Abstract
Based on a variety of genetic, biochemical, and neuropathological evidence, amyloid-β peptide (Aβ) has been suggested to be causal in Alzheimer's disease (AD). Aβ has been shown to mediate neurodegenerative and inflammatory changes associated with amyloid plaques, as well as exert direct neurotoxicity through oligomeric forms of Aβ. The mechanism of Aβ toxicity, however, remains largely unknown. In this work, we show that an early event after exposure of postmitotic neurons to Aβ is tyrosine phosphorylation of FISH adapter protein. FISH binds to and potentially regulates certain ADAM family members. We present evidence that FISH and ADAM12 mediate the neurotoxic effect of Aβ. Expression of an ADAM12 protease-deficient mutant (ADAM12ΔMP) blocks Aβ-induced neuronal death, and expression of an N-terminal fragment of FISH reduces Aβ toxicity. The C-terminal fragment of FISH containing the ADAMs binding region is found to be sufficient for induction of neuronal death, which is prevented by coexpression of the ADAM12ΔMP. Aβ treatment, as well as expression of the C-terminal toxic FISH fragment, induces accumulation of ADAM12 N-terminal cleavage product in conditioned medium, demonstrating activation of the ADAM metalloprotease/sheddase activity. ADAM12 protein is reduced in AD brains, pointing to a possible increase in ADAM12 proteolytic activity. These data suggest that Aβ toxicity is mediated by FISH and ADAM12 and may provide insights into therapeutic strategies for AD treatment.
Keywords: Alzheimer's disease
Alzheimer's disease (AD) is characterized by accumulation of amyloid-β peptide (Aβ), tau phosphorylation, paired helical filament formation, and massive neurodegeneration (1), which is thought to result from the deposition of Aβ in the cortex and hippocampus. It has been shown that Aβ induces neuronal death in vitro (2), and several proteins have been proposed for the role of the specific Aβ receptor (3–6), although little information is available about early steps of the signaling pathway that mediates Aβ neurotoxicity (7). c-Jun N-terminal kinase as well as the p38 kinase cascades have been implicated as downstream effectors mediating the Aβ-induced neuronal death (8–11). Several reports have suggested that activation of tyrosine kinases may play a role in Aβ-induced neuronal death (12, 13) or may have a neuroprotective function, as was shown for nicotinic receptor-dependent Jak2 kinase activation, which is induced by nicotine, but not Aβ, binding to the receptor (14). Some other putative Aβ receptors are thought to signal through G proteins (5, 15), which might implicate tyrosine kinase activity in downstream signaling (reviewed in ref. 16), although the kinase substrates functionally involved in Aβ toxicity are not known. In an attempt to identify these substrates in an in vitro model of Aβ toxicity, we used a panel of anti-phosphotyrosine site-specific antibodies to probe a whole-cell lysate (WCL) of Aβ-treated and control human cortical cultures (HCC). Because our search for proteins phosphorylated on tyrosine residues was limited to commercially available anti-phosphotyrosine-specific antibodies, many potentially interesting proteins were excluded, such as tau, for which Aβ-induced tyrosine phosphorylation in neuronal culture has been reported previously (13). c-Jun N-terminal kinase and p38 stimulation was detected in our model (data not shown) in accordance with previous reports (8, 10).
Among the antibodies tested, anti-phospho-epidermal growth factor receptor (EGFR) detected phosphorylation of a protein distinct from EGFR. No other phosphospecific anti-receptor tyrosine kinase antibodies detected increased levels of phosphorylation. We isolated the phosphoprotein and identified it as the FISH (five SH3 domains) adapter protein. FISH was initially cloned in a screen for Src kinase substrates (17); it contains an N-terminal PX domain and five SH3 domains distributed throughout the rest of the protein. The most C-terminal SH3 domain (SH3#5) has been shown to bind the cytoplasmic domain of three members of the ADAMs family (a disintegrin and metalloprotease): ADAM12, -15, and -19 (18). Tyrosine phosphorylation of FISH plays a regulatory role in the protein localization, because it was shown that FISH is maintained in the cytoplasm by intramolecular interaction of the PX domain with the third SH3 domain (SH3#3). Phosphorylation disrupts this interaction, resulting in redistribution of the protein from cytoplasmic to perimembrane localization, thus enabling FISH binding to ADAMs (18).
We demonstrate that FISH can stimulate ADAMs metalloprotease activity and therefore propose that increased proteolytic activity of ADAMs is an intermediate step in the Aβ neurotoxicity signaling pathway.
Materials and Methods
Materials. Rabbits were immunized with bacterially produced GSTFISH723–1105 to obtain anti-FISH antibodies, which were purified by protein A chromatography. The reagents used from commercial sources were as follows: anti-phospho-EGFR, phospho-ErbB2 antibodies, and other site-specific anti-phosphotyrosine antibodies (Cell Signaling Technology, Beverly, MA), mouse anti-EGFR (Upstate Biotechnology, Lake Placid, NY), rabbit anti-EGFR (Cell Signaling Technology), goat anti-FISH antibody (Santa Cruz Biotechnology), anti-phosphotyrosine antibodies (US Biological, Swampscott, MA and Sigma–Aldrich), anti-ADAM12 and ADAM19 antibodies (Sigma–Aldrich; BioDesign, Kennebunk, ME; Chemicon; and US Biological), anti-80 K-H (BD Biosciences), anti-His tag antibodies (Novagen), PD153035 (Calbiochem), SP FF cation exchange and PD-10 columns (Amersham Pharmacia Biologicals), Alamar Blue (BioSource International, Camarillo, CA), protease inhibitor set (Roche Molecular Biology), and leukocyte antigen-related phosphatase (BioMol, Plymouth Meeting, PA).
HCC Preparation and Treatment. Human fetal cerebral cortical tissue (Advanced Bioscience Resources, Alameda, CA) between 13 and 16 weeks of gestation was processed as described in ref. 13, and cultures were plated in modified minimal essential medium with Eagle's salts (MMEM) supplemented with B27 (Invitrogen) on poly(d-lysine)-coated plates (VWR Scientific) at 75,000 cells per well in 96-well plates, 2.5 × 106 cells per well in 6-well plates, or 5 × 107 cells in T75 flasks. Two- to 3-week-old cultures were used in all experiments.
The cultures were treated with 16 pg/ml EGF for 15 min or with various forms of Aβ for indicated periods of time. Aggregated (seed) Aβ was generated by adding distilled deionized H2O to Aβ powder [Aβ(1–40), California Peptide Research, Napa, CA] to make up a 1mM stock. This solution was incubated for 72 h at 37°C, aliquoted, and stored frozen at -20°C. Soluble Aβ was prepared by adding DMSO to Aβ powder to make a 7.5 mM stock, sonicated for 30 min, and frozen at -20°C. HCC were treated with 1 μM “seed” Aβ, followed by 20 μM “soluble” Aβ in MMEM, as described in ref. 19. This two-component treatment was generally used for the experiments described, although similar results were obtained with one-component Aβ treatment, in which Aβ stock was prepared by adding water to Aβ powder to make a 1 mM solution, which was snap-frozen at -20°C. HCC were treated with 20 μMAβ in MMEM. Aβ(1–42) (California Peptide Research) was prepared as a 7.5 mM DMSO stock and used at 10 μM final concentration. Toxicity of different Aβ preparations was tested in viability assays (Fig. 5A, which is published as supporting information on the PNAS web site).
Viability was determined by incubating cells in 10% Alamar Blue in MMEM for 1 h, and fluorescence levels were determined as described in ref. 20 after 2 days of Aβ exposure. In all cases, cell treatments were performed in triplicate wells.
ADAM12 shedding was evaluated by collecting conditioned medium from 2.5 × 106 cells infected with adenovirus constructs and treated with Aβ for 18 h where indicated. Samples were concentrated on Amicon ultracentrifugation units (10-kDa size exclusion) from 2-ml to 100-μl final volume, and 20 μl was used for Western blot analysis.
Immunoprecipitation and Western Blots. Cells were lysed in 20 mM Hepes (pH 7.6), 150 mM NaCl, 1% Nonidet P-40, 10% glycerol, 1 mM Na3VO4, 10 mM NPP, and protease inhibitors (1 ml of buffer per 7.5 × 106 cells), and proteins of interest were immunoprecipitated from cleared lysates (15,000 × g centrifugation for 5 min at 4°C) by using 5 μg of antibodies and 30 μl of 50% suspension of protein G Sepharose for 1 h at 4°C. Sepharose beads were washed twice with 1.5 ml of lysis buffer and proteins eluted in Laemmli sample buffer (50 mM Tris·HCl, pH 6.8/2% SDS/10% glycerol/100 mM DTT). Samples were resolved on 4–12% gradient SDS/PAGE and transferred on poly(vinylidene difluoride) membrane (Millipore). Western blots were performed according to the manufacturer's protocols for phosphospecific antibodies; for Western blot detection of nonphosphorylated proteins, the membranes were blocked in TBST (10 mM Tris·HCl, pH 8.0/150 mM NaCl/0.1% Tween 20) plus 5% nonfat milk and incubated with antibodies in TBST plus 1% nonfat milk for 1 h at room temperature followed by three TBST washes and incubation with horseradish peroxidaseconjugated secondary antibodies at 1:10,000 dilution for1hat room temperature. Chemiluminescence was visualized by using the ECL kit (Amersham Pharmacia Biosciences).
Immunocytochemistry. After6hofAβ treatment, cells were fixed in 4% paraformaldehyde/PBS for 15 min at room temperature, washed in PBS, and permeabilized in 0.1% Triton X-100 in PBS for 30 min. After blocking in 1% BSA/PBS for 2 h, samples were incubated with 1 μg/ml affinity-purified rabbit anti-FISH polyclonal antibodies in PBS for 1 h at room temperature. Incubation with preimmune serum or omitting primary antibodies were used as specificity controls. Bound antibodies were visualized by incubation with Cy3-coupled goat anti-rabbit Ig secondary antibody. Cells were analyzed by using a Nikon TE300 microscope, chargecoupled device camera (Roper Scientific, Trenton, NJ), and metamorph software (Universal Imaging, Downington, PA).
p165 Purification. HCC (5 × 107 cells) were treated for 6 h with Aβ, lysed in 10 ml of immunoprecipitation buffer (pH 7.6), and cleared by centrifugation at 15,000 × g for 15 min at 4°C followed by 150,000 × g centrifugation for 1 h. The supernatant was diluted 3× with 20 mM Hepes·KOH (pH 7.6) and 10 mM NaCl and loaded onto a 5-ml SP-FF column equilibrated with 20 mM Hepes·KOH (pH 7.6) and 50 mM NaCl. The column was washed with 50 ml of similar buffer with 175 mM NaCl, and p165-containing fraction was eluted in 10 ml of buffer with 275 mM NaCl. The salt concentration in the sample then was decreased to100 mM by using a PD-10 desalting column. The sample was incubated with monoclonal anti-phosphotyrosine antibody beads (O.T.162, US Biological, or PT-66, Sigma) for 16 h at 4°C (50 μg/ml), and beads were washed with 50 ml of buffer of the same composition. Bound proteins were eluted with 10% SDS and separated by SDS/PAGE, followed by silver staining for p165 band visualization. Protein identification was performed at the Harvard Microchemistry Facility (Cambridge, MA) by microcapillary RP-HPLC nano-electrospray tandem MS on a LCQ DECA XP quadripole ion trap mass spectrometer (Finnigan-MAT, San Jose, CA).
FISH and ADAM12 Constructs. Most of the human FISH mRNA sequence was present in Integrated Molecular Analysis of Genomes and Their Expression (IMAGE) Consortium clone 6155848 (GenBank database accession no. BQ424230). The missing 5′ fragment was cloned by PCR of human brain cDNA library (BD Biosciences) by using the primers gggcgccactccgctttgtggggggaagatg and cgaaagctgatgccatcggaaatgcacgac. The additional sequence matched putative exons on chromosome 10 between base pairs 105030140 and 105279611 (GenBank accession no. NC_000010.7), adjacent to the remainder of the gene (between base pairs 105028135 and 105026160). Deletion mutants of FISH were cloned as His-tagged constructs by subcloning into pcDNA3.1/His (N-terminal tag) or pcDNA3.1/V5-His (C-terminal tag) vectors, using restriction sites KpnI/BamHI for FISH348–1105, KpnI/EcoRV for FISH348–911, and EcoRI/ApaI for FISH1–388; fusions were recloned into pADTrack-CMV plasmid by using HindIII/RV or HindIII/XbaI sites, followed by recombination with adenoviral backbone construct as described in ref. 21. ADAM12 deletion mutant was obtained by PCR of human brain cDNA (BD Bioscience) using primers aagcttatggcagcgcgcccgctgcccg(I), atcgggtggtggaaacacattctttgcagc, gctctagatcacttaatataggcggtgtgg(II), and atcgaagtcagggagtctttcgggggccag. PCR products annealed to each other and a new PCR reaction performed with primers I and II, and the resulting cDNA was cloned in pADTrack-CMV HindIII/XbaI. BglII and XbaI sites flanking the ORF of full-length ADAM12 cDNA were introduced by PCR using Invitrogen clone (ID 19600411894933) as a template. BglII/XbaI and BglII/BglII fragments were consequently cloned in pADTrack-CMV. All constructs were verified by DNA sequencing.
Adenoviral Constructs. Adenovirus constructs were produced by using the AdenoEasy system kindly provided by Bert Vogelstein (Johns Hopkins Medical Institutions, Baltimore). The system allows expression of GFP and a protein of interest from the same adenoviral construct as described in ref. 21. Puresyn Inc. (Malvern, PA) adenovirus purification kit and production services were used for the viral purification. On average, an infection rate of 50 viral particles per cell resulted in 25–30% GFP positive cells. Efficiency of infection was calculated as the percent of GFP-positive cells among at least 1,000 DAPI-stained cells counted in random microscope fields 24 h after infection. Viral mediated expression of the proteins of interest was confirmed by Western blot. Coinfection of two different viral constructs of the same titer resulted in coexpression of proteins of interest in almost all GFP-positive cells as verified by immunostaining in pilot experiments (data not shown).
AD Sample Preparation. Control and AD inferior frontal gyrus gray matter, with postmortem intervals of <3 h (a generous gift from Joe Rogers, Sun Health Research Institute, Sun City, AZ), were homogenized in immunoprecipitation buffer (pH 7.6) plus 0.1% SDS. Homogenates were cleared by 10,000 × g centrifugation for 5 min at 4°C, equal amounts of total protein were probed on Western blot with anti-ADAM12 and reprobed with anti-80K-H (cytoplasmic protein) antibodies, and the ratios of optical densities ADAM12/80K-H were determined by using an Si Densitometer and imagequant software (Molecular Dynamics).
Results and Discussion
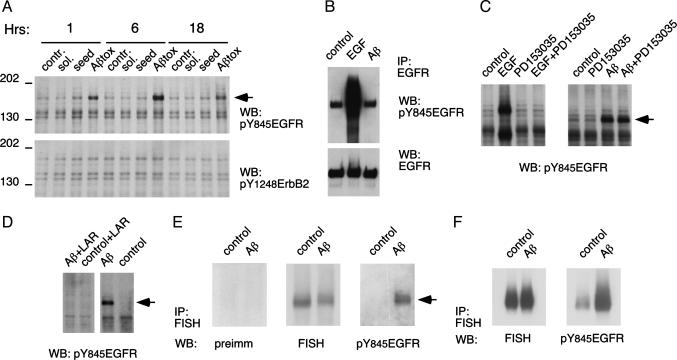
p165 Is Phosphorylated upon Aβ Stimulation of Primary Cortical Neuronal Cultures. By using a panel of anti-phosphotyrosine site-specific antibodies, proteins phosphorylated in response to the Aβ treatment of HCC were identified. Anti-phosphotyrosine-845 EGFR antibody (α-pY845EGFR), but no other anti-receptor tyrosine kinase antibodies, detected a phosphoprotein of an apparent molecular mass of 165 kDa (Fig. 1A). Notably, toxic forms of Aβ(1–40) induced the p165 phosphorylation, whereas nontoxic aggregated seed and DMSO-solubilized Aβ alone did not. Treatment with Aβ(1–42) also resulted in p165 phosphorylation (Fig. 5B). Aβ-induced neuronal death in this model requires a minimum of 24 h of exposure to the peptide (data not shown) and is preceded by phosphorylation of p165, which is detected within1hof peptide application and persists for at least 18 h (Fig. 1A).
Fig. 1.
Neurotoxic Aβ treatment results in phosphorylation of FISH protein. (A)pY845EGFR and pY1248ErbB2 WCL Western blot. “Tox” is the neurotoxic Aβ form that is described in Materials and Methods. Seed, aggregated Aβ; sol., DMSO-solubilized Aβ. Arrow indicates p165. (B and C) p165 is distinct from EGFR. (B) EGFR immunoprecipitated from cultures treated with EGF or Aβ probed with α-pY845EGFR and reprobed with α-EGFR antibodies. (C) WCL Western blot of HCC treated with EGF or Aβ, with or without 2.8 μM PD153035. (D) p165 is phosphorylated on tyrosine residue(s). Shown is a Western blot of HCC lysate incubated with leukocyte antigen-related phosphatase (20 units/ml for 30 min at 37°C). (E and F) p165 is FISH protein. (E) Immunoprecipitated FISH probed on Western blot as indicated. (F) FISH immunoprecipitated from cultures expressing exogenous FISH protein probed with α-pY845EGFR and reprobed with anti-FISH antibodies. FISH and α-pY845EGFR expression was 5- to 10-fold higher than that of endogenous protein.
Direct immunoprecipitation of EGFR did not identify the Aβ-induced phosphoband as EGFR (Fig. 1B), and treatment of cultured neurons with PD153035, a small molecule inhibitor of EGFR kinase activity, did not block the phosphorylation of the p165 protein (Fig. 1C). Additionally, the apparent molecular mass of p165 is smaller than that of EGFR. Treating the lysate of Aβ-exposed neurons with leukocyte antigen-related phosphotyrosine phosphatase resulted in complete disappearance of the Aβ-induced α-pY845EGFR signal on Western blot, confirming that the antibody was detecting a phosphotyrosine-specific epitope (Fig. 1D). We concluded that the α-pY845EGFR antibody was crossreacting with an unknown protein that is tyrosine phosphorylated in response to Aβ treatment.
p165 Is the FISH Adapter Protein. After preliminary characterization of p165 as a Nonidet P-40-soluble, nonglycosylated protein (data not shown), a protocol for p165 isolation was established to identify this protein. The procedure included differential centrifugation, followed by cation exchange chromatography and anti-phosphotyrosine immunoprecipitation (Fig. 5). Two independent MS analyses of the protein identified p165 as the FISH adapter protein, which was initially characterized in a screen for Src kinase substrates (17).
α-pY845EGFR Western blot of immunoprecipitated FISH protein demonstrated that p165 phosphoprotein is recognized by anti-FISH antibodies, and the electrophoretic mobility of the p165 is identical to that of FISH protein (Fig. 1E). To further confirm Aβ-induced FISH phosphorylation, FISH protein was expressed in HCC by using an adenoviral vector. Aβ treatment resulted in phosphorylation of immunoprecipitated exogenously expressed FISH, providing additional evidence of Aβ-induced tyrosine phosphorylation of this protein (Fig. 1F).
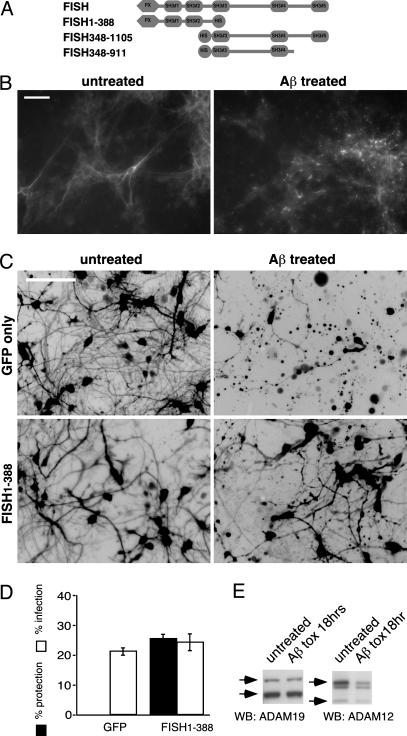
As shown in Fig. 2A, the domain architecture of FISH is defined by an N-terminal PX domain followed by five SH3 domains. The C-terminal SH3 (SH3#5) domain is able to interact with the C-terminal tail of three members of the ADAM family, and the SH3#3 is able to interact intramolecularly with the N-terminal PX domain of FISH, thus keeping the protein in an inactive state (18). The described regulation of FISH localization through tyrosine phosphorylation (18) prompted us to test whether Aβ application induces a change in FISH distribution.
Fig. 2.
Aβ induces FISH subcellular relocalization, and FISH1–388 provides neuroprotection from Aβ toxicity. (A) Schematic representation of FISH deletion mutants. (B) HCC were treated with Aβ for 6 h and immunostained with anti-FISH antibodies. (Scale bar: 50 μm.) (C and D) FISH1–388 expression results in increased viability of Aβ treated neurons. (C) Inverted image of GFP-positive cells expressing deletion mutant as shown. (D) Cytotoxicity assay of Aβ-treated cultures infected with adenoviruses as in C. Error bars in A–D represent SD. (E) Aβ treatment results in ADAM12, but not ADAM19, degradation. ADAM12 and ADAM19 WCL Western blot of control and 18-h Aβ-treated cultures is shown. Arrows indicate specific ADAMs proform (Upper) and mature protein (Lower); unmarked is nonspecific band in ADAM12 Western blot.
Immunostaining of control and Aβ-treated HCC demonstrated profound changes in the localization of FISH from diffuse cytoplasmic staining in the untreated cells to punctate staining in cells treated with Aβ for 6 h (Fig. 2B). These data show that FISH phosphorylation results in redistribution of the protein in Aβ-treated neurons similar to the Src-dependent FISH redistribution previously described in the NIH 3T3 cell line (18) and suggest that FISH function is stimulated by Aβ treatment.
Expression of N-Terminal Moiety of FISH Is Neuroprotective in Aβ Toxicity. If FISH has a role in mediating Aβ-induced neurotoxicity, then interference with the protein function should result in a neuroprotective effect. Because the intramolecular interaction of the PX domain with SH3#3 controls FISH localization (18), which is changed upon Aβ treatment (Fig. 2B), expression of FISH N-terminal moiety alone (FISH1–388, Fig. 2A) might result in a dominant-negative effect on the function of the endogenous protein. As shown in Fig. 2 C and D, neurons infected with adenovirus expressing FISH1–388 had increased resistance to Aβ-induced cell death compared with neurons infected with control adenovirus (GFP). This result indicates that endogenous FISH is involved in the neurotoxic signaling induced by Aβ. It is not clear how FISH1–388 interferes with the function of the endogenous protein. The fragment could compete with full-length FISH for a kinase, for SH3#3 domain binding to the PX domain, for PX-dependent interaction at the perimembrane locale, or for binding to an as-yet-unidentified protein.
ADAM12 Activity Is Increased in Aβ Treatment. Considering that ADAMs may be downstream targets for regulation by FISH (18), we looked for evidence of altered ADAMs function in the HCC model of Aβ toxicity. ADAM family members are known to act as “sheddases” (22) that cleave extracellular domains in a number of transmembrane proteins. The metalloproteases also are reported to undergo autocatalytic self-processing (23), which can be used as an indication of their proteolytic activity. ADAM12 and ADAM19 are both known to interact with FISH (18) and are expressed in HCC (Fig. 2E). WCL Western blots of untreated or Aβ-treated (18 h) cultures showed an Aβ-dependent decrease in ADAM12 protein level but no significant change in ADAM19 level (Fig. 2E). It cannot be ruled out that ADAM19 protease activity also is increased by Aβ treatment, but it is undetectable in this model. After 18 h of Aβ treatment, levels of other proteins tested remained virtually unchanged (data not shown), suggesting that the decreased level of ADAM12 is the result of self-cleavage due to stimulation of proteolytic activity.
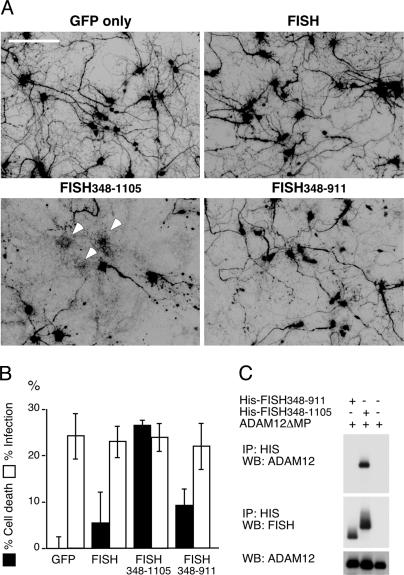
Expression of FISH348–1105 Induced Neuronal Death in an ADAM12-Dependent Manner. To further study the role of FISH and ADAM12 proteins in Aβ toxicity, we created new deletion mutants of FISH and ADAM12. In contrast to the inhibitory effect of FISH1–388 on Aβ-mediated neurotoxicity, removal of the N-terminal region from the full-length protein, creating a deletion mutant FISH348–1105 (Fig. 2A), causes cell death even without application of Aβ when FISH348–1105 is expressed in cortical neurons (Fig. 3 A and B). It is possible that FISH348–1105 is acting as a “constitutively active” FISH mutant, because it does not contain the PX domain, necessary to keep FISH in an inactive state. The toxicity was not observed upon full-length FISH protein expression (Fig. 3 A and B).
Fig. 3.
Neurotoxicity of FISH348–1105 deletion mutant depends on interaction with ADAMs. (A and B) Expression of FISH348–1105 results in neuronal death. Expression of either full-length FISH or a C-terminal fragment lacking the ADAMs binding region is not cytotoxic. (A) Inverted image of GFP-positive cells expressing FISH constructs as indicated, 3 days after infection. (B) Cytotoxicity assay of cultures expressing full-length FISH and deletion mutants. (C) FISH348–1105, but not FISH348–911, binds ADAM12ΔMP. Anti-His immunoprecipitates from HCC expressing ADAM12ΔMP (Top) and His-tagged deletion mutants of FISH (Middle) were probed with anti-ADAM12 and anti-FISH antibodies as shown. (Bottom) WCL Western blot probed for ADAM12.
Neuronal death induced by FISH348–1105 might result from the unhindered activation of ADAMs by the deletion mutant. In this case, deletion of the ADAMs binding region in FISH348–1105 or coexpression of a protease-deficient ADAM12 mutant would be predicted to block the toxicity. To test whether FISH348–1105 induces neuronal death in an ADAMs-dependent manner, the C-terminal region was removed from the FISH348–1105 to generate FISH348–911 (Fig. 2A). Unlike FISH348–1105, the new deletion mutant did not bind ADAM12 (Fig. 3C), and FISH348–911 expression in HCC did not result in neuronal death as compared with FISH348–1105 (Fig. 3 A and B).
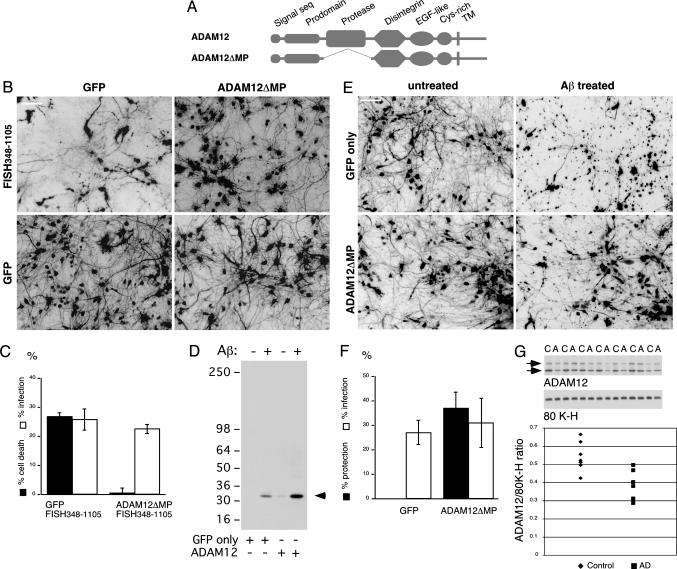
To further substantiate the possibility that FISH348–1105 neurotoxicity is mediated through ADAM12 activation, the effect of full-length ADAM12 expression was compared with that of a metalloprotease-deficient ADAM12 (ADAM12ΔMP, Fig. 4A) on FISH348–1105-induced neuronal death. Coexpressing ADAM12ΔMP with FISH348–1105 resulted in protection from FISH348–1105-induced toxicity (Fig. 4 B and C), whereas coexpression of full-length ADAM12 did not result in a protective effect (Fig. 6, which is published as supporting information on the PNAS web site). Furthermore, longer-term expression of ADAM12 on its own had a neurotoxic effect, which was not observed for ADAM12ΔMP or control virus (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 4.
Neurotoxicity induced by FISH348–1105 or Aβ is prevented by expression of ADAM12ΔMP. (A) Schematic representation of ADAM12 and the deletion mutant. (B) Inverted image of GFP-positive cells expressing FISH and/or ADAM12 deletion mutants as indicated, 3 days after infection, with no Aβ treatment. (C) Cytotoxicity assay of cultures expressing proteins as in B. (D) An 18-h Aβ treatment induces ADAM12 shedding. Shown is an N-terminal ADAM12 Western blot of conditioned medium from cortical cultures infected with adenoviruses and treated as indicated. (E and F) ADAM12ΔMP rescues neurons from Aβ cytotoxicity. (E) Inverted image of GFP-positive neurons expressing ADAM12ΔMP and treated with Aβ as indicated. (F) Cytotoxicity assay of cultures as in E.(G) ADAM12 level is decreased in AD cortex. ADAM12 Western blot and densitometry of AD (A) and control (C) samples are shown. An 80K-H (intracellular protein) Western blot is shown for comparison to demonstrate equal protein loading. Arrow indicates proform (Upper) and mature form (Lower) of ADAM12. The mature form was imaged for densitometry. Difference between the two groups is statistically significant (Mann–Whitney test, P = 0.0268).
Activation of the protease function and ADAM12 self-cleavage, as opposed to down-regulated expression, should result in accumulation of the cleavage product(s) in the conditioned medium from the cortical cultures. Indeed, FISH348–1105 expression induces accumulation of a 32-kDa ADAM12 cleavage product in conditioned medium that is recognized by N-terminal specific anti-ADAM12 antibodies. Coexpression of ADAM12ΔMP prevents this shedding (Fig. 8, which is published as supporting information on the PNAS web site).
These data demonstrate that FISH348–1105 neurotoxicity is dependent on its ability to interact with metalloprotease-competent ADAM12 and suggest that Aβ toxicity also may be mediated through FISH-dependent induction of ADAMs activity.
Aβ-Induced Neuronal Death Is ADAM12 Metalloprotease Activity-Dependent. The decrease of ADAM12 cellular level upon Aβ treatment, as well as the dependence of FISH348–1105 toxicity on ADAM12, led us to test whether Aβ application, similar to FISH348–1105 expression, would result in accumulation of the ADAM12 32-kDa soluble fragment in the conditioned medium. As shown in Fig. 4D, Aβ treatment results in accumulation of the ADAM12 soluble fragment in the conditioned medium, and the amount is increased when ADAM12 is overexpressed (correlating with overexpressed ADAM12 toxicity). These data and toxicity resulting from full-length ADAM12 expression (Fig. 7) allowed us to put forward a prediction that Aβ neurotoxicity in our in vitro model can be prevented by expression of protease-deficient ADAM12. HCC infected with adenovirus expressing ADAM12ΔMP or control virus were treated with Aβ. As shown in Fig. 4 E and F, expression of ADAM12ΔMP blocks neuronal death induced by Aβ in vitro, further supporting the hypothesis of FISH and ADAM12 mediating cytotoxicity of the peptide. Protection of ADAM12ΔMP against Aβ toxicity might result from a few possible effects of the mutant expression: sequestration of FISH protein or competition with endogenous ADAM12 for a protein binding to the other structural motifs present in ADAM12ΔMP.
Protein Level of ADAM12 Is Reduced in AD Cortex. To determine whether a similar pathological process might occur in AD, we compared levels of ADAM12 protein in AD and control cortical samples. Western blot analysis of ADAM12 indicated lower levels of ADAM12 in AD samples vs. controls (Fig. 4G). Although other causes, such as down-regulated transcription, cannot be ruled out, the data correlate with the in vitro observation, thereby supporting the possibility of ADAM12 activation in AD.
The presented data demonstrate that neurons respond to toxic Aβ treatment by phosphorylation of the FISH adapter protein and changes in its localization, which correlate with cleavage of ADAM12. Interfering with FISH and ADAM12 metalloprotease function results in a protective effect against Aβ-induced neurotoxicity. Thus, we conclude that an intermediate step in Aβ-induced cell death is activation of ADAM12, which is triggered by FISH phosphorylation enabling the adapter protein to bind to the cytoplasmic tail of the metalloprotease. Activation of the other two ADAMs family members known to interact with FISH, ADAM15 and ADAM19, also cannot be ruled out. It remains to be seen whether the observed phenomenon of ADAM(s)-mediated neuronal death is specific to Aβ toxicity, or whether it is part of an “executioner mechanism,” which can be stimulated by other neurotoxic agents. These findings show an unusual pathway of Aβ-induced neurotoxicity through FISH-mediated regulation of ADAM(s) and suggest a previously undescribed strategy for AD treatment by targeting the metalloprotease activity of ADAM(s).
Supplementary Material
Acknowledgments
We thank Russell Rydel for contributions in establishing the in vitro model of Aβ toxicity, Tamie Chilcote and Jason Goldstein for helpful discussions, Anke Meyer-Franke and Zhen Jin for help with cortical culture preparation, and Robin Barbour and Linnea Diep for help with antibody production.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Aβ, amyloid-β peptide; AD, Alzheimer's disease; EGFR, EGF receptor; HCC, human cortical culture; MMEM, modified minimal essential medium with Eagle's salts; WCL, whole-cell lysate; FISH, five SH3 domains; ADAM, a disintegrin and metalloprotease; ADAM12ΔMP, ADAM12 deleted metalloprotease domain.
References
- 1.Selkoe, D. J. & Schenk, D. (2003) Annu. Rev. Pharmacol. Toxicol. 43, 545-584. [DOI] [PubMed] [Google Scholar]
- 2.Lorenzo, A. & Yankner, B. A. (1994) Proc. Natl. Acad. Sci. USA 91, 12243-12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorenzo, A., Yuan, M., Zhang, Z., Paganetti, P. A., Sturchler-Pierrat, C., Staufenbiel, M., Mautino, J., Vigo, F. S., Sommer, B. & Yankner, B. A. (2000) Nat. Neurosci. 3, 460-464. [DOI] [PubMed] [Google Scholar]
- 4.Yaar, M., Zhai, S., Fine, R. E., Eisenhauer, P. B., Arble, B. L., Stewart, K. B. & Gilchrest, B. A. (2002) J. Biol. Chem. 277, 7720-7725. [DOI] [PubMed] [Google Scholar]
- 5.Kajkowski, E. M., Lo, C. F., Ning, X., Walker, S., Sofia, H. J., Wang, W., Edris, W., Chanda, P., Wagner, E., Vile, S., et al. (2001) J. Biol. Chem. 276, 18748-18756. [DOI] [PubMed] [Google Scholar]
- 6.Wang, H. Y., Lee, D. H., D'Andrea, M. R., Peterson, P. A., Shank, R. P. & Reitz, A. B. (2000) J. Biol. Chem. 275, 5626-5632. [DOI] [PubMed] [Google Scholar]
- 7.Mattson, M. P. (2004) Nature 430, 631-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morishima, Y., Gotoh, Y., Zieg, J., Barrett, T., Takano, H., Flavell, R., Davis, R. J., Shirasaki, Y. & Greenberg, M. E. (2001) J. Neurosci. 21, 7551-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bozyczko-Coyne, D., O'Kane, T. M., Wu, Z. L., Dobrzanski, P., Murthy, S., Vaught, J. L. & Scott, R. W. (2001) J. Neurochem. 77, 849-863. [DOI] [PubMed] [Google Scholar]
- 10.Savage, M. J., Lin, Y. G., Ciallella, J. R., Flood, D. G. & Scott, R. W. (2002) J. Neurosci. 22, 3376-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuperstein, F. & Yavin, E. (2003) J. Neurochem. 86, 114-125. [DOI] [PubMed] [Google Scholar]
- 12.Lambert, M. P., Barlow, A. K., Chromy, B. A., Edwards, C., Freed, R., Liosatos, M., Morgan, T. E., Rozovsky, I., Trommer, B., Viola, K. L., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 6448-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson, R., Scales, T., Clark, B. R., Gibb, G., Reynolds, C. H., Kellie, S., Bird, I. N., Varndell, I. M., Sheppard, P. W., Everall, I. & Anderton, B. H. (2002) J. Neurosci. 22, 10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw, S., Bencherif, M. & Marrero, M. B. (2002) J. Biol. Chem. 277, 44920-44924. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto, Y., Kaneko, Y., Tsukamoto, E., Frankowski, H., Kouyama, K., Kita, Y., Niikura, T., Aiso, S., Bredesen, D. E., Matsuoka, M. & Nishimoto, I. (2004) J. Neurochem. 90, 549-558. [DOI] [PubMed] [Google Scholar]
- 16.Shah, B. H. & Catt, K. J. (2004) Trends Neurosci. 27, 48-53. [DOI] [PubMed] [Google Scholar]
- 17.Lock, P., Abram, C. L., Gibson, T. & Courtneidge, S. A. (1998) EMBO J. 17, 4346-4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abram, C. L., Seals, D. F., Pass, I., Salinsky, D., Maurer, L., Roth, T. M. & Courtneidge, S. A. (2003) J. Biol. Chem. 278, 16844-16851. [DOI] [PubMed] [Google Scholar]
- 19.Wogulis, M., Wright, S., Cunningham, D., Chilcote, T., Powell, K. & Rydel, R. E. (2005) J. Neurosci., in press. [DOI] [PMC free article] [PubMed]
- 20.Estus, S., Tucker, H. M., van Rooyen, C., Wright, S., Brigham, E. F., Wogulis, M. & Rydel, R. E. (1997) J. Neurosci. 17, 7736-7745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W. & Vogelstein, B. (1998) Proc. Natl. Acad. Sci. USA 95, 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seals, D. F. & Courtneidge, S. A. (2003) Genes Dev. 17, 7-30. [DOI] [PubMed] [Google Scholar]
- 23.Chesneau, V., Becherer, J. D., Zheng, Y., Erdjument-Bromage, H., Tempst, P. & Blobel, C. P. (2003) J. Biol. Chem. 278, 22331-22340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.