ABSTRACT
The Bacillus subtilis trpEDCFBA operon is regulated by a transcription attenuation mechanism in which tryptophan-activated TRAP binds to the nascent transcript and blocks the formation of an antiterminator structure such that the formation of an overlapping intrinsic terminator causes termination in the 5′ untranslated region (5′ UTR). In the absence of bound TRAP, the antiterminator forms and transcription continues into the trp genes. RNA polymerase pauses at positions U107 and U144 in the 5′ UTR. The general transcription elongation factors NusA and NusG stimulate pausing at both positions. NusG-stimulated pausing at U144 requires sequence-specific contacts with a T tract in the nontemplate DNA (ntDNA) strand within the paused transcription bubble. Pausing at U144 participates in a trpE translation repression mechanism. Since U107 just precedes the critical overlap between the antiterminator and terminator structures, pausing at this position is thought to participate in attenuation. Here we carried out in vitro pausing and termination experiments to identify components of the U107 pause signal and to determine whether pausing affects the termination efficiency in the 5′ UTR. We determined that the U107 and U144 pause signals are organized in a modular fashion containing distinct RNA hairpin, U-tract, and T-tract components. NusA-stimulated pausing was affected by hairpin strength and the U-tract sequence, whereas NusG-stimulated pausing was affected by hairpin strength and the T-tract sequence. We also determined that pausing at U107 results in increased TRAP-dependent termination in the 5′ UTR, implying that NusA- and NusG-stimulated pausing participates in the trp operon attenuation mechanism by providing additional time for TRAP binding.
IMPORTANCE The expression of several bacterial operons is controlled by regulated termination in the 5′ untranslated region (5′ UTR). Transcription attenuation is defined as situations in which the binding of a regulatory molecule promotes transcription termination in the 5′ UTR, with the default being transcription readthrough into the downstream genes. RNA polymerase pausing is thought to participate in several attenuation mechanisms by synchronizing the position of RNA polymerase with RNA folding and/or regulatory factor binding, although this has only been shown in a few instances. We found that NusA- and NusG-stimulated pausing participates in the attenuation mechanism controlling the expression of the Bacillus subtilis trp operon by increasing the TRAP-dependent termination efficiency. The pause signal is organized in a modular fashion containing RNA hairpin, U-tract, and T-tract components.
KEYWORDS: NusA, NusG, RNA polymerase pausing, TRAP, gene regulation, transcription attenuation, transcription factors
INTRODUCTION
The Bacillus subtilis trpEDCFBA operon is regulated by transcription attenuation and translation repression mechanisms in response to tryptophan by the trp RNA-binding attenuation protein (TRAP) (1, 2). Three RNA secondary structures that form in the 5′ untranslated region (5′ UTR) participate in the attenuation mechanism (Fig. 1). The TRAP binding target consists of the 5′ stem-loop (3) and 11 equivalently spaced (G/U)AG repeats, six of which are present within the antiterminator structure (4, 5). The binding of tryptophan-activated TRAP to the triplet repeats results in the RNA wrapping around the periphery of the protein (6, 7). Thus, bound TRAP prevents the formation of the antiterminator structure, which allows the formation of the overlapping intrinsic terminator hairpin and hence the termination of transcription in the 5′ UTR. In the absence of TRAP binding, the formation of the antiterminator allows for transcription of the entire operon (8–10). Since TRAP must bind before RNA polymerase (RNAP) transcribes past the terminator, the timing of TRAP binding is crucial for this regulatory decision. Transcripts that fail to terminate in the 5′ UTR are subject to TRAP-dependent translation repression; TRAP binding to readthrough transcripts promotes the formation of the trpE Shine-Dalgarno (SD) sequestering hairpin, thereby reducing TrpE synthesis by inhibiting ribosome binding (11, 12).
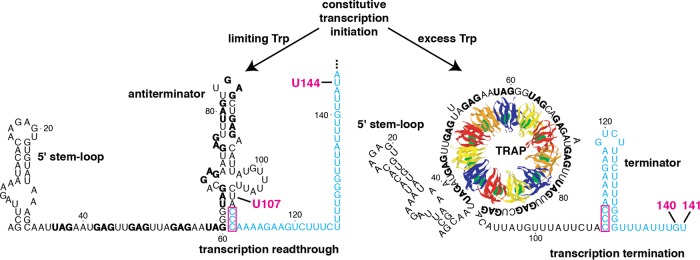
FIG 1.
Model of the B. subtilis trp operon transcription attenuation mechanism. During transcription, NusA and NusG stimulate RNAP pausing at U107. Under limiting tryptophan conditions, TRAP is not activated and does not bind to the nascent transcript. RNAP eventually overcomes the pause and resumes transcription, resulting in the formation of the antiterminator and transcription readthrough into the trp operon structural genes. Under excess tryptophan conditions, tryptophan-activated TRAP binds to the 5′ stem-loop and the (G/U)AG repeats soon after they are synthesized. Pausing at U107 provides additional time for TRAP binding. Bound TRAP prevents the formation of the antiterminator, which allows for the formation of the overlapping terminator hairpin such that transcription terminates at G140 or U141. The 11 TRAP subunits are shown in alternating colors with bound tryptophan shown in green. The 11 (G/U)AG repeats are indicated in boldface type, while the critical overlapping nucleotides between the antiterminator and terminator structures are boxed. The U144 pause site is also shown. Numbering is from the start of transcription.
In addition to regulating the trp operon, TRAP regulates the translation of trpG (a tryptophan biosynthesis gene), trpP (a tryptophan transport gene), and ycbK (encoding a putative efflux protein) by binding to triplet repeats that surround and overlap the cognate SD sequences (13–16). Thus, TRAP coordinately regulates tryptophan biosynthesis and transport in response to changing tryptophan levels. Another protein, called anti-TRAP (AT), antagonizes the activity of tryptophan-activated TRAP by competing with mRNA for TRAP's RNA binding surface (17–19). Since the transcription and the translation of the gene encoding AT (rtpA) is regulated by uncharged tRNATrp, B. subtilis regulates tryptophan biosynthesis by sensing the levels of both tryptophan and uncharged tRNATrp in the cell (20).
Even though transcription elongation by RNAP is highly processive, it is punctuated by transient pausing events. Pausing can allow for synchronization of the RNAP position with RNA folding and/or regulatory factor binding (21–25). Pause signals within the nascent RNA and DNA template cause isomerization of RNAP such that it enters into an elemental pause state in which elongation is reversibly inhibited (26). Hairpin-stimulated pause signals include an RNA pause hairpin and the downstream RNA sequence (27, 28). The interaction of transcription factors with the transcription elongation complex (TEC) can dramatically alter the efficiency and duration of pausing. NusA and NusG are two general transcription elongation factors that affect transcription, both positively and negatively, in response to specific signals in the nascent transcript and/or DNA template (29, 30). These two proteins exhibit remarkable flexibility in their ability to affect RNAP's response to pause signals. NusA from Escherichia coli increases the pause half-life by stimulating an interaction between the pause hairpin and the β flap of RNAP (31, 32). As NusA proteins from B. subtilis and E. coli can substitute for one another in stimulating pausing in vitro (33), it is apparent that their mechanisms of action are essentially identical. In contrast to NusA, NusG proteins from these organisms have opposite functions on pausing. Whereas E. coli NusG increases the transcription elongation rate by suppressing pausing (34, 35), B. subtilis NusG stimulates pausing by making sequence-specific contacts with the nontemplate DNA (ntDNA) strand within the paused transcription bubble (28, 33, 36).
RNAP pauses at positions U107 and U144 in the 5′ UTR of the B. subtilis trp operon (22). U107 just precedes the critical overlap between the antiterminator and terminator structures. Thus, pausing at U107 is thought to participate in the attenuation mechanism by providing additional time for TRAP to bind to the nascent transcript and promote termination. The U144 pause site is just downstream from the G140 and U141 termination sites (Fig. 1). Pausing at this position participates in the trpE translation repression mechanism by providing a second opportunity for TRAP binding (23). NusA and NusG stimulate pausing at both of these positions. As the U144 pause half-life is much longer in vitro and in vivo, our previous mechanistic pausing studies focused on the U144 site (23, 28, 33, 36). Based on the mechanistic insight gained from these prior studies, we conducted experiments to determine the features of the U107 pause site that distinguish it from the long-lived U144 pause. We found that features of the RNA hairpin, the sequence of the RNA between the hairpin and the 3′ end of the RNA (i.e., U107), and the sequence of the ntDNA within the paused transcription bubble all contribute to the disparate pause half-lives at these two sites. We further show that pausing at U107 increases the efficiency of TRAP-dependent termination in the trp 5′ UTR, consistent with pausing at this position providing additional time for TRAP to bind and interfere with the formation of the antiterminator structure.
RESULTS
Sequence and structural features of the U107 pause signal.
Single-round in vitro transcription assays were used to monitor pausing at U107 and U144 in the B. subtilis trp leader. Nascent transcripts were labeled at their 5′ ends during the first step in which a 29-nucleotide (nt) halted TEC was formed by the omission of CTP. In the second step, elongation was resumed by the addition of CTP together with heparin to prevent further RNAP initiation events. When added, NusA and NusG were included in the second step of the reaction. Since positions 108 and 145 are both A residues, a limiting concentration of ATP (10 μM) was used to increase the dwell time of RNAP at each pause site. The hallmark feature of a pause site is that the corresponding paused transcript initially increases in abundance and then chases to longer transcripts. This feature is evident for the U107 and U144 pause sites in the B. subtilis trp operon 5′ UTR (Fig. 2B). Note that some terminated products were also observed under these conditions. The identity of the 3′ nucleotide in the active site of RNAP (U107 and U144) is critical for basal and NusA- and NusG-stimulated pausing (33). Our previous studies of the U144 pause site also identified a T-rich tract of the ntDNA strand within the paused transcription bubble as being critical for NusG-stimulated pausing, with residues T135, T137, T138, and T139 being particularly important (28, 33, 36). Note that the corresponding U-rich tract in the nascent RNA would be within the RNA-DNA hybrid at the U144 pause site. The finding that the wild-type (WT) NusG N-terminal domain (NGN) cross-linked to T138, while the cross-linking was greatly reduced or absent in pause-defective NGN mutants, established that NusG makes sequence-specific contacts with the ntDNA strand within the paused transcription bubble (36).
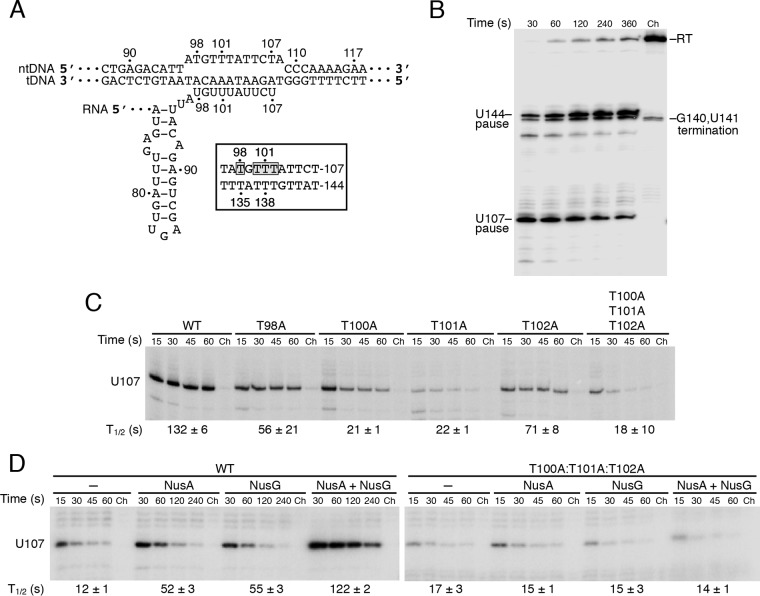
FIG 2.
Mutations in the T tract/U tract interfere with NusA- and NusG-stimulated pausing. (A) Diagram of the U107 paused transcription bubble showing the RNA-DNA hybrid in the pretranslocated state and the RNA hairpin. The nontemplate DNA (ntDNA) and template DNA (tDNA) strands are indicated. Numbering is from the start of transcription. (Inset) Comparison of the ntDNA strands at the U107 and U144 pause sites. The four T residues that were mutagenized in this study are boxed and shaded. (B) Single-round in vitro transcription reactions were performed with a DNA template containing the U107 and U144 pause sites in the presence of 1 μM NusA and 1 μM NusG. Positions of the U107 and U144 pause, terminated, and readthrough (RT) transcripts are marked. (C) Single-round in vitro transcription reactions were performed with wild-type (WT) and mutant DNA templates, as indicated, in the presence of 1 μM NusA and 1 μM NusG. (D) Single-round in vitro transcription reactions were performed with WT and T100A-T101A-T102A triple mutant DNA templates in the absence or presence of 1 μM NusA and/or 1 μM NusG, as indicated. (B to D) Reactions were stopped at the times shown above the lanes. Chase reactions (Ch) were extended for an additional 10 min at 37°C in the presence of 500 μM (each) NTPs. Each experiment was performed at least twice and representative gels are shown. (C, D) The position of the U107 pause transcript is marked. Pause half-life (t1/2) values and standard deviations are shown at the bottom of each set of lanes.
The precise positions of T135, T137, T138, and T139 within the U144 pause signal are conserved within the U107 pause signal (T98, T100, T101, and T102) (Fig. 2A). Note that all four of these U107 pause site residues are present within an internal loop of the predicted antiterminator structure involved in attenuation (Fig. 1). Thus, we introduced T-to-A substitutions to assess their role in RNAP pausing with the expectation that these changes would have a minimal impact on the antiterminator structure. In the presence of both NusA and NusG, the T98A and T102A mutations reduced the pause half-life 2-fold, whereas the T100A and T101A substitutions had stronger 6-fold pausing defects (Fig. 2C). Pausing was reduced to a similar extent in the T100A-T101A-T102A triple mutant. Since both NusA and NusG stimulate pausing at U107 (28, 33), we performed pausing experiments with the triple mutant template with or without NusA and/or NusG. Whereas NusA and NusG both stimulated pausing at the WT U107 pause site, neither protein stimulated pausing of the triple mutant (Fig. 2D). Because the substitution of the equivalent T-tract/U-tract residues at the U144 pause site also resulted in the loss of pause stimulation by NusA and NusG (28), combined with the prior demonstration that NusG cross-links to T138 of the ntDNA strand at the U144 pause site (36), we infer that NusG also makes sequence-specific contacts with the ntDNA strand at the U107 pause site. Furthermore, since it is known that NusA reduces the rate of transcription through U-rich tracts (28, 37), our results suggest that the loss of NusA-stimulated pausing in the T100A-T101A-T102A triple mutant is a consequence of the reduced number of U residues in the RNA-DNA hybrid.
Another critical component of hairpin-dependent pause signals is the hairpin itself. Although previous in vitro pausing studies using oligonucleotide competitors provided evidence for a U107 pause hairpin (22), the presence of a pause hairpin had not been demonstrated directly. Thus, we deleted nucleotides 73 to 94 encompassing the presumed U107 pause hairpin (ΔHp) (Fig. 1 and 3A) and found that pausing was reduced approximately 2-fold compared with that of the WT pause site in the absence of Nus factors (Fig. 2D and 3B). However, NusA- and NusG-stimulated pausing was virtually eliminated with the ΔHp template (Fig. 3B). Since Mfold (38) did not predict any RNA structural changes other than the removal of the pause hairpin, we conclude that U107 represents a hairpin-dependent pause site and that the hairpin contributes to pause stimulation by both Nus factors.
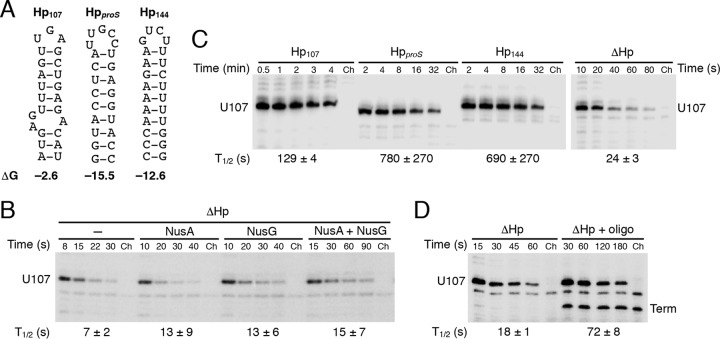
FIG 3.
The presence and strength of the pause hairpin greatly affect pausing at U107. (A) Structures of the U107 (Hp107), proS (HpproS), and U144 (Hp144) hairpins predicted by Mfold (38). (B) Single-round in vitro transcription reactions were performed with a ΔHp DNA template in the absence or presence of 1 μM NusA and/or NusG, as indicated. (C) Single-round in vitro transcription reactions were performed in the presence of 1 μM NusA and 1 μM NusG with templates containing the U107 pause hairpin (Hp107), a hairpin derived from the proS terminator (HpproS), the U144 pause hairpin (Hp144), or no hairpin (ΔHp). Note that the time scale for the ΔHp template is in seconds while those for the other samples are in minutes. (D) Single-round in vitro transcription reactions were performed in the presence of 1 μM NusA and 1 μM NusG with a ΔHp template in the absence or presence of a DNA oligonucleotide complementary to the region just upstream of the U tract. The resulting RNA-DNA hybrid generated a synthetic U107 pause hairpin. The oligonucleotide also resulted in an apparent termination event at position 102 (term). (B to D) Reactions were stopped at the times shown above the lanes. Chase reactions (Ch) were extended for an additional 10 min at 37°C in the presence of 500 μM (each) NTPs. The position of the U107 pause transcript is marked. Pause half-life (t1/2) values and standard deviations are shown at the bottom of each set of lanes. Each experiment was performed at least twice and representative gels are shown.
The U144 pause hairpin consists of 10 uninterrupted base pairs with a calculated ΔG value of −12.6 kcal/mol. By contrast, the 9-bp U107 pause hairpin is disrupted by a 2-by-1 internal loop with a ΔG value of only −2.6 kcal/mol. Thus, we compared the pause hairpin strengths of these two hairpins by replacing the U107 hairpin (Hp107) with that from the U144 pause site (Hp144). For comparison, we also replaced the U107 hairpin with the strong hairpin from the B. subtilis proS terminator (HpproS, ΔG = −15.5 kcal/mol) (Fig. 3A). The pause half-lives in the presence of both Nus factors were correlated with hairpin strength, with the strong Hp144 and HpproS resulting in pause half-lives that were approximately 6-fold longer than that with Hp107 (Fig. 3C).
Pairing of antisense DNA and RNA oligonucleotides to the appropriate position of the nascent transcript was previously shown to mimic a pause RNA hairpin at the E. coli his pause site (39). Hence, we tested whether pausing with the ΔHp template could be restored by annealing a DNA oligonucleotide to the nascent RNA just upstream of the U tract. Inclusion of a 15-nt antisense DNA oligonucleotide complementary to positions 57 to 71 of the trp 5′ UTR (Fig. 1) increased the pause half-life 4-fold in the presence of both Nus factors (Fig. 3D). Although the natural U107 hairpin resulted in a longer pause half-life than what was observed with the antisense oligonucleotide (Fig. 3C and D), these results indicate that a DNA-RNA duplex is capable of mimicking the U107 RNA hairpin in NusA- and NusG-stimulated pausing. The annealing of the antisense oligonucleotide also resulted in some termination within the U tract upstream of the pause site, presumably by mimicking a terminator hairpin upstream of a U-rich sequence, a common feature of intrinsic terminators (Fig. 3D). This phenomenon of oligonucleotide-mediated transcript release was observed previously (40). In addition, the antisense oligonucleotide caused nearly 100% transcription termination at residues G140 and U141, because it disrupted the base of the antiterminator structure by hybridization to residues 60 to 65, thereby allowing the formation of the attenuator terminator hairpin (Fig. 1 and data not shown).
Modular arrangement of the U107 and U144 pause signals.
The pause half-life at the U144 pause site is much longer than that at U107 (Fig. 2B) (22, 23, 33). Our results indicate that the relative strengths of the two pause hairpins are at least partly responsible for this difference (Fig. 3C). Our results also indicate that the conserved T/U residues downstream of the U107 and U144 pause hairpins are critical for NusA- and NusG-stimulated pausing. However, the identities of the non-T/U residues downstream of the U107 and U144 pause hairpins differ among the two pause sites, and these differences could contribute to the difference in pause half-lives (Fig. 4A). Thus, we compared all combinations of hairpins and downstream T-tract/U-tract regions from the U107 and U144 pause sites. Compared with that of the WT U107 pause site, the U144 pause half-life is 2.5-fold longer in the absence of Nus factors, 3.5-fold longer in the presence of NusA, 32-fold longer in the presence of NusG, and 37-fold longer in the presence of both Nus factors (Fig. 4B and E). Thus, it is apparent that the two pause sites differ most drastically in their response to NusG. When a hybrid pause site containing the U144 hairpin and the U107 T tract/U tract was tested (Fig. 4C), the basal and NusA-stimulated pausing were similar to that for the WT U144 pause site (Fig. 4C and E) and about 2.5-fold longer than that for the WT U107 pause site (Fig. 4B and C). However, NusG-stimulated pausing was greatly affected at this hybrid pause site; the pause half-life was increased ∼5-fold compared with that for the WT U107 site and reduced ∼7-fold compared with that for the WT U144 site. When a hybrid pause site containing the U107 hairpin and the U144 T tract/U tract was tested (Fig. 4D), the basal and NusA-stimulated pausing was slightly elevated compared with that for the WT U107 pause site (Fig. 4B and D) and reduced approximately 2-fold relative to that for the WT U144 pause site (Fig. 4D and E). As for the Hp144 plus T tract/U tract107 hybrid pause site (Fig. 4C), NusG-stimulated pausing was greatly affected by the Hp107 plus T tract/U tract144 hybrid pause site; the pause half-life was increased ∼6-fold compared with that for the WT U107 site and reduced ∼6-fold compared with that for the WT U144 site. From these data we conclude that the U107 and U144 pause signals are organized in a modular fashion containing distinct hairpin and T-tract/U-tract components. Moreover, our results indicate that both NusA and NusG respond to each of these modular components.
FIG 4.
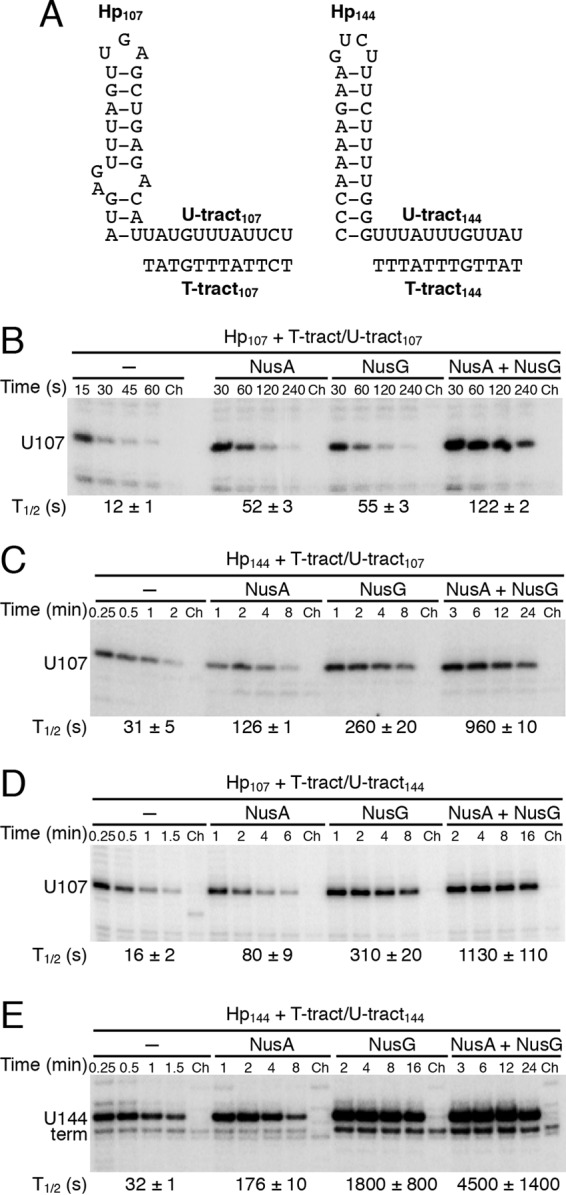
Relative contributions of the pause hairpin and T tract/U tract for pausing. (A) Sequence comparison of the 107 and 144 pause hairpins, RNA U tracts, and ntDNA T tracts. (B to E) Single-round in vitro transcription reactions were performed with various DNA templates in the absence or presence of 1 μM NusA and/or 1 μM NusG, as indicated. Reactions were stopped at the times shown above the lanes. Chase reactions (Ch) were extended for an additional 10 min at 37°C in the presence of 500 μM (each) NTPs. The position of the U107 pause transcript is marked. Pause half-life (t1/2) values and standard deviations are shown at the bottom of each set of lanes. Each experiment was performed at least twice and representative gels are shown.
Pausing at U107 increases TRAP-dependent termination in the trp 5′ UTR.
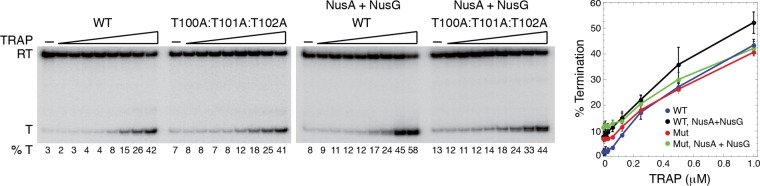
In the transcription attenuation mechanism, the formation of the antiterminator structure promotes transcription readthrough into the trpEDCFBA operon structural genes by preventing the formation of the overlapping terminator hairpin (Fig. 1). Since U107 just precedes the critical overlap between the antiterminator and terminator structures, pausing at U107 is assumed to provide additional time for TRAP binding. Thus, pausing at U107, which is stimulated ∼10-fold by the NusA-NusG combination (Fig. 2D and 4B), is predicted to increase the TRAP-dependent termination efficiency. To test this prediction, we performed single-round in vitro termination assays using WT and T100A-T101A-T102A triple mutant templates. Although the T-to-A substitutions were predicted to have a minimal impact on antiterminator function, in the absence of Nus factors, we observed a 2-fold increase in basal termination efficiency (i.e., in the absence of TRAP) (Fig. 5, left). As the TRAP concentration was gradually increased, the termination efficiency with the WT and mutant templates both reached ∼40%. In the presence of NusA and NusG, the basal termination efficiency was also elevated with the T100A-T101A-T102A triple mutant template (Fig. 5, right). As was observed in the absence of Nus factors, the termination efficiency of the mutant template reached ∼40%. Importantly, the termination efficiency reached approximately 55% with the WT template at the highest TRAP concentration, indicating that pausing results in increased TRAP-dependent termination at the trp leader attenuator. These data are consistent with a model in which NusA- and NusG-stimulated pausing at U107 participates in the trp operon attenuation mechanism in vitro. We further infer that pausing provides additional time for TRAP binding. Unfortunately, the increase in basal termination efficiency of the pause-defective mutant template prevented us from demonstrating that pausing at U107 participates in the transcription attenuation mechanism in vivo.
FIG 5.
Effect of T-tract/U-tract mutations on TRAP-dependent termination at the trp attenuator. Single-round in vitro transcription reactions were performed with WT and T100A-T101A-T102A triple mutant DNA templates in the absence or presence of 1 μM NusA, 1 μM NusG, and various concentrations of TRAP (0, 0.008, 0.016, 0.031, 0.062, 0.125, 0.25, 0.5, and 1.0 μM), as indicated. Positions of the terminated (T) and readthrough (RT) transcripts are marked. Termination efficiency (%T) is shown at the bottom of each lane. Quantification of results from four experiments and standard deviations are shown at the right.
DISCUSSION
An in vivo consensus sequence for elemental pause sites was identified for E. coli and B. subtilis, with pausing occurring every 200 nt on average (41, 42). Structural information at elemental pause sites revealed a relaxed open-clamp RNAP conformation, suggesting that the failure of RNAP to reestablish contacts with downstream DNA contributes to pausing (43). Although entry into an elemental pause state is a fundamental property of RNAP, the majority of these pause events are probably not regulatory. Other than a pyrimidine at the pause site and an A residue at the position immediately downstream of the pause site, the two trp pause sites bear little resemblance to the elemental pause site consensus.
Our previous studies demonstrated that NusA and NusG stimulate RNAP pausing at positions U107 and U144 of the B. subtilis trp leader (22, 28, 33, 36). Although NusA and NusG stimulate pausing at both of these sites, stimulation at U144 is highly synergistic (Fig. 2D and 4B and E). Pausing at U144 represses the translation of trpE by providing an opportunity for TRAP to bind and promote the formation of the trpE SD-sequestering hairpin (23). Although pausing is generally assumed to provide an opportunity for regulatory factor binding in attenuation mechanisms (26), this had not been established for the B. subtilis trp operon.
Both of the trp pause sites are preceded by a 12-nt T-rich sequence in the ntDNA strand, corresponding to a U-rich sequence in the nascent transcript, most of which would be part of the RNA-DNA hybrid in the main channel of RNAP. The relative positions of eight T residues are conserved in both pause signals (Fig. 2A). We previously demonstrated that several of these T residues in the U144 pause signal constitute a sequence-specific NusG recognition element (28, 36). Our current work indicates that the T residues play a similar role in pausing at U107. For example, the relative importance of the U144 pause site T residues is in the order T137≈T138>T135>T139 (28), which is similar to the order of the U107 pause site (T100≈T101>T98≈T102) (Fig. 2A and C). However, the relative contributions of these conserved T residues differ between the two pause sites. Whereas simultaneous substitution of T137, T138, and T139 was required to eliminate NusG-stimulated pausing (28), the substitution of just T100 or T101 had an effect that was similar to the T100A-T101A-T102A triple substitution (Fig. 2).
In contrast to the conserved T residues, the non-T residues at the U107 and U144 pause sites differ considerably (Fig. 2A). A comparison of the U107 and U144 T tracts indicates that the non-T residues of the U144 pause site provide a much stronger NusG-stimulated pausing signal (compare Fig. 4B with D and C with E). The simple absence of T at position 97 near the U107 pause site cannot explain this difference, because the T134C substitution at the U144 pause site had little effect on pausing (28). Instead, the sequence differences between the U107 and U144 T tracts (Fig. 2A), combined with our extensive mutagenesis study of the U144 pause site (28), suggest that the A102 and C106 residues negatively impact pausing at U107, whereas positions A97 and G99 are unlikely to be important for pausing.
Pause stimulation by the RNA hairpin depends on the relative strength of the secondary structure but not on its specific sequence (Fig. 3A and C). Mutagenesis of the U144 pause hairpin revealed a sharp drop from full to little pause stimulation in the narrow range of hairpin stability between ΔG values −8.6 and −6.0 kcal/mol (28), whereas our current work indicates that a hairpin with a ΔG value as low as −2.6 kcal/mol can stimulate pausing (Fig. 3). Thus, the modular combination of the hairpin and T tract/U tract leads to a wide range in pause strengths (Fig. 4).
The relative influence of the hairpin and T tract/U tract on pausing depends on the presence of two general transcription elongation factors. NusA-stimulated pausing is highly sensitive to the strength of the pause hairpin (Fig. 4) and to the U richness of the U tract (Fig. 2D) (28). Although NusA is known to stimulate the interaction of the pause hairpin with the β-flap domain of RNAP (31, 32), our results suggest that NusA may also assist with hairpin folding, as appears to be the case during intrinsic termination (44). Since results from previous studies indicate that NusA slows transcription through U tracts of intrinsic terminators (28, 37), the deficiency of NusA-stimulated pausing with the T100A-T101A-T102A mutant template may be due to the reduction in the U richness of the nascent transcript, which would abrogate its effect on slowing the rate of transcription as RNAP approaches the U107 pause site.
NusG-stimulated pausing is also highly sensitive to the strength of the pause hairpin (Fig. 4) and the sequence of the T tract (Fig. 2 and 4) (28). NusG makes sequence-specific contacts with T residues in the ntDNA strand within the U144 paused transcription bubble (28, 36). Since the U107 pause site mutations altered T residues corresponding to T residues that are critical for NusG-stimulated pausing at U144, the NusG-stimulated pausing defects at U107 are probably caused by the loss of ntDNA contacts within the U107 paused transcription bubble. Structure modeling of the paused elongation complex places the RNA exit channel and ntDNA strand far away from each other (36), excluding the possibility of direct contact between NusG and the pause hairpin. Therefore, the pause hairpin likely affects NusG-stimulated pausing via allosteric communication between two remote regions of the TEC.
The U107 pause hairpin contains 4 of the 11 (G/U)AG repeats that constitute the TRAP binding target, while the T-tract/U-tract portion of the U107 pause signal is positioned 4 nt downstream from the last repeat (Fig. 1 and 4A). Therefore, the disruption of the pause hairpin is the likely mechanism responsible for the previously observed release of paused RNAP by TRAP binding (22). Once RNAP resumes transcription, the terminator hairpin can form, resulting in transcript release at the attenuator (Fig. 1). Our observation that the T100A-T101A-T102A mutant trp leader template is defective in pausing (Fig. 2) and TRAP-dependent termination (Fig. 5) is consistent with a model in which pause stimulation by NusA and NusG provides additional time for tryptophan-activated TRAP to bind to the nascent transcript such that termination in the trp leader is increased. Thus, our results provide compelling evidence that NusA- and NusG-stimulated pausing synchronize the position of RNAP with TRAP binding in the B. subtilis trp operon attenuation mechanism.
The only function of TRAP binding was long thought to be to prevent antiterminator formation during attenuation (2). However, recent evidence indicates that TRAP is capable of inducing efficient termination to the otherwise weak intrinsic terminator in the absence of a functional antiterminator structure (45–47). In addition to TRAP, NusA is required to achieve efficient termination in the trp leader (44, 46). One model posits that in addition to antiterminator disruption, bound TRAP is capable of promoting forward translocation of RNAP, thereby leading to more efficient termination (45). The identification of mutant TRAP proteins that are still capable of binding tryptophan and RNA but are defective in termination is consistent with this model (47); however, studies will be required to fully define the interrelationship between pausing, TRAP binding, and termination.
MATERIALS AND METHODS
DNA templates and proteins.
All templates for the U107 pausing and termination assays were obtained by PCR amplification of the B. subtilis trp leader, in which a cryptic promoter located between +2 and +30 was converted into a strong consensus promoter along with an extended −10 region to drive transcription initiation from residue +37. Use of this modified cryptic promoter rather than the natural trp promoter alleviated the requirement for using an RNA trimer oligonucleotide for initiation to obtain a C-less halted complex and avoided the problems associated with transcription initiation from two promoters (23). Mutations were introduced using the QuikChange protocol (Agilent Technologies). Briefly, 185-nt PCR fragments (containing −1 to +182) of the wild-type (WT) and mutant templates were used as the templates for in vitro pausing and termination assays. B. subtilis TRAP (48), His-tagged NusA (22), and His-tagged NusG (33) proteins were overproduced and purified as described previously. His-tagged B. subtilis RNAP was purified as described previously (23, 49).
In vitro pausing assays.
Pausing assays and data analysis were performed as described previously (22, 23, 33). Halted TECs containing a 29-nt transcript were formed in a reaction mixture containing ATP and GTP (8 μM each), 2 μM UTP, and 1 mCi · ml−1 [α-32P]UTP at 37°C. Transcription elongation was halted at position 65 of the trp leader due to the absence of CTP. The elongation of halted transcription complexes was resumed by the addition of all four nucleoside triphosphates ([NTPs] 150 μM GTP, CTP, and UTP and 10 μM ATP), together with heparin at room temperature (23 to 25°C). When included, NusA (1 μM), NusG (1 μM), and/or a DNA oligonucleotide (50 μM) were added in the second step of the reaction. Aliquots of the elongation reaction mixtures were removed at various times, and the last aliquot was chased for an additional 10 min at 37°C in the presence of 500 μM NTPs. The reactions were stopped by mixing with gel loading solution, and samples were fractionated through standard 6% sequencing gels. The pause half-lives were calculated by plotting the relative intensities of the U107 pause bands against the incubation times and fitting the data with a single exponential equation, as described previously (22, 50).
In vitro termination assays.
In vitro termination assays were performed as described previously (22, 23, 33, 44). Halted TECs containing a 29-nt transcript were formed in a reaction mixture containing ATP and GTP (8 μM each), 2 μM UTP, and 1 mCi · ml−1 [α-32P]UTP at 37°C. The elongation of halted complexes was resumed by the addition of 150 μM of all four NTPs for 5 min at 37°C. When included in the reaction mixtures, NusA (1 μM), NusG (1 μM), and TRAP (various concentrations) together with 1 mM l-tryptophan were added in the second step of the reaction. TRAP concentrations were 0 nM, 7.5 nM, 15 nM, 30 nM, 60 nM, 125 nM, 250 nM, 500 nM, and 1 μM. The reactions were stopped by mixing with gel loading solution, and samples were fractionated through standard 6% sequencing gels. The efficiency of termination was calculated as the fraction of a terminated transcript relative to the sum of all terminated and readthrough transcripts, as described previously (33).
ACKNOWLEDGMENTS
We thank Helen Yakhnin for technical assistance.
This work was supported by a National Institutes of Health grant (no. GM098399) to Paul Babitzke.
REFERENCES
- 1.Babitzke P. 2004. Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: the Bacillus subtilis TRAP protein. Curr Opin Microbiol 7:132–139. doi: 10.1016/j.mib.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Gollnick P, Babitzke P, Antson A, Yanofsky C. 2005. Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis. Annu Rev Genet 39:47–68. doi: 10.1146/annurev.genet.39.073003.093745. [DOI] [PubMed] [Google Scholar]
- 3.McGraw AP, Mokdad A, Major F, Bevilacqua PC, Babitzke P. 2009. Molecular basis of TRAP-5′SL RNA interaction in the Bacillus subtilis trp operon transcription attenuation mechanism. RNA 15:55–66. doi: 10.1261/rna.1314409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babitzke P, Stults JT, Shire SJ, Yanofsky C. 1994. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J Biol Chem 269:16597–16604. [PubMed] [Google Scholar]
- 5.Babitzke P, Yealy J, Campanelli D. 1996. Interaction of the trp RNA-Binding attenuation protein (TRAP) of Bacillus subtilis with RNA: effects of the number of GAG repeats, the nucleotides separating adjacent repeats, and RNA secondary structure. J Bacteriol 178:5159–5163. doi: 10.1128/jb.178.17.5159-5163.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antson AA, Otridge J, Brzozowski AM, Dodson EJ, Dodson GG, Wilson KS, Smith TM, Yang M, Kurecki T, Gollnick P. 1995. The structure of trp RNA-binding attenuation protein. Nature 374:693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- 7.Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X, Gollnick P. 1999. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature 401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 8.Babitzke P, Gollnick P, Yanofsky C. 1992. The mtrAB operon of Bacillus subtilis encodes GTP cyclohydrolase I (MtrA), an enzyme involved in folic acid biosynthesis, and MtrB, a regulator of tryptophan biosynthesis. J Bacteriol 174:2059–2064. doi: 10.1128/jb.174.7.2059-2064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otridge J, Gollnick P. 1993. MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner. Proc Natl Acad Sci U S A 90:128–132. doi: 10.1073/pnas.90.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babitzke P, Yanofsky C. 1993. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc Natl Acad Sci U S A 90:133–137. doi: 10.1073/pnas.90.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merino E, Babitzke P, Yanofsky C. 1995. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J Bacteriol 177:6362–6370. doi: 10.1128/jb.177.22.6362-6370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du H, Babitzke P. 1998. trp RNA-binding attenuation protein-mediated long distance RNA refolding regulates translation of trpE in Bacillus subtilis. J Biol Chem 273:20494–20503. doi: 10.1074/jbc.273.32.20494. [DOI] [PubMed] [Google Scholar]
- 13.Du H, Tarpey R, Babitzke P. 1997. The trp RNA-binding attenuation protein regulates TrpG synthesis by binding to the trpG ribosome binding site of Bacillus subtilis. J Bacteriol 179:2582–2586. doi: 10.1128/jb.179.8.2582-2586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarsero JP, Merino E, Yanofsky C. 2000. A Bacillus subtilis gene of previously unknown function, yhaG, is translationally regulated by tryptophan-activated TRAP and appears to be involved in tryptophan transport. J Bacteriol 182:2329–2331. doi: 10.1128/JB.182.8.2329-2331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yakhnin H, Zhang H, Yakhnin AV, Babitzke P. 2004. The trp RNA-binding attenuation protein of Bacillus subtilis regulates translation of the tryptophan transport gene trpP (yhaG) by blocking ribosome binding. J Bacteriol 186:278–286. doi: 10.1128/JB.186.2.278-286.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yakhnin H, Yakhnin AV, Babitzke P. 2006. The trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis regulates translation initiation of ycbK, a gene encoding a putative efflux protein, by blocking ribosome binding. Mol Microbiol 61:1252–1266. doi: 10.1111/j.1365-2958.2006.05278.x. [DOI] [PubMed] [Google Scholar]
- 17.Valbuzzi A, Yanofsky C. 2001. Inhibition of the B. subtilis regulatory protein TRAP by the TRAP-inhibitory protein, AT. Science 293:2057–2059. doi: 10.1126/science.1062187. [DOI] [PubMed] [Google Scholar]
- 18.Valbuzzi A, Gollnick P, Babitzke P, Yanofsky C. 2002. The anti-trp RNA-binding attenuation protein (anti-TRAP), AT, recognizes the tryptophan-activated RNA binding domain of the TRAP regulatory protein. J Biol Chem 277:10608–10613. doi: 10.1074/jbc.M111813200. [DOI] [PubMed] [Google Scholar]
- 19.Ihms EC, Zhou M, Zhang Y, Kleckner IR, McElroy CA, Wysocki VH, Gollnick P, Foster MP. 2014. Gene regulation by substoichiometric heterocomplex formation of undecameric TRAP and trimeric anti-TRAP. Proc Natl Acad Sci U S A 111:3442–3447. doi: 10.1073/pnas.1315281111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen G, Yanofsky C. 2003. Tandem transcription and translation regulatory sensing of uncharged tryptophan tRNA. Science 301:211–213. doi: 10.1126/science.1084902. [DOI] [PubMed] [Google Scholar]
- 21.Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. 1999. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc Natl Acad Sci U S A 96:9545–9450. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yakhnin AV, Babitzke P. 2002. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism in vitro. Proc Natl Acad Sci U S A 99:11067–11072. doi: 10.1073/pnas.162373299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yakhnin AV, Yakhnin H, Babitzke P. 2006. RNA polymerase pausing participates in the Bacillus subtilis trpE translation control mechanism by providing additional time for TRAP to bind to the nascent trp leader transcript. Mol Cell 24:547–557. doi: 10.1016/j.molcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 24.Wong TN, Sosnick TR, Pan T. 2007. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc Natl Acad Sci U S A 104:17995–18000. doi: 10.1073/pnas.0705038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perdue SA, Roberts JW. 2011. σ70-dependent transcription pausing in Escherichia coli. J Mol Biol 412:782–792. doi: 10.1016/j.jmb.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Landick R. 2016. A two-way street: regulatory interplay between RNA polymerase and nascent RNA structure. Trends Biochem Sci 41:293–310. doi: 10.1016/j.tibs.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan CL, Landick R. 1993. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol 233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- 28.Yakhnin AV, Babitzke P. 2010. Mechanism of NusG-stimulated pausing, hairpin-dependent pause site selection and intrinsic termination at overlapping pause and termination sites in the Bacillus subtilis trp leader. Mol Microbiol 76:690–705. doi: 10.1111/j.1365-2958.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Lewis PJ. 2010. The interaction between bacterial transcription factors and RNA polymerase during the transition from initiation to elongation. Transcription 1:66–69. doi: 10.4161/trns.1.2.12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yakhnin AV, Babitzke P. 2014. NusG/Spt5: are there common functions of this ubiquitous transcription elongation factor? Curr Opin Microbiol 18:68–71. doi: 10.1016/j.mib.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Artsimovitch I, Landick R. 2000. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci U S A 97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toulokhonov I, Artsimovitch I, Landick R. 2001. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science 292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 33.Yakhnin AV, Yakhnin H, Babitzke P. 2008. Function of the Bacillus subtilis transcription elongation factor NusG in hairpin-dependent RNA polymerase pausing in the trp leader. Proc Natl Acad Sci U S A 105:16131–16136. doi: 10.1073/pnas.0808842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mooney RA, Schweimer K, Rösch P, Gottesman M, Landick R. 2009. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol 391:341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herbert KM, Zhou J, Mooney RA, Porta AL, Landick R, Block SM. 2010. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol 399:17–30. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yakhnin AV, Murakami KS, Babitzke P. 2016. NusG is a sequence-specific RNA polymerase pause factor that binds to the non-template DNA within the paused transcription bubble. J Biol Chem 291:5299–5308. doi: 10.1074/jbc.M115.704189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gusarov I, Nudler E. 2001. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell 107:437–449. doi: 10.1016/S0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- 38.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolb KE, Hein PP, Landick R. 2014. Antisense oligonucleotide-stimulated transcriptional pausing reveals RNA exit channel specificity of RNA polymerase and mechanistic contributions of NusA and RfaH. J Biol Chem 289:1151–1163. doi: 10.1074/jbc.M113.521393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yarnell WS, Roberts JW. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 41.Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. 2014. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vvedenskaya I, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, Ebright RH, Nickels BE. 2014. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science 344:1285–1289. doi: 10.1126/science.1253458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weixlbaumer A, Leon K, Landick R, Darst SA. 2013. Structural basis of transcriptional pausing in bacteria. Cell 152:431–441. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mondal S, Yakhnin AV, Sebastien A, Albert I, Babitzke P. 2016. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat Microbiol 1:15007. doi: 10.1038/nmicrobiol.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Potter KD, Merlino NM, Jacobs T, Gollnick P. 2011. TRAP binding to the Bacillus subtilis trp leader region RNA causes efficient transcription termination at a weak intrinsic terminator. Nucleic Acids Res 39:2092–2102. doi: 10.1093/nar/gkq965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McAdams NM, Gollnick P. 2014. The Bacillus subtilis TRAP protein can induce transcription termination in the leader region of the tryptophan biosynthetic (trp) operon independent of the trp attenuator RNA. PLoS One 9:e88097. doi: 10.1371/journal.pone.0088097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McAdams NM, Patterson A, Gollnick P. 2017. Identification of a residue (Glu60) in TRAP required for inducing efficient transcription termination at the trp attenuator independent of binding tryptophan and RNA. J Bacteriol 199:e00710-16. doi: 10.1128/JB.00710-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yakhnin AV, Trimble JJ, Chiaro CR, Babitzke P. 2000. Effects of mutations in the l-tryptophan binding pocket of the trp RNA-binding attenuation protein of Bacillus subtilis. J Biol Chem 275:4519–4524. doi: 10.1074/jbc.275.6.4519. [DOI] [PubMed] [Google Scholar]
- 49.Qi Y, Hulett FM. 1998. PhoP-P and RNA polymerase sigmaA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP-P activator sites within the coding region stimulate transcription in vitro. Mol Microbiol 28:1187–1197. doi: 10.1046/j.1365-2958.1998.00882.x. [DOI] [PubMed] [Google Scholar]
- 50.Landick R, Wang D, Chan CL. 1996. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: his leader pause site as paradigm. Methods Enzymol 274:334–353. doi: 10.1016/S0076-6879(96)74029-6. [DOI] [PubMed] [Google Scholar]