicates that motif III is crucial for the correct biological function of Has1p.
A mutation in the putative second Walker A motif (K389A) has no effect on growth
Upon analysis of the Has1p protein sequence, we found a sequence with strong similarity to the Walker A motif (383GtkGkGKS390) downstream of motif VI. In general, the Walker A motif (consensus A/GxxxxGKS/T) is part of the nucleotide-binding site, and it is found in many NTPases (10). We decided to test the significance of this motif adjacent to motif VI for the function of Has1p. In analogy to the first Walker A motif (motif I in RNA helicases), we changed the lysine at position 389 to alanine (388GKS390→388GAS390) to study the effects of this substitution on the functional and biochemical properties of Has1p (Figure 1A). The K389A mutant was able to form colonies on 5-FOA-containing plates similar to wild-type cells (Figure 1B), which indicated that altering this motif was not deleterious to growth. We performed a polysome analysis to analyze this mutant further, but no differences from the wild-type profile were observed (data not shown). For biochemical studies, we also prepared a double mutant bearing the mutations in motif I and in the downstream putative Walker A motif (K92A–K389A). As expected, this double mutant was not able to complement the growth of the ΔHAS1 strain (Figure 1B).
Recombinant Has1p is an RNA-dependent ATPase
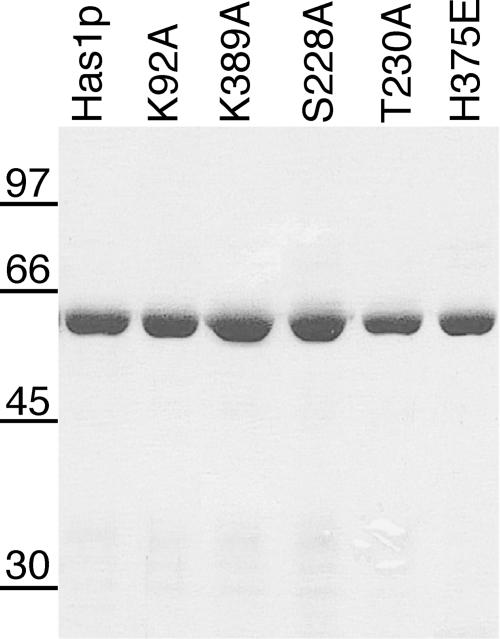
To facilitate the biochemical characterization of Has1p, His6-tagged Has1p was expressed in bacteria and purified from soluble bacterial lysates by nickel–agarose chromatography. IPTG induction led to the accumulation of a protein of ∼56 kDa that was consistent with the calculated molecular weight of His6-Has1p. The identity of the protein was confirmed both with anti-His6 and anti-Has1p antibodies (data not shown). The protein bound to nickel–agarose was eluted with 100 mM imidazole. Protein was eluted in 1 ml of total volume at a concentration between 1 and 3 mg/ml. The quality of the protein preparations was verified on an SDS–polyacrylamide gel (Figure 2).
Figure 2.
Aliquots (1 μg) of purified wild-type (Has1p) and mutant proteins were analyzed by SDS–PAGE, and the polypeptides were visualized by staining with Coomassie blue. The positions of marker proteins (in kDa) are indicated at the left. Proteins K92A–K389A and R376A are not shown; they were purified to the comparable purity and homogeneity.
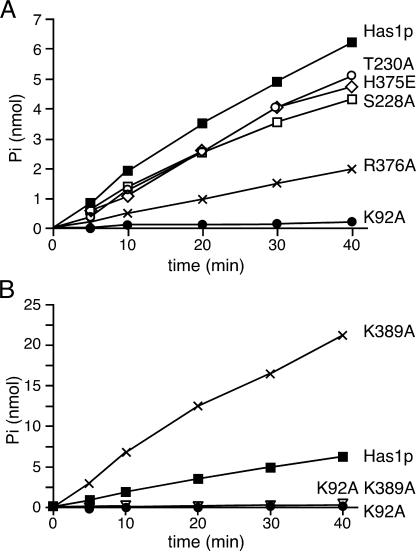
The presence of DEAD-box motifs in Has1p led to the prediction that this protein is an ATPase. Also, the finding that the K92A mutation in motif I (Walker A motif) has a lethal phenotype indicated that the ATPase activity is important for the biological function of Has1p (36). The rate of ATP hydrolysis was measured in the presence of total yeast RNA (Type III; Sigma) to test whether purified Has1p behaves as an ATPase in vitro (Figure 3). The reaction velocity was proportional to the enzyme concentration (data not shown), confirming that the observed activity was the consequence of protein input in the assay. As a control, we constructed the plasmid expressing a mutant protein with a motif I substitution (91GKT93→91GAT93). This mutant protein showed only 2–5% of the ATPase activity of the wild-type (Figure 3). This strongly suggests that the observed ATPase activity is directly linked to Has1p.
Figure 3.
ATP hydrolysis by the wild-type Has1p and the mutants was measured over time. Reactions were carried out at pH 6.5 with 1 mM ATP, 2 mM MgCl2 and 390 nM proteins at 30°C in the presence of 400 ng/μl of total yeast RNA. No ATPase activity was observed without RNA (data not shown). (A) ATPase activity of Has1p (filled square) and mutant proteins K92A (filled circle), S228A (open square), T230A (open circle), H375E (open diamond) and R376A (x). (B) ATPase activity of Has1p (filled square) and mutant proteins K92A (filled circle), K389A (x) and the double mutant K92A–K389A (inverted open triangle).
We then tested whether the ATPase activity was dependent on an RNA cofactor, as was reported for other proteins from the DExH/D-box families. The amount of ATP hydrolysis increased in a time-dependent manner in the presence of saturating concentrations of total yeast RNA. The ATP hydrolysis was abolished either when RNA was omitted or when DNA was added instead of RNA, which showed that the ATPase activity of Has1p is strictly dependent on the presence of the RNA cofactor (data not shown).
The requirement for divalent cations was also measured using increasing concentrations of MgCl2. The optimal activity was observed between 1.5 and 2 mM MgCl2. The optimal pH was established to be pH 6.5; no activity was detected below pH 5 and above pH 8. A 20% higher activity was observed when using potassium acetate than when using KCl at the optimal pH. Both ATP and dATP were efficiently hydrolyzed in the presence of RNA, whereas for other nucleotide triphosphates <3% background was observed.
The Michaelis–Menten kinetic parameters were determined with variable concentrations of ATP at saturating concentrations of RNA. Using the reciprocal plot (Hanes–Woolf), we calculated KM (ATP) of ∼450 μM and kcat of ∼5.5 min−1 (Table 1). Data were also fitted directly to the Michaelis–Menten equation with the program GraphPad Prism 4 (GraphPad Software) and essentially the same values were obtained. Note that some of our mutants could not be fully saturated for ATP in our assays; therefore, these results should be considered as kinetic parameters rather than true thermodynamic values.
Table 1.
Kinetic analysis of ATP binding and hydrolysis for wild-type and mutant Has1pa
| Enzyme | KM (μM) | kcat (min−1) | Relative kcat/KM |
|---|---|---|---|
| Has1p | 440 ± 90 | 5.4 ± 0.9 | 1 |
| K92A | ND | 0.17 ± 0.05 | ND |
| K389A | 450 ± 200 | 17 ± 2 | 3.06 |
| K92AK389A | ND | 0.7 ± 0.1 | ND |
| S228A | 760 ± 410 | 3.8 ± 1 | 0.41 |
| T230A | 810 ± 420 | 3.9 ± 0.9 | 0.38 |
| H375E | 920 ± 380 | 4.2 ± 0.6 | 0.37 |
| R376A | ND | 1.6 ± 0.5 | ND |
aValues derived from nonlinear fits to the Michaelis–Menten equation using the mean values of three independent experiments. The standard errors of estimation were derived from the curve fits.
ND, not determined.
ATPase activities of the Has1p mutant proteins
Conserved residues in motifs I, III and VI are important for Has1p function in vivo (36), whereas the altered downstream Walker A-like motif did not change the ability of the protein to support growth on 5-FOA plates. To study the biochemical consequences of the mutations, we produced His6-tagged Has1 proteins K92A (motif I), S228A and T230A (motif III), H375E and R376A (motif VI), K389A (putative second Walker A motif) and the double mutant K92A–K389A. Mutant proteins were purified from bacteria using the same method as for the wild-type protein with comparable yield and purity (Figure 2). ATPase activities of the mutants are shown in Figure 3 and the kinetic analysis is summarized in Table 1. The K92A mutant showed ∼2–5% of the wild-type activity and an almost 20 times higher KM for ATP. Mutant proteins S228A, T230A and H375E exhibited 70–80% of the wild-type activity and a slight decrease (about two times) in affinity for ATP. The lethal motif VI mutant R376A exhibited <30% of the wild-type activity (Figure 3A). Surprisingly, the K389A mutant in the putative downstream Walker A motif showed a 3-fold increase in the ATPase activity with no actual difference in ATP binding (Figure 3B and Table 1). The double mutant K92A–K389A had ∼13% of the wild-type activity, which was more than three times above the marginal activity observed for the motif I mutant K92A. Both wild-type Has1p and the mutants could be inhibited by the presence of ADP; when ATP and ADP were present at equimolar amounts, then the activities of Has1p, S228A, T230A, H375E and K389A were reduced 5-fold (data not shown).
Has1p ATPase activity is stimulated by ribosomal RNAs
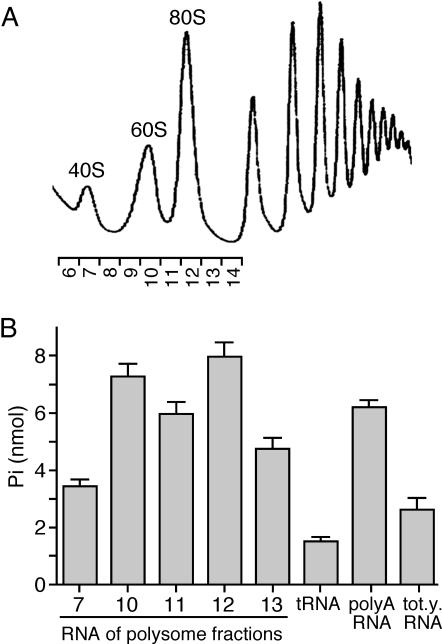
In general, the in vitro ATPase activity of DEAD-box proteins is dependent on the presence of RNA. In some cases, such as Prp5p, which is required for pre-mRNA splicing, this activity can be particularly stimulated with specific RNA species that are believed to be the substrates of those enzymes (46). In the case of the E.coli DbpA protein, the ATPase activity is increased three orders of magnitude in the presence of a specific 23S rRNA loop when compared with nonspecific RNA (47). In order to screen the main types of cellular RNAs for the stimulation of ATPase activity of Has1p, we prepared total yeast RNA, poly(A) RNA, tRNA and rRNAs. The purity and quantity of the RNAs were checked on agarose gels and by measuring the A260/A280 (data not shown). Ribosomal RNA was considered to be the most likely substrate for Has1p and it was prepared from different fractions of a polysome gradient (Figure 4). The fractions presumably contained different pre-rRNAs that could potentially be substrates of Has1p (36). The ATPase activity of Has1p was significantly stimulated when rRNA fractions or poly(A) RNA were used compared with that for total yeast RNA or tRNA. In the case of poly(A) RNA, we could not exclude the possibility that a small amount of rRNA was present in the samples and that it contributed to the activation. Stimulation by 25S rRNA (fraction 10) was two times higher than with 18S RNA (fraction 7). 80S fractions (fractions 11 and 12) showed similar activation as fractions containing 25S RNA; those fractions contained 18S and 25S rRNA, and presumably the large 35S precursors (36).
Figure 4.
ATPase stimulation of Has1p by various RNAs. (A) rRNA was recovered from the polysome fractions as shown. (B) ATPase activity was measured at pH 6.5 in the presence of 1 mM ATP, 2 mM MgCl2 and 10 nM rRNA, tRNA, poly(A) RNA or total yeast RNA after 40 min of incubation at 30°C. Stimulation with rRNAs and poly(A) RNA was observed, with the highest values obtained with fractions 10 and 12 presumably containing 60S/pre-60S and 80S/95S complexes, respectively (36). The RNA amount in the reaction was normalized to 10 μg/ml.
Has1p unwinds RNA/DNA heteroduplexes in vitro
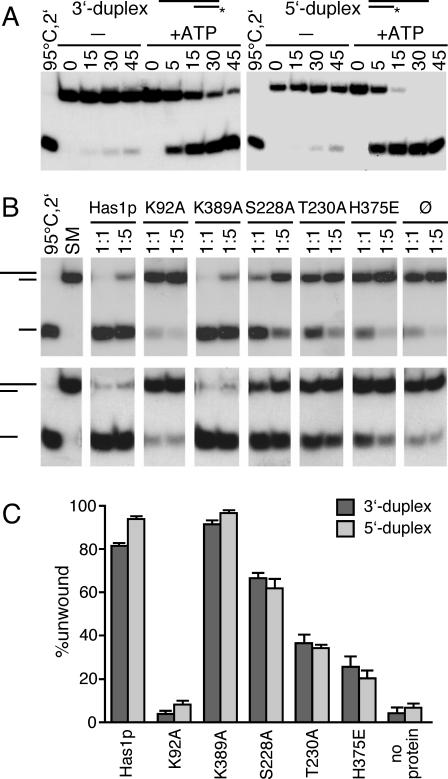
To date, no yeast DEAD-box protein involved in eukaryotic ribosome biogenesis has been shown to have an ATP-dependent RNA unwinding activity in vitro. In order to test whether Has1p has such an activity, we constructed two RNA/DNA heteroduplexes. It was previously shown that RNA/DNA duplexes can serve as substrates where the DNA strand is not interfering with the helicase activity (45,48). The first substrate (3′ duplex) was a 44mer RNA transcript with a 16mer DNA oligonucleotide complementary to the 3′end of the RNA. The second substrate (5′ duplex) was a 48mer RNA with a 17mer DNA complementary to the 5′ region of RNA. Reproducible disruption of the duplex was detected with both substrates in the presence of purified Has1p and ATP (Figure 5A). Interestingly, under the same conditions, the extent of unwinding was higher with the 5′ duplex. Although both duplexes have similar stabilities (ΔG = −19.8 kcal/mol for 3′ duplex and ΔG = −19.1 kcal/mol for 5′ duplex), these results indicate that intrinsic properties of the duplexes contribute to the properties of the unwinding reaction. Under our assay conditions, the efficient unwinding was observed with concentrations of Has1p that were either equal to or lower than the concentration of the duplex. This is in contrast with previously reported unwinding properties of other DEAD-box proteins, where a large excess of enzyme was required for efficient unwinding (18,49,50).
Figure 5.
Unwinding of RNA/DNA duplexes by wild-type and Has1p mutants. (A) Time course for ATP-dependent unwinding of 3′ duplex and 5′ duplex by wild-type Has1p. In short, 50 nM duplex was incubated with 50 nM protein with or without 1 mM ATP, at 30°C for the time indicated in minutes. To prevent reannealing of the displaced 32P-labeled oligonucleotide, 1 μM cold DNA oligonucleotide was added as a competitor. Products were separated on a 15% polyacrylamide gel, which was then subjected to autoradiography. (B) Unwinding activities of the wild-type and Has1p mutants. An aliquot of 50 nM duplex (3′ top panel, 5′ bottom panel) was incubated either with 50 nM protein (molar ratio 1:1, as indicated) or 10 nM protein (molar ratio 1:5, as indicated). The reactions were carried out for 60 min at 30°C in the presence of 3 mM ATP. To prevent reannealing of the displaced 32P-labeled oligonucleotide, 1 μM cold DNA oligonucleotide was added as a competitor. Products were separated on a 15% polyacrylamide gel, which was then subjected to autoradiography. As control, the duplex was either heat-denatured for 2 min at 95°C, quick-cooled on ice and then loaded, or starting material (SM) containing the same amount of duplex was loaded. (C) Quantification of the experiments that were performed with 50 nM duplex and 50 nM enzyme, and incubated for 60 min at 30°C in the presence of 3 mM ATP. The standard errors of estimation were derived from the curve fits based on three independent sets of experiments.
The motif I mutant with abolished ATPase activity was not able to unwind heteroduplexes (Figure 5B and C); the marginal unwinding was identical to the extent of unwinding without protein, and we consider it as a background unwinding in our experiments. We also tested whether ADP can stimulate unwinding; like in the reaction without nucleotide, no unwinding activity was observed (data not shown).
RNA binding and unwinding of RNA/DNA heteroduplexes by Has1p mutants
Since the ATPase activity of Has1p was dependent on an RNA cofactor, we wanted to test whether the Has1p mutants display differences in RNA binding and in unwinding properties compared with the wild-type protein. Unwinding activities of the mutants are shown in the Figure 5B and C. Although displaying similar ATPase activities, the mutant S228A showed a slightly reduced unwinding activity, while T230A and H375E mutants unwound the duplexes with significantly lower efficiencies. Because of its lower ATPase activity, the mutant protein R376A was not used in these assays. After 1 h of incubation at 30°C of both enzyme and duplex at concentrations of 50 nM (Figure 5B and C), S228A showed ∼60% of the wild-type activity, whereas T230A and H375E were ∼20–30% of the wild-type (when the background unwinding was subtracted). In agreement with its increased ATPase activity, the Walker A-like motif mutant (K389A) unwound duplexes with higher efficiency than the wild-type. Together with our in vivo data, these results are consistent with the unwinding activity of Has1p being necessary for its in vivo function, since the impaired unwinding coincided with the loss of ability to complement the ΔHAS1 strain.
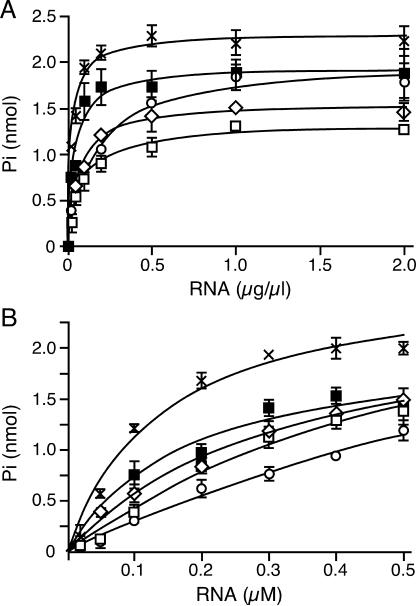
In order to determine whether the differences in the helicase activity were due to an altered RNA binding, we attempted a gel-mobility assay to see directly the formation of a Has1p–RNA complex. Unfortunately, under several different conditions used, purified Has1p always precipitated in the wells of the gel, and we were not able to obtain reproducible and reliable results. Therefore, we performed kinetic analysis of RNA binding through the stimulation of ATPase activity by the RNA (Figure 6). We used both total yeast RNA (Type III; Sigma) and ssRNA that was used as an RNA strand for the 3′ duplex in the unwinding assay (see Materials and Methods). It has been shown for other helicases that ssRNA overhangs are required for protein loading onto the partial duplex and for unwinding activity (18); therefore, if binding of this particular ssRNA is altered, we should be able to correlate it directly to the 3′ duplex unwinding. Note that in the case of ssRNA, proteins were not fully saturated for ssRNA. Although the obtained values were successfully fit to the Michaelis–Menten equation, they should be considered as kinetic parameters, and they were used for comparisons between the wild-type and mutant Has1p. We defined K1/2 as the concentration of RNA at which the velocity of ATP hydrolysis reaches half of the maximal value (half-activity). For both the RNA substrates, the mutants showed the same trend when compared with the wild-type values (Figure 6 and Table 2). The differences were less pronounced when total yeast RNA was used. Affinities for ssRNA were lower for the motif III mutants, as well as for the motif VI mutant H375E (Table 2). Although S228A and H375E mutants exhibited similar ATPase activities, H375E displayed significantly lower unwinding than S228A and consequently it was not able to support growth. Interestingly, the affinity for ssRNA was slightly higher in H375E; this means that this mutant, despite having 80% of the wild-type ATPase activity and its ability to bind RNA, somehow lost the ability to use the energy from ATP hydrolysis to perform productive unwinding. It is possible that this failure also contributed to the lower activities of S228A and T230A, but to a lesser extent.
Figure 6.
ATPase activities of Has1p (filled square) and mutant proteins S228A (open square), T230A (open circle), H375E (open diamond) and K389A (x) in the presence of the increasing concentrations of total yeast (Type III) RNA (A), and RNA transcript (RNA strand of 3′ duplex used in unwinding assay) (B). Reactions were carried out for 40 min at 30°C with 1 mM ATP, 2 mM MgCl2 and 390 nM proteins. The curves were fit to the nonlinear fit (one binding site) using GraphPad Prism 4. The standard errors of estimation were derived from the curve fits based on three independent sets of experiments.
Table 2.
Kinetic analysis of RNA binding through the stimulation of ATP hydrolysis
| Enzyme | K1/2 (RNA transcript) (nM) | Relative K1/2 | K1/2 (total yeast RNA) (μg/μl) | Relative K1/2 |
|---|---|---|---|---|
| Has1p | 160 ± 20 | 1 | 28 ± 16 | 1 |
| K389A | 150 ± 20 | 0.92 | 25 ± 5 | 0.89 |
| S228A | 750 ± 160 | 4.58 | 62 ± 12 | 2.21 |
| T230A | 1500 ± 520 | 9.18 | 130 ± 20 | 4.64 |
| H375E | 430 ± 80 | 2.60 | 60 ± 15 | 2.14 |
K1/2 represents the concentration of RNA at which the velocity of ATP hydrolysis reaches the half of maximal value (half-activity). Values derived from nonlinear fits to the Michaelis–Menten equation using the mean values of three independent experiments. The standard errors of estimation were derived from the curve fits.
DISCUSSION
The yeast DEAD-box protein Has1p is involved in 40S r-subunit biogenesis, but is also found associated with the pre-60S pathway (36). DEAD-box proteins share nine conserved motifs, and most of them are also conserved in other members of the DExH-box superfamily. As for many other DEAD-box proteins involved in ribosome biogenesis, the importance of the conserved helicase motifs for the function of Has1p has not been analyzed. Mutational studies of several DExH/D-box proteins, including yeast eIF4A, Ded1p, Prp2p, Prp16p, Prp22p as well as viral helicases NS3 and NPH-II, revealed that conserved amino acids in motifs I (AxxGxGKT), II (DExH/D), III (S/TAT) and VI (Q/HRxGRxGR) are important for in vivo function and in vitro ATPase activity and unwinding (11,14,18,51–56).
The biochemical characterization of Has1p reveals that this protein is an RNA-dependent ATPase in vitro and that it can unwind RNA/DNA heteroduplexes in an ATP-dependent manner. Our results suggest that both the activities are essential in vivo, as mutants that lost one of these activities were not able to support the growth of a ΔHAS1 strain. Mutations in the conserved motifs I, III and VI, as well as in the putative Walker A motif downstream of motif VI, reveal the importance of the corresponding residues for Has1p function.
The observation that Has1p exhibits the capacity to hydrolyze ATP is not surprising, as many proteins that possess a helicase core domain previously have been shown to be ATPases (1,57,58). ATP hydrolysis of Has1p is strictly dependent on the presence of RNA, which is also true for many other DEAD-box proteins characterized so far (18,59,60). The kcat for ATP hydrolysis by Has1p is ∼5.5 min−1, which is comparable with kcat values for yeast eIF4A [6.8 min−1, (61)], Xenopus An3 [6 min−1, (62)] and E.coli SrmB [1.2 min−1, (63)], but is much lower than kcat measured for E.coli DbpA [600 min−1, (64)], yeast Prp22p [400 min−1, (65)] or Ded1p [300 min−1, (18)]. The affinity of Has1p for ATP is comparable with the affinities of other DEAD-box proteins, with a KM value of ∼400 μM, which is still well below the cellular concentration of ATP (5–10 mM). The relatively weak ATPase activity of Has1p could reflect low intrinsic catalytic activity. Alternatively, the absence of a specific substrate or protein cofactors, or the lack of post-translational modifications in the recombinant protein may contribute to the low activity. Nevertheless, in the absence of RNA, Has1p exhibits no ATPase activity, whereas in the presence of total yeast RNA or ssRNA transcript, there is a substantial level of ATPase activity.
The exciting result of our studies is that recombinant Has1p exhibits ATP-dependent duplex unwinding activity in vitro. Recently, it was reported that Dbp9p unwinds DNA duplexes in an ATP-dependent manner, but the biological relevance of this finding is unclear (66). Here, we show that Has1p is able to unwind RNA/DNA heteroduplexes that have either a 3′ duplex with a 5′ RNA overhang or a 5′ duplex with a 3′ RNA overhang. Despite similar stabilities of the substrates, Has1p unwinds the 5′ duplex with a higher efficiency. Although this may imply directionality in the Has1p-mediated unwinding, it is at present more probable that intrinsic characteristics of the two duplexes cause this difference (N. K. Tanner, J. Banroques, O. Cordin and P. Linder, unpublished data). As we have not tested a blunt-end duplex, we cannot exclude the possibility that Has1p is able to bind to a duplex region per se. At this point, we believe that the mechanism of duplex unwinding by Has1p is a result of nonprocessive duplex destabilization through the interaction of the protein with the RNA rather than a processive unwinding of the duplex that involves translocation along the RNA strand. The relatively weak ATPase activity and the ability to unwind both 3′ and 5′ duplexes point to such a conclusion. The majority of the helicases studied so far display directional unwinding; nevertheless, weaker helicases, such as eIF4A and p68, were reported to unwind duplexes in both directions in vitro (49,60). For that reason, we avoid using the term helicase in the case of Has1p, although with the substrates used the resulting activity is indistinguishable from the activity of processive helicases [e.g. NPH-II, (67)].
For the mutation of the conserved lysine (91GKT93→ 91GAT93) in motif I, there is a clear correlation between lack of growth and the abolished ATPase activity. This is consistent with studies for other NTPases including yeast eIF4A, Prp16p, Prp22p and Ded1p (55,65,68). Crystal structures of eIF4A (5,7) and NS3 (8,13) show that the lysine of motif I contacts the α, β and sometimes γ phosphates of the bound NTP.
The motif III hydroxyls (SAT) seem not to be important for ATP hydrolysis, although they are considered necessary for RNA unwinding by eIF4A (51), NPH-II (14), Prp22 (69) and Prp43 (70). A motif III mutant exhibited a dominant-negative phenotype that abolished splicing in Prp2p (71). As ATPase activity of this mutant was reduced only slightly, it was proposed that the dominant-negative phenotype is due primarily to a defect in the putative RNA helicase activity. Proteins mutated in motif III retain ATPase activity, but they do not unwind duplexes, suggesting that there is an uncoupling of the ATPase and unwinding activities. In agreement with that, S228A and T230A mutants of Has1p hydrolyze ATP with ∼70% of the wild-type activity, but the duplex unwinding is significantly impaired, particularly for T230A (Table 1 and Figure 5B and C). The severity of the growth phenotype is proportional to the loss of unwinding activity in vitro; S228A displays cold-sensitive growth, while the T230A substitution is lethal for the cell. The low level of unwinding by T230A probably comes from its inability to bind RNA strong enough, since it requires almost 10 times more RNA than the wild-type to reach half of the maximal velocity of ATP hydrolysis (Table 2). Additionally, besides lower ATPase activities and RNA-binding affinities, impaired transmission of conformational changes upon ATP hydrolysis may also contribute to the loss of unwinding activity. As predicted from solved helicase crystal structures with and without a bound nucleotide, the relative orientation of two helicase domains changes with ATP hydrolysis, and this movement is what probably leads to strand separation (72). Although there are no comparable data for DEAD-box proteins, in the structures of eIF4A (7) and MjDEAD (6) both serine and threonine from motif III (SAT) form hydrogen bonds with the aspartate from motif II (DEAD), thus providing intramolecular communication between motifs.
The significance of the proper transmission of the conformational changes for the function of Has1p is even more obvious from our analysis of the motif VI mutant H375E. Although motif VI was described as important for ATP hydrolysis because it forms part of the ATP-binding cleft (4), substitution of histidine at the beginning of the motif by a negatively charged glutamate decreased the ATPase activity by only 20%. Although the RNA binding was slightly affected (two times higher K1/2 than for the wild-type), this mutant retained only ∼20% of the unwinding activity (Figure 5B and C). The solved crystal structure of eIF4A (7) indicated that this histidine (HRxGRxGR) could make an intramolecular interaction with an aspartate in motif II (DEAD). The equivalent interaction between glutamine in motif VI (QxxGRxGR) and histidine in motif II (DExH) was seen in both UvrB (73–75) and NS3 helicases (8,13). Notably, the two motifs are part of two distinct structural domains of the helicase core. A functional linkage of the histidine in motif VI and the aspartate in motif II was also suggested by biochemical analyses of eIF4A (51), where DEAD/HRxGRxGR to DEAH/QrxGRxGR changes resulted in a doubling of ATPase activity but eliminated unwinding. Altogether, these data indicate a functional crosstalk between these two motifs. However, it cannot only be a question of amino acid interactions, since the reciprocal exchanges abolish helicase activity. As motifs II and VI are important for ATP binding and hydrolysis, this intramolecular interaction may be needed to sense the release of Pi upon ATP hydrolysis and, as a consequence, to transmit conformational changes throughout the molecule. In support for that view, the H375E mutation dissociates ATP hydrolysis and unwinding activities without a drastic decrease of ATP and RNA binding, which reflects a more complex role for motif VI that is not limited to ATP hydrolysis. The second mutation in motif VI, R376A, reduces the ATP hydrolysis of Has1p to <30% of the wild-type activity and is lethal in vivo. Although this arginine residue seems to be less crucial for the interaction with ATP than in other helicases (76), in Has1p it is important for ATPase activity. It is possible that a threshold ATPase activity is necessary for supporting the growth of the ΔHAS1 strain, as was suggested for various Prp16 mutants (53). The equivalent arginine was also essential for eIF4A (56), but in contrast in Ded1p it diminishes ATPase activity but it is not lethal (J. Banroques and P. Linder, unpublished data). Similarly, an equivalent substitution in Dbp8p resulted in a wild-type in vivo phenotype (24).
Downstream of motif VI in Has1p, there is a motif that has a consensus sequence of a Walker A motif (383GtkGkGKS390). Although we could show that the mutation of lysine 389, which is equivalent to the conserved lysine in motif I, was not deleterious in vivo, we found that this mutant protein displays more than three times higher ATPase activity and higher unwinding activity in vitro compared with the wild-type. Moreover, the ATPase activity of the K92A–K389A mutant was 13% of wild-type value, yet almost 4-fold higher than that of the K92A single mutant. Although it is difficult to interpret this finding without further analysis, there are several possibilities. One possibility is that the effect we observe is indirectly due to the distortion of motif VI caused by mutagenesis of the adjacent region. Alteration of ATP binding may result in faster dissociation of the hydrolysis products leading to an increase in kcat. A second and more intriguing possibility is that this motif is able to participate in ATP binding, since it is in close proximity to the ATP-binding site that is formed by motifs Q, I, II and VI. In that case, this could be a way to regulate the ATPase activity; once ATP is bound, it can be accommodated in the active site in two ways. One is productive binding (through motifs Q, I, II and VI) that leads to hydrolysis, whereas the other is nonproductive or less productive binding in which the putative Walker A motif downstream of motif VI also interacts with ATP. When the lysine in this motif is not available (as in the case of the K389A mutant), the ATP is bound only in the productive way and hydrolysis occurs faster. It is noteworthy that the affinities for ATP are the same for the wild-type and the K389A mutant, which implies that the increase in activity results from the increased velocity of the reaction. A biological role can be envisioned for this type of regulation: once the protein encounters a proper substrate or interacting partner, the resulting conformational changes may preclude the interaction of ATP with the putative second Walker A motif. Consequently, this may lead to derepression of enzymatic activity. Detailed structural information for DEAD-box proteins that contain this second Walker-A motif is needed to clarify this idea.
In general, the lack of specific substrates and interacting proteins make in vitro studies difficult to interpret. It is likely that the in vivo activities of Has1p are higher than suggested from the in vitro results, and several ways to achieve that higher activity can be envisioned. It is possible that a Has1p-specific substrate exists, but it still needs to be found. We looked for candidates among rRNA precursors (36), and we could observe 4–8 times higher stimulation with the rRNAs and poly(A) RNA compared with tRNA. In general, the less compact RNA molecules seem to serve as better substrates for Has1p. The highest stimulation observed with the rRNAs isolated from 60S or pre-60S, 80S and 90S ribosomal particles is in agreement with the postulated biological role of Has1p (36). Bearing in mind that the specific activity of Has1p (∼5.5 min−1) was measured in the presence of total yeast RNA (Sigma), it is clear that Has1p would display higher activity if only rRNA is provided. Nevertheless, several different rRNA pools, as well as poly(A) RNA, activated Has1p to a similar extent, which indicates a lack of high substrate specificity in vitro. Another possibility is that specific substrates cannot be found in our current in vitro conditions; rather, the substrate RNA becomes presented to Has1p only at a particular point during ribosome biogenesis, and that this happens only in the context of the larger complex of proteins, snoRNAs and pre-rRNAs. Finally, it is possible that the activity of Has1p becomes derepressed in vivo, probably through its interaction with another protein. In this case, a mode of regulation through the putative second Walker A motif can be imagined; at the proper time and in the proper complex, Has1p becomes derepressed due to slight conformational changes that make the lysine of this motif no longer exposed to the ATP-binding pocket.
Acknowledgments
We are grateful to all members of our laboratory for useful discussions and support. We thank Monique Doère for excellent technical help with the polysome profiles. We are grateful to Josette Banroques for critical reading of the manuscript, as well as to the referees for their comments and suggestions. S.R. was supported by the Novartis and the E. & L. Schmidheiny foundations. This work was supported by a grant from the Swiss National Science Foundation and by the canton of Geneva. Funding to pay the Open Access publication charges for this article was provided by the Swiss National Science Foundation.
REFERENCES
- 1.Tanner N.K., Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 2.Schwer B. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nature Struct. Biol. 2001;8:113–116. doi: 10.1038/84091. [DOI] [PubMed] [Google Scholar]
- 3.Linder P., Tanner N.K., Banroques J. From RNA helicases to RNPases. Trends Biochem. Sci. 2001;26:339–341. doi: 10.1016/s0968-0004(01)01870-9. [DOI] [PubMed] [Google Scholar]
- 4.Rocak S., Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nature Rev Mol Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 5.Benz J., Trachsel H., Baumann U. Crystal structure of ATPase domain of translation initiation factor eIF4A from Saccharomyces cerevisiae—the prototype of the DEAD box protein family. Structure Fold. Des. 1999;7:671–679. doi: 10.1016/s0969-2126(99)80088-4. [DOI] [PubMed] [Google Scholar]
- 6.Story R.M., Li H., Abelson J.N. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc. Natl Acad. Sci. USA. 2001;98:1465–1470. doi: 10.1073/pnas.98.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caruthers J.M., Johnson E.R., McKay D.B. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl Acad. Sci. USA. 2000;97:3080–3085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao N., Hesson T., Cable M., Hong Z., Kwong A.D., Le H.V., Weber P.C. Structure of the hepatitis C virus RNA helicase domain. Nature Struct. Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 9.Tanner N.K., Cordin O., Banroques J., Doère M., Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 10.Walker J.E., Saraste M., Runswick M.J., Gay N.J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pause A., Méthot N., Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol. Cell. Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross C.H., Shuman S. The QRxGRxGRxxxG motif of the vaccinia virus DExH box RNA helicase NPH-II is required for ATP hydrolysis and RNA unwinding but not for RNA binding. J Virol. 1996;70:1706–1713. doi: 10.1128/jvi.70.3.1706-1713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J.L., Morgenstern K.A., Griffith J.P., Dwyer M.D., Thomson J.A., Murcko M.A., Lin C., Caron P.R. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 14.Gross C.H., Shuman S. The nucleoside triphosphate and helicase activities of vaccinia virus NPH-II are essential for virus replication. J. Virol. 1998;72:4729–4736. doi: 10.1128/jvi.72.6.4729-4736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang T.-H., Latus L.J., Liu Z., Abbott J.M. Genetic interactions of conserved regions in the DEAD-box protein Prp28p. Nucleic Acids Res. 1998;25:5033–5040. doi: 10.1093/nar/25.24.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svitkin Y.V., Pause A., Haghighat A., Pyronnet S., Witherell G., Belsham G.J., Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider S., Campodonico E., Schwer B. Motifs IV and V in the DEAH box splicing factor Prp22 are important for RNA unwinding, and helicase-defective Prp22 mutants are suppressed by Prp8. J. Biol. Chem. 2004;279:8617–8626. doi: 10.1074/jbc.M312715200. [DOI] [PubMed] [Google Scholar]
- 18.Iost I., Dreyfus M., Linder P. Ded1p, a DEAD-box protein required for translation initation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 19.O'Day C.L., Chavanikamannil F., Abelson J. 18S rRNA processing requires the RNA helicase-like protein Rrp3. Nucleic Acids Res. 1996;24:3201–3207. doi: 10.1093/nar/24.16.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kressler D., de la Cruz J., Rojo M., Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang W.-Q., Clark J.A., Fournier M.J. The rRNA-processing function of the yeast U14 small nucleolar RNA can be rescued by a conserved RNA helicase-like protein. Mol. Cell. Biol. 1997;17:4124–4132. doi: 10.1128/mcb.17.7.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venema J., Bousquet-Antonelli C., Gelugne J.-P., Caizergues-Ferrer M., Tollervey D. Rok1p is a putative RNA helicase required for rRNA processing. Mol. Cell. Biol. 1997;17:3398–3407. doi: 10.1128/mcb.17.6.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colley A., Beggs J., Tollervey D., Lafontaine D.L.J. Dhr1p, a putative DEAH-box RNA helicase is associated with the box C+D snoRNA U3. Mol. Cell. Biol. 2000;20:7238–7246. doi: 10.1128/mcb.20.19.7238-7246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daugeron M.C., Linder P. Characterization and mutational analysis of yeast Dbp8p, a putative RNA helicase involved in ribosome biogenesis. Nucleic Acids Res. 2001;29:1144–1155. doi: 10.1093/nar/29.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ripmaster T.L., Vaughn G.P., Woolford J.L., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc. Natl Acad. Sci. USA. 1992;89:11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weaver P.L., Sun C., Chang T.-H. Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol. Cell. Biol. 1997;17:1354–1365. doi: 10.1128/mcb.17.3.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daugeron M.C., Linder P. Dbp7p, a putative ATP-dependent RNA helicase of Saccharomyces cerevisiae is required for 60S ribosomal subunit assembly. RNA. 1998;4:566–581. doi: 10.1017/s1355838298980190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de la Cruz J., Kressler D., Rojo M., Tollervey D., Linder P. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA. 1998;4:1268–1281. doi: 10.1017/s1355838298981158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de la Cruz J., Kressler D., Tollervey D., Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kressler D., de la Cruz J., Rojo M., Linder P. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:1855–1865. doi: 10.1128/mcb.18.4.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burger F., Daugeron M.-C., Linder P. Dbp10p, a putative RNA helicase from Saccharomyces cerevisiae, is required for ribosome biogenesis. Nucleic Acids Res. 2000;28:2315–2323. doi: 10.1093/nar/28.12.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bond A.T., Mangus D.A., He F., Jacobson A. Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol. Cell. Biol. 2001;21:7366–7379. doi: 10.1128/MCB.21.21.7366-7379.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zagulski M., Kressler D., Becam A.M., Rytka J., Herbert C.J. Mak5p, which is required for the maintenance of the M1 dsRNA virus, is encoded by the yeast ORF YBR142w and is involved in the biogenesis of the 60S subunit of the ribosome. Mol. Genet. Genomics. 2003;270:216–224. doi: 10.1007/s00438-003-0913-4. [DOI] [PubMed] [Google Scholar]
- 34.Daugeron M.C., Kressler D., Linder P. Dbp9p, a putative ATP-dependent RNA helicase involved in 60S-ribosomal-subunit biogenesis, functionally interacts with Dbp6p. RNA. 2001;7:1317–1334. doi: 10.1017/s1355838201010640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oh J.Y., Kim J. ATP hydrolysis of the DEAD box protein Rok1p is required for in vivo ROK1 function. Nucleic Acids Res. 1999;27:2753–2759. doi: 10.1093/nar/27.13.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emery B., De La Cruz J., Rocak S., Deloche O., Linder P. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 2004;52:141–158. doi: 10.1111/j.1365-2958.2003.03973.x. [DOI] [PubMed] [Google Scholar]
- 37.Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schafer T., Kuster B., Tschochner H., Tollervey D., et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 38.Harnpicharnchai P., Jakovljevic J., Horsey E., Miles T., Roman J., Rout M., Meagher D., Imai B., Guo Y., Brame C., et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 39.Nissan T.A., Bassler J., Petfalski E., Tollervey D., Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bassler J., Grandi P., Gadal O., Lessmann T., Petfalski E., Tollervey D., Lechner J., Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;88:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 41.Muhlrad D., Hunter R., Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 42.Boeke J.D., Trueheart J., Natsoulis G., Fink G.R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 43.Company M., Arenas J., Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 44.Pugh G.E., Nicol S.M., Fuller-Pace F.V. Interaction of the Escherichia coli DEAD box protein DbpA with 23 S ribosomal RNA. J. Mol. Biol. 1999;292:771–778. doi: 10.1006/jmbi.1999.3112. [DOI] [PubMed] [Google Scholar]
- 45.Cordin O., Tanner N.K., Doere M., Linder P., Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Day C.-L., Dalbadie-McFarland G., Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J. Biol. Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- 47.Fuller-Pace F.V., Nicol S.M., Reid A.D., Lane D.P. DbpA: a DEAD box protein specifically activated by 23S rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogers G.W.J., Lima W.F., Merrick W.C. Further characterization of the helicase activity of eIF4A. Substrate specificity. J. Biol. Chem. 2001;276:12598–12608. doi: 10.1074/jbc.M007560200. [DOI] [PubMed] [Google Scholar]
- 49.Rozen F., Edery I., Meerovitch K., Dever T.E., Merrick W.C., Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogers G.W., Jr, Komar A.A., Merrick W.C. eIF4A: the godfather of the DEAD box helicases. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 51.Pause A., Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edwalds-Gilbert G., Kim D.H., Kim S.H., Tseng Y.H., Yu Y., Lin R.J. Dominant negative mutants of the yeast splicing factor Prp2 map to a putative cleft region in the helicase domain of DExD/H-box proteins. RNA. 2000;6:1106–1119. doi: 10.1017/s1355838200992483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider S., Hotz H.R., Schwer B. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J. Biol. Chem. 2002;277:15452–15458. doi: 10.1074/jbc.M112473200. [DOI] [PubMed] [Google Scholar]
- 54.Heilek G.M., Peterson M.G. A point mutation abolishes the helicase but not the nucleoside triphosphatase activity of hepatitis C virus NS3 protein. J. Virol. 1997;71:6264–6266. doi: 10.1128/jvi.71.8.6264-6266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blum S., Schmid S.R., Pause A., Buser P., Linder P., Sonenberg N., Trachsel H. ATP hydrolysis by initiation factor 4A is required for translation initiation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1992;89:7664–7668. doi: 10.1073/pnas.89.16.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmid S.R., Linder P. Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol. Cell. Biol. 1991;11:3463–3471. doi: 10.1128/mcb.11.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gorbalenya A.E., Koonin E.V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 58.Schmid S.R., Linder P. D-E-A-D protein family of putative RNA helicases. Mol. Microbiol. 1992;6:283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 59.Diges C.M., Uhlenbeck O.C. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hirling H., Scheffner M., Restle T., Stahl H. RNA helicase activity associated with the human p68 protein. Nature. 1989;339:562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- 61.Lorsch J.R., Herschlag D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 62.Gururajan R., Weeks D.L. An3 protein encoded by a localized maternal mRNA in Xenopus laevis is an ATPase with substrate-specific RNA helicase activity. Biochim. Biophys. Acta. 1997;1350:169–182. doi: 10.1016/s0167-4781(96)00155-8. [DOI] [PubMed] [Google Scholar]
- 63.Nishi K., Morel-Deville F., Hershey J.W.B., Leighton T., Schnier J. An eIF-4A-like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly [Erratum (1989) Nature, 340, 246.] Nature. 1988;336:496–498. doi: 10.1038/336496a0. [DOI] [PubMed] [Google Scholar]
- 64.Tsu C.A., Kossen K., Uhlenbeck O.C. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wagner J.D., Jankowsky E., Company M., Pyle A.M., Abelson J.N. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kikuma T., Ohtsu M., Utsugi T., Koga S., Okuhara K., Eki T., Fujimori F., Murakami Y. Dbp9p, a member of the DEAD box protein family, exhibits DNA helicase activity. J. Biol. Chem. 2004;279:20692–20698. doi: 10.1074/jbc.M400231200. [DOI] [PubMed] [Google Scholar]
- 67.Jankowsky E., Gross C.H., Shumann S., Pyle A.M. The DexH protein NPH-II is a processive and directional molecular motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 68.Schwer B., Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 69.Schwer B., Meszaros T. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 2000;19:6582–6591. doi: 10.1093/emboj/19.23.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin A., Schneider S., Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- 71.Plumpton M., McGarvey M., Beggs J.D. A dominant negative mutation in the conserved RNA helicase motif ‘SAT’ causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 1994;13:879–887. doi: 10.1002/j.1460-2075.1994.tb06331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Velankar S.S., Soultanas P., Dillingham M.S., Subramanya H.S., Wigley D.B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 73.Machius M., Henry L., Palnitkar M., Deisenhofer J. Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc. Natl Acad. Sci. USA. 1999;96:11717–11722. doi: 10.1073/pnas.96.21.11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Theis K., Chen P.J., Skorvaga M., Van Houten B., Kisker C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J. 1999;18:6899–6907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakagawa N., Sugahara M., Masui R., Kato R., Fukuyama K., Kuramitsu S. Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J. Biochem. 1999;126:986–990. doi: 10.1093/oxfordjournals.jbchem.a022566. [DOI] [PubMed] [Google Scholar]
- 76.Caruthers J.M., McKay D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]