Abstract
Characterization of the conformational landscapes for proteins with different secondary structures is important in elucidating the mechanism of protein folding. The folding trajectory of single-chain monellin composed of a five-stranded β-sheet and a helix was investigated by using a pH-jump from the alkaline unfolded to native state. The kinetic changes in the secondary structures and in the overall size and shape were measured by circular dichroism spectroscopy and small-angle x-ray scattering, respectively. The formation of the tertiary structure was monitored by intrinsic and extrinsic fluorescence. A significant collapse was observed within 300 μs after the pH-jump, leading to the intermediate with a small amount of secondary and tertiary structures but with an overall oblate shape. Subsequently, the stepwise formation of secondary and tertiary structures was detected. The current observation was consistent with the theoretical prediction that a more significant collapse precedes the formation of secondary structures in the folding of β-sheet proteins than that of helical proteins [Shea, J. E., Onuchic, J. N. & Brooks, C. L., III (2002) Proc. Natl. Acad. Sci. USA 99, 16064–16068]. Furthermore, it was implied that the initial collapse was promoted by the formation of some specific structural elements, such as tight turns, to form the oblate shape.
Keywords: energy landscape, protein folding, submillisecond dynamics, x-ray scattering
Characterization of the folding dynamics for proteins and their free energy landscapes offers a key to understanding protein folding phenomena (1, 2). The coarse-grained approximation of intraprotein interactions (a “funneled” landscape) has been proposed (3, 4) and has succeeded in explaining the structures of proteins in the folding transition state (5–8). However, the molecular basis of the funneled landscape, which is realized by a complex interplay of various intraprotein interactions and polypeptide dynamics, is largely unknown. For example, although the hydrophobic interaction and hydrogen bond (H-bond) are the major stabilizing interactions for the specific folded conformations of proteins, their contribution to the energy landscape, including the unfolded conformations, is controversial (9). Furthermore, the relationship between protein folds and the characteristics of landscapes has not been well investigated. Therefore, the observation of the entire folding process of proteins with various folds is required to characterize the free energy landscapes.
We previously observed the folding dynamics of α-helical proteins, cytochrome c (cyt c) and apomyoglobin (apoMb), based on helical content and compactness, which mainly reflect the formation of the H-bonds and hydrophobic interactions, respectively (10–12). The characterized landscapes of these proteins demonstrated the cooperative acquisition of the helical structure and compactness after the initial collapse and suggested that the hydrophobic environment created by the collapse facilitates the subsequent conformational search. Our observations were consistent with recent molecular dynamics calculations, in which helix formation and native tertiary packing occur simultaneously in the folding trajectories of helical proteins, including cyt c (13–15). Interestingly, Brooks and coworkers (15–17) further predicted that the initial collapse is more pronounced for proteins composed of β-sheets (β-sheet proteins). However, the prediction has not been experimentally verified, because the processes of compaction of β-sheet proteins have not been characterized at ambient temperature except for proteins with disulfide bonds (18). Thus, the purpose of this study is to characterize the folding landscapes of a β-sheet protein as an experimental test of the prediction by Brooks (15) and to investigate the implications of the collapse for the folding of β-sheet proteins.
We chose single-chain monellin (SMN) to research β-sheet proteins without a disulfide bond. SMN is genetically created by fusing two chains of monellin to retain its topology and activity (19). SMN is composed of an N-terminal strand (strand I), segregated helix, and four-stranded β-meander (strands II–V) separated by three tight β-turns (turns 1–3), as shown in Fig. 5, which is published as supporting information on the PNAS web site (20). To multilaterally observe the folding dynamics of SMN in the time domain from 300 μs to 5 s, we used solution-mixing devices combined with various methods. Circular dichroism (CD) spectroscopy can detect the amount of secondary structure. Small-angle x-ray scattering (SAXS) can monitor the compactness and shape of macromolecules that reflect the tertiary structure. The transient clustering of hydrophobic residues can be followed by monitoring the fluorescence from 1-anilinonaphthalene-8-sulfonate (ANS). Intrinsic fluorescence mainly from single Trp-4 is a good indicator of the environment of the N-terminal strand. By analyzing the kinetic information obtained from multiple structural probes, we characterized in detail the features of the folding of SMN.
Materials and Methods
Preparation of SMN. The SMN used in this study is slightly different from that reported by Lee et al. [SCM (20)] and contains Met at the N-terminal and a mutation of Cys-45 to Ser-45. SMN was prepared as described by Konno (19). Lyophilized SMN was dissolved in water and centrifuged to remove insoluble components. Unfolded SMN was prepared by adding appropriate amounts of 1 M NaOH to adjust the pH of the solution to 13.0. The reactions were initiated by mixing the solution of alkaline-unfolded SMN and 300 mM glycine buffer at a volume ratio of 5:1 to a final pH of 9.4. The protein concentrations were confirmed by measuring the absorption at 277 nm (ε277 = 1.46 × 104 M–1·cm–1) (19). All measurements were conducted at 20 ± 1°C except for the SAXS measurements, which were performed at 26 ± 1°C. The amount of SMN consumed for the entire measurements was ≈4 g.
CD Measurements. CD spectra were measured by using a CD spectrometer (J-720, Jasco, Easton, MD). The static samples contained 12 μM SMN and 50 mM glycine adjusted at various pHs between 7.0 and 13.0. The midpoint of unfolding and the number of protons involved are calculated from the Hill equation (21). Time-resolved CD spectra from 500 μs to 15 ms were collected by using the continuous-flow (CTF) mixer, as reported (12, 22), whose mixing efficiency was 98% at 130 μs. At least five scans were averaged, and the deformation of CD spectra was corrected as described (10). The refolding kinetics from 20 ms to 4 s was followed by using a stopped-flow (STF) mixer (Unisoku, Osaka) and the same condition used in the CTF experiments. At least 30 shots were performed and averaged. The concentration of SMN after mixing was 30 μM. The secondary structure contents were roughly evaluated by fitting the CD spectra to a linear combination of the standard spectra for the α-helix of poly(Glu), β-sheet of poly(Lys), and random structure of poly(Pro)II helix, with an error of ±8% for each.
Intrinsic Fluorescence of Trp-4. The kinetics of the intrinsic fluorescence intensity was obtained by excitation at 280 nm and by monitoring the emission passed through a 320-nm sharp-cut filter. The refolding kinetics from 1 to 10 ms was monitored with a CTF mixer used in a previous Raman study (23). The fluorescence from the channel was imaged onto a charge-coupled device (Princeton Instruments, Trenton, NJ). The mixing time of the apparatus estimated based on the fluorescence quench of N-acetyltryptophanamide by N-bromosuccinimide was 520 μs. The refolding kinetics from 15 ms to 5 s was monitored with a STF mixer (Unisoku, Hirakata, Japan) coupled with a photon-counting detector (Hamamatsu Photonics, Hamamatsu, Japan). The final concentration of SMN was 30 μM. The fluorescence intensities were normalized with respect to 30 μM SMN in 50 mM glycine buffer at pH 9.4.
Extrinsic Fluorescence of ANS. The kinetics of the fluorescence from ANS was monitored by excitation at 344 nm and by selecting the emission by using a 420-nm sharp-cut filter. ANS was contained in the refolding buffer, giving the final concentrations between 30 and 200 μM after mixing. The intensities were normalized relative to the fluorescence of the solution containing 80 μM ANS and 40 μM native SMN in 50 mM glycine buffer at pH 9.4. To confirm the negligible effect of the ANS binding to the folding of SMN, the following control experiments were conducted by monitoring the whole kinetic traces by using the same CTF (from 520 μs to 8 ms) and STF (from 10 ms to 5 s) mixers used for the Trp fluorescence measurements. First, the existence of ANS either with the refolding buffer or with unfolded SMN before the mixing did not alter the kinetic traces. Second, the time constants for the first and second kinetic phases obtained at the two SMN concentrations of 40 and 80 μM did not depend on the ANS concentration between 30 and 200 μM. Third, the time constants were invariant on the SMN concentrations at 40, 60, and 80 μM in the presence of 2.5 times excess of ANS.
SAXS Measurements. All SAXS measurements were performed at RIKEN beamline I (BL45XU) at SPring-8 (24). The refolding kinetics from 300 μs to 10 ms was monitored with the CTF mixer whose path length was 1 mm with 50-μm-thick kapton windows. The experimental setup was basically the same as reported (11). The consecutive observation of the sample and baseline scatterings without stopping the flow was essential to avoid artifacts in the baseline subtraction caused by small variations in the path length. The kinetics after 12 ms was monitored with the STF mixer as reported (11, 12). The final concentration of SMN was 3.7 mg·ml–1.
The radius of gyration (Rg) and the zero-angle scattering intensity (I(0)) were estimated based on the Guinier approximation (25):
 |
where S = (2sinθ)/λ; 2θ is the scattering angle; λ is the wavelength of the incident x-ray; and I(S) is the scattering intensity at S. The maximum S value for the Guinier fitting was determined by the criterion 2π RgS < 1.4. The Rg values for native and unfolded states described in the text are the values extrapolated to the infinite dilution. The scattering intensities were scaled by the data collection time, x-ray intensity, SMN concentration, and scattering intensity of the standard protein solution.
The pair-distribution function, P(r), was calculated by using the gnom package (26). In brief, a series of trial P(r)s was calculated with various values of maximum r (Dmax) at 1-Å resolution for a scattering profile whose S range was limited from 0.0052 to 0.040 Å–1. Among the trial P(r)s that can reproduce the original scattering with a reasonably small rms difference, the P(r) giving the smallest Dmax was selected. The reliability of the selected P(r) was confirmed by comparing the Rg estimated from the Guinier fitting of the original scattering with the Rg calculated from P(r). The errors in the ordinate values of P(r) were within 3% at each data point.
Results
Equilibrium Unfolding of SMN. We first characterized the equilibrium unfolding properties of SMN to determine the initial and final conditions for the kinetic folding experiments. A negative peak at 213 nm in the CD spectrum of SMN at pH 9.4 (Fig. 1A, black line) is characteristic of β-sheet proteins. A positive band ≈233 nm likely reflects the contribution of aromatic side chains (27). SMN at pH 9.4 retains the compact conformation because of a small Rg (15.8 ± 0.2 Å at the infinite dilution) in the static SAXS measurements. In contrast, SMN at pH 13.0 possesses the expanded and unfolded conformation as indicated in the CD spectrum devoid of secondary structures (Fig. 1 A, blue line) and by the expanded Rg (25.5 ± 0.5 Å at the infinite dilution). The alkaline-induced unfolding of SMN monitored by CD at 216 nm ([θ216]) indicates the midpoint of transition is pH 11.1 ± 0.03 and the number of protons involved is 3.2 (not shown). Because the unfolding transition is fully reversible, the kinetic process of the transition from the unfolded to the native state can be initiated by a pH-jump from 13.0 to 9.4.
Fig. 1.
The time-dependent changes in the CD data for the refolding of SMN initiated by the pH-jump from 13.0 to 9.4. The concentration of SMN after the mixing was 30 μM. (A) The time-resolved CD spectra of the refolding of SMN. For clarity, only two time points, 300 μs (red circles) and 100 ms (green diamonds), are presented. The CD spectra of the native (black line) and unfolded (blue line) conformations measured at pH 13.0 and 9.4, respectively, are also presented. The α-helix contents estimated from the spectra for the unfolded SMN, at 300 μs and 100 ms and for the native SMN were 3%, 2%, 9%, and 9%, respectively. The β-sheet contents were 0%, 4%, 13%, and 43% in the same order. (B) The refolding kinetics of SMN measured by [θ216]. Data obtained by the CTF and STF methods are represented by open circles and a thin line, respectively. The open and filled squares represent [θ216] for the unfolded and native states, respectively. The kinetic data were fitted by a double exponential with time constants of 14 ± 3 ms and 1.3 ± 0.4 s (thick line). The amplitudes of the first and second kinetic phases were –2,930 ± 230 and –2,220 ± 170 deg·cm2·dmol–1, respectively.
Stepwise Secondary Structure Formation Monitored by CD Spectroscopy. To observe the overall dynamics of the folding of SMN, we next monitored the kinetic changes of secondary structure upon the refolding of alkaline-unfolded SMN. We followed the kinetic changes of [θ216] after the pH-jump from 13.0 to 9.4 (Fig. 1B). The CTF and STF systems were used to cover the time windows from 500 μs to 15 ms and from 20 ms to 4 s, respectively. The two traces obtained independently were connected seamlessly and exhibited two distinct phases, the first with a time constant of 14 ± 3 ms (amplitude of 57% of the total change) and the second with a time constant of 1.3 ± 0.4 s (43%). The observed kinetics therefore accounts for the total change in [θ216]; however, a close examination of the kinetic CD spectrum immediately after the mixing suggests an additional conformational change. As shown in Fig. 1 A, the kinetic CD spectrum at 300 μs (red circles) is distinct from that of the initial unfolded state (blue line) beyond the experimental errors in the regions around 233 nm and under 215 nm. The rapid changes in the CD spectrum suggest changes in the main chain conformation. Thus, the kinetic process of the folding likely consists of three phases: the burst phase occurring within the mixing dead time (<300 μs) and the first and second phases occurring with the time constants of ≈14 ms and 1.3 s, respectively. These results correspond to the stepwise formation of secondary structures. We observed that the kinetic trace after 20 ms is invariant upon changes in the SMN concentration from 20 to 50 μM (not shown), suggesting that the second phase is not caused by a formation of multimeric components.
Folding Kinetics Monitored by Intrinsic and Extrinsic Fluorescence. To obtain information on the conformational changes of N-terminal strand, changes in the intrinsic fluorescence intensity from Trp-4 were observed. Unfolded SMN at pH 13.0 exhibits a weaker fluorescence intensity (open square, Fig. 2A) than native SMN (filled square), indicating the exposure of Trp-4 to solvent and the deprotonation of the indole group in the alkaline condition. In contrast, the stronger intensity for the native state likely corresponds to the protonation of Trp-4 and its partial burial in the nearby residues (20). The kinetic fluorescence trace for the folding of SMN can be fitted by a double exponential function with time constants of 8.0 ± 0.2 ms and 0.89 ± 0.06 s (Fig. 2 A), which correspond to those of the first and second phases observed in the kinetic CD trace, respectively. Thus, although the burst-phase change could not be detected, the first and second phases were confirmed and demonstrated the stepwise formation of the tertiary contacts around Trp-4.
Fig. 2.
The refolding kinetics of SMN induced by the pH-jump from 13.0 to 9.4. (A) A representative trace measured by the intrinsic fluorescence from Trp-4. The final concentration of SMN was 30 μM. Data obtained by the CTF and STF methods are represented by open circles and a thin line, respectively. The fluorescence intensities for the initial (open square) and final (filled square) conformations were measured at pH 13.0 and 9.4, respectively. The kinetic data were fitted by a double exponential with time constants of 8.0 ± 0.2 ms and 0.89 ± 0.06 s (thick line). (B) A representative trace measured by the extrinsic fluorescence from ANS. The final concentrations of SMN and ANS were 40 and 100 μM, respectively. The intensities for the initial and final states (open and filled squares) were measured at pH 13.0 and 9.4, respectively. The kinetic data obtained by the CTF and STF methods are represented by open circles and a thin line, respectively. The kinetic data were fitted by a double exponential with time constants of 19 ± 1 ms and 1.3 ± 0.1 s (thick line). (C) The titration of the kinetic ANS fluorescence intensities at 100 ms against the concentration of ANS. Open circles and filled triangles indicate measurements for the SMN concentrations of 40 and 80 μM, respectively, with various concentrations of ANS.
The first and second phases in the folding of SMN and their structural changes were also reflected in the changes in ANS fluorescence. The fluorescence from ANS is amplified upon its binding to hydrophobic surfaces or clusters that are characteristics of partially structured proteins (28, 29). We observed a small amplification of the ANS fluorescence with both the native (Fig. 2B, filled square) and unfolded (Fig. 2B, open square) SMN and confirmed that the binding of ANS with these conformations is weak. In contrast, the kinetic experiments exhibited an increase in intensity with a time constant of 19 ± 1 ms and a decrease with a time constant of 1.3 ± 0.1 s (Fig. 2B). These time constants were essentially independent of the ANS concentration between 30 and 200 μM, indicating that both the first and second phases are not rate-limited by the ANS binding. The similarity in the time constants obtained in CD, Trp, and ANS fluorescence measurements strongly supports that the ANS binding does not perturb the folding of SMN. In addition, the time constants for first and second phases were not affected by the concentration of SMN between 40 and 80 μM (not shown), supporting that the two kinetic phases cannot be explained by a formation of multimeric components. The kinetic titration of ANS with SMN monitored at the transient fluorescence intensity at 100 ms evidences the transient binding of ANS to SMN in a 1:1 manner (Fig. 2C), excluding the possibility of a nonspecific binding of ANS to SMN. We interpret that the changes in ANS fluorescence correspond to the formation and burial of the single ANS-binding site with the time constant of 19 ms and 1.3 s, respectively.
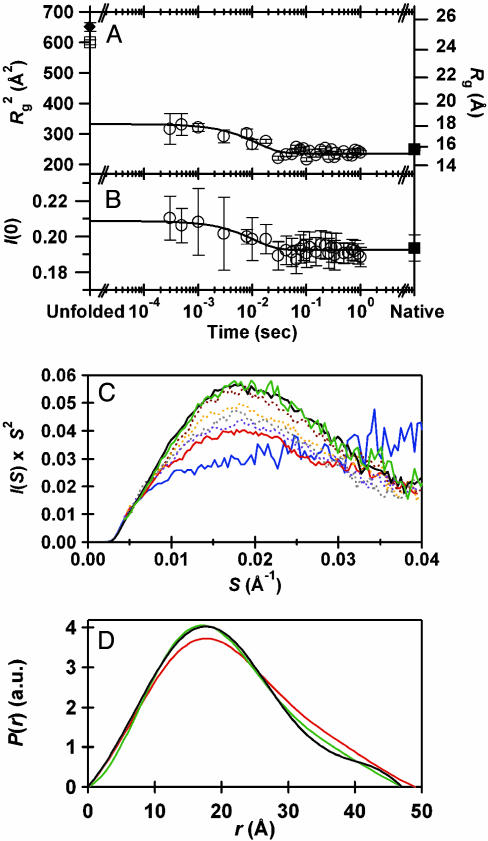
Rapid Chain Condensation Monitored by SAXS. Finally, to monitor the compaction and overall shape of the main chain in the folding of SMN, we conducted time-resolved SAXS measurements by using the synchrotron x-ray source. We estimated the kinetic changes in Rg and I(0) based on a Guinier analysis of the scattering data, which were plotted in Fig. 3 A and B, respectively. The plot for Rg demonstrates the burst phase contraction of the main chain within 300 μs, followed by the slower contraction, which can be fitted by a single exponential with a time constant of 12 ± 3 ms (Fig. 3A). The obtained time constant is comparable to the first phase observed in the CD measurements (14 ms). Extrapolations of the fitted trace to zero and infinite times yield a Rg of 18.2 ± 0.4 Å and 15.4 ± 0.1 Å, respectively. Although the former value is ≈6 Å smaller than that of the initial unfolded state (24.3 Å), the latter value is similar to that of the native conformation (15.8 Å). Thus, the kinetic SAXS data demonstrated the burst and first kinetic phases involving ≈6 and ≈3 Å of compaction, respectively. The change in I(0) can also be fitted by an exponential function with a time constant of 10 ms, consistent with that of the first kinetic phase (Fig. 3B).
Fig. 3.
The time-dependent changes in the SAXS data during the folding of SMN initiated by the pH-jump from 13.0 to 9.4. The final concentration of SMN was 3.7 mg·ml–1. (A) The time course of the change in the Rg (open circles) obtained from the Guinier analysis of the SAXS profiles. The filled diamond and square represent the Rgs for the unfolded and native states at an infinite dilution, respectively. The Rg value for the unfolded SMN determined at 3.7 mg·ml–1 was 24.3 ± 0.4 Å (open square). The line represents a single exponential fitting (13 ± 6 ms) of the kinetic data from 300 μs to 1 s.(B) The time course of the change in the I(0) (open circles). The line represents a single exponential fitting with a time constant of 10 ± 6 ms. The filled square denotes I(0) for the native conformation obtained in the same solution. (C) The time-resolved Kratky profiles for the same folding process. For clarification, eight representative profiles are presented, which were obtained for the native (black line) and unfolded (blue) conformations and for the time-resolved data at 300 μs (red), 1 ms (dotted purple), 3 ms (dotted gray), 8 ms (dotted orange), 10 ms (dotted brown), and 100 ms (green). (D) The pair distribution functions [P(r)] calculated from the scattering profiles. The black line corresponds to P(r) for the native state, whose maximum r (Dmax) is 47Å. The red and green lines correspond to P(r)s for 300 μs and 100 ms, respectively. Dmax at 300 μs and 100 ms are 49 and 47 Å, respectively. P(r)s were normalized to have a constant area.
The molecular shape of SMN can be deduced from the Kratky plots of the scattering profiles (Fig. 3C). The Kratky plot of unfolded SMN possesses a plateau in the moderate angle region (dotted blue line), which is indicative of a random coil (30). In contrast, a peak found in the scattering at 300 μs (red line) demonstrates the formation of a globular domain (30). The single peak of the plot also excludes the accumulation of the putative dimer that should exhibit a shoulder at a shorter value of S. Interestingly, the plot at 100 ms (green line) is very similar in shape to that of SMN in the native state, suggesting that the overall molecular shape of SMN at 100 ms is similar to the native conformation. The changes in shape are also seen in the pair distribution function, P(r), which represents the distribution of distances between every pair of atoms (Fig. 3D). The P(r) function for native SMN (black line) shows a peak at ≈18 Å and a shoulder above ≈30 Å that reflect the oblate shape for SMN in the native state. The P(r) function at 100 ms (green line) is similar to that of native SMN, confirming the similarity in overall structure at 100 ms and for the native conformation. The P(r) function at 300 μs (red line) suggests a collapse, because the most frequent r value and Dmax are identical with and ≈2 Å larger than those of the native structure, respectively. The small difference in Dmax at 300 μs and for the native state excludes a possible presence of expanded loops. Therefore, it is most likely that SMN at 300 μs is slightly expanded compared with the native conformation but already possesses a oblate overall shape.
Discussion
Sequential Folding Scheme of SMN. We observed three kinetic phases in the folding of SMN by using CD, SAXS, Trp, and ANS fluorescence. The burst phase was demonstrated in the SAXS data and completed within 300 μs. The first and second phases detected by the other methods occurred with time constants of ≈14 ms and ≈1.3 s, respectively. These observations can be explained by the sequential folding scheme (Scheme 1), in which the formations of the two intermediates, I1 and I2, and the native state, N, correspond to the burst, first, and second phases, respectively. As explained in Supporting Text, which is published as supporting information on the PNAS web site, the scheme is the most consistent and simplest model to explain the current observations. Because the apparent rate constants for the phases are well separated, the kinetic data obtained at 300 μs and 100 ms can be considered to represent the data for I1 and I2, respectively.
Scheme 1.
L-Shaped Conformational Landscape of SMN. The folding dynamics of SMN can be separated into distinct structural events based on the sequential folding scheme. The burst phase occurs within 300 μs and forms I1 possessing no binding site for ANS and no tertiary contacts around Trp-4. The total amount of secondary structure for I1, roughly estimated as the sum of α-helix and β-sheet contents from the CD spectrum at 300 μs, is ≈6% and is comparable to that of U (≈3%). In contrast, the SAXS data demonstrated that I1 possesses a collapsed main chain and suggested that its overall shape is oblate. The analysis of the scattering data based on the triaxial ellipsoid approximation was also consistent with an oblate shape for I1, which is slightly expanded compared with N (Fig. 6, which is published as supporting information on the PNAS web site). Thus, the burst phase of the folding can be described as a significant collapse to form the loosely packed and oblate shape by using a small number of secondary and tertiary structures. Such a rapid collapse has been observed in the formation of intermediates for some β-sheet proteins (31–33). For example, the burst-phase intermediates of dihydrofolate reductase and IL-1β show significant protection of the main chain hydrogens in the hydrophobic core of the β-sheet, suggesting an initial collapse (31, 32). A rapid collapse without significant tertiary structures was observed in the refolding of ubiquitin conducted at subzero temperature (33), although the protein is the two-state folder at room temperature (34). The current study unequivocally and quantitatively demonstrated that a significant collapse is the initial folding phase for a β-sheet protein without S–S bonds.
The second component, I2, is formed with a time constant of ≈14 ms and possesses a further collapsed conformation whose overall shape is very similar to that of the native SMN. I(0) decreases to the native level in the first phase, which suggests a decrease in the amount of hydrated waters in the transition from I1 to I2 (12, 35–37). I2 possesses partial tertiary contacts demonstrated by the specific binding site for ANS and the increased Trp-4 fluorescence. The total content of secondary structures estimated from the CD spectrum at 100 ms is ≈22% and is smaller than that of N (≈52%). Thus, the transition from I1 to I2 can be described as the process whereby the partial tertiary structure and the overall structural framework of SMN are formed. We suggest that the overall fold of I2 might be very similar to that of the native conformation; however, the H-bond network for the β-sheet has not been correctly developed.
The final phase, corresponding to the transition from I2 to N, occurs with a time constant of ≈1.3 s. Because [θ216] and the fluorescence intensity of Trp-4 develop in this phase, the formation of main chain H-bonds and the complete side chain packing characterize the final phase in the folding of SMN. The final folding phase for β-sheet proteins is frequently the simultaneous formation of the β-sheet and tertiary structure (38–40). Furthermore, the formation of the native structure is generally slower for β-sheet proteins than for helical proteins, as demonstrated by the correlation found for the rates of formation of native states from intermediates against the absolute contact orders (41). The significant energy barrier confirmed for the final phase in the folding of SMN might be explained by a breakage of the potential nonnative contacts formed in I2 having the collapsed conformation without the correct H-bond network.
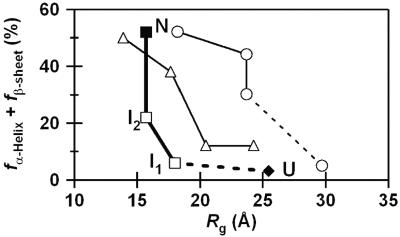
To demonstrate the remarkable differences between helical and β-sheet proteins, the folding trajectories of SMN, apoMb, and cyt c were compared in the 2D space defined by Rg and the amount of secondary structure (Fig. 4) (11, 12). The L-shaped trajectory of SMN contrasts with the more diagonal trajectories of helical proteins, indicating that the folding landscapes are distinct for proteins composed of different secondary structures. Although both cyt c and apoMb exhibit the cooperative acquisition of helices and Rg after the initial collapse, SMN shows extensive collapse (≈70% of the total change) with a small increase in the content of secondary structure in the initial stage and subsequently builds up its β-sheet in the collapsed conformation. Brooks and coworkers (14, 15) found for the helical proteins that native tertiary and secondary structures are formed more or less commensurately. In contrast, their calculations on β-sheet proteins showed L-shaped energy surfaces (15–17). Our results are consistent with the theoretical prediction and suggest the importance of the initial collapse for the β-sheet formation.
Fig. 4.
Comparison of the folding trajectories for SMN, apoMb, and cyt c in the 2D conformational space defined by secondary structural content (the sum of the helix and β-sheet contents) and Rg. The filled diamond and square represent the unfolded (U) and native (N) conformations of SMN, respectively. The open squares correspond to the kinetic intermediates of SMN (I1 and I2). The open circles and triangles correspond to the folding trajectories for apoMb and cyt c, respectively, determined previously (11, 12).
Origins of the L-Shaped Landscape of SMN. To understand the origins of the rapid collapse in SMN, we first noticed that I1 possesses no binding site for ANS and only a small content of secondary structure. These observations seemingly suggest that the significant collapse reflects the nonspecific condensation of hydrophobic elements (42) and the possibility that the hydrophobicity of SMN might be significantly higher than that of apoMb or cyt c. However, the average hydrophobicity of SMN (–0.85) calculated with the Kyte–Doolittle scale (43) is comparable to that of cyt c (–0.85) and smaller than that of apoMb (–0.37). In fact, the primary sequence of SMN is less hydrophobic than those of two-state proteins that fold without an initial collapse, such as the IgG-binding domain of protein L (–0.53), ubiquitin (–0.60), and common-type acylphosphatase (–0.62). Accordingly, it is difficult to interpret the initial collapse only in terms of a nonspecific and random event. In addition, the oblate shape of I1 is less likely to be formed as the result of a nonspecific random collapse. The collapse dynamics of SMN should involve the formation of specific structural elements to create the oblate shape of I1.
We propose the following mechanisms to explain the initial folding dynamics of SMN. The native-like overall shape of I1 can be naturally explained if we assume the formation of tight turns in SMN and an extended conformation between the turns. In line with this possibility, turn structures have been demonstrated as a part of folding nucleation sites for many β-sheet proteins that fold in the two-state manner (44–48). In addition, β-sheet proteins generally possess many middle-range contacts separated by 10–30 residues in the primary sequence due to neighboring strands extending from tight turns (49). Thus, once the tight turns are formed, they might induce the formation of the middle-range contacts without the correct H-bond network. The intrinsic length limits of the two-stranded sheet extending from a tight turn might also explain the compactness of I1 (50). The initial formation of tight turns is reminiscent of the classical proposal that two-stranded β-ribbons might be formed initially in Greek key β-barrels (51). We propose that the specific collapse observed in SMN would be important for the construction of the topology of other β-sheet proteins.
Supplementary Material
Acknowledgments
This study was supported by Grants-in-Aid for Scientific Research (to S.T., K.I., and I.M.) and Special Coordination Funds for Promoting Science and Technology (to T.F.) from the Ministry of Education, Culture, Sports, Science and Technology. T.K. is supported by the fellowship of the Japan Society for the Promotion of Science to Young Scientists. S.A. is supported by the Special Postdoctoral Researchers Program of RIKEN.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SMN, single-chain monellin; CD, circular dichroism; SAXS, small-angle x-ray scattering; Rg, radius of gyration; I(0), zero-angle scattering intensity; ANS, 1-anilinonaphthalene-8-sulfonate; apoMb, apomyoglobin; cyt c, cytochrome c; H-bond, hydrogen bond; CTF, continuous-flow; STF, stopped-flow.
References
- 1.Fersht, A. R. (1999) Structure and Mechanism in Protein Science (Freeman, New York).
- 2.Kuhlman, B. & Baker, D. (2004) Curr. Opin. Struct. Biol. 14, 89–95. [DOI] [PubMed] [Google Scholar]
- 3.Go, N. (1983) Annu. Rev. Biophys. Bioeng. 12, 183–210. [DOI] [PubMed] [Google Scholar]
- 4.Bryngelson, J. D., Onuchic, J. N., Socci, N. D. & Wolynes, P. G. (1995) Proteins 21, 167–195. [DOI] [PubMed] [Google Scholar]
- 5.Daggett, V. & Fersht, A. R. (2003) Trends Biochem. Sci. 28, 18–25. [DOI] [PubMed] [Google Scholar]
- 6.Onuchic, J. N. & Wolynes, P. G. (2004) Curr. Opin. Struct. Biol. 14, 70–75. [DOI] [PubMed] [Google Scholar]
- 7.Munoz, V. & Eaton, W. A. (1999) Proc. Natl. Acad. Sci. USA 96, 11311–11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koga, N. & Takada, S. (2001) J. Mol. Biol. 313, 171–180. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin, R. L. (2003) J. Biol. Chem. 278, 17581–17588. [DOI] [PubMed] [Google Scholar]
- 10.Akiyama, S., Takahashi, S., Ishimori, K. & Morishima, I. (2000) Nat. Struct. Biol. 7, 514–520. [DOI] [PubMed] [Google Scholar]
- 11.Akiyama, S., Takahashi, S., Kimura, T., Ishimori, K., Morishima, I., Nishikawa, Y. & Fujisawa, T. (2002) Proc. Natl. Acad. Sci. USA 99, 1329–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uzawa, T., Akiyama, S., Kimura, T., Takahashi, S., Ishimori, K., Morishima, I. & Fujisawa, T. (2004) Proc. Natl. Acad. Sci. USA 101, 1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardenas, A. E. & Elber, R. (2003) Proteins 51, 245–257. [DOI] [PubMed] [Google Scholar]
- 14.Guo, Z., Brooks, C. L., III, & Boczko, E. M. (1997) Proc. Natl. Acad. Sci. USA 94, 10161–10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brooks, C. L., III (2002) Acc. Chem. Res. 35, 447–454. [DOI] [PubMed] [Google Scholar]
- 16.Shea, J. E., Onuchic, J. N. & Brooks, C. L., III (2002) Proc. Natl. Acad. Sci. USA 99, 16064–16068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheinerman, F. B. & Brooks, C. L., III (1998) Proc. Natl. Acad. Sci. USA 95, 1562–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segel, D. J., Bachmann, A., Hofrichter, J., Hodgson, K. O., Doniach, S. & Kiefhaber, T. (1999) J. Mol. Biol. 288, 489–499. [DOI] [PubMed] [Google Scholar]
- 19.Konno, T. (2001) Protein Sci. 10, 2093–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, S. Y., Lee, J. H., Chang, H. J., Cho, J. M., Jung, J. W. & Lee, W. T. (1999) Biochemistry 38, 2340–2346. [DOI] [PubMed] [Google Scholar]
- 21.Cantor, C. R. & Schimmel, P. R. (1980) Biophysical Chemistry, Part III (Freeman, New York).
- 22.Kimura, T., Takahashi, S., Akiyama, S., Uzawa, T., Ishimori, K. & Morishima, I. (2002) J. Am. Chem. Soc. 124, 11596–11597. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi, S., Yeh, S. R., Das, T. K., Chan, C. K., Gottfried, D. S. & Rousseau, D. L. (1997) Nat. Struct. Biol. 4, 44–50. [DOI] [PubMed] [Google Scholar]
- 24.Fujisawa, T., Inoue, K., Oka, T., Iwamoto, H., Uruga, T., Kumasaka, T., Inoko, Y., Yagi, N., Yamamoto, M. & Ueki, T. (2000) J. Appl. Crystallogr. 33, 797–800. [Google Scholar]
- 25.Guinier, A. & Fournet, G. (1955) Small Angle Scattering of X-Rays (Wiley, New York).
- 26.Semenyuk, A. V. & Svergun, D. I. (1991) J. Appl. Crystallogr. 24, 537–540. [Google Scholar]
- 27.Vuilleumier, S., Sancho, J., Loewenthal, R. & Fersht, A. R. (1993) Biochemistry 32, 10303–10313. [DOI] [PubMed] [Google Scholar]
- 28.Semisotnov, G. V., Rodionova, N. A., Kutyshenko, V. P., Ebert, B., Blanck, J. & Ptitsyn, O. B. (1987) FEBS Lett. 224, 9–13. [DOI] [PubMed] [Google Scholar]
- 29.Maki, K., Cheng, H., Dolgikh, D. A., Shastry, M. C. & Roder, H. (2004) J. Mol. Biol. 338, 383–400. [DOI] [PubMed] [Google Scholar]
- 30.Glatter, O. & Kratky, O. (1982) Small Angle X-Ray Scattering (Academic, London).
- 31.Jones, B. E. & Matthews, C. R. (1995) Protein Sci. 4, 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heidary, D. K., Gross, L. A., Roy, M. & Jennings, P. A. (1997) Nat. Struct. Biol. 4, 725–731. [DOI] [PubMed] [Google Scholar]
- 33.Larios, E., Li, J. S., Schulten, K., Kihara, H. & Gruebele, M. (2004) J. Mol. Biol. 340, 115–125. [DOI] [PubMed] [Google Scholar]
- 34.Jacob, J., Krantz, B., Dothager, R. S., Thiyagarajan, P. & Sosnick, T. R. (2004) J. Mol. Biol. 338, 369–382. [DOI] [PubMed] [Google Scholar]
- 35.Svergun, D. I., Richard, S., Koch, M. H., Sayers, Z., Kuprin, S. & Zaccai, G. (1998) Proc. Natl. Acad. Sci. USA 95, 2267–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, L., Wildegger, G., Kiefhaber, T., Hodgson, K. O. & Doniach, S. (1998) J. Mol. Biol. 276, 225–237. [DOI] [PubMed] [Google Scholar]
- 37.Arai, M., Ito, K., Inobe, T., Nakao, M., Maki, K., Kamagata, K., Kihara, H., Amemiya, Y. & Kuwajima, K. (2002) J. Mol. Biol. 321, 121–132. [DOI] [PubMed] [Google Scholar]
- 38.Jones, B. E., Beechem, J. M. & Matthews, C. R. (1995) Biochemistry 34, 1867–1877. [DOI] [PubMed] [Google Scholar]
- 39.Parker, M. J., Dempsey, C. E., Lorch, M. & Clarke, A. R. (1997) Biochemistry 36, 13396–13405. [DOI] [PubMed] [Google Scholar]
- 40.Mizuguchi, M., Kroon, G. J., Wright, P. E. & Dyson, H. J. (2003) J. Mol. Biol. 328, 1161–1171. [DOI] [PubMed] [Google Scholar]
- 41.Kamagata, K., Arai, M. & Kuwajima, K. (2004) J. Mol. Biol. 339, 951–965. [DOI] [PubMed] [Google Scholar]
- 42.Krantz, B. A., Mayne, L., Rumbley, J., Englander, S. W. & Sosnick, T. R. (2002) J. Mol. Biol. 324, 359–371. [DOI] [PubMed] [Google Scholar]
- 43.Kyte, J. & Doolittle, R. F. (1982) J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- 44.Walkenhorst, W. F., Edwards, J. A., Markley, J. L. & Roder, H. (2002) Protein Sci. 11, 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferguson, N. & Fersht, A. R. (2003) Curr. Opin. Struct. Biol. 13, 75–81. [DOI] [PubMed] [Google Scholar]
- 46.Viguera, A. R. & Serrano, L. (2003) Proc. Natl. Acad. Sci. USA 100, 5730–5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krantz, B. A., Dothager, R. S. & Sosnick, T. R. (2004) J. Mol. Biol. 337, 463–475. [DOI] [PubMed] [Google Scholar]
- 48.Deechongkit, S., Nguyen, H., Powers, E. T., Dawson, P. E., Gruebele, M. & Kelly, J. W. (2004) Nature 430, 101–105. [DOI] [PubMed] [Google Scholar]
- 49.Gromiha, M. M. & Selvaraj, S. (2004) Prog. Biophys. Mol. Biol. 86, 235–277. [DOI] [PubMed] [Google Scholar]
- 50.Stanger, H. E., Syud, F. A., Espinosa, J. F., Giriat, I., Muir, T. & Gellman, S. H. (2001) Proc. Natl. Acad. Sci. USA 98, 12015–12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richardson, J. S. (1981) Adv. Protein Chem. 34, 167–339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.