ABSTRACT
Nitrite-oxidizing bacteria (NOB) are responsible for the second step of nitrification in natural and engineered ecosystems. The recently discovered genus Nitrotoga belongs to the Betaproteobacteria and potentially has high environmental importance. Although environmental clones affiliated with Nitrotoga are widely distributed, the limited number of cultivated Nitrotoga spp. results in a poor understanding of their ecophysiological features. In this study, we successfully enriched the nonmarine cold-adapted Nitrotoga sp. strain AM1 from coastal sand in an eelgrass zone and investigated its physiological characteristics. Multistep-enrichment approaches led to an increase in the abundance of AM1 to approximately 80% of the total bacterial population. AM1 was the only detectable NOB in the bacterial community. The 16S rRNA gene sequence of AM1 was 99.6% identical to that of “Candidatus Nitrotoga arctica,” which was enriched from permafrost-affected soil. The highest nitrogen oxidation rate of AM1 was observed at 16°C. The half-saturation constant (Km) and the generation time were determined to be 25 μM NO2− and 54 h, respectively. The nitrite oxidation rate of AM1 was stimulated at concentrations of <30 mM NH4Cl but completely inhibited at 50 mM NH4Cl. AM1 can grow well under specific environmental conditions, such as low temperature and in the presence of a relatively high concentration of free ammonia. These results help improve our comprehension of the functional importance of Nitrotoga.
IMPORTANCE Nitrite-oxidizing bacteria (NOB) are key players in the second step of nitrification, which is an important process of the nitrogen cycle. Recent studies have suggested that the organisms of the novel NOB genus Nitrotoga were widely distributed and played a functional role in natural and engineered ecosystems. However, only a few Nitrotoga enrichments have been obtained, and little is known about their ecology and physiology. In this study, we successfully enriched a Nitrotoga sp. from sand in a shallow coastal marine ecosystem and undertook a physiological characterization. The laboratory experiments showed that the Nitrotoga enrichment culture could adapt not only to low temperature but also to relatively high concentrations of free ammonia. The determination of as-yet-unknown unique characteristics of Nitrotoga contributes to the improvement of our insights into the microbiology of nitrification.
KEYWORDS: Nitrospira, Nitrotoga, ammonia, coastal sand, cultivation, enrichment, microbial communities, nitrification, nitrite-oxidizing bacteria, physiology
INTRODUCTION
In nitrification, ammonia is oxidized into nitrate via nitrite by phylogenetically different chemolithoautotrophic microorganisms, and this is an integral part of the global nitrogen cycle. The reactions are catalyzed by ammonia-oxidizing bacteria (AOB), ammonia-oxidizing archaea (AOA), and nitrite-oxidizing bacteria (NOB). Recent investigations have also demonstrated complete nitrification by a single cell (1, 2). Biological nitrite oxidation prevents an accumulation of this highly toxic intermediate and produces nitrate as a nitrogen source for microbes, fungi, and plants (3, 4). Therefore, nitrite oxidation is an important process of the nitrogen cycle. The phylogenetically diverse NOB are, to date, classified into seven genera (Nitrobacter, Nitrococcus, Nitrospina, Nitrospira, Nitrotoga, Nitrolancea, and “Candidatus Nitromaritima”) in four phyla (5). The uncultured new marine NOB “Candidatus Nitromaritima,” which is a Nitrospina-like bacterium, was suggested by single-cell genomics in the brine-seawater interface layer of the Red Sea (6).
During the 3 decades since the first description of Nitrospira (7), cultivation-independent methods have shown that this genus is widely distributed and predominant in many natural ecosystems and engineered systems (8–11). Recently, “Candidatus Nitrotoga arctica” was enriched from permafrost-affected soil in the Siberian Arctic, and it is the only known cold-adapted nitrite-oxidizing betaproteobacterium (12). Nitrotoga-like 16S rRNA genes were identified in other cold habitats, such as deglaciated soils (13) and periglacial soils at 5,400 m elevation (14). Moreover, the 16S rRNA gene clone libraries obtained from a subglacial Antarctic lake showed a significant level of Nitrotoga sp. (13% and 7.8% of the sequences obtained in the water column and sediment, respectively) (15). Nitrotoga-related clones were also detected in oligotrophic ecosystems, such as cave biofilms (16), groundwater seep (17), river water (18), Yellow Sea intertidal beach seawater (19), and salt marsh sediments (20). Surprisingly, Nitrotoga was the only detectable NOB in two out of 20 full-scale wastewater treatment plants (WWTPs) analyzed in Germany and Switzerland (21). Subsequent analysis of microbial communities in 13 WWTPs in Denmark demonstrated that Nitrotoga had a higher transient read abundance than Nitrospira in several activated sludge samples (22). These investigations suggest that Nitrotoga is the primary nitrite oxidizer in some ecosystems.
To understand the ecological roles of widely distributed Nitrotoga organisms, it is necessary to elucidate their physiological properties by the use of enriched or pure cultures. So far, only two Nitrotoga enrichment cultures, one from permafrost soil (12) and one from a biofilter of a cold-freshwater aquaculture plant (23), have been characterized in detail. The affinity for nitrite of “Ca. Nitrotoga arctica” was higher than that of Nitrobacter spp., and the half-saturation constant for nitrite (Km) of “Ca. Nitrotoga arctica” was 2.1 to 6.4 times higher than that of Nitrospira (24). Hüpeden and colleagues demonstrated that Nitrotoga could adapt not only to low temperature but also to a moderately low pH environment (23). However, the cultivation of Nitrotoga-like bacteria under laboratory conditions is difficult, and thus, most of their physiological properties remain unknown.
The purpose of this study was to obtain and characterize a highly enriched culture of Nitrotoga from a novel source, a sandy sediment of eelgrass (Zostera marina). The eelgrass zones in the coastal shallows of Japan assume a key role as primary producers and contribute to the conservation of biodiversity (25). Z. marina enhances nitrogen transformations, including ammonification by discharge of organic nitrogen and nitrification by release of O2 from the roots (26). Previously, the number of crenarchaeotal and betaproteobacterial ammonia monooxygenase alpha subunit gene copies was estimated by quantitative PCR in sand of the eelgrass zone at Tanoura Bay, Shizuoka, Japan, where nitrification occurs throughout the year (27). Considering that clones related to Nitrotoga were detected in coastal ecosystems (19, 20), Nitrotoga was likely to contribute to nitrite oxidation in the sand of the eelgrass zone during winter, although nitrite in the sand was estimated to be below the quantitative limit (27). Here, we successfully performed selective enrichment of Nitrotoga sp. from sand of the eelgrass zone by incubating at low temperature and controlling the NaCl concentration. Then, physiological characterization of the enrichment culture was conducted to reveal its optimum temperature for nitrite oxidation, the influence of NH4Cl, its tolerance for NaCl, and its kinetic parameters, which suggested as-yet-unknown unique characteristics of Nitrotoga.
RESULTS
Enrichment of Nitrotoga-like bacteria.
Combined incubation in batch cultures and in a bioreactor was applied to enrich Nitrotoga from coastal sand over 6 years (Fig. 1). A Nitrotoga-specific primer pair (NTG200F/840R) did not amplify the 16S rRNA gene from the sand of the eelgrass zone. After incubation in artificial seawater (ASW) medium at 10°C for several months, using type-specific primers (see Table 1), only PCR product amplified by NTG200F/840R was detected by agarose gel electrophoresis. Nitrotoga-like denaturing gradient gel electrophoresis (DGGE) bands were detected from the first enrichment culture after a serial dilution step (see Fig. S2 in the supplemental material); however, the cells corresponding to Nitrotoga could not be observed with a microscope. Microscopic observation was performed without cell concentration.
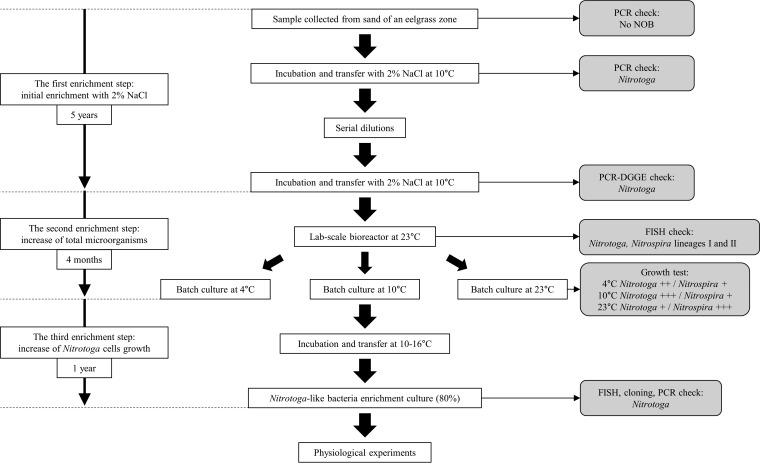
FIG 1.
Flow chart of the procedure to enrich Nitrotoga-like bacteria from sand in an eelgrass zone. All cultures for enrichment were incubated in mineral medium containing 0.5 to 1.4 mM NaNO2 in the dark without shaking. The cultures were transferred with inocula of 1 or 10% (vol/vol), except when serial dilutions were used. In growth tests, + indicates the relative cell growth rate in batch cultures.
TABLE 1.
Oligonucleotide primers and probes used in this study
| Enrichment step | Primer or probe name | Use | Sequence (5′ to 3′) | Target | Reference or source |
|---|---|---|---|---|---|
| First step | |||||
| NTG200F | PCR | CTC GCG TTT TCG GAG CGG | Nitrotoga 16S rRNA gene | 12 | |
| NTG840R | PCR | CTA AGG AAG TCT CCT CCC | 12 | ||
| Ntspmar62F | PCR | GCC CCG GAT TCT CGT TCG | Nitrospira marina-related Nitrospira 16S rRNA gene | 11 | |
| Ntspa662R | PCR | GGA ATT CCG CGC TCC TCT | 10 | ||
| Ntspa81F | PCR | TTR TAA RGC GGC GAA CGG GT | Nitrospira lineage I and II 16S rRNA genes | This study | |
| Ntspa662R | PCR | GGA ATT CCG CGC TCC TCT | 10 | ||
| NitSSU_130F | PCR | GGG TGA GTA ACA CGT GAA TAA | Nitrospina 16S rRNA gene | 60 | |
| NitSSU_282R | PCR | TCA GGC CGG CTA AMC A | 60 | ||
| Nbacter1050F | PCR | CAC CTG TGC TCC ATG CTC CG | Nitrobacter 16S rRNA gene | 61 | |
| Nbacter1433R | PCR | CGG GTT AGC GCA CCG CCT | 62 | ||
| Eub341F-GC | PCR-DGGE | CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC GCC TAC GGG AGG CAG CAG | All bacterial 16S rRNA genes | 52 (modified) | |
| Univ907R | PCR-DGGE | CCG TCA ATT CCC TTT RAG TTT | 53 | ||
| Second or third step | |||||
| 27f | Cloning | AGA GTT TGA TCM TGG CTC AG | All bacterial 16S rRNA genes | 53 | |
| 1492r | Cloning | TAC GGY TAC CTT GTT ACG ACT T | 53 | ||
| nxrB169f | PCR | TAC ATG TGG TGG AAC A | Nitrospira and Nitrospina nxrB genea | 63 | |
| nxrB638r | PCR | CGG TTC TGG TCR ATC A | 63 | ||
| F1norA | PCR | CAG ACC GAC GTG TGC GAA AG | Nitrobacter nxrA gene | 64 | |
| R1norA | PCR | TCY ACA AGG AAC GGA AGG TC | 64 | ||
| EUB338 | FISH | GCT GCC TCC CGT AGG AGT | Most bacteria | 57 | |
| EUB338 II | FISH | GCA GCC ACC CGT AGG TGT | Most Planctomycetales | 65 | |
| EUB338 III | FISH | GCT GCC ACC CGT AGG TGT | Most Verrucomicrobiales | 65 | |
| NTG840 | FISH | CTA AGG AAG TCT CCT CCC | Nitrotoga | 12 | |
| Ntspa1431 | FISH | TTG GCT TGG GCG ACT TCA | Nitrospira lineage I | 66 | |
| Ntspa1151 | FISH | TTC TCC TGG GCA GTC TCT CC | Nitrospira lineage II | 66 |
The reverse primer has three mismatches to the nxrB gene of Nitrospina toward the 5′ end of the primer.
Subsequently, a bioreactor at room temperature (23°C), which had been successfully used for enrichment of NOB in our laboratory (28), was applied to increase the abundance of Nitrotoga cells. Modified medium without NaCl was fed to the bioreactor. Overgrown microorganisms attached to the polyester nonwoven fabric materials were regularly discharged. The ratios of NOB cells to the total bacterial population were estimated by fluorescence in situ hybridization (FISH) analysis by microscopic direct counting throughout the second and third enrichment steps. As expected, after incubation in the bioreactor for 4 months, Nitrotoga cells increased to 12% of the total bacteria. The abundances of Nitrospira lineages I and II reached 24% and 4%, respectively, although these Nitrospira spp. were not detected by Ntspa81F/Ntspa662R after the first enrichment step (see Fig. S1 in the supplemental material). The second enrichment step with the bioreactor was certainly effective for the growth of Nitrotoga. Nevertheless, the population ratio of NOB in the bioreactor showed that room temperature (23°C) placed Nitrospira in a more advantageous position than Nitrotoga.
To test whether temperature enhanced the cell growth of Nitrotoga bacteria, bioreactor samples were incubated in batch cultures at 4, 10, and 23°C. The batch test revealed that the ratio of Nitrotoga bacteria increased selectively at 10°C, conditions under which the growth of Nitrospira was inhibited. To operate the bioreactor at low temperature was difficult, and thus the microorganisms attached to the nonwoven fabric materials were transferred to batch culture and incubated at 10 to 16°C for 1 year to selectively enrich Nitrotoga cells. Consequently, the abundance of Nitrotoga increased to approximately 80% of the total microbial cells. The primer pairs F1norA/R1norA and nxrB169f/638r, targeting nxrA of Nitrobacter and nxrB of Nitrospira and Nitrospina, respectively, were used to check the possibility of contamination. The genes possessed by these NOB were not amplified. Therefore, no Nitrobacter, Nitrospina, or Nitrospira bacteria were detected by agarose gel electrophoresis. However, considering that the source sample was sand of the eelgrass zone, these marine NOB might be still present in the high-Nitrotoga enrichment below the detection limit of PCR. The presence of very few other NOB cells did not affect the physiological analysis of the Nitrotoga-enriched culture.
Phylogenetic analysis.
A partial 16S rRNA gene of Nitrotoga amplified by NTG200F/840R in the early stage culture of the first enrichment step was directly sequenced. The closest environmental clone was an uncultured bacterium clone from salt mash sediments (accession no. HQ272404, 99.6% identity). We named the initial Nitrotoga clone AM0.
In the high-Nitrotoga enrichment culture after the third enrichment step, the microbial community structure was analyzed based on 16S rRNA gene sequences (Table S1). A total of 29 clones picked for plasmid isolation were grouped into 16 operational taxonomic units (OTUs). The most abundant OTU, OTU1 (8/29 clones), defined as Nitrotoga sp. strain AM1, showed a high level of identity with the sequence of “Ca. Nitrotoga arctica” (99.6%) (Fig. 2). The closest isolate, based on the 16S rRNA gene sequence, was the Fe-oxidizing bacterium Sideroxydans lithotrophicus strain ES-1 (95.0%) (29). OTU1 belongs to a different cluster than the Nitrotoga clones detected in Yellow Sea intertidal beach seawater and salt marsh sediments. The 16S rRNA gene sequence of AM1 was different from that of AM0 (99.2%). The second largest OTU in the Nitrotoga-enriched culture (4/29 clones) was assigned to the recently proposed novel genus Pseudorhodoferax, which is in the class Betaproteobacteria and the family Comamonadaceae (30). The total number of bacterial species was estimated by Chao1 to be 30 (31).
FIG 2.
Phylogenetic tree based on 16S rRNA gene sequences of the genus Nitrotoga and representatives of the family Gallionellaceae as the outgroup. The tree was constructed using the maximum likelihood algorithm. Bootstrap values at the branch nodes were iterated 1,000 times. Sequences of Nitrotoga obtained in this study and other enriched cultures are in bold. The scale bar corresponds to 1% estimated sequence divergence. Accession numbers are shown to the right of the organism names/descriptions. DO, dissolved oxygen.
Morphology.
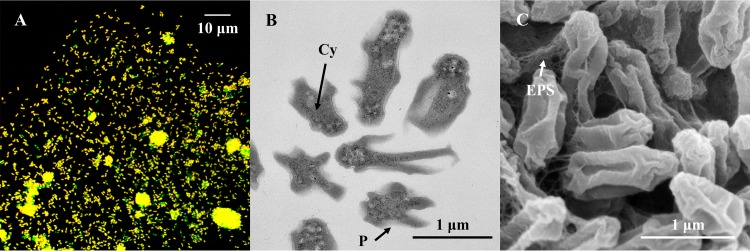
Nitrifying bacteria have genus-specific morphology and ultrastructure, for example, the presence or absence of intracytoplasmic membranes. The cell size and tendency to aggregation vary between species. Although other AOB and NOB isolated from activated sludge by a cell sorting system form single-species microcolonies (32–34), such microcolonies of Nitrotoga were not found in this study. The enriched Nitrotoga cells were planktonic or aggregated with heterotrophs (Fig. 3A). The multiple-species aggregates were mainly composed of Nitrotoga cells.
FIG 3.
Morphology of AM1. (A) Confocal laser scanning microscopy image of FISH-stained Nitrotoga cells (yellow) by Cy3-labeled NTG840, and other microorganisms (green) by fluorescein isothiocyanate (FITC)-labeled EUB338 mixture. (B) Overview of ultrathin sections of Nitrotoga-like bacteria observed by TEM. An extraordinarily wide periplasmic space (P) surrounded the cytoplasm (Cy). (C) SEM image of Nitrotoga-like bacterial cells loosely coupled by thin layers of extracellular polymeric substances (EPS).
We regarded dominant bacteria forming aggregates as Nitrotoga in the electron microscopic images. As shown by electron microscopy, a coccoid or short straight cytoplasm was surrounded by an extraordinarily wide periplasmic space, especially toward the long axis of the cells (Fig. 3B). Cross-sections of irregular star-shaped Nitrotoga cells enriched in this study resembled Nitrotoga sp. strain HAM-1 from activated sludge (35). The cells were loosely coupled by thin layers of extracellular polymeric substances, as well as other Nitrotoga cells (Fig. 3C) (23, 35). Planktonic cells of Nitrotoga-like bacteria ranged from 0.3 to 0.5 μm in the short axis and from 0.5 to 1.2 μm in the long axis. The overall appearance was a wrinkled rod shape.
Optimum temperature.
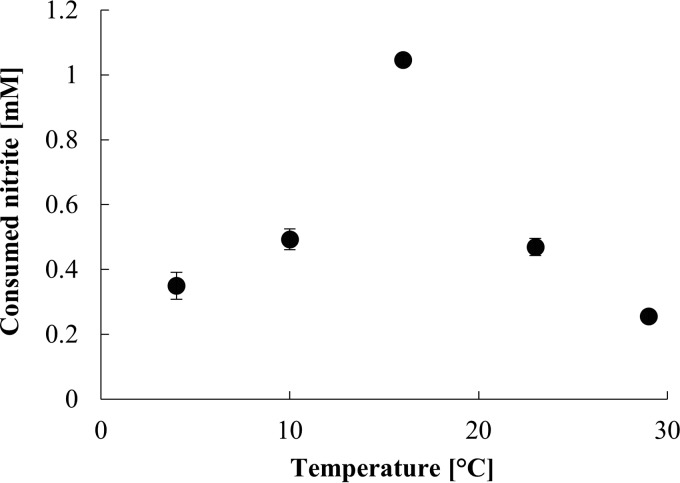
AM1 consumed nitrite within the temperature range of 4 to 29°C, as monitored over 3 days. The optimum temperature for nitrite consumption was 16°C, and nitrite oxidation activity at 10 to 23°C was >40% of the maximal activity (Fig. 4).
FIG 4.
Nitrite oxidation activity of AM1 during 3 days of incubation at different temperatures. Experiments were performed with transferred cells in early stationary phase. Error bars show standard deviations of biological triplicate measurements.
Kinetic parameters of nitrite oxidation.
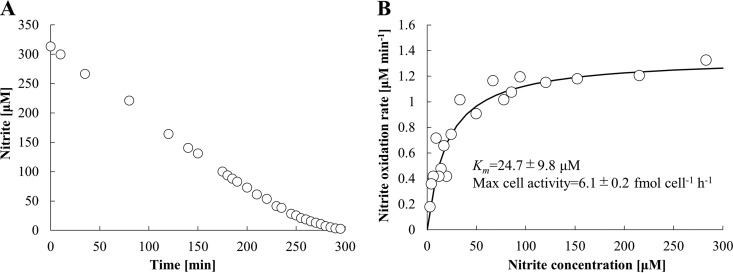
AM1 cells in the exponential-growth phase oxidized nitrite immediately after the addition of nitrite. Since the nitrite oxidation rate of a single Nitrotoga cell was too low to measure, the Nitrotoga enrichment culture was concentrated by centrifugation. Short-time centrifugation did not negatively affect the nitrite oxidation activity of Nitrotoga (Fig. S3). The half-saturation constant (apparent Km value) and maximum cell activity were calculated by manually fitting the data obtained to the Michaelis-Menten equation. The Km and maximum cell activity were determined to be 24.7 ± 9.8 μM NO2− and 6.1 ± 0.2 fmol NO2− · cell−1 · h−1, respectively (Fig. 5A and B). AM1 cells in exponential phase produced a little more nitrate than oxidized nitrite (data not shown). The concentration of excess nitrate had no influence on determination of the Km value.
FIG 5.
Representative data of half-saturation constants (apparent Km) for nitrite of exponential-growth-phase cells of AM1 at 16°C without shaking. (A) The initial nitrite concentration was adjusted to 0.3 mM. The samples were taken out every 2.5 to 20 min. (B) Michaelis-Menten plots were fitted by the least-squares method. The Km value and maximum cell activity were calculated from biological 3-fold measurements.
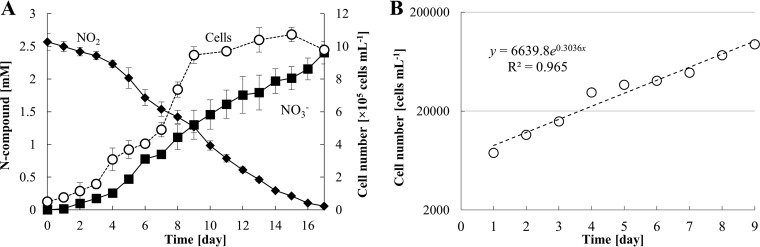
The generation time was also determined. AM1 stoichiometrically oxidized nitrite to nitrate and grew over time in nitrite-limited mineral medium (Fig. 6A). To facilitate single-cell counting with a microscopy, the samples were sonicated for an appropriate time. The Nitrotoga aggregates were completely disrupted to single planktonic cells without the loss of too many cells (Fig. S4). Generation time was calculated by fitting the cell growth curve to an exponential equation (Fig. 6B). Based on the change in cell number between days 1 and 9, the average generation time was calculated to be 54 h.
FIG 6.
Growth and nitrite oxidation activities of AM1. (A) The plots indicate nitrite concentration (diamonds), nitrate concentration (squares), and cell numbers (circles) in nitrite-limited mineral medium at 16°C. Each experiment was performed with transferred cells (10% inoculum) in early stationary phase. (B) Changes in the cell number from day 1 to day 9 were fitted to the exponential-growth curve. All error bars show the standard deviations of biological triplicate measurements.
Influence of stress on nitrite oxidation rate.
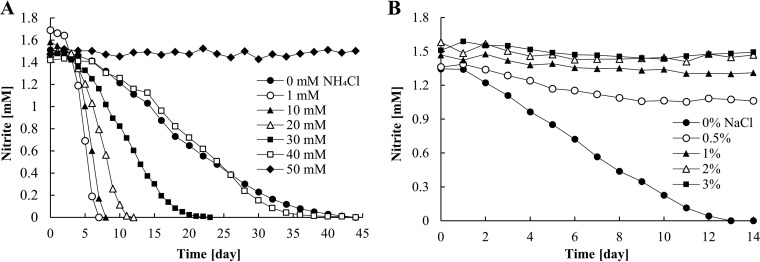
To investigate the influences of increasing concentrations of NH4Cl and NaCl on nitrite oxidation activity, AM1 was incubated in batch culture with medium containing NH4Cl or NaCl. Compared to a control without NH4Cl, the nitrite oxidation rates of AM1 were accelerated at concentrations below 30 mM NH4Cl (Fig. 7A). The highest nitrite oxidation rates were recorded in 1 or 10 mM NH4Cl. No nitrite oxidation occurred in the presence of 50 mM NH4Cl.
FIG 7.
Nitrite consumption of the enrichment culture of AM1 at 16°C. Incubations were performed with transferred cells in early stationary phase and in mineral medium supplemented with various NH4Cl concentrations (A) or NaCl concentrations (B). The experiments were started with inocula of 1% (A) and 10% (B). Noticeable results were obtained; thus, these experiments were performed once.
NaCl strongly affected the activity of AM1. Complete depletion of nitrite occurred only in medium without NaCl (Fig. 7B). About 0.3 mM NO2− was consumed over 2 weeks at 0.5% NaCl, but no more oxidation was observed after that. AM1 could not oxidize nitrite in NaCl-supplemented medium, although the salinity in standard seawater is generally about 3.5% (36).
DISCUSSION
Enrichment.
The 16S rRNA genes of the genus Nitrotoga are distributed in a variety of habitats, but only two highly enriched cultures have been obtained, one from permafrost-affected soil (12) and one from a biofilter of a cold-freshwater aquaculture plant (23). In this study, we successfully enriched a new member of the genus Nitrotoga from coastal sand. During the enrichment procedures, the cultures were checked regularly by type-specific primers and FISH analysis to monitor the growth of Nitrotoga. This routine allowed us to establish a protocol for enrichment of Nitrotoga. As a result, multiple enrichment steps led to the enrichment of Nitrotoga sp. AM1.
In the first enrichment step, an initial sample from coastal sand was incubated in 2% NaCl-amended mineral medium at 10°C, but the growth of AM0 was limited in ASW medium. Similarly, this condition was not suitable for enrichment of AM1. In fact, later physiological experiments revealed that AM1 cannot tolerate concentrations higher than 0.5% NaCl (Fig. 7B). Therefore, the abundance of Nitrotoga was below the microscopic detection limit, even after long-term incubation at low temperature in ASW medium. This suggested that AM1 enriched from coastal sand might have originated on land and has low activity in the seafloor.
Since Nitrotoga was not enriched any more despite extended incubation for 5 years, the culture conditions were renewed. In the second enrichment step, modified medium without NaCl was continuously fed into the bioreactor, which allowed the growth of Nitrotoga. This is consistent with a previous study (37) where an influx of synthetic medium with 4% NaCl in a sequencing batch reactor resulted in a decreased nitrogen consumption-specific rate by non-salt-adapted nitrifiers from nitrifying activated sludge (genera Nitrosomonas, Nitrobacter, and Nitrospira), but their activities were partially restored after salt stress was removed.
In the last enrichment step, the culture was transferred from the bioreactor into batches incubated at 10 to 16°C to selectively enrich Nitrotoga. On incubation for 4 months in the bioreactor at 23°C, Nitrospira outcompeted Nitrotoga. This was simply because the culture conditions were more suitable for Nitrospira than Nitrotoga. Growth tests using microorganisms attached to the fabric materials within the bioreactor showed that Nitrotoga grew well at 4 or 10°C. In contrast, the optimum temperature for the culture of Nitrospira cells is 28 to 32°C (23, 32, 33). Thus, temperature was a key driver regulating the competition between Nitrotoga and Nitrospira. Previous studies demonstrated that Nitrotoga outgrew Nitrospira during long-term cultivation at 5 and 10°C (35, 38). Consistent with these studies, a final enrichment step in batch culture at 10 to 16°C resulted in AM1 enrichment to approximately 80% of the total bacterial population. Incubation in low temperature without NaCl was an important process to obtain the new high level of Nitrotoga enrichment.
The sampling site was a freshwater-saltwater interface, so nonhalophilic Nitrotoga sp. was enriched depending on the culture conditions. For the same reason, the ratios of Nitrospira lineages I and II increased in the second enrichment step. AM1 and these Nitrospira lineages probably survive as very minor members during the initial 5 years.
Response to free ammonia.
NOB and ammonia-oxidizing microbes coexist in environments where nitrification occurs (39). The two coupled steps imply that NOB are exposed to NH3/NH4+. Free ammonia, i.e., unionized ammonia, affects potential nitrite oxidation rates (40).
According to Blackburne and colleagues, Nitrospira had an inhibition threshold for free ammonia between 0.04 and 0.08 mg of NH3-N · liter−1 (41). Furthermore, a recent study based on Nitrospira isolates demonstrated that the nitrite oxidation activity of Nitrospira strains ND1 and NJ1 was inhibited at concentrations above 0.85 and 4.3 mg of NH3-N · liter−1, respectively (42). With respect to thermophilic NOB related to Nitrospira calida derived from a nutrient-rich composting fertilizer, its sensitivity for free ammonia was lower than those of other reported Nitrospira spp., characterized by half-maximal inhibition of 5.0 mg of NH3-N · liter−1 (43). In this study, the presence of free ammonia at 0.04 to 1.30 mg of NH3-N · liter−1 (1 to 30 mM NH4Cl [pH 8.1]) prompted the nitrite oxidation by AM1. Thus, AM1 had a much higher tolerance for free ammonia than Nitrospira, although the nitrite oxidation activity of AM1 was completely inhibited at 30.2 mg of NH3-N · liter−1 (50 mM NH4Cl). These results suggest that free ammonia can be a selective factor for the enrichment of Nitrotoga versus Nitrospira.
Interestingly, the nitrite oxidation rate of AM1 was higher at concentrations between 1 and 30 mM NH4Cl than in the control (0 mM NH4Cl). Nitrobacter winogradskyi grew well in 1 mM NH4Cl-amended medium, and the expression level of Nwi_0718, which is the associated hypothetical assimilatory nitrite reductase NirBC encoded by Nwi_0719 and Nwi_0720, was 5.0-log-fold lower than that in unamended culture (44). Similarly, AM1 could save energy for assimilatory nitrite reduction using ammonium in the 1 to 30 mM NH4Cl-augmented treatments, so that complete depletion of nitrite was faster than in other treatments.
Physiological characteristics compared with other Nitrotoga and Nitrospira.
This cultivation-based study presents further physiological information on Nitrotoga. In contrast to the relatively high 16S rRNA gene sequence similarity of 99.0 to 99.6% among “Ca. Nitrotoga arctica,” Nitrotoga sp. strains HW29 and AM1, their physiological properties, such as optimum temperature for nitrite oxidation, pH for cultivation, and kinetic parameters, are different (Table 2).
TABLE 2.
Comparison of characteristics among Nitrotoga-enriched cultures investigated in detail
| Characteristic | AM1 | “Ca. Nitrotoga arctica”a | HW29b |
|---|---|---|---|
| Sample source | Coastal sand | Permafrost-affected soil | RAS (fish tank) |
| Tendency to aggregate | Weak | Weak | Weak |
| Overall appearance | Wrinkled rod shape | Irregular coccoid | Irregular coccoid |
| Optimum temp (°C) | 16 | 13 | 22 |
| Applied pH for cultivation | 8.0–8.3 (7.8)c | 7.4–7.6 | 6.8–7.4 |
| Km (μM) | 24.7 ± 9.8 | 58 ± 28 | ND |
| Maximum cell activity (fmol · cell−1 · h−1) | 6.1 ± 0.2 | 2 | ND |
| Generation time (h) | 54 | 44 | ND |
Low temperature enhanced the activities of the three Nitrotoga-enriched cultures, but their optimum temperatures ranged from 13°C to 22°C. Slightly acidic pH provided an advantage for Nitrotoga sp. HW29 (23), while the other Nitrotoga enrichment cultures were incubated in media with pH values of 7.4 to 8.3 (Table 2). The affinities for nitrite also differed among the enrichment cultures; this is an important factor that can determine the community structure of NOB (24, 45, 46). The Km value of AM1 (25 μM NO2−) indicated better adaptation to low nitrite concentration than that of “Ca. Nitrotoga arctica” (58 μM NO2−), although its affinity was still lower than or equal to those of Nitrospira defluvii (9 μM NO2−), Nitrospira moscoviensis (9 μM NO2−), and Nitrospira lenta BS10 (27 μM NO2−) (24). The generation times of the Nitrotoga-enriched cultures were 54 h (Nitrotoga sp. AM1) and 44 h (“Ca. Nitrotoga arctica”), which were ≥7 h longer than that of Nitrospira cultures (24). Diversity of physiological characteristics is thus observed in the genus Nitrotoga as well as Nitrobacter and Nitrospira, whereas Nitrotoga and Nitrobacter form a monophyletic clade, and Nitrospira consists of deeply branching lineages (10, 47–49).
Initially, AM0 was detected in the early stage culture of the first enrichment step, which was potentially halophilic Nitrotoga bacteria. The growth of AM0 was confirmed by PCR-DGGE, although the cells were below the detection limit of microscopic observation without cell concentration. Not all halophilic NOB grow in artificial marine medium prepared with NaCl (7, 50), indicating that AM0 might be enriched in natural seawater at low temperature. Interestingly, the partial 16S rRNA gene sequence of AM0 was closely related to clones of Nitrotoga retrieved from coastal ecosystems (accession numbers GU061253 and HQ272404 in Fig. 2). This suggests the possibility that these clones related to Nitrotoga are marine NOB. The abundance of AM0 was decreased in the second enrichment step. Probably, this is because the mineral medium without NaCl fed to the bioreactor collapsed ion homeostasis of AM0. After culture conditions were changed, AM1 (which is phylogenetically different from AM0) was successfully enriched.
In conclusion, this study revealed previously unknown physiological characteristics of Nitrotoga. AM1 showed adaptation to low temperature and relatively high concentrations of free ammonia. Considering that the few Nitrotoga cultures characterized to date are stimulated by specific environmental conditions, such as low temperature and slightly acidic pH, Nitrotoga might grow under such conditions which are unfavorable for other NOB and play an important role in the nitrogen cycle in natural and engineered systems. The availability of pure cultures and genomic analyses of Nitrotoga will shed further light on their microbiology and unique physiological properties.
MATERIALS AND METHODS
Sampling and enrichment.
Sand samples from an eelgrass (Z. marina) zone were collected 0 to 5 cm below the seafloor at a water depth of about 4 m on 16 July 2009. The seawater temperature above the eelgrass at the time of sampling was 19°C. The outline of the enrichment procedures is shown in Fig. 1. The first enrichment before the serial dilution was started using the following artificial seawater (ASW) medium: 34.5 mg · liter−1 NaNO2 as the sole energy and nitrogen source, 20 g · liter−1 NaCl, 10.64 g · liter−1 MgCl2·6H2O, 1.1 g · liter−1 CaCl2, 660 mg · liter−1 KCl, 10 mg · liter−1 K2HPO4, 3.91 g · liter−1 Na2SO4, 3 mg · liter−1 EDTA-Fe(III), 119 mg · liter−1 NaHCO3, and 1 ml · liter−1 nonchelated trace element mixture (51). The cultures incubated at 10°C were transferred (10% inoculum) to fresh medium when the absence of nitrite was confirmed. Subsequent serial dilutions (10−1 to 10−10) were performed in test tubes. For the serial dilution step and incubation after the serial dilutions, 22.3 ml of the filtered 2.5% NaHCO3 solution and 1 ml of the trace element mixture (51) were added to 1 liter of autoclaved ASW medium. The pH was adjusted to 7.8 with 1 M HCl or 3 M NaOH. After the serial dilutions, nitrite-oxidizing cultures were inoculated in a 100-ml flask including 30 ml of the modified ASW medium and then incubated at 10°C in the dark without agitation. Detailed information on the initial enrichment step is shown in Fig. S1.
As the second enrichment step to increase total microorganisms, the culture incubated at 10°C was inoculated into a continuous feeding bioreactor with nonwoven fabric materials (0.7-cm thickness; Japan Vilene) as biomass carriers (28). The bioreactor was operated at room temperature (23°C) with aeration. The effluent nitrite concentration was maintained below 0.07 mM. The influent nitrite concentration was controlled at 0.7 to 1.4 mM. The composition of mineral medium fed into the bioreactor was modified from that in a previous report (28). The modified inorganic medium comprised 25.4 mg · liter−1 K2HPO4, 40.6 mg · liter−1 MgSO4·7H2O, 6.6 mg · liter−1 CaCl2·2H2O, 3.2 mg · liter−1 FeSO4·7H2O, 54.2 μg · liter−1 MnSO4·5H2O, 49.4 μg · liter−1 H3BO3, 43.1 μg · liter−1 ZnSO4·7H2O, 27.6 μg · liter−1 Na2Mo4O4, and 25 μg · liter−1 CuSO4·5H2O. Unless otherwise noted, the modified mineral medium was used for enrichment and physiological experiments. Microbial community structures were observed routinely using fluorescence in situ hybridization (FISH) probes. The ratios of FISH-stained NOB to total bacteria were quantitatively measured by a microscopic direct counting method (see below). To determine the effect of temperature on Nitrotoga growth, aliquots of the bioreactor culture were initially incubated in Erlenmeyer flasks at 4, 10, and 23°C. Subsequently, biomass attached to the nonwoven fabric materials was transferred to Erlenmeyer flasks, and selective enrichment in batch culture was performed in the dark at 10 to 16°C without shaking. The initial nitrite concentration in the flasks was 1.4 mM NaNO2. The culture was transferred to fresh medium regularly to avoid any influence of metabolite accumulation.
PCR and phylogenetic analyses.
For the first enrichment culture, DNA was extracted using a DNeasy blood and tissue kit (Qiagen, Valencia, CA), and DNA fragments containing NOB 16S rRNA genes were amplified in TaKaRa Ex Taq (TaKaRa Bio, Otsu, Japan) using type-specific primer pairs (listed in Table 1). The Nitrotoga-PCR product was visualized on an agarose gel stained with ethidium bromide, purified with a Qiagen II gel extraction kit (Qiagen), and then cloned with a TOPO TA cloning kit (Life Technologies, Carlsbad, CA). The M13F/M13R PCR product was sequenced with a BigDye Terminator version 3.1 (Applied Biosystems, Foster City, CA) on a 3130xl genetic analyzer (Applied Biosystems). Denaturing gradient gel electrophoresis (DGGE) was used to assess the absence of Nitrotoga in the first enrichment culture after the serial dilutions. For DGGE analysis, DNA fragments encoding bacterial 16S rRNA genes were amplified in TaKaRa Ex Taq (TaKaRa Bio) PCR cocktail with the primers Eub341F with GC-clamp and Univ907R (52). Several DGGE bands on the gel image were purified and used as the template DNA for sequencing.
DNA was extracted from the high Nitrotoga enrichment culture in the third enrichment step with the Isoplant II extraction kit (Nippon Gene, Tokyo, Japan), as per the manufacturer's instructions. Bacterial 16S rRNA genes were amplified with the eubacterial primers 27f and 1492r (53). PCR products were purified using the Wizard SV gel and PCR cleanup system (Promega, Tokyo, Japan) and cloned into the pGEM-T Easy vector system (Promega), as per the manufacturer's instructions. The cloned 16S rRNA genes were sequenced by Fasmac (Kanagawa, Japan). The nucleotide sequences obtained were combined into operational taxonomic units (OTUs) based on 16S rRNA gene similarities of ≥98.7% (54). OTUs were assigned to the lowest possible Silva taxonomy (55). OTUs belonging to the genus Nitrotoga and related clone sequences were aligned using MEGA 6.06 (56). A phylogenetic tree was constructed based on the maximum likelihood algorithm in MEGA.
FISH.
Microbial community structures during the second and third enrichment processes were checked regularly by FISH analysis. FISH was conducted based on a previous protocol (57). The oligonucleotide probes used in this study are listed in Table 1. FISH-stained cells were observed and recorded using a fluorescence microscope (Axioskop 2 Plus; Carl Zeiss, Oberkochen, Germany) or confocal laser scanning microscope (FV1000-IX81-S; Olympus, Tokyo, Japan) after dropping SlowFade antifade reagents (Thermo Fisher Scientific, Waltham, MA).
Disruption of Nitrotoga aggregates for cell counting.
Since Nitrotoga formed aggregates with heterotrophic bacteria, single planktonic cells were counted after ultrasonic treatment (model Q55; QSonica, Newtown, CT, USA) for 30 s at an amplitude of dial 30 on ice. Cell numbers were counted directly by microscopic observation. The average cell numbers were determined from at least 10 representative microscopic images of enrichment samples.
Electron microscopy.
Ultrathin sections of enriched Nitrotoga cells were analyzed with a transmission electron microscope (TEM) (JEM-1200EX; JEOL, Tokyo, Japan) at 80 kV. The cell appearance was viewed in a scanning electron microscope (SEM) (JSM-6320F; JEOL) at 5 kV. These analyses were conducted at the Hanaichi Ultrastructure Research Institute, Okazaki, Japan, as previously described (32).
Physiological analyses.
Physiological characterization was conducted in the modified mineral medium using early stationary-phase cells, which were incubated for 1 to 2 days after nitrite was completely depleted, at a temperature of 16°C, unless otherwise noted. The physiological experiments were performed at low nitrite concentrations (≤2.6 mM NO2−) to avoid inhibition of Nitrotoga cell growth.
Optimum temperature for nitrite oxidation, and tolerance for NH4Cl and NaCl.
To determine the optimum temperature for nitrite oxidation, enrichment cultures were incubated at 4 to 29°C for 3 days. Samples (10 ml) were inoculated into 50-ml test tubes closed with rubber stoppers and shaken (100 rpm). The experiment was performed in biological triplicates. To test the tolerance limits, NH4Cl or NaCl was added in different concentrations, with the pH maintained at 8.1. Nitrotoga-enriched culture was inoculated into Erlenmeyer flasks. Samples were taken regularly, and nitrite concentrations were measured colorimetrically with the Griess reagent (58). The tolerance experiments against NH4Cl and NaCl were performed independently once.
The free ammonia concentration was calculated from the NH4Cl concentration, pH value, and temperature, as follows (59):
| (1) |
where Kb is the ionization constant of the ammonia equilibrium equation and Kw is the ionization constant of water. Kb/Kw may be related to temperature in the following manner:
| (2) |
Also, the proportion of free ammonia depends on salinity. However, we neglected the effect of salinity, because the mineral medium used in the tolerance experiment for free ammonia contained negligible amounts of salts. The composition of the mineral medium was described above.
Determination of kinetic parameters.
The half-saturation constant Km was calculated based on the relationship between nitrite concentration and the nitrite oxidation rate. Cells in the exponential-growth phase, which were incubated for 2 to 4 days after nitrite addition, were centrifuged (2,900 × g, 30 min), and then the supernatant was removed. Determination of the exponential-growth phase referred to the experiment for generation time. Fresh mineral medium containing 0.3 mM nitrite was added to a 5-ml glass chamber in a final volume of 3 ml. The glass chamber was incubated at 16°C without shaking until all nitrite was completely depleted. During incubation, 50-μl aliquots of the samples were taken out every 2.5 to 20 min and immediately heated at 95°C for 5 min to inactivate the Nitrotoga cells. After measurement for nitrite concentration with the Griess reagent (58), apparent Km values were determined by the least-squares method. Here, we defined the half-saturation constant for nitrite concentration (in micromoles) as the apparent Km value, because purified enzymes were not used in these experiments. However, cell growth can be neglected in a short-term experiment. The difference between the nitrite oxidation rate (V) obtained from the experiment described above and V estimated by the Michaelis-Menten equation was minimized. Maximum cell activity (in femtomoles NO2− per cell per hour) was calculated from the maximum nitrite oxidation rate per hour and per single cell.
To measure generation time, early stationary-phase cells were transferred into Erlenmeyer flasks, and 2-ml samples were taken regularly to analyze nitrite and nitrate concentrations using ion chromatography. These experiments for determination of kinetics parameters were performed in biological triplicates.
Chemical analysis.
The concentrations of nitrite and nitrate were determined by ion chromatography with a TSKgel SuperIC-Anion HS (IC-2010; Tosoh, Tokyo, Japan). Nitrite concentration was alternatively checked colorimetrically with Griess reagent (58). All samples measured were filtered through 0.22-μm-membrane filters (SLGP033NB; Merck Millipore, Billerica, MA) before analysis.
Accession number(s).
All 16S rRNA gene sequences in this study were deposited to the DNA Data Bank of Japan under accession numbers LC190436 to LC190451 and LC217313. No chimeric sequences were submitted.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful for many helpful discussions with members of the Tsuneda laboratory. We also thank Norisuke Ushiki for technical support and the staff of the General Research Institute with their support in the sequencing at the Nihon University.
This research was supported by a grant-in-aid for young scientists (B) from the Japan Society for the Promotion of Science (JSPS) (16K18609) (to H.F.), by a Scientific Research Grant from the Mayekawa Houonkai Foundation (to H.F.), and by the Large Research Projects program from the Institute for Fermentation, Osaka (to S.T.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00549-17.
REFERENCES
- 1.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. 2015. Complete nitrification by Nitrospira bacteria. Nature 528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Kessel MA, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJ, Kartal B, Jetten MS, Lücker S. 2015. Complete nitrification by a single microorganism. Nature 528:555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin JT, Stewart V. 1998. Nitrate assimilation by bacteria. Adv Microb Physiol 39:1–30. doi: 10.1016/S0065-2911(08)60014-4. [DOI] [PubMed] [Google Scholar]
- 4.Philips S, Laanbroek HJ, Verstraete W. 2002. Origin, causes and effects of increased nitrite concentrations in aquatic environments. Rev Environ Sci Biotechnol 1:115–141. doi: 10.1023/A:1020892826575. [DOI] [Google Scholar]
- 5.Daims H, Lücker S, Wagner M. 2016. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol 24:699–712. doi: 10.1016/j.tim.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngugi DK, Blom J, Stepanauskas R, Stingl U. 2016. Diversification and niche adaptations of Nitrospina-like bacteria in the polyextreme interfaces of Red Sea brines. ISME J 10:1383–1399. doi: 10.1038/ismej.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watson SW, Bock E, Valois FW, Waterbury JB, Schlosser U. 1986. Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch Microbiol 144:1–7. doi: 10.1007/BF00454947. [DOI] [Google Scholar]
- 8.Hovanec TA, Taylor LT, Blakis A, Delong EF. 1998. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl Environ Microbiol 64:258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juretschko S, Timmermann G, Schmid M, Schleifer KH, Pommerening-Roser A, Koops HP, Wagner M. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol 64:3042–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol 67:5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foesel BU, Gieseke A, Schwermer C, Stief P, Koch L, Cytryn E, de la Torre JR, van Rijn J, Minz D, Drake HL, Schramm A. 2008. Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol Ecol 63:192–204. doi: 10.1111/j.1574-6941.2007.00418.x. [DOI] [PubMed] [Google Scholar]
- 12.Alawi M, Lipski A, Sanders T, Pfeiffer EM, Spieck E. 2007. Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J 1:256–264. doi: 10.1038/ismej.2007.34. [DOI] [PubMed] [Google Scholar]
- 13.Sattin SR, Cleveland CC, Hood E, Reed SC, King AJ, Schmidt SK, Robeson MS, Ascarrunz N, Nemergut DR. 2009. Functional shifts in unvegetated, perhumid, recently-deglaciated soils do not correlate with shifts in soil bacterial community composition. J Microbiol 47:673–681. doi: 10.1007/s12275-009-0194-7. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt SK, Nemergut DR, Miller AE, Freeman KR, King AJ, Seimon A. 2009. Microbial activity and diversity during extreme freeze-thaw cycles in periglacial soils, 5400 m elevation, Cordillera Vilcanota, Peru. Extremophiles 13:807–816. doi: 10.1007/s00792-009-0268-9. [DOI] [PubMed] [Google Scholar]
- 15.Christner BC, Priscu JC, Achberger AM, Barbante C, Carter SP, Christianson K, Michaud AB, Mikucki JA, Mitchell AC, Skidmore ML, Vick-Majors TJ, WISSARD Science Team. 2014. A microbial ecosystem beneath the West Antarctic ice sheet. Nature 512:310–313. doi: 10.1038/nature13667. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Wu L, Boden R, Hillebrand A, Kumaresan D, Moussard H, Baciu M, Lu Y, Colin Murrell J. 2009. Life without light: microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J 3:1093–1104. doi: 10.1038/ismej.2009.57. [DOI] [PubMed] [Google Scholar]
- 17.Roden EE, McBeth JM, Blöthe M, Percak-Dennett EM, Fleming EJ, Holyoke RR, Luther GW, Emerson D, Schieber J. 2012. The microbial ferrous wheel in a neutral pH groundwater seep. Front Microbiol 3:172. doi: 10.3389/fmicb.2012.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li D, Qi R, Yang M, Zhang Y, Yu T. 2011. Bacterial community characteristics under long-term antibiotic selection pressures. Water Res 45:6063–6073. doi: 10.1016/j.watres.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Na H, Kim OS, Yoon SH, Kim Y, Chun J. 2011. Comparative approach to capture bacterial diversity of coastal waters. J Microbiol 49:729–740. doi: 10.1007/s12275-011-1205-z. [DOI] [PubMed] [Google Scholar]
- 20.Martiny JB, Eisen JA, Penn K, Allison SD, Horner-Devine MC. 2011. Drivers of bacterial beta-diversity depend on spatial scale. Proc Natl Acad Sci U S A 108:7850–7854. doi: 10.1073/pnas.1016308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lücker S, Schwarz J, Gruber-Dorninger C, Spieck E, Wagner M, Daims H. 2015. Nitrotoga-like bacteria are previously unrecognized key nitrite oxidizers in full-scale wastewater treatment plants. ISME J 9:708–720. doi: 10.1038/ismej.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saunders AM, Albertsen M, Vollertsen J, Nielsen PH. 2016. The activated sludge ecosystem contains a core community of abundant organisms. ISME J 10:11–20. doi: 10.1038/ismej.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hüpeden J, Wegen S, Off S, Lucker S, Bedarf Y, Daims H, Kuhn C, Spieck E. 2016. Relative abundance of Nitrotoga spp. in a biofilter of a cold-freshwater aquaculture plant appears to be stimulated by slightly acidic pH. Appl Environ Microbiol 82:1838–1845. doi: 10.1128/AEM.03163-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowka B, Daims H, Spieck E. 2015. Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl Environ Microbiol 81:745–753. doi: 10.1128/AEM.02734-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herbert R. 1999. Nitrogen cycling in coastal marine ecosystems. FEMS Microbiol Rev 23:563–590. doi: 10.1111/j.1574-6976.1999.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 26.Caffrey JM, Kemp WM. 1990. Nitrogen cycling in sediments with estuarine populations of Potamogeton perfoliatus and Zostera marina. Mar Ecol Prog Ser 66:147–160. doi: 10.3354/meps066147. [DOI] [Google Scholar]
- 27.Ando Y, Nakagawa T, Takahashi R, Yoshihara K, Tokuyama T. 2009. Seasonal changes in abundance of ammonia-oxidizing archaea and ammonia-oxidizing bacteria and their nitrification in sand of an eelgrass zone. Microbes Environ 24:21–27. doi: 10.1264/jsme2.ME08536. [DOI] [PubMed] [Google Scholar]
- 28.Fujitani H, Aoi Y, Tsuneda S. 2013. Selective enrichment of two different types of Nitrospira-like nitrite-oxidizing bacteria from a wastewater treatment plant. Microbes Environ 28:236–243. doi: 10.1264/jsme2.ME12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emerson D, Field EK, Chertkov O, Davenport KW, Goodwin L, Munk C, Nolan M, Woyke T. 2013. Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front Microbiol 4:254. doi: 10.3389/fmicb.2013.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruland N, Bathe S, Willems A, Steinbuchel A. 2009. Pseudorhodoferax soli gen. nov., sp. nov. and Pseudorhodoferax caeni sp. nov., two members of the class Betaproteobacteria belonging to the family Comamonadaceae. Int J Syst Evol Microbiol 59:2702–2707. doi: 10.1099/ijs.0.006791-0. [DOI] [PubMed] [Google Scholar]
- 31.Chao A. 2005. Species estimation and applications, p 7907–7916. In Balakrishnan N, Read CB, Vidakovic B (ed), Encyclopedia of statistical sciences, 2nd ed, vol 12 John Wiley & Sons, New York, NY. [Google Scholar]
- 32.Fujitani H, Ushiki N, Tsuneda S, Aoi Y. 2014. Isolation of sublineage I Nitrospira by a novel cultivation strategy. Environ Microbiol 16:3030–3040. doi: 10.1111/1462-2920.12248. [DOI] [PubMed] [Google Scholar]
- 33.Ushiki N, Fujitani H, Aoi Y, Tsuneda S. 2013. Isolation of Nitrospira belonging to sublineage II from a wastewater treatment plant. Microbes Environ 28:346–353. doi: 10.1264/jsme2.ME13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujitani H, Kumagai A, Ushiki N, Momiuchi K, Tsuneda S. 2015. Selective isolation of ammonia-oxidizing bacteria from autotrophic nitrifying granules by applying cell-sorting and sub-culturing of microcolonies. Front Microbiol 6:1159. doi: 10.3389/fmicb.2015.01159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alawi M, Off S, Kaya M, Spieck E. 2009. Temperature influences the population structure of nitrite-oxidizing bacteria in activated sludge. Environ Microbiol Rep 1:184–190. doi: 10.1111/j.1758-2229.2009.00029.x. [DOI] [PubMed] [Google Scholar]
- 36.Millero FJ, Feistel R, Wright DG, McDougall TJ. 2008. The composition of standard seawater and the definition of the reference-composition salinity scale. Deep-Sea Res Pt I 55:50–72. doi: 10.1016/j.dsr.2007.10.001. [DOI] [Google Scholar]
- 37.Moussa M, Sumanasekera D, Ibrahim S, Lubberding H, Hooijmans C, Gijzen H, van Loosdrecht M. 2006. Long term effects of salt on activity, population structure and floc characteristics in enriched bacterial cultures of nitrifiers. Water Res 40:1377–1388. doi: 10.1016/j.watres.2006.01.029. [DOI] [PubMed] [Google Scholar]
- 38.Karkman A, Mattila K, Tamminen M, Virta M. 2011. Cold temperature decreases bacterial species richness in nitrogen-removing bioreactors treating inorganic mine waters. Biotechnol Bioeng 108:2876–2883. doi: 10.1002/bit.23267. [DOI] [PubMed] [Google Scholar]
- 39.Arp DJ, Bottomley PJ. 2006. Nitrifiers: more than 100 years from isolation to genome sequences. Microbe 1:229–234. [Google Scholar]
- 40.Kim D, Lee D, Keller J. 2006. Effect of temperature and free ammonia on nitrification and nitrite accumulation in landfill leachate and analysis of its nitrifying bacterial community by FISH. Bioresour Technol 97:459–468. doi: 10.1016/j.biortech.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 41.Blackburne R, Vadivelu VM, Yuan Z, Keller J. 2007. Kinetic characterisation of an enriched Nitrospira culture with comparison to Nitrobacter. Water Res 41:3033–3042. doi: 10.1016/j.watres.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 42.Ushiki N, Jinno M, Fujitani H, Suenaga T, Terada A, Tsuneda S. 2017. Nitrite oxidation kinetics of two Nitrospira strains: the quest for competition and ecological niche differentiation. J Biosci Bioeng 123:581–589. doi: 10.1016/j.jbiosc.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 43.Courtens EN, Spieck E, Vilchez-Vargas R, Bodé S, Boeckx P, Schouten S, Jauregui R, Pieper DH, Vlaeminck SE, Boon N. 2016. A robust nitrifying community in a bioreactor at 50°C opens up the path for thermophilic nitrogen removal. ISME J 10:2293–2303. doi: 10.1038/ismej.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sayavedra-Soto L, Ferrell R, Dobie M, Mellbye B, Chaplen F, Buchanan A, Chang J, Bottomley P, Arp D. 2015. Nitrobacter winogradskyi transcriptomic response to low and high ammonium concentrations. FEMS Microbiol Lett 362:1–7. doi: 10.1093/femsle/fnu040. [DOI] [PubMed] [Google Scholar]
- 45.Schramm A, de Beer D, van den Heuvel J, Ottengraf S, Amann R. 1999. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol 65:3690–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim DJ, Kim SH. 2006. Effect of nitrite concentration on the distribution and competition of nitrite-oxidizing bacteria in nitratation reactor systems and their kinetic characteristics. Water Res 40:887–894. doi: 10.1016/j.watres.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 47.Sorokin DY, Muyzer G, Brinkhoff T, Kuenen JG, Jetten MS. 1998. Isolation and characterization of a novel facultatively alkaliphilic Nitrobacter species, N. alkalicus sp. nov. Arch Microbiol 170:345–352. doi: 10.1007/s002030050652. [DOI] [PubMed] [Google Scholar]
- 48.Lebedeva EV, Alawi M, Maixner F, Jozsa PG, Daims H, Spieck E. 2008. Physiological and phylogenetic characterization of a novel lithoautotrophic nitrite-oxidizing bacterium, ‘Candidatus Nitrospira bockiana.’ Int J Syst Evol Microbiol 58:242–250. doi: 10.1099/ijs.0.65379-0. [DOI] [PubMed] [Google Scholar]
- 49.Lebedeva EV, Off S, Zumbragel S, Kruse M, Shagzhina A, Lucker S, Maixner F, Lipski A, Daims H, Spieck E. 2011. Isolation and characterization of a moderately thermophilic nitrite-oxidizing bacterium from a geothermal spring. FEMS Microbiol Ecol 75:195–204. doi: 10.1111/j.1574-6941.2010.01006.x. [DOI] [PubMed] [Google Scholar]
- 50.Spieck E, Keuter S, Wenzel T, Bock E, Ludwig W. 2014. Characterization of a new marine nitrite oxidizing bacterium, Nitrospina watsonii sp. nov., a member of the newly proposed phylum “Nitrospinae.” Syst Appl Microbiol 37:170–176. doi: 10.1016/j.syapm.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 51.Widdel F, Bak F. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p 3352–3378. In Balows A, Tr̈uper HG, Dworkin M, Harder W, Schleifer KH (ed), The prokaryotes, 2nd ed, vol 4 Springer-Verlag, New York, NY. [Google Scholar]
- 52.Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified gene coding for 16S rRNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley, Chichester, United Kingdom. [Google Scholar]
- 54.Stackebrandt E, Ebers J. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155. [Google Scholar]
- 55.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hewitt EJ, Nicholas DJD. 1964. Enzymes of inorganic nitrogen metabolism, p 167–172. In Linskens HF, Sanwal BD, Tracey MV (ed), Modern methods of plant analysis, vol 7 Springer, Heidelberg, Germany. [Google Scholar]
- 59.Anthonisen AC, Loehr RC, Prakasam TB, Srinath EG. 1976. Inhibition of nitrification by ammonia and nitrous acid. J Water Pollut Control Fed 48:835–852. [PubMed] [Google Scholar]
- 60.Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF. 2007. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific subtropical gyre. Environ Microbiol 9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- 61.Freitag TE, Chang L, Clegg CD, Prosser JI. 2005. Influence of inorganic nitrogen management regime on the diversity of nitrite-oxidizing bacteria in agricultural grassland soils. Appl Environ Microbiol 71:8323–8334. doi: 10.1128/AEM.71.12.8323-8334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagner M, Rath G, Koops HP, Flood J, Amann R. 1996. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci Technol 34:237–244. doi: 10.1016/0273-1223(96)00514-8. [DOI] [Google Scholar]
- 63.Pester M, Maixner F, Berry D, Rattei T, Koch H, Lucker S, Nowka B, Richter A, Spieck E, Lebedeva E, Loy A, Wagner M, Daims H. 2014. nxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ Microbiol 16:3055–3071. doi: 10.1111/1462-2920.12300. [DOI] [PubMed] [Google Scholar]
- 64.Poly F, Wertz S, Brothier E, Degrange V. 2008. First exploration of Nitrobacter diversity in soils by a PCR cloning-sequencing approach targeting functional gene nxrA. FEMS Microbiol Ecol 63:132–140. doi: 10.1111/j.1574-6941.2007.00404.x. [DOI] [PubMed] [Google Scholar]
- 65.Daims H, Ramsing NB, Schleifer KH, Wagner M. 2001. Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescence in situ hybridization. Appl Environ Microbiol 67:5810–5818. doi: 10.1128/AEM.67.12.5810-5818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M, Daims H. 2006. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol 8:1487–1495. doi: 10.1111/j.1462-2920.2006.01033.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.