Abstract
The production of cholera toxin (CT) during Vibrio cholerae infection results in the hallmark diarrhea that characterizes the disease cholera. The transmembrane protein ToxR was originally identified as a functional transcriptional activator of ctxAB in a heterologous Escherichia coli system. However, direct ToxR activation of the ctxAB promoter in V. cholerae has not been previously demonstrated. Instead, a regulatory cascade has been defined in which the activators ToxRS and TcpPH modulate ctxAB expression by acting in concert to transcriptionally activate another regulator, ToxT. ToxT, in turn, directly activates ctxAB expression as well as expression of the tcp genes and other virulence-associated genes. In this study, we show that ToxRS directly activates ctxAB in a ToxT-independent manner in a classical biotype V. cholerae, and that this activation requires the presence of bile acids. Although the levels of CT induced by this mechanism are lower than levels induced under other in vitro conditions, the bile-dependent conditions described here are more physiologic, being independent of pH and temperature. We further show that the inability of bile acids to stimulate ToxRS-dependent expression of CT in El Tor biotype strains is related to the differences between classical and El Tor ctxAB promoters, which differ in the number of heptad TTTTGAT repeats in their respective upstream regions. The ability of bile acids to stimulate direct activation of ctxAB by ToxRS depends upon the transmembrane domain of ToxR, which may interact with bile acids in the inner membrane of V. cholerae.
The Gram-negative bacterium Vibrio cholerae is the infectious agent that causes the severe human diarrheal disease cholera (1). Two major virulence factors, cholera toxin (CT) and the toxin-coregulated pilus (TCP), play major roles in the pathogenesis of this infection. Secretion of CT, an ADP-ribosylating toxin encoded in the genome of the filamentous, lysogenic CTXΦ phage, results in elevated cAMP levels in intestinal epithelial cells and subsequent secretory diarrhea (2). The regulation of CT and TCP expression has been studied intensely and has been reported to respond to temperature, pH, osmolarity, bile salts, and certain amino acids in vitro (3, 4); nevertheless, a clear understanding of the in vivo environmental stimuli that trigger CT and TCP production remains elusive.
The first transcriptional regulator of CT production identified was ToxR, a 32-kDa inner-membrane protein that activates the ctx promoter in Escherichia coli and whose deletion in V. cholerae results in the inability to produce CT (5). Deletion analysis of the ctxAB promoter localized the binding site of ToxR to multiple heptad repeats, TTTTGAT, upstream of ctxAB (6). However, ToxR activation of ctxAB transcription in V. cholerae has not been demonstrated (7). Instead, ToxR in V. cholerae binds with its downstream enhancer ToxS to the upstream region of the toxT gene, which encodes another transcriptional activator (8). ToxT belongs to the AraC family of transcriptional activators, activates multiple virulence genes [including ctxAB, acfA, the tcp genes (9)], and autoregulates itself, presumably by controlling read-through transcription from the upstream tcpA promoter (10, 11). In cascade-like fashion, ToxRS participates in the activation of the toxT promoter, which subsequently activates other virulence genes.
ToxR forms homodimers via its 100-aa C-terminal periplasmic domain and heterodimers with ToxS (12, 13). However, the in vitro activity of ToxR is neither dependent on nor regulated by this phenomemon, as demonstrated by ToxR fusion protein analysis (14). Its activity in vitro does require the ability of the 180-aa N-terminal cytoplasmic domain to bind DNA and activate transcription (15) and the localization of ToxR to the membrane by the 16- to 19-aa transmembrane domain (16). Substitutions of the periplasmic and transmembrane domains do not completely abrogate ToxR activity but do appear to affect the conditions under which ToxR is active in vitro. In contrast, ToxR dimerization is required for full colonization in an in vivo infant mouse model (16).
An additional inner-membrane regulator, TcpP (17), along with its downstream enhancer, TcpH, is essential for in vitro ToxRS activation of toxT. TcpPH alone can activate toxT transcription in vitro (18). TcpPH appears to play a major role in mediating the signals that couple environmental stimuli to virulence regulation. It is responsible for in vitro induction of CT in a biotype-specific manner, for a significant albeit incomplete response to pH and temperature (19), and for regulation of virulence gene expression in response to a quorum-sensing system (20). These effects are mediated by AphA and AphB, transcriptional regulators that directly activate tcpPH transcription and ultimately regulate ToxT and CT (21).
ToxRS regulates multiple genes other than ToxT (22), including the ompU and ompT genes (3, 7, 23). ToxR activity, independent of ToxT, results in the reciprocal expression of OmpU and repression of OmpT, two outer-membrane proteins that may act as adhesins (24) or porins (25), and that may affect virulence factor expression, intestinal colonization, and resistance to bile acids (26).
Bile is present in the lumen of the human intestine as a necessary element of the digestive process. Bile acids, as a mixture of the sodium salts of taurocholic, glycocholic, deoxycholic, chenodeoxycholic, and cholic acid, are a major component of crude bile and are secreted by the gallbladder into the proximal portion of the duodenum to an estimated concentration of ≈0.2–2% for individual bile acids. They aid in lipid solubilization and also serve to protect the host against enteric pathogens, presumably due to bacteriocidal activity related to their detergent-like properties (27).
Numerous effects of crude bile on V. cholerae have been described. Gupta and Chowdhury (4) reported that, in classical biotype O395 and 569B strains, crude ox bile extract (0.2–0.4%) decreased expression of the virulence factors CT and TcpA and increased motility in a ToxR-independent manner but had no effect on OmpU/OmpT expression. Schumacher and Klose (28) reported that, in O395 expressing ToxT from a plasmid-based, heterologous Plac promoter, sodium choleate (0.4%) also repressed CT and TCP production by inhibiting ctx and tcpA transcription. These results led to the hypothesis that perhaps bile acids inhibit ToxT activity, preventing their premature expression during infection. However, in contrast to Gupta and Chowdhury's earlier study, Provenzano et al. (29) reported that bile increases OmpU expression and transcription in a ToxR-dependent manner in both O395 and the El Tor strain E7946. They also reported that increased OmpU expression conferred increased resistance to bile and OmpT conferred decreased virulence factor expression and decreased colonization of the infant mouse (26). Collectively, these results imply that the relationship between bile and V. cholerae is complex because of the heterogenous composition of bile as well as its pleiotropic effects on the bacteria. Furthermore, the in vivo effects of bile on infection and virulence are not clear.
In this study, we report that purified bile acids do not inhibit CT or TcpA expression in the classical biotype O395 strain. Instead, they activate CT expression in a ToxRS-dependent, ToxT-independent manner. This activation is indifferent to pH or temperature and thus can be achieved in vitro under conditions that more closely mimic in vivo environmental conditions (alkaline pH and 37°C) (30). This report of the direct ability of ToxRS to activate the ctxAB promoter in V. cholerae mirrors the original discovery of ToxR as an activator of ctxAB transcription in E. coli and allows consideration of a direct role of ToxRS in in vivo CT expression.
Materials and Methods
Bacterial Strains and Plasmids. The V. cholerae classical strain O395 (31) was used in all experiments except for the generation of growth curves, where strain BGD4 was used (32). BGD4 is an O395 derivative carrying an in-frame deletion of tcpA. All V. cholerae mutants used in this study were derived from O395. The construction of in-frame deletions of toxRS, tcpPH, and toxT in O395 is described in ref. 17. The method of Skorupski and Taylor (33) was used to construct the double and triple deletion mutants. Strains JJM3.10, JJM3.16, and JJM3.13 are derivatives of El Tor strains N16961 (34), E7946 (6), and P27459, respectively, that underwent ctxA promoter exchange with the ctxA promoter region from classical biotype strain 569B (J.J.M., unpublished data). The construction of the ToxRS fusion proteins in O395 is described in ref. 16. Lab stocks of E. coli DH5αλpir (35) and SM10λpir (3) were used for cloning and mating, respectively, into V. cholerae.
Growth Conditions. CT induction of O395 and its derivatives was examined in LB with an initial pH of 6.5 at 30°C or pH 8.5 at 37°C. Stock solutions of sodium cholate (10% in water; Sigma), sodium deoxycholate (4% in water; Sigma), sodium taurocholate (10% in water; Sigma), or SDS (1% in water) were added at the appropriate dilutions to 5 ml of LB in 150 mm × 15 mm test tubes. A stock solution of crude bile was obtained by making a 10% suspension of crude ox bovine bile (B3883, Sigma) in water, centrifuging the suspension at 16,000 × g for 10 min, and filtering the supernatant through a 5-μm filter. Overnight cultures were subcultured at a 1:1,000 dilution unless otherwise specified. OD600 was measured at the appropriate time points.
Detection of CT. GM1 ganglioside enzyme-linked immunosorbent CT assays (36) were performed after incubation of V. cholerae strains overnight with shaking under the conditions described. CT expression was normalized for OD600 of the growing culture. All samples were performed, minimally, in triplicate.
Western Blot Analysis of TcpA. Cells were grown in LB (initial pH 6.5) at 30°C overnight in the absence or presence of 0.1% sodium cholate. Cell extracts were subjected to SDS/PAGE, transferred to nitrocellulose, probed with anti-TcpA Ab (37), and visualized by enhanced chemiluminescence (Amersham Pharmacia).
Results
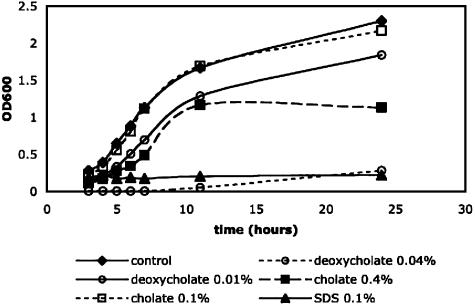
Bile Acids Do Not Inhibit CT Production Independent of Toxicity to V. cholerae O395. Because crude bile has been reported to inhibit CT production, we were interested in examining the ability of the individual components of crude bile, specifically bile acids, to inhibit CT production. Bile acids are bacteriocidal, so to determine the concentrations at which the individual bile acids demonstrated evidence of growth inhibition or delay, we generated growth curves at 30°C by using the strain BGD4, which is O395 strain deleted for tcpA, to remove the confounding factor of agglutination in measuring OD600 (Fig. 1). Significant growth defects were noted in the presence of SDS (0.1%) and sodium deoxycholate (0.04%), but no detectable defect was noted in the presence of sodium cholate (0.1%). Sodium deoxycholate (0.04%) was >10-fold more toxic to cells relative to sodium cholate (0.4%). Similar results were obtained for growth at pH 8.5 and 37°C (data not shown).
Fig. 1.
Growth curves of classical V. cholerae strain BGD4 in the presence of varying detergents. LB (5 ml), initial pH 6.5, was inoculated with a 1:1,000 dilution of an overnight culture of BGD4. Cultures were grown at 30°C, and the OD600 was measured at various time points in the presence of no detergent, 0.1% sodium cholate, 0.4% sodium cholate, 0.01% sodium deoxycholate, 0.04% sodium deoxycholate, or 0.01% SDS.
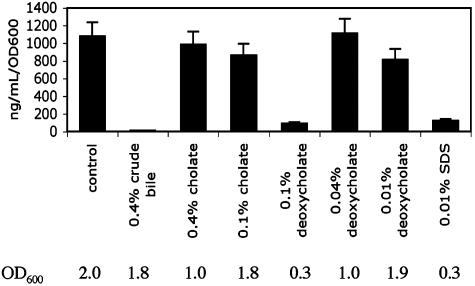
We then measured the amount of CT expressed by O395 in the presence of varying concentrations of the bile acids under standard inducing conditions (LB pH 6.5 and 30°C), compared with the amount made in the presence of crude bovine bile (Fig. 2). Because growth in the presence of bile acids is sensitive to the amount of inoculum, we subcultured an overnight culture of O395 at the lower dilution of 1:200 to attain higher final culture density for the strain, even in the presence of the more toxic concentrations of the bile acid. Whereas inhibition by crude bile was observed to completely inhibit CT expression, no inhibition of CT expression was seen by pure bile acids. Relative to the control where no bile acid or detergent was added, the presence of 0.4% or 0.1% sodium cholate and 0.04% or 0.01% sodium deoxycholate had no statistically significant effect on the amount of CT expressed when normalized to OD600 as a surrogate for cell number. These same results were observed when other purified bile acids were tested, including lithocholate, dehydrocholate, chenodeoxycholate, and the conjugated bile acids glycocholate and taurocholate (results not shown).
Fig. 2.
CT expression in O395 in the presence of various detergents. LB (pH 6.5) with no detergent, 0.4% crude bile, 0.1% sodium cholate, 0.4% sodium cholate, 0.1% sodium deoxycholate, 0.04% sodium deoxycholate, 0.01% sodium deoxycholate, or 0.01% SDS was inoculated with overnight cultures of the classical biotype V. cholerae strain O395 at a dilution of 1:200. They were grown for 12 h at 30°C, and the cultures were subsequently assayed for toxin production by ELISA. The amount of CT produced was normalized by OD600. The OD600 of the corresponding culture from which the CT was assayed in the supernatant is shown below.
Inhibition of CT expression was seen when cells were grown in the presence of 0.1% sodium deoxycholate. However, growth was also significantly inhibited, with a final OD600 of only 0.28, compared with the growth in the other samples in which all final OD600 values were >1. Similar inhibition of both CT expression and growth was also seen when cells were grown in the presence of a detergent (0.01% SDS with a final OD600 of 0.30). The implication is therefore that expression of CT by O395 is sensitive to growth conditions and that, unless growth is maintained at a certain level (in our case, the ability to reach an OD600 of 1.0 in 12 h), CT is not expressed. This conclusion is consistent with the previous report that TcpPH is expressed in a growth-dependent manner, i.e., only expressed in mid-logarithmic phase (38). These findings also suggest that the major bile acids are not the component within bile that accounts for the previously described inhibition of CT expression but rather that some other component within the crude mixture accounts for that activity.
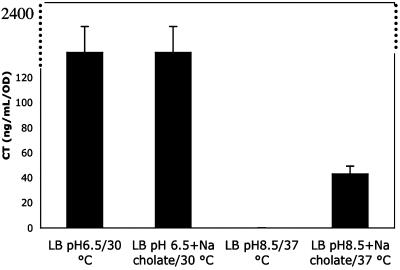
Bile Acids Activate CT Expression Under Standard Noninducing Conditions in a ToxT-Independent Manner. We discovered that under standard noninducing conditions (LB pH 8.5, 37°C), the addition of 0.1% sodium cholate to the medium resulted in activation of CT expression in O395 (Fig. 3). Although the amount of CT expressed (43 ng/ml) is 50-fold less than the amount made under standard inducing conditions (2,050 ng/ml), these conditions may more closely resemble the physiological conditions during infection than the standard inducing conditions (pH 6.5, 30°C). Although 0.1% sodium cholate was used because it has the least effect on growth, this same activation was attained with sodium deoxycholate (0.01%) or sodium taurocholate (0.1%). However, the activation is not the result of a detergent effect because no CT expression was seen when O395 was grown similarly in the presence of SDS (0.05%), Triton X-100 (0.1%), Tween (2%), and Nonidet P-40 (0.5%) (results not shown).
Fig. 3.
CT expression in O395 under various conditions in the presence of sodium cholate. LB at a starting pH of 6.5 with or without 0.1% sodium cholate was inoculated with an overnight culture of O395. These cultures were grown with shaking overnight at 30°C. These samples were compared with cultures grown in LB at a starting pH of 8.5, with or without 0.1% sodium cholate, grown overnight with shaking at 37°C. The resulting supernatants were assayed for CT production by ELISA.
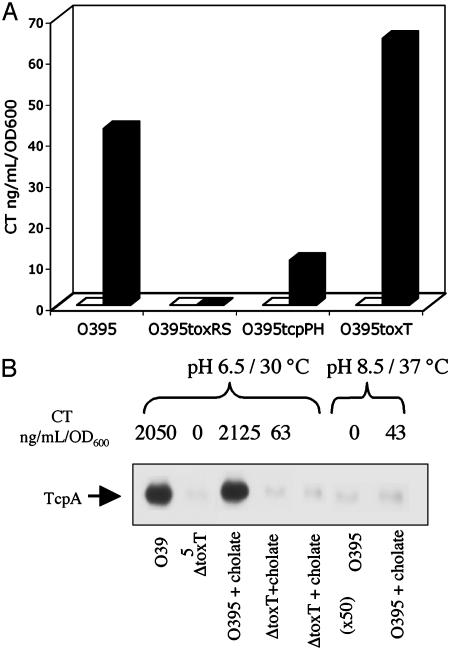
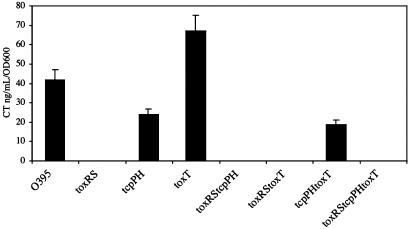
The ability of sodium cholate to activate CT expression was next tested in mutants of O395 that were deleted for the known virulence regulators ToxRS, TcpPH, and ToxT (Fig. 4A). Induction of CT expression by bile under the conditions of LB pH 8.5 and 37°C was ToxRS-dependent and ToxT-independent. Induction was partially reduced in the tcpPH deletion mutant, implying that TcpPH may enhance the activity of ToxRS. The inability of ToxRS alone to induce tcpA transcription, even in the presence of bile acids, was confirmed by Western blot analysis of cell extracts, within the detection sensitivity of this method (Fig. 4B), and by autoagglutination (data not shown). This finding is not surprising in light of the established model whereby ToxT is required for tcpA activation, and no direct role has been shown for ToxR (9, 39). A similar profile of bile acid induction of CT expression in the deletion strains was observed in LB pH 6.5 and 30°C and in AKI media (data not shown), suggesting that this induction is insensitive to pH, temperature, and growth media. It is likely that induction in the wild-type strain occurs under these conditions as well, but the effect is masked by the over-whelming amount of CT already expressed via the ToxT-dependent mechanism.
Fig. 4.
Sodium cholate induction of CT expression is toxRS-dependent. (A) O395 and the deletion mutants O395ΔtoxRS, O395ΔtcpPH, and O395ΔtoxT were grown overnight in LB (initial pH 8.5) at 37°C in the presence or absence of 0.1% sodium cholate. The resulting supernatants were assayed for CT expression by ELISA. (□, without sodium cholate; ▪, with 0.1% sodium cholate.) Errors were <10% in all samples. (B) Sodium cholate does not activate TcpA expression. O395 and O395ΔtoxT were grown in LB (initial pH 6.5) at 30°C in the absence or presence of 0.1% sodium cholate. The cell pellets were analyzed by Western blot and probed with anti-TcpA Ab (Upper), and the supernatants were assayed for CT expression (Lower). Lane 5 contains 50 times the amount of total protein, compared with the other lanes, to demonstrate that there is no TcpA expressed, even in proportion to the 50-fold lower CT that is expressed relative to standard inducing conditions.
That ToxRS is necessary and sufficient for CT expression in the presence of bile acids was confirmed by examining the ability of sodium cholate to activate CT expression in the double and triple mutants ΔtoxRSΔtcpPH, ΔtoxRSΔtoxT, ΔtcpPHΔtoxT, and ΔtoxRSΔtcpPHΔtoxT (Fig. 5). In all mutants, the absence of ToxRS was sufficient to completely eliminate CT activation by bile acids. In contrast, CT expression was independent of the presence of ToxT under all conditions tested. This is the first demonstration in V. cholerae of the ability of ToxRS to directly activate CT expression, a phenomenon that was originally described in the heterologous E. coli system.
Fig. 5.
Sodium cholate induction of CT production is ToxRS-dependent but ToxT-independent. O395, the deletion mutants O395ΔtoxRS, O395ΔtcpPH, and O395ΔtoxT, and the double and triple mutants O395ΔtoxRSΔtcpPH, O395ΔtoxRSΔtoxT, O395ΔtcpPHΔtoxT, and O395ΔtoxRSΔtcpPHΔtoxT were grown overnight in LB (initial pH 8.5) at 37°C in the presence of 0.1% sodium cholate. The resulting supernatants were assayed for CT expression by ELISA.
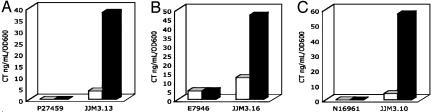
ToxRS Can Activate the CT Promoter from Classical Biotype Strains in the Presence of Bile Acids but Does Not Activate the Promoter from El Tor Biotype Strains. We tested the ability of bile acids to induce CT expression in the El Tor biotype strains N16961, E7946, and PA27459 (Fig. 6). Although each of these strains is able to make CT under AKI conditions (40), they are typical of El Tor biotype strains in that they make very little or no CT in LB at 37°C. The addition of 0.1% sodium cholate did not induce any CT expression by these wild-type strains. This result recalls the observation that constitutive expression of ToxR in an El Tor strain results in considerably lower production of CT than in a classical strain (6). In that study, exchange of the El Tor promoter in strain RV79 for the classical promoter from strain 569B, resulted in increased production of CT, suggesting that it is the promoters themselves, rather than some strain-specific regulatory step, that accounts for the difference in CT production. The most glaring dissimilarity that could account for the difference in promoter responsiveness is the number of heptad repeats (TTTTGAT) that appear in the promoters. El Tor strains typically carry two to four copies, whereas classical strains typically have eight repeats.
Fig. 6.
Sodium cholate induced CT expression in El Tor biotype strains of V. cholerae. The ability of sodium cholate to induce CT production was examined in the El Tor biotype strains P27459 (A), E7946 (B), and N16961 (C). The strains were inoculated from an overnight culture into LB, initial pH 8.5, and grown overnight in the absence or presence of sodium cholate at 30°C. CT expression was assayed by ELISA. (□, without sodium cholate; ▪, with 0.1% sodium cholate). Sodium cholate (0.1%) did not induce CT expression when the CT genes ctxA and ctxB were under the control of their native El Tor promoters; this is in contrast to sodium cholate induced CT expression in strains JJM3.13, JJM3.16, and JJM3.10 in which ctxA and ctxB are driven by the classical CT promoter from 569B in P27459, E7946, and N16961, respectively. Errors were all within 12% of their respective mean values.
To examine whether the ability of bile acids to induce ToxRS-activated CT expression in classical but not El Tor strains is also due to a difference in promoters, we measured CT expression in mutant El Tor biotype strains JJM3.10, JJM3.16, and JJM3.13, in which the native ctxAB promoter was replaced by the ctxAB promoter from classical biotype strain 569B (Fig. 6 A–C). The promoters differ by four copies of the heptad repeat (6). In all strain backgrounds, the classical ctxAB promoter in the El Tor background was activated by sodium cholate in LB at 37°C, conditions under which there is typically no CT expression in either classical or El Tor strains. Thus, it seems to be a difference in promoter that accounts for the apparent resistance of El Tor ctxAB expression to bile activation. This same phenomenon was first described in the original descriptions of ToxR activation of CT expression (6).
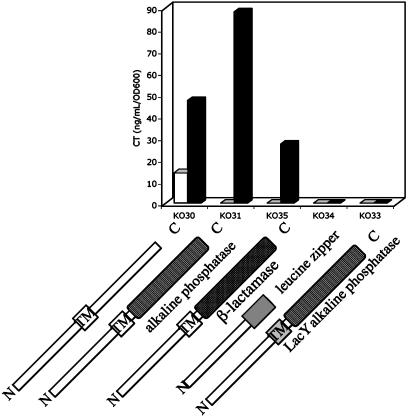
Bile Acids Require the ToxR Transmembrane Domain to Activate CT Expression. To examine more closely the relationship between ToxR and bile acids, we examined the ability of sodium cholate to activate CT expression in a series of previously described ToxR fusion proteins (16) (Fig. 7). The fusion constructs are controlled by the tet promoter and are integrated into the chromosomal lacZ locus of JJM43 (O395-N1 ΔtoxR43). KO30 expresses wild-type ToxR. KO31 expresses a ToxR-phoA fusion with ToxR truncated at amino acid 212 and its C-terminal periplasmic domain replaced by alkaline phosphatase. This fusion preserves the ability to dimerize through the alkaline phosphatase domains. KO35 expresses a ToxR-Bla fusion that cannot dimerize through the periplasmic domains. In both these cases, relative to wild-type ToxR, sodium cholate was able to induce CT expression in LB pH 8.5 and 37°C. The KO34 fusion (ToxR-GCN4-C), which does not localize to the membrane but can dimerize, was unable to induce CT expression in either the absence or presence of sodium cholate. KO33, a ToxR-LacY-PhoA fusion that can dimerize by its periplasmic PhoA domain and can localize to the membrane via its LacY domain, could not be activated by sodium cholate to induce CT. This result suggests that the integrity of the ToxR transmembrane domain must be maintained in order for sodium cholate to exert its effect. Given that sodium cholate likely inserts into the membrane because of its detergent-like properties, the transmembrane region of ToxR may be the site of direct or indirect interaction with bile acids.
Fig. 7.
Sodium cholate induced CT expression in mutant V. cholerae bearing ToxR fusion proteins. The ability of sodium cholate to induce CT production was examined in O395 derivatives bearing ToxR fusion proteins in the presence or absence of sodium cholate. The strains were inoculated from an overnight culture into LB, initial pH 8.5, and grown overnight in the absence or presence of sodium cholate at 37°C. CT expression was assayed by ELISA. (□, without sodium cholate; ▪, with 0.1% sodium cholate.) ToxR fusion proteins are schematically depicted from amino to carboxyl termini. TM boxes indicate transmembrane domains. The different fusion domains are designated beneath the portion of each protein. The ToxR-PhoA and ToxR-Bla fusion proteins have the periplasmic C terminus of ToxR (residues 212–294) replaced by their respective fused domains. In ToxR-GCN4-C, the transmembrane and C terminus of ToxR (residues 182–294) have been replaced by a leucine zipper, which preserves the ability to dimerize. In ToxR-lacY-PhoA, the transmembrane and C terminus of ToxR have also been eliminated, but a heterologous transmembrane domain and periplasmic carboxyl domain have been substituted.
Bile acids were unable to enhance the activity of the same ToxR fusion proteins in the heterologous E. coli system containing a ctx-lacZ reporter (5, 16) (data not shown). ToxR and many of its derived fusion proteins are already sufficient to activate the ctx promoter by themselves, and thus, the presence of bile acids may be superfluous in this system. One possible explanation for the differences observed between the native V. cholerae and E. coli systems could be the presence of some factor in V. cholerae that sequesters ToxR in an inactive form. Bile acids could function to liberate and thus activate ToxR in V. cholerae, whereas in E. coli, this factor is simply absent, resulting in bile acid insensitivity.
Discussion
ToxR was first discovered as a central virulence regulator due to its in vitro ability in E. coli to activate CT expression, but the apparent lack of ToxR activity at the ctx promoter in V. cholerae has remained a conundrum. Instead, a cascade model for virulence regulation has arisen, involving ToxRS, AphAB, TcpPH, and ToxT, with ToxT acting as the critical linchpin that directly activates expression of CT and other virulence factors.
Similarly, the in vitro study of CT regulation in classical V. cholerae strains has relied on an unusual paradox whereby CT expression is induced under the nonphysiological conditions of pH 6.5 and 30°C but inhibited under the physiological conditions that mimic infection of the human intestine (pH 8.5 and 37°C). Although this clearly illustrates the limitations of in vitro systems, significant insight has nevertheless been gained by these studies, leading to the elucidation of pathways and regulators that appear to be significant in vivo, as demonstrated in the infant mouse colonization model (41), in a rabbit ileal loop model (42), and in microarray data obtained from human stool samples (22, 43). For example, a toxT deletion mutant clearly results in a significant colonization defect in the infant mouse model, relative to wild-type O395 (8). In contrast however, in the El Tor biotype strain C6709-1, requirements for infant mouse colonization clearly differ, compared with in vitro induction of ctx and tcpA (41).
In this study, we demonstrate that bile acids can induce in vitro CT expression under conditions that more closely resemble the physiological conditions during infection, and that this induction is due to direct ToxR activity at the ctx promoter and not through ToxT. Although the induction level is small relative to levels measured under standard in vitro inducing conditions, it reconciles previously conflicting data with regard to ToxR activity at the ctx promoter in V. cholerae. Previous evidence suggests that transcriptional induction of tcpA and ctxA differ during in vivo infection, both temporally and with regards to upstream, required regulators, raising the possibility of alternative pathways of activation rather than the central dogma established for in vitro induction (41). Our findings suggest that the case for direct ToxR activity should be reconsidered, especially as progress is made in further characterizing the in vivo signals that govern virulence regulation.
Bile modulates CT expression, outer membrane protein expression, motility, and induction of efflux systems (44). Its pleiotropic effects are complex given that it is a heterogenous mixture of electrolytes, bile acids, phospholipids, cholesterol, and bilirubin. Furthermore, each individual component may have multiple effects. The major components of bile are bile acids, which at higher concentrations are bacteriocidal, presumably due to their detergent-like properties. Thus, they may play a role in host defense against infection. However, the bacterium may combat this defense by its virulence response in the presence of bile acids. We demonstrate that bile acids at sub-bacteriocidal concentrations can actually induce ctx expression but not other virulence factors such as TCP. Provenzano et al. (26) have also demonstrated that bile may play a role in bacterial resistance mechanisms by up-regulating OmpU expression in a ToxR-dependent manner. Further evidence for a bacterial response includes the stimulation of biofilm formation in response to bile acids (D.T.H. and J.J.M., unpublished data). Together, these data suggest that a complex and as yet ill-defined in vivo relationship may exist between bile and V. cholerae.
The role of the various domains of ToxR has remained unclear, with contradictions observed between in vitro and in vivo requirements. ToxR activity in vitro is not dependent on the specific transmembrane or periplasmic domains as long as the ability to localize to the membrane is preserved (16). However, in vivo activity in the mouse colonization model depends on the identity of the periplasmic domain. This observation led to speculation that perhaps the specific transmembrane and periplasmic domains play a more critical role in vivo than in vitro. Our data demonstrate that under appropriate environmental conditions (in this case, the presence of bile acids), the specific transmembrane domain is required for activation, thus supporting this hypothesis. Whether these conditions mimic those experienced by the bacteria during infection is not known. Additionally, the role of ToxS in this model is not known.
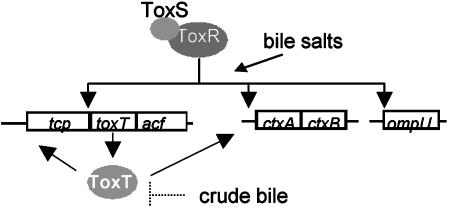
This demonstration of the ability of ToxRS to activate ctx expression directly in O395, with some enhancement by TcpPH, expands the possible roles that ToxRS plays in virulence, with both ToxT-dependent and ToxT-independent functions (Fig. 8). Direct ToxRS activation of ctx in strains of the classical but not El Tor biotypes raises interesting questions in light of the more severe diarrheal disease caused by classical strains. It also suggests a potentially more prominent role for this mechanism in vivo than has been previously recognized in vitro. Clearly, much remains to be understood about the in vivo significance of pathways that have been defined in vitro, about the in vivo environment itself, and about the signals that allow V. cholerae to interact with this environment during infection.
Fig. 8.
Model for the role of ToxR in V. cholerae. ToxRS plays a role in the activation of toxT, ctxAB, and ompU. Crude bile inhibits CT expression, possibly by inhibition of ToxT. Bile acids activate the ToxT-independent branch of ToxRS.
Acknowledgments
We thank Drs. Jun Zhu, Michelle Dziejman, and Su Chiang for helpful discussion and review of the manuscript. D.T.H. was supported by a Howard Hughes Medical Institute fellowship for physician-scientists and a K08 award from the National Institutes of Health. This study was supported by National Institutes of Health Grant AI18045 and National Science Foundation Grant MCB-0094447 (to J.J.M.).
Abbreviations: CT, cholera toxin; TCP, toxin-coregulated pilus.
References
- 1.Faruque, S. M., Albert, M. J. & Mekalanos, J. J. (1998) Microbiol. Mol. Biol. Rev. 62, 1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waldor, M. K. & Mekalanos, J. J. (1996) in Enteric Infections and Immunity, ed. Paradise, L. J. (Plenum, New York), pp. 37-55.
- 3.Miller, V. L. & Mekalanos, J. J. (1988) J. Bacteriol. 170, 2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta, S. & Chowdhury, R. (1997) Infect. Immun. 65, 1131-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller, V. L. & Mekalanos, J. J. (1984) Proc. Natl. Acad. Sci. USA 81, 3471-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mekalanos, J. J., Swartz, D. J., Pearson, G. D., Harford, N., Groyne, F. & de Wilde, M. (1983) Nature 306, 551-557. [DOI] [PubMed] [Google Scholar]
- 7.Champion, G. A., Neely, M. N., Brennan, M. A. & DiRita, V. J. (1997) Mol. Microbiol. 23, 323-331. [DOI] [PubMed] [Google Scholar]
- 8.Higgins, D. E. & DiRita, V. J. (1994) Mol. Microbiol. 14, 17-29. [DOI] [PubMed] [Google Scholar]
- 9.DiRita, V. J., Parsot, C., Jander, G. & Mekalanos, J. J. (1991) Proc. Natl. Acad. Sci. USA 88, 5403-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, R. C. & Taylor, R. K. (1995) Mol. Microbiol. 16, 425-439. [DOI] [PubMed] [Google Scholar]
- 11.Yu, R. R. & DiRita, V. J. (1999) J. Bacteriol. 181, 2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dziejman, M. & Mekalanos, J. J. (1994) Mol. Microbiol. 13, 485-494. [DOI] [PubMed] [Google Scholar]
- 13.DiRita, V. J. & Mekalanos, J. J. (1991) Cell 64, 29-37. [DOI] [PubMed] [Google Scholar]
- 14.Ottemann, K. M. & Mekalanos, J. J. (1996) J. Bacteriol. 178, 156-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, V. L., Taylor, R. K. & Mekalanos, J. J. (1987) Cell 48, 271-279. [DOI] [PubMed] [Google Scholar]
- 16.Ottemann, K. M. & Mekalanos, J. J. (1995) Mol. Microbiol. 15, 719-731. [DOI] [PubMed] [Google Scholar]
- 17.Hase, C. C. & Mekalanos, J. J. (1998) Proc. Natl. Acad. Sci. USA 95, 730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krukonis, E. S., Yu, R. R. & DiRita, V. J. (2000) Mol. Microbiol. 38, 67-84. [DOI] [PubMed] [Google Scholar]
- 19.Murley, Y. M., Carroll, P. A., Skorupski, K., Taylor, R. K. & Calderwood, S. B. (1999) Infect. Immun. 67, 5117-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu, J., Miller, M. B., Vance, R. E., Dziejman, M., Bassler, B. L. & Mekalanos, J. J. (2002) Proc. Natl. Acad. Sci. USA 99, 3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovacikova, G. & Skorupski, K. (2002) Mol. Microbiol. 46, 1135-1147. [DOI] [PubMed] [Google Scholar]
- 22.Bina, J., Zhu, J., Dziejman, M., Faruque, S., Calderwood, S. & Mekalanos, J. (2003) Proc. Natl. Acad. Sci. USA 100, 2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crawford, J. A., Kaper, J. B. & DiRita, V. J. (1998) Mol. Microbiol. 29, 235-246. [DOI] [PubMed] [Google Scholar]
- 24.Sperandio, V., Giron, J. A., Silveira, W. D. & Kaper, J. B. (1995) Infect. Immun. 63, 4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakrabarti, S. R., Chaudhuri, K., Sen, K. & Das, J. (1996) J. Bacteriol. 178, 524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Provenzano, D., Schuhmacher, D. A., Barker, J. L. & Klose, K. E. (2000) Infect. Immun. 68, 1491-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofmann, A. F. (1998) in Gastrointestinal and Liver Disease, eds. Feldman, M., Scharschmidt, B. F. & Sleisenger, M. H. (Saunders, Philadelphia), pp. 937-948.
- 28.Schuhmacher, D. A. & Klose, K. E. (1999) J. Bacteriol. 181, 1508-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Provenzano, D. & Klose, K. E. (2000) Proc. Natl. Acad. Sci. USA 97, 10220-10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakrabarti, S., Sengupta, N. & Chowdhury, R. (1999) Infect. Immun. 67, 1025-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mekalanos, J. J., Collier, R. J. & Romig, W. R. (1979) J. Biol. Chem. 254, 5855-5861. [PubMed] [Google Scholar]
- 32.Chiang, S. L., Taylor, R. K., Koomey, M. & Mekalanos, J. J. (1995) Mol. Microbiol. 17, 1133-1142. [DOI] [PubMed] [Google Scholar]
- 33.Skorupski, K. & Taylor, R. K. (1996) Gene 169, 47-52. [DOI] [PubMed] [Google Scholar]
- 34.Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., Haft, D. H., Hickey, E. K., Peterson, J. D., Umayam, L., et al. (2000) Nature 406, 477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. (1995) J. Bacteriol. 177, 4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardel, C. L. & Mekalanos, J. J. (1994) Methods Enzymol. 235, 517-526. [DOI] [PubMed] [Google Scholar]
- 37.Sun, D., Seyer, J. M., Kovari, I., Sumrada, R. A. & Taylor, R. K. (1991) Infect. Immun. 59, 114-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carroll, P. A., Tashima, K. T., Rogers, M. B., DiRita, V. J. & Calderwood, S. B. (1997) Mol. Microbiol. 25, 1099-1111. [DOI] [PubMed] [Google Scholar]
- 39.Higgins, D. E., Nazareno, E. & DiRita, V. J. (1992) J. Bacteriol. 174, 6974-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwanaga, M., Yamamoto, K., Higa, N., Ichinose, Y., Nakasone, N. & Tanabe, M. (1986) Microbiol. Immunol. 30, 1075-1083. [DOI] [PubMed] [Google Scholar]
- 41.Lee, S. H., Hava, D. L., Waldor, M. K. & Camilli, A. (1999) Cell 99, 625-634. [DOI] [PubMed] [Google Scholar]
- 42.Xu, Q., Dziejman, M. & Mekalanos, J. J. (2003) Proc. Natl. Acad. Sci. USA 100, 1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merrell, D. S., Butler, S. M., Qadri, F., Dolganov, N. A., Alam, A., Cohen, M. B., Calderwood, S. B., Schoolnik, G. K. & Camilli, A. (2002) Nature 417, 642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee, A., Chaudhuri, S., Saha, G., Gupta, S. & Chowdhury, R. (2004) J. Bacteriol. 186, 6809-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]