Abstract
Key to the remarkable ability of vestimentiferan tubeworms to thrive in the harsh conditions of hydrothermal vents are hemoglobins that permit the sequestration and delivery of hydrogen sulfide and oxygen to chemoautotrophic bacteria. Here, we demonstrate that zinc ions, not free cysteine residues, bind sulfide in vestimentiferan hemoglobins. The crystal structure of the C1 hemoglobin from the hydrothermal vent tubeworm Riftia pachyptila has been determined to 3.15 Å and revealed the unexpected presence of 12 tightly bound Zn2+ ions near the threefold axes of this D3 symmetric hollow sphere. Chelation experiments on R. pachyptila whole-coelomic fluid and purified hemoglobins reveal a role for Zn2+ ions in sulfide binding. Free cysteine residues, previously proposed as sulfide-binding sites in vestimentiferan hemoglobins, are found buried in surprisingly hydrophobic pockets below the surface of the R. pachyptila C1 molecule, suggesting that access of these residues to environmental sulfide is restricted. Attempts to reduce the sulfide-binding capacities of R. pachyptila hemoglobins by addition of a thiol inhibitor were also unsuccessful. These findings challenge the currently accepted paradigm of annelid hemoglobin evolution and adaptation to reducing environments.
Keywords: protein crystallography, protein assembly, cysteine, deep sea, vestimentiferan
Deep-sea hydrothermal vents are remarkable for their geology, chemistry, and biology. Hot anoxic water is mixed to various degrees with cold, oxic, deep-sea bottom water beneath the sea floor and then expelled along mid-ocean ridges (1). The result is a wide variety of chemical and thermal habitats that are colonized by organisms with various degrees of tolerance for these abiotic factors (2). Of particular interest has been the ability of hydrothermal vent organisms to cope with toxic levels of hydrogen sulfide.
Vestimentiferan tubeworms are the visually dominant organism living around many deep-sea hydrothermal vents. These worms lack a mouth or digestive system and rely on a symbiosis with sulfide-oxidizing bacteria as their source of nutrition (3). Interestingly, vestimentiferan mitochondria are inhibited by moderate sulfide concentrations, as are those of most metazoans (4). To avoid the toxic effects of sulfide while supplying it to their internal chemoautotrophic symbionts, vestimentiferan tubeworms possess several large extracellular hemoglobins (Hbs) that can simultaneously and reversibly bind sulfide and oxygen (5). The binding characteristics of these Hbs not only protect against the inhibition of aerobic respiration but also facilitate the delivery of sulfide and oxygen to the chemoautotrophic endosymbiotic bacteria (3, 4).
The multi-Hb system of vestimentiferan tubeworms consists of a hexagonal bilayer (HBL) Hb dissolved in the vascular blood, designated V1, and two smaller Hb forms, one in the vascular blood and the other dissolved in the coelomic fluid, designated V2 and C1 respectively (6). As with other annelid HBL Hbs, tubeworm V1 Hb has globin and nonglobin polypeptide chains and a mass of ≈3,600 kDa (7). The smaller vestimentiferan Hb forms, each with a mass of ≈400 kDa, are assembled from globin chains only (6). It is worth noting that no crystal structures of vestimentiferan Hbs have previously been described.
Sulfide binding to these Hbs has been thought to involve the numerous cysteine residues (both free and disulfide bonded) found in the polypeptide chains of these Hbs (8–10). Specifically, free cysteine (free-Cys) residues not involved in disulfide bonds have been proposed as the sole location of sulfide binding in the 400-kDa (V2 and C1) Hbs, whereas it has been hypothesized that the V1 Hb has multiple sulfide-binding mechanisms involving free-Cys residues and the abundant intrachain disulfide bonds in the nonglobin linker chains (8, 9).
The C1 Hb from the deep-sea hydrothermal vent tubeworm, Riftia pachyptila, provides an opportunity to study a stable alternative to the HBL assembly of annelid globin polypeptide chains. This paper reports the crystal structure of the C1 Hb from R. pachyptila to 3.15-Å resolution. The structure reveals an Hb arrangement of 24 globin chains tightly associated to form a hollow sphere with 12 previously unknown Zn2+ ions ligated to histidine and glutamate residues near the threefold axes of the molecule. Moreover, results are presented that demonstrate these Zn2+ ions play a major role in sulfide binding in both C1 and V1 Hbs, whereas results from thiol-inhibition experiments question any role for free-Cys residues in sulfide binding.
Materials and Methods
Sample Collection and Purification. Specimens of the hydrothermal vent tubeworm R. pachyptila were collected at a depth of 2,500 m by the deep submergence vehicle Alvin from hydrothermal vents at 9°50′ N longitude along the East Pacific Rise. Whole-fluid samples were collected and purified into their constituent Hb fractions by gel filtration chromatography (6, 10). Before structural or functional analyses, all samples of purified Hb were checked for degradation by gel filtration chromatography.
Crystallization and Data Collection. Samples of the C1 Hb from R. pachyptila were transferred to a solution of 0.01 M Hepes buffer (pH 7.5 at 20°C). Oxy-crystals were grown at 20°C by using the sitting-drop method and equilibrated against a reservoir solution consisting of 1.6 M ammonium sulfate, 0.1 M Hepes sodium salt (pH 7.5 at 20°C), and 0.1 M NaCl. Diffraction data were collected at BioCARS beamline 14-BMC, Advanced Photon Source at the Argonne National Laboratory (Argonne, IL), on the Area Detector Systems Company Quantum 4 charge-coupled device detector by using a crystal to detector distance of 300 mm. The data were processed and scaled with the hkl package (11), and the crystallographic statistics are given in Table 1.
Table 1. Crystallographic statistics.
| Space group | P41212 |
| Unit cell dimensions, Å | |
| a | 195.84 |
| b | 195.84 |
| c | 308.82 |
| Resolution, Å (highest shell) | 50.0-3.15 (3.26-3.15) |
| No. of unique reflections | 104,444 |
| Rmerge | 0.065 (0.316) |
| I/σ(I) | 11.3 (2.4) |
| Completeness, % | 99.9 (99.9) |
| Multiplicity | 4.6 |
| Refinement | |
| R factor, % | 24.4 |
| Rfree, % | 27.1 |
| Rms deviations | |
| Bonds, Å | 0.009 |
| Angles, ° | 1.45 |
Structure Determination and Refinement. Structure determination by molecular replacement used a Hb dodecamer from Lumbricus terrestris HBL Hb (12) as a search molecule. Molecular replacement calculations were carried out with the software package cns (13) at 4.5-Å resolution. Translation function searches identified the correct space group as P41212 (rather than the alternative P43212) and provided a solution for two dodecamers per asymmetric unit. Rigid body refinement for all 24 subunits resulted in an initial R = 0.473 and free residual index (Rfree) = 0.477 at 4.5-Å resolution. This model was used to generate the matrices and mask for six-fold averaging by using one L. terrestris tetramer (14) and to calculate phases only to 5.5-Å resolution, to minimize phase bias from the L. terrestris Hb model. Molecular averaging carried out with rave (15) to extend the resolution from 5.5 to 3.15 Å resulted in a readily interpretable electron density map, with a residual index (R factor) of 20.5% between the observed structure factors and those calculated from the averaged map.
A molecular model for the R. pachyptila C1 Hb was fit in o (16) by using the published sequence for A2 (8), partial sequences for the A1, B1, and B2 chains (17), and previously unpublished terminal amino acid sequences determined by nucleotide sequencing of R. pachyptila genomic DNA (see Table 3, which is published as supporting information on the PNAS web site).
Five percent of randomly selected reflections were designated as test reflections for use in the Rfree cross-validation method and used throughout refinement. All simulated annealing and minimization calculations were carried out in cns (13) with either strict or tight noncrystallographic restraints (weight – ncs = 300). After refinement of the A1A2B1B2 tetramer model by using strict noncrystallographic symmetry, the conventional and free R factors were 26.5% and 27.7%, respectively. Refinement of the whole 24-mer by using noncrystallographic restraints improved the model only slightly. Figures were generated with bobscript (18), raster3d (19), and midas (20).
Inductively coupled plasma MS was used to determine the identity of bound metals revealed in the electron density. The samples were heat digested in concentrated nitric acid and then redissolved into 2% nitric acid before analyses. A Finnigan MAT element high-resolution inductively coupled plasma mass spectrometer (Thermo Electron) was used.
Examination of the noncovalent interactions within the C1 Hb was performed by using electrospray ionization TOF MS in a manner similar to that described in Green et al. (21). The mass of the assembly was derived from the highest points of the smoothed multiply charged peaks assuming multiple protonation (see Supporting Results and Discussion, which is published as supporting information on the PNAS web site).
Sulfide-Binding Analyses. Purified V1 and C1 Hb was analyzed by electrospray ionization MS under denaturing conditions to quantify the total and free-Cys content of the globin and nonglobin polypeptide chains. Sample preparation and analyses was performed similarly to the methods described in Green et al. (22). Although some of these data have been published previously (6), there was little information available in that study from the nonglobin linker chains [proposed to play a major role in sulfide binding (9)] of the V1 Hb. Results from all of the individual polypeptide chains and proposed whole molecule models are presented in Table 2.
Table 2. The role of Zn2+ in sulfide binding by R. pachyptila Hbs.
| V1 | C1 | |
|---|---|---|
| Controls | ||
| Sulfide-bound* | 2.65 ± 0.16 | 0.79 ± 0.15 |
| Sites occupied† | (381) | (19) |
| Experimental (EDTA) | ||
| Sulfide-bound* | NA | 0.33 ± 0.04 |
| Sites occupied† | NA | (8) |
| Experimental (TPEN) | ||
| Sulfide-bound* | 2.16 ± 0.21 | 0.34 ± 0.15 |
| Sites occupied† | (312) | (8) |
| Control — experimental* | 0.49 | 0.46 |
| Chelated Zn2+ sites† | 71 | 11 |
| Zn2+ ions per Hb‡ | 72 | 12 |
| Free-Cys per Hb‡ | 96 | 12 |
| Linker Cys per Hb‡ | 336 | NA |
Binding data are presented as HS-/heme ± SD.
The number of binding sites occupied by sulfide and the number of chelated Zn2+ ions estimated, using 144 and 24 heme groups for the V1 and C1 Hbs, respectively.
Determined analytically.
The effect of Zn2+ chelation on the sulfide-binding capacities of purified V1 and C1 Hb was analyzed per the protocol of Arp and Childress (23) and Childress and coworkers (23, 24). Experimental Hb samples at 2 mM concentration (based on heme) were pretreated for 30 min at room temperature with either 1 mM N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN) or 1 mM EDTA and dialyzed against 1 mM Na2S. Untreated V1 and C1 Hb served as controls in all experiments. All bound sulfide measurements were standardized to the concentration of heme in the Hb sample determined at the end of each experiment by the method of Drabkin and Austin (25). Sulfide-binding measurements are presented as moles of sulfide bound per mole of heme (HS–/heme).
Thiol inhibition experiments were conducted on R. pachyptila whole-coelomic fluid and purified V1 Hb to assess the contribution of free-Cys residues to the sulfide-binding capacities of vestimentiferan Hbs (9). Following the protocol of Zal et al. (9), we pretreated the Hb samples with 3 mM N-ethylmaleimide (NEM) for 30 min at room temperature and then dialyzed these samples in a saline buffer (3) along with untreated controls against 2 mM Na2S for 18 h. Samples were analyzed per the protocol of Arp and Childress (23) and Childress and coworkers (23, 24).
Results and Discussion
The crystal structure of the R. pachyptila 400-kDa C1 Hb was determined by a combination of molecular replacement, using a model for the Hb dodecamer from L. terrestris (12), and six-fold molecular averaging. A portion of the six-fold-averaged 3.15-Å electron density map is shown in Fig. 1. R. pachyptila C1 Hb is assembled from four distinct globin subunits designated A1, A2, B1, and B2 (17). The electron density allowed fitting a model for the entire sequence of the A2 chain, all but the first six N-terminal residues for chain B2, and all but the first two N-terminal residues for chain A1, which were not modeled and presumed disordered. The electron density for the B1 chain is consistent with a mixture of B1a and B1b isomers and was modeled as such. The C1 crystal structure also enabled us to align the nomenclature assigned to the various globin polypeptide chains from previous studies with the globin polypeptide chains of L. terrestris (see Table 4, which is published as supporting information on the PNAS web site). The atomic model has been refined by using tight noncrystallographic restraints and group B factors (two per residue) to a conventional R factor of 24.4%, and an Rfree of 27.1% (Table 1).
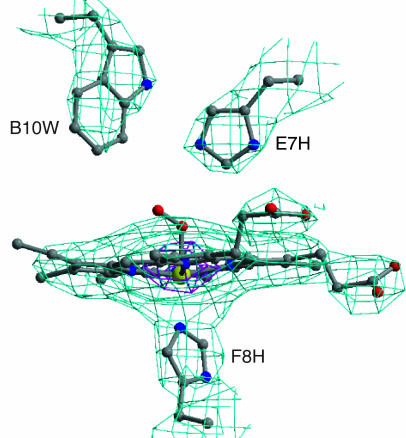
Fig. 1.
Six-fold-averaged electron density map for the R. pachyptila C1 Hb. The electron density map derived from phase extension by using molecular averaging of the six A1A2B1B2 tetramers in R. pachyptila C1 is shown for the heme region of the A2 subunit contoured at 3 σ (light blue) and 15 σ (purple). The quality of this 3.15-Å map is excellent and allows confident placement of side chains and clear definition of the heme iron position.
R. pachyptila Hb subunits exhibit the standard myoglobin fold, including seven helices designated A-H (but lacking a D helix) that are connected by corners designated as AB-GH. In presenting the structure, we have identified individual residues by their helical or corner designation by using standard myoglobin nomenclature. The proximal histidine is designated as F8, because it is the eighth residue in the F-helix in myoglobin, even though it is actually the 15th residue in the F-helix of R. pachyptila Hb chains.
Functional Binding Sites. The electron density maps from the C1 Hb crystal structure clearly reveal the presence of 12 very strong peaks (>10 σ) located in depressions near the molecular three-fold axes (Fig. 2). These peaks are found in groups of six, forming a triangular ring at each pole of the molecule (Figs. 2 and 3). Results from samples of R. pachyptila C1 Hb analyzed by inductively coupled plasma MS confirmed the presence of 12 Zn2+ ions per C1 Hb (0.5 moles of Zn2+ per mole of heme), which is consistent with an assignment of Zn2+ to these features in the density map.
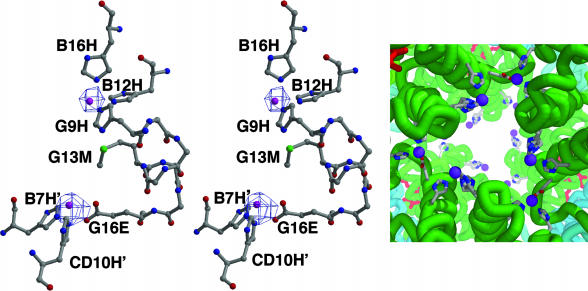
Fig. 2.
Zn2+-binding sites. (Left and Center) Stereoview of two Zn2+-binding sites with protein ligands along with a 2Fo – Fc map contoured at 6 σ. Each A2 subunit provides one complete Zn2+-binding site involving three His ligands and shares two others at interfaces with neighboring A2 subunits. The refined Zn2+ positions are shown as purple balls. (Right) The arrangement of Zn2+-binding sites near the molecular threefold axis of R. pachyptila C1 Hb. Zn2+ ions are purple, along with protein ligands and main-chain traces in the vicinity of the Zn2+ ions.
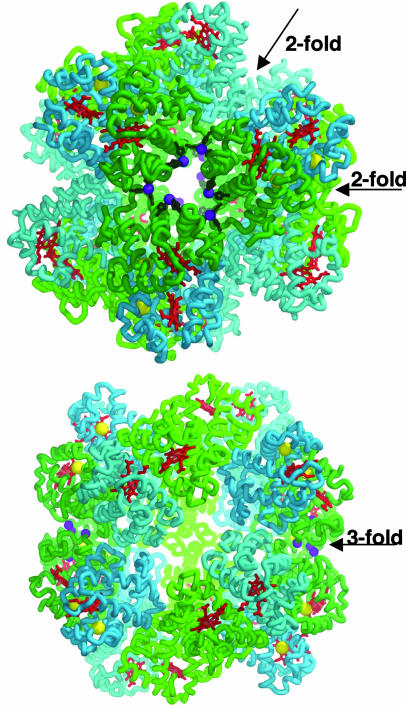
Fig. 3.
Overview of the R. pachyptila C1 Hb structure. The subunit trace for A1 is light green, A2 is dark green, B1 is light blue, and B2 is dark blue. In addition, the heme groups are red, the Zn2+ ions are purple, free-Cys residues are yellow, and the protein side-chain ligands are dark green. (Upper) The view along the threefold axis. (Lower) The view along the twofold axis. Note the location of the Zn2+ atoms clustered in groups of six near the threefold axis at both poles of the molecule.
Each A2 chain contains one complete Zn2+-binding site and shares two others with adjoining A2 subunits. The Zn2+-binding site contained within chain A2 is composed of three His residues (B12, B16, and G9), and the site shared between two A2 subunits is created by Glu G16 of one A2 chain, and His B7 and His CD10 of the adjoining A2 chain (Fig. 2). The refined Zn2+ positions are 9.5 Å apart. Between pairs of Zn2+ sites is a Met residue, whose sulfur atom is 4.5 and 5.9 Å, respectively, from each Zn2+ site.
Zn2+ plays a key role in sulfide binding in the vesicomyid clam, Calyptogena elongata (26). In C. elongata, the Zn2+-based, serum-borne sulfide-binding protein fosters uptake and transport of sulfide from the reducing environment to chemoautotrophic endosymbionts (26). To investigate the role of Zn2+ ions in sulfide binding by R. pachyptila Hbs, samples of both C1 and V1 Hbs from R. pachyptila were treated with metal chelators and dialyzed against a saturating concentration of Na2S (Table 2). Samples (n = 8) of the C1 Hb were exposed to the general chelator, EDTA. When compared with control data, the sulfide-binding capacity of the treated C1 Hb was reduced by 58%. Next, samples of both the V1 (n = 6) and C1 Hbs (n = 11) were exposed to the Zn2+-specific chelator, TPEN. When compared with control fluids, the sulfide-binding capacity of the treated C1 Hb was reduced 57%, whereas purified V1 Hb showed an 18% decrease. The difference between the control data and the inhibitor data were significant for each Hb under the respective experimental conditions (P < 0.001; Student's t test). For both the V1 and C1 Hbs, it is unclear from our data whether the reduction in sulfide binding after treatment with TPEN is due to in situ binding or complete removal of the Zn2+ ions from the protein.
The average sulfide-binding capacity for the C1 controls was 0.79 HS–/heme (Table 2). A 58% decrease in sulfide binding by the chelated C1 Hb corresponds to a loss of 11 sulfide-binding sites (0.46 HS–/heme) and agrees well with the 12 Zn2+ ions observed in the C1 Hb crystal structure (Table 2). The remaining sulfide, 0.33 HS–/heme, would occupy approximately eight sulfide-binding sites, indicating the presence of an additional mechanism for sulfide binding in these Hbs.
Chelation of the Zn2+ ions in the V1 Hb shows that Zn2+ accounts for only 18% of bound sulfide (Table 2). The average sulfide-binding capacity in these experiments for the untreated V1 Hb controls was 2.65 HS–/heme. An 18% decrease in sulfide binding would account for a loss of 71 sulfide-binding sites (0.49 HS–/heme). By using a similar molar ratio to that found in the C1 Hb, this value is in good agreement with the 72 Zn2+ sites in this molecule.
The sulfide that remains bound after chelation of the Zn2+-binding sites in tubeworm Hbs suggests that there are multiple mechanisms for sulfide binding in both Hbs. Arp et al. (10) present unpublished evidence suggesting that capping the Cys residues resulting from disulfide bond reduction in vestimentiferan Hbs can lead to decreased sulfide-binding capacities. If this mechanism can be substantiated, then the 336 disulfide-bonded Cys in the linker chains of the V1 Hb, determined by electrospray ionization MS (Table 2), provide adequate binding space for the remaining hydrogen sulfide as has been suggested (9, 10). Because linker chains are not incorporated into the assembly of the R. pachyptila C1 Hb, this mechanism does not seem feasible for this Hb. A sulfide-binding mechanism involving the numerous free-Cys residues in this Hb has been suggested (9); however, further examination of this mechanism is warranted.
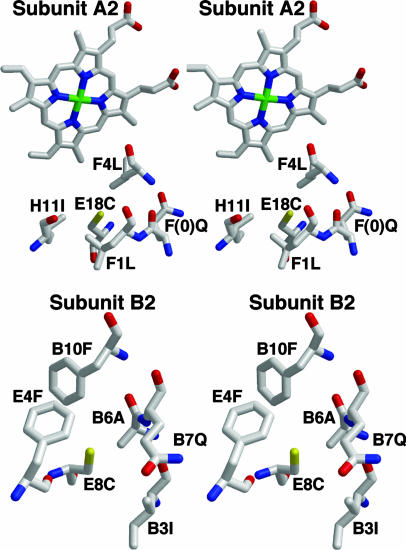
The relatively high Cys content of vestimetniferan Hbs led to the hypothesis that these residues play a role in sulfide binding (9, 10). Cys residues at positions E18 (A2 subunit) and E8 (B2 subunit), that are not involved in disulfide bonds have been implicated as the specific residues in the globin chains involved in the binding of hydrogen sulfide by the formation of S-sulfohemoglobin (9) (Fig. 4). The crystal structure reveals that the E18 and E8 free-Cys residues are buried 5–6 Å beneath the subunit surface in a hydrophobic pocket, which is created by Leu (F1 and F4), Ile (H11), and the base of the heme group in the A2 chain, and Phe (B10 and E4), Ile (B3), and Ala (B6) on the B2 subunit. In addition, a hydrophilic Gln is present near both free-Cys residues, with an approach of 5.5 Å from Gln F (0) Oε to the sulfur of Cys E18 in subunit A2, and 5.3 Å from Gln B7 Oε to the sulfur of Cys E8 in subunit B2.
Fig. 4.
Stereoviews of the hydrophobic pockets surrounding the two free-Cys residues that are proposed as sites of sulfide binding. (Upper) Cys E18 in subunit A2. (Lower) Cys E8 in subunit B2. In both cases, the free-Cys is buried ≈5 Å beneath the protein surface in a hydrophobic pocket, with a nearby (≈5.5 Å) glutamine residue.
The surrounding hydrophobic residues are likely to restrict access of the free-Cys to environmental sulfide. This would be especially true for HS–, the sulfide species that predominates at physiological pH and the species reported to bind to R. pachyptila Hbs (24, 27). Zal et al. (9) reported a significant decrease in the sulfide-binding capacity of V1 and C1 Hbs upon treatment of the free thiol groups with NEM. Those experiments revealed a 75% inhibition of sulfide binding in V1 Hb and complete sulfide-binding inhibition in C1 Hb after NEM pretreatment, indicating that free-Cys was the sole source of sulfide binding in the latter Hbs (9). Based on the stoichiometry of the free-Cys in V1 globin and nonglobin linker chains (Table 2), and by using the sulfide-binding capacity of the Zal et al. (9) experiments (2.85 HS–/heme), sulfide binding by free-Cys should only have accounted for 23% of the sulfide-binding capacity of the V1 Hb.
We repeated the Zal et al. (9) experiments because our structure of the R. pachyptila C1 Hb calls into question whether the free-Cys are capable of binding sulfide, and the results of those experiments did not fit with the discovery of a Zn2+-sulfide-binding mechanism. After five experiments using NEM-treated whole-coelomic fluid (n = 12) and purified V1 Hb (n = 3), we have determined that the combination of 3 mM NEM and 2 mM Na2S in the dialysate has the effect of oxidizing all of the sulfide in the experimental system. An inability to detect sulfide in the dialysate at the end of each experiment precludes the calculation of bound sulfide (24), the central component of the experimental results. An additional six experiments were conducted on NEM-treated whole-coelomic fluid (n = 16) and purified C1 Hb (n = 2), but without NEM in the dialysate solution. In these experiments, there was no indication of sulfide oxidation or an inhibition of sulfide binding in either the whole fluid (controls: 0.92 ± 0.17 HS–/heme; NEM-treated whole-coelomic fluid: 0.90 ± 0.12 HS–/heme; P > 0.80, Student's t test) or purified C1 Hb (Controls: 0.58 ± 0.02 HS–/heme; NEM-treated C1 Hb: 0.62 ± 0.06 HS–/heme; P > 0.30, Student's t test). Incidental to the experimental findings, Zal et al. (9) observed that NEM added to solution with Na2S transformed the color of the buffer solution from clear to pink. In none of the five experiments where NEM was added to the dialysate did any visible color change to the buffer occur. Subsequent analysis found that a light-pink coloration was only present when both NEM and Na2S concentrations were >10 mM, and a strong pink coloration was visible at >20 mM. Because we were unable to reproduce the experimental results of Zal et al. (9), we must therefore conclude based on the structural data presented and our subsequent functional experiments with NEM-treated Hbs that free-Cys residues do not play a significant role in sulfide binding in the V1 and C1 Hbs as has been proposed. Interestingly, a hydrothermal vent polychaete that lives on the sides of active chimneys in a higher-sulfide regime than R. pachyptila, Alvinella pompejana, maintains several free-Cys residues in its extracellular HBL Hb but does not have the ability to bind sulfide (28, 29).
R. pachyptila Hbs have a relatively high affinity for oxygen (< 1 torr at 3°C) (30–32). The distal heme pockets of the C1 Hb differ between the A and B chains in subtle ways that are likely to affect oxygen-binding properties (See Fig. 5, which is published as supporting information on the PNAS web site). Each maintains histidines at the classical positions E7 and F8 (the distal and proximal His residues). The B chain pockets have a Phe at position B10, whereas the A chains have a Trp at B10, making the A-chain heme pockets somewhat more crowded than the B chain pockets. In all cases, the edges of the aromatic rings appear to be positioned close enough (3.5–3.6 Å) to directly impact the oxygen-binding affinity of these subunits. The presence of aromatic amino acids at position B10 is a noteworthy aspect of annelid extracellular Hbs. The size and electrostatic properties of the B10 residue can significantly impact the ligand-binding properties in Hbs (33), with aromatic residues stabilizing bound oxygen molecules and lowering autooxidation of the heme iron (34). Thus, large aromatic residues at position B10 are likely to be an important factor in the success of these hemoglobins as extracellular oxygen carriers.
Structure of R. pachyptila C1 Hb. The 24 subunits of R. pachyptila C1 Hb are arranged into a 130-Å diameter hollow sphere composed of two tightly associated dodecamers (Fig. 3). Each dodecamer has a threefold axis of symmetry (C3) that relates three A1A2B1B2 heterotetramers, creating a similar arrangement to that in the L. terrestris Hb dodecamer subassemblage (14). Two dodecamers then come together with the threefold axes aligned to form a complex with dihedral D3 symmetry. This association results in an approximately spherical protein shell that has an inner radius of ≈25 Å.
Each dodecamer is an assembly of three tetramers, formed from one A1B1 noncovalent dimer, and one A2B2 noncovalent dimer. These two dimers have contacts primarily involving residues in the E and F helices in a similar arrangement to that observed in cooperative Hb dimers found in mollusks and echinoderms (35). These dimers have the most extensive interactions between pairs of subunits in the assemblage, with the A1B1 interface burying a total surface area of 1,940 Å2, whereas the A2B2 interface buries a total surface area of 2,140 Å2. Specific contacts between subunits of the dimers are very similar to those observed in the Hb dodecamer from L. terrestris erythrocruorin (see Fig. 6, which is published as supporting information on the PNAS web site), including key interactions proposed to be important in the allosteric function (12). The A1B1-A2B2 tetramer involves contacts from all four subunits, forming an interface that buries a total of 1,940 Å2 between the two dimers along a quasi-twofold axis. The contacts between neighboring tetramers of the dodecamer form a somewhat smaller interface that buries a surface area of 1,270 Å2. The 12 subunits of the dodecamer are assembled by using a hierarchy of interactions between unlike subunits.
In contrast, the assembly of two dodecamers into a hollow sphere (Fig. 3) results from isologous contacts between chemically identical subunits at three dyad symmetry axes. A total surface area of 6,900 Å2 is buried between the two dodecamers, with each A1 chain contributing ≈900 Å2 and each B2 chain contributing ≈250 Å2 of the buried surface area (for more details on the stability of this protein, see Supporting Results and Discussion).
In light of our discovery of a Zn2+-sulfide-binding mechanism in vestimentiferan V1 and C1 Hbs, our inability to recreate the results from Zal et al. (9) is especially significant because it had been reported that treatment of the C1 Hb with a thiol-binding agent completely extinguished sulfide binding. The loss of free-Cys by annelids living in nonreducing environments may still have been driven by selection against reactive thiol groups as has been suggested (36). Kenesi et al. (37) found that the presence of buried free-Cys residues in trypsinogen, such as those in R. pachyptila Hbs, may be more deleterious than surface free-Cys. Nonetheless, it is unclear how unreacted free thiols would negatively impact the oxygen-binding function of Hbs in annelids from nonreducing habitats, especially when the two functions are known to be mutually exclusive in vestimentiferan Hbs (10). Therefore, their absence in the Hbs of annelids in nonreducing environments does not necessarily confirm their involvement in sulfide binding as previously proposed (36). Our findings challenge the current hypothesis on the mechanism of sulfide binding in annelid Hbs and question the paradigm proposed to drive annelid evolution (36).
Supplementary Material
Acknowledgments
We thank the captains and crews of the research vessels Atlantis and New Horizon and the pilots and crew of the deep submergence vessel Alvin for assistance with sample collection. J. J. Childress (University of California, Santa Barbara) provided essential logistical support during the 2001 and 2002 research cruises. We thank John Kittleson of the Penn State Materials Characterization Laboratory and the staff of BioCARS at the Advanced Photon Source for providing data collection assistance. We thank R. Smith for assistance with protein separation. We thank J. T. Lecomte for her comments and suggestions that improved this manuscript and Eric Fontano (Brandeis University, Waltham, MA) for his work on Fig. 3. This work was supported by the Alfred P. Sloan Foundation (to J.F.F.), National Oceanic and Atmospheric Administration/National Oceanic and Atmospheric Administration's Undersea Research Program Grant UAF01-0042 (to S.W.S., C.R.F., and S. Hourdez), National Institutes of Health Grant DK43323 (to W.E.R.), and National Science Foundation Grant OCE-0002729 (to C.R.F.). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, Contract No. W-31–109-Eng-38. Use of the BioCARS Sector 14 was supported by National Institutes of Health–National Center for Research Resources Grant RR07707.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HBL, hexagonal bilayer; free-Cys, free cysteine; HS–/heme, moles of sulfide per mole of heme; NEM, N-ethylmaleimide; TPEN, N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1YHU). The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY885670–AY885672).
References
- 1.Johnson, K. S., Childress, J. J., Hessler, R. R., Sakamoto-Arnold, C. M. & Beehler, C. L. (1988) Deep-Sea Res. 35, 1723–1744. [Google Scholar]
- 2.Childress, J. J. & Fisher, C. R. (1992) Oceanogr. Mar. Biol. Ann. Rev. 30, 337–441. [Google Scholar]
- 3.Fisher, C. R., Childress, J. J. & Sanders, N. K. (1988) Symbiosis 5, 229–246. [Google Scholar]
- 4.Powell, M. A. & Somero, G. N. (1986) Biol. Bull. (Woods Hole, Mass.) 171, 274–290. [Google Scholar]
- 5.Arp, A. J., Childress, J. J. & Fisher, C. R. (1985) Bull. Biol. Soc. Wash. 6, 289–300. [Google Scholar]
- 6.Zal, F., Lallier, F. H., Green, B. N., Vinogradov, S. N. & Toulmond, A. (1996) J. Biol. Chem. 271, 8875–8881. [DOI] [PubMed] [Google Scholar]
- 7.Vinogradov, S. N., Sharma, P. K. & Walz, D. A. (1991) Comp Biochem Physiol B Biochem. Mol. Biol. 98, 187–194. [DOI] [PubMed] [Google Scholar]
- 8.Zal, F., Suzuki, T., Kawasaki, Y., Childress, J. J., Lallier, F. H. & Toulmond, A. (1997) Proteins Struct. Funct. Genet. 29, 562–574. [PubMed] [Google Scholar]
- 9.Zal, F., Leize, E., Lallier, F. H., Toulmond, A., Dorsselaer, A. V. & Childress, J. J. (1998) Proc. Natl. Acad. Sci. USA 95, 8997–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arp, A. J., Childress, J. J. & Vetter, R. D. (1987) J. Exp. Biol. 128, 139–158. [Google Scholar]
- 11.Otwinowski, Z. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 12.Strand, K., Knapp, J. E., Bhyravbhatla, B. & Royer, W. E. (2004) J. Mol. Biol. 244, 119–134. [DOI] [PubMed] [Google Scholar]
- 13.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 14.Royer, W. E., Jr., Strand, K., van Heel, M. & Hendrickson, W. A. (2000) Proc. Natl. Acad. Sci. USA 97, 7107–7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleywegt, G. J. & Jones, T. A. (1994) in From First Map to Final Model, eds. Bailey, S., Hubbard, R. & Waller, D. A. (Science and Engineering Research Council Daresbury Lab, Warrington, U. K.), pp. 59–66.
- 16.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 17.Bailly, X., Jollivet, D., Vanin, S., Deutsch, J., Zal, F., Lallier, F. & Toulmond, A. (2002) Mol. Biol. Evol. 19, 1421–1433. [DOI] [PubMed] [Google Scholar]
- 18.Esnouf, R. M. (1999) Acta Crystallogr. D 55, 938–940. [DOI] [PubMed] [Google Scholar]
- 19.Merritt, E. & Bacon, D. (1997) Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]
- 20.Ferrin, T. E., Huang, C. C., Jarvis, L. E. & Langridge, R. (1988) J. Mol. Graphics 6, 13–27. [Google Scholar]
- 21.Green, B. N., Gotoh, T., Suzuki, T., Zal, F., Lallier, F. H., Toulmond, A. & Vinogradov, S. N. (2001) J. Mol. Biol. 309, 553–560. [DOI] [PubMed] [Google Scholar]
- 22.Green, B. N., Kuchimov, A. R., Waltz, D. A., Moens, L. & Vinogradov, S. N. (1998) Biochemistry 37, 6598–6605. [DOI] [PubMed] [Google Scholar]
- 23.Arp, A. J. & Childress, J. J. (1983) Science 219, 295–297. [DOI] [PubMed] [Google Scholar]
- 24.Childress, J. J., Arp, A. J. & Fisher, C. R. (1984) Mar. Biol. (Berlin) 83, 109–124. [Google Scholar]
- 25.Drabkin, D. L. & Austin, J. H. (1935) J. Biol. Chem. 112, 51–65. [Google Scholar]
- 26.Childress, J. J., Fisher, C. R., Favuzzi, J. A., Arp, A. J. & Oros, D. R. (1993) J. Exp. Biol. 179, 131–158. [Google Scholar]
- 27.Goffredi, S. K., Childress, J. J., Desauliniers, N. T. & Lallier, F. H. (1997) J. Exp. Biol. 200, 2609–2616. [DOI] [PubMed] [Google Scholar]
- 28.Zal, F., Green, B. N., Lallier, F. H. & Toulmond, A. (1997) Biochemistry 36, 11777–11786. [DOI] [PubMed] [Google Scholar]
- 29.Martineu, P., Juniper, S. K., Fisher, C. R. & Massoth, G. J. (1997) Physiol. Zool. 70, 578–588. [DOI] [PubMed] [Google Scholar]
- 30.Arp, A. J. & Childress, J. J. (1981) Science 213, 342–344. [DOI] [PubMed] [Google Scholar]
- 31.Arp, A. J., Doyle, M., Di Cera, E. & Gill, S. (1990) Respir. Physiol. 80, 323–334. [DOI] [PubMed] [Google Scholar]
- 32.Wittenberg, J., Morris, R., Gibson, Q. & Jones, M. (1981) Science 213, 344–346. [DOI] [PubMed] [Google Scholar]
- 33.Yang, J., Kloek, A. P., Goldberg, D. E. & Mathews, F. S. (1995) Proc. Natl. Acad. Sci. USA 92, 4224–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carver, T. E., Brantley, R. E., Jr., Singleton, E. W., Arduini, R. M., Quillin, M. L., Phillips, G. N., Jr., & Olson, J. S. (1992) J. Biol. Chem. 267, 14443–14450. [PubMed] [Google Scholar]
- 35.Royer, W. E., Jr., Knapp, J. E., Strand, K. & Heaslet, H. A. (2001) Trends Biochem. Sci. 26, 297–304. [DOI] [PubMed] [Google Scholar]
- 36.Bailly, X., Leroy, R., Carney, S., Collin, O., Zal, F., Toulmond, A. & Jollivet, D. (2003) Proc. Natl. Acad. Sci. USA 100, 5885–5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenesi, E., Katona, G. & Szilagyi, L. (2003) Biochem. Biophys. Res. Commun. 309, 749–754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.