How cell–cell signaling coordinates cell movement, gene expression, and differentiation during the development of multicellular organisms is a fundamental question. At the molecular level, the answer is complex, even in the simplest cases. Myxococcus xanthus is one of the simplest organisms that undergo development to produce a multicellular structure of uniform size and shape. Cells of this bacterium sense nutrient limitation and alter their gliding movements to produce mounds (Fig. 1). Within these nascent fruiting bodies, cells differentiate into spores. Driving this developmental process is a program of gene expression that is temporally and spatially regulated by intracellular and extracellular signals. Are the typical prokaryotic signaling and gene regulatory mechanisms sufficient for multicellular development of M. xanthus? Previous work has indicated that eukaryotic-like mechanisms play a role. The work of Jelsbak et al. (1) in this issue of PNAS extends this observation by describing a group of 12 M. xanthus genes that seem to encode enhancer-binding proteins (EBPs) with a forkhead-associated (FHA) domain. The authors show that the FHA domain of one of the EBPs is required for normal development. Because FHA domains interact with phosphothreonine residues, the results suggest a crucial link to eukaryotic-like Ser/Thr protein kinases (STPKs), which are abundant in M. xanthus.
Fig. 1.
Cartoon depicting starved M. xanthus cells forming a mound, then a fruiting body, which would contain ≈105 spores.
Bacterial EBPs derive their name from the fact that they activate transcription by σ54-RNA polymerase (σ54-RNAP) in much the same way some EBPs activate transcription in eukaryotes. σ54-RNAP requires interaction with an EBP to form the transcriptionally competent open promoter complex. Bacterial EBPs bind to DNA enhancer elements typically located 70–150 bp upstream of the transcription start site (reviewed in ref. 2). Bending of DNA allows the EBP to contact σ54-RNAP, forming a DNA loop. ATP hydrolysis by the conserved central domain of the EBP enables it to convert the σ54-RNAP closed promoter complex to the open complex. Although eukaryotic EBPs also bind DNA and in some cases cause a DNA loop to form by contacting RNAP or a general transcription factor, they do not perform the ATP hydrolysis necessary for open complex formation [e.g., transcription factor IIH (TFIIH) does this in the case of RNAP II transcription].
The work of Jelsbak et al. (1) builds on previous efforts to identify and characterize EBPs of M. xanthus (3–5). By using the conserved central ATPase domain to search the recently completed M. xanthus genome sequence, Jelsbak et al. (1) found 52 putative EBPs. This is the largest number so far found in a bacterial genome. At least 16 of these EBPs have been shown to be required for normal fruiting body development or motility (refs. 3–5 and references therein). Clearly, this form of regulation is critical for M. xanthus development. Moreover, σ54 is essential for growth of this bacterium (6), unlike other bacteria so far tested. The situation in M. xanthus is strikingly different from that in Streptomyces coelicolor, another prokaryotic model for development, which is devoid of σ54 and EBPs (7).
Jelsbak et al. (1) recognize FHA domains in 12 of the M. xanthus EBPs. The FHA domain mediates phosphorylation-dependent, protein–protein interactions by recognizing a phosphothreonine-containing epitope in a protein partner (reviewed in ref. 8). Originally recognized in a subset of eukaryotic forkhead-type transcription factors, the FHA domain has been found in eukaryotic proteins with diverse functions, including signal transduction, protein transport, and DNA repair. Likewise, FHA domains are found in a variety of prokaryotic proteins, implicating them in many bacterial processes (9). However, until now, only two putative FHA domain-containing EBPs (FHA-EBPs) have been described in bacteria (10). The finding of 12 such proteins in M. xanthus (1) suggests abundant connections between EBP-dependent transcription by σ54-RNAP and phosphorylation of Thr residues in proteins.
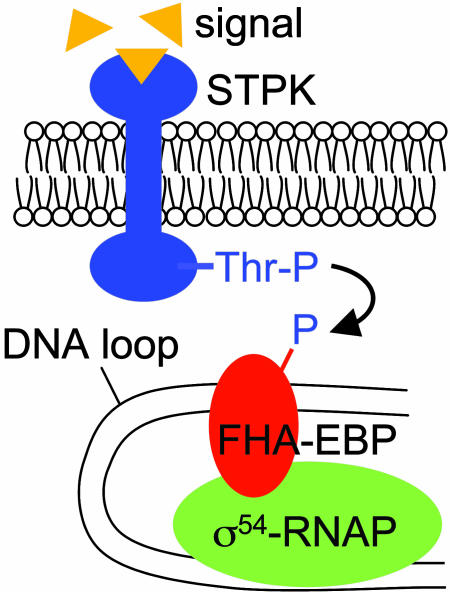
There is ample opportunity for phosphothreonine formation in proteins of M. xanthus. The first eukaryotic-like STPK found in bacteria was discovered in M. xanthus (11), and subsequent work revealed a large family of STPKs, many required for normal development (12). Half of the 12 FHA-EBP genes are next to or near STPK genes in the M. xanthus genome (1). This proximity suggests that the FHA domain of these EBPs might interact with the STPK encoded nearby. According to this model (Fig. 2), Thr autophosphorylation of the STPK in response to a signal would promote interaction with the FHA-EBP. Transfer of phosphate from the STPK to the FHA-EBP would allow it to activate transcription by σ54-RNAP.
Fig. 2.
Model showing how an STPK/FHA-EBP signal transduction system might work.
A precedent for parts of this model has been described recently (13). PknH, an STPK, and EmbR, an FHA domain-containing transcription factor, are encoded by adjacent genes in Mycobacterium tuberculosis. PknH can autophosphorylate on Ser and Thr residues in vitro, then interact with the FHA domain of EmbR and transfer phosphate to one or more of its Thr residues. Presumably, phosphorylation of EmbR enables it to activate transcription. Although EmbR lacks the conserved central ATPase domain of EBPs, the N-terminal two-thirds of EmbR is similar to AfsR, a transcriptional activator of S. coelicolor. AfsR does not have a recognizable FHA domain, but it is phosphorylated by an STPK, and this phosphorylation enhances its ability to bind to the –35 region of its target promoter, where its ATPase activity is proposed to isomerize the RNAP closed promoter complex to the open complex (reviewed in ref. 14). Although this mechanism is still speculative, and AfsR differs from EBPs in that it binds closer to the start site of transcription and its ATPase domain does not share all of the same sequence features, the theme of signal transduction by means of STPK phosphorylation of a transcriptional activator whose function depends on ATP hydrolysis is preserved in S. coelicolor, despite its lack of EBPs and σ54 (7).
As in eukaryotes, the signaling and regulatory proteins in prokaryotes are modular and seem to contain nearly every conceivable combination of domains. The finding of two FHA-EBPs in Pirellula (10) and now 12 in the phylogenetically distant M. xanthus (1) raises interesting questions about the evolutionary history of this domain combination. Further analysis is needed to assess whether lateral transfer likely played a role in creating the combination, and genomic sequencing of more bacterial species will reveal its prevalence and perhaps its evolution. It will be interesting to compare the FHA domains of the 12 M. xanthus EBPs with that of EspA, a putative protein kinase that delays M. xanthus sporulation until mounds have formed (15). In addition to its N-terminal FHA domain, EspA has a histidine protein kinase (HPK) domain followed by a receiver domain at its C terminus. Because a receiver domain is more often found in response regulator proteins of two-component signal transduction systems (16), EspA is considered a hybrid kinase. Given EspA's FHA domain, its lack of a predicted transmembrane domain, and the fact that it is encoded next to a putative STPK gene (15), it is possible that EspA is phosphorylated by the STPK, activating its HPK. Hence, EspA might directly link STPK and HPK signaling. It seems likely that mining the M. xanthus genome sequence for FHA domains will reveal more proteins with interesting domain combinations.
Of the 12 M. xanthus genes predicted to encode FHA-EBPs, 9 had been mutated previously and 2 of the mutations caused a developmental defect (3, 4). Jelsbak et al. (1) disrupted the remaining 3 genes, and one of the mutants had a developmental defect. Disruption of the Mx4885 gene caused a delay in mound formation, and the mounds never became as compact and uniform in shape as those produced by wild type. Sporulation was reduced 500-fold compared with wild type. An in-frame deletion of the part of the gene predicted to encode the FHA domain produced the same phenotype. Analysis of the mutant suggests a partial defect in the response to C-signal.
The C-signal is a 17-kDa protein believed to be processed from a 25-kDa precursor at the cell surface (reviewed in ref. 17). Transmission of the C-signal requires that cells move into alignment and make contact. The level of C-signal increases during development, and a higher level is needed for sporulation than for mound formation. Low level C-signaling produces rippling behavior, a pattern of cell movement in which cells accumulate in transient ridges between nascent fruiting bodies. Expression of most genes induced after 6 h into development depends partly or completely on C-signaling. FruA, a putative two-component response regulator, mediates responses to C-signal, presumably after being phosphorylated by an unidentified HPK (18–20).
The developmental defects of the Mx4485 mutant are not as severe as those of a fruA mutant. A fruA mutant, like a mutant unable to produce C-signal, fails to ripple, make compact mounds, or form spores (18, 20). The Mx4485 mutant was able to ripple (1), indicating some ability to respond to C-signal. Also, expression of two late developmental reporter fusions, created by Tn5 lac insertions Ω4414 and Ω4403, was much less severely impaired in the Mx4485 mutant (1) than in a fruA mutant (18, 19). Jelsbak et al. (1) show that the Mx4485 mutant makes normal levels of C-signal and FruA, so they propose that Mx4485 affects the phosphorylation step believed to activate FruA.
How could this work? In the context of the model shown in Fig. 2, an STPK might phosphorylate Mx4485 in response to an early developmental signal. The phosphorylated FHA-EBP would activate transcription by σ54-RNAP of one or more genes whose products facilitate phosphorylation of FruA. For example, increased transcription of a gene coding for the unidentified HPK thought to phosphorylate FruA would be a simple way to link an early STPK/Mx4485 signaling pathway to a later HPK/FruA pathway that responds to C-signal. Of course, the connection might be less direct.
It will be interesting to see just how eukaryotic-like the signaling and gene regulatory network is during the “simple” developmental process of M. xanthus.
See companion article on page 3010.
References
- 1.Jelsbak, L., Givskov, M. & Kaiser, D. (2005) Proc. Natl. Acad. Sci. USA 102, 3010–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buck, M., Gallegos, M. T., Studholme, D. J., Guo, Y. & Gralla, J. D. (2000) J. Bacteriol. 182, 4129–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caberoy, N. B., Welch, R. D., Jakobsen, J. S., Slater, S. C. & Garza, A. G. (2003) J. Bacteriol. 185, 6083–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorski, L. & Kaiser, D. (1998) J. Bacteriol. 180, 5896–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakobsen, J. S., Jelsbak, L., Welch, R. D., Cummings, C., Goldman, B., Stark, E., Slater, S. & Kaiser, D. (2004) J. Bacteriol. 186, 4361–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keseler, I. & Kaiser, D. (1997) Proc. Natl. Acad. Sci. USA 94, 1979–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentley, S. D., Chater, K. F., Cerdeno-Tarraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., Harris, D. E., Quail, M. A., Kieser, H., Harper, D., et al. (2002) Nature 417, 141–147. [DOI] [PubMed] [Google Scholar]
- 8.Durocher, D. & Jackson, S. P. (2002) FEBS Lett. 513, 58–66. [DOI] [PubMed] [Google Scholar]
- 9.Pallen, M., Chaudhuri, R. & Khan, A. (2002) Trends Microbiol. 10, 556–563. [DOI] [PubMed] [Google Scholar]
- 10.Studholme, D. J. & Dixon, R. (2004) FEMS Microbiol. Lett. 230, 215–225. [DOI] [PubMed] [Google Scholar]
- 11.Munoz-Dorado, J., Inouye, S. & Inouye, M. (1991) Cell 67, 995–1006. [DOI] [PubMed] [Google Scholar]
- 12.Inouye, S., Jain, R., Ueki, T., Nariya, H., Xu, C. Y., Hsu, M. Y., Fernandez-Luque, B. A., Munoz-Dorado, J., Farez-Vidal, E. & Inouye, M. (2000) Microb. Comp. Genomics 5, 103–120. [DOI] [PubMed] [Google Scholar]
- 13.Molle, V., Kremer, L., Girard-Blanc, C., Besra, G. S., Cozzone, A. J. & Prost, J. F. (2003) Biochemistry 42, 15300–15309. [DOI] [PubMed] [Google Scholar]
- 14.Umeyama, T., Lee, P. C. & Horinouchi, S. (2002) Appl. Microbiol. Biotechnol. 59, 419–425. [DOI] [PubMed] [Google Scholar]
- 15.Cho, K. & Zusman, D. R. (1999) Mol. Microbiol. 34, 714–725. [DOI] [PubMed] [Google Scholar]
- 16.Stock, A. M., Robinson, V. L. & Goudreau, P. N. (2000) Annu. Rev. Biochem. 69, 183–215. [DOI] [PubMed] [Google Scholar]
- 17.Kaiser, D. (2004) Annu. Rev. Microbiol. 58, 75–98. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa, M., Fujitani, S., Mao, X., Inouye, S. & Komano, T. (1996) Mol. Microbiol. 22, 757–767. [DOI] [PubMed] [Google Scholar]
- 19.Ellehauge, E., Norregaard-Madsen, M. & Sogaard-Andersen, L. (1998) Mol. Microbiol. 30, 807–817. [DOI] [PubMed] [Google Scholar]
- 20.Sogaard-Andersen, L., Slack, F., Kimsey, H. & Kaiser, D. (1996) Genes Dev. 10, 740–754. [DOI] [PubMed] [Google Scholar]