Abstract
Primate genomes encode a variety of innate immune strategies to defend themselves against retroviruses. One of these, TRIM5α, can restrict diverse retroviruses in a species-specific manner. Thus, whereas rhesus TRIM5α can strongly restrict HIV-1, human TRIM5α only has weak HIV-1 restriction. The biology of TRIM5α restriction suggests that it is locked in an antagonistic conflict with the proteins encoding the viral capsid. Such antagonistic interactions frequently result in rapid amino acid replacements at the protein–protein interface, as each genetic entity vies for evolutionary dominance. By analyzing its evolutionary history, we find strong evidence for ancient positive selection in the primate TRIM5α gene. This selection is strikingly variable with some of the strongest selection occurring in the human lineage. This history suggests that TRIM5α evolution has been driven by antagonistic interactions with a wide variety of viruses and endogenous retroviruses that predate the origin of primate lentiviruses. A 13-aa “patch” in the SPRY protein domain bears a dense concentration of positively selected residues, potentially implicating it as an antiviral interface. By using functional studies of chimeric TRIM5α genes, we show that this patch is generally essential for retroviral restriction and is responsible for most of the species-specific antiretroviral restriction activity. Our study highlights the power of evolutionary analyses, in which positive selection identifies not only the age of genetic conflict but also the interaction interface where this conflict plays out.
Keywords: capsid, human endogenous retroviruses, HIV type 1, SPRY
Innate immune strategies that defend primates against retroviruses are of considerable medical and evolutionary importance. Two distinct cellular antiviral defense systems, APOBEC3G/F (1–3) and TRIM5α (4), that limit HIV infection have been described recently. APOBEC3G and APOBEC3F are cytidine deaminases that can cause hypermutation in the viral genome, but a viral accessory protein, Vif, can counteract their antiviral effect. TRIM5α is a postentry restriction factor that accounts for the resistance to HIV type 1 (HIV-1) observed in rhesus monkey cells. It is not yet known how TRIM5α mediates viral restriction, although a shorter, alternate transcript of the TRIM5 gene has been shown to be an ubiquitin ligase (5). TRIM5α restriction depends on the viral capsid, and its effect is saturable (4), although direct physical interaction between TRIM5α and capsid has not been demonstrated. TRIM5α from human and nonhuman primates also can restrict other lentiviruses and some strains of murine leukemia virus, a distantly related gammaretrovirus (6–9).
Although host genomes benefit from TRIM5α's recognition of viruses, it is in the best interest of the virus to evade recognition. Such antagonistic interactions have been formalized as the “Red Queen” hypothesis (10) and lead to the rapid fixation of amino acid replacements (positive selection), most likely at the interaction interface. The history of positive selection is thus informative for determining how long genes have been participants in genetic conflict, for identifying the likely sources of this conflict, and even for defining interaction domains involved. We previously have performed such an analysis on the APOBEC genes to show that APOBEC3G's role in genome defense predates the origin of primate lentiviruses (11, 12) and that many other APOBEC cytidine deaminase genes likely participate in defending the primate genome against retroviruses.
Here, we show that the TRIM5α restriction factor has undergone multiple episodes of positive selection that predate the origin of primate lentiviruses. Selection pressures on TRIM5α vary widely among primate lineages, suggesting that distinct episodes of retroviral infection have dominated TRIM5α evolution. The positive selection in TRIM5α appears to be concentrated largely in the α-isoform-specific SPRY domain. We use this concentration of positively selected residues to propose and validate the presence of a SPRY “patch” that is essential for TRIM5α's general and species-specific restriction of retroviruses.
Materials and Methods
Primate Genomic DNA Sources. Genomic DNA samples obtained from Coriell Cell Repositories (Camden, NJ) are as follows (including Coriell repository numbers): Pan troglodytes (chimpanzee, NAO3448A), Gorilla gorilla (gorilla, NG05251B), Pongo pygmaeus (orangutan, NAO4272), Erythrocebus patas (patas monkey, NG06254), Lagothrix lagotricha (common woolly monkey, NG05356), Ateles geoffroyi (black-handed spider monkey, NGO5352), and Saguinus labiatus (red-chested mustached tamarin, NG05308). Genomic DNA from Hylobates syndactylus (island siamang, KB11539), Colobus guereza kikuyuensis (kikuyu colobus, OR160), Pygathrix nemaeus (douc langur, OR259), Callithrix pygmaea (pygmy marmoset, OR690), Saimiri sciureus sciureus (squirrel monkey, KB4544), Callicebus donacophilus donacophilus (Bolivian gray titi, OR1522), Pithecia pithecia pithecia (white-faced saki, KB5932), and Alouatta sara (Bolivian red howler, OR749) was obtained from the Center for Reproduction of Endangered Species FrozenZoo Project (San Diego Zoo, San Diego). Papio anubis (baboon) DNA was a personal gift from Trent Colbert (Fred Hutchinson Cancer Research Center). Cercopithecus aethiops (African green monkey) genomic DNA was prepared from Cos-7 cells with the QIAamp-DNA kit (Qiagen, Valencia, CA).
Sequencing of TRIM5α Exons from Primate Genomic DNA. TRIM5α was amplified and sequenced exon by exon from genomic DNA with PCR Supermix High Fidelity (Invitrogen) by using the PCR and sequencing primers shown in Table 2, which is published as supporting information on the PNAS web site. PCR products were sequenced directly, except in a few cases where they were first cloned into the TOPO TA cloning vector (Invitrogen), followed by sequencing of three independent clones. Exon reads were spliced together to create virtual transcripts for each primate, and have been entered into the GenBank database under the accession nos. AY843504–AY843520. The TRIM5α cDNA sequences for human (AY625000), Macaca mulatta (Rhesus macaque, AY523632.1), and Cercopithecus tantalus (tantalus monkey, AY593973.2) were obtained from the GenBank database. A phylogeny constructed by using the isolated TRIM5α genes is in good agreement with the accepted primate phylogeny, indicating that all sequences isolated by our PCR strategy are truly orthologous.
Confirmation of TRIM5 Transcripts by Sequencing from RNA. The following monkey fibroblast cell lines were obtained from Coriell: Ateles geoffroyi (AG05352), Lagothrix lagotricha (AG05356), Callicebus moloch (AG06115A), and Saguinus labiatus (AG05308A). RNA was prepared from ≈1 million cells with the RNeasy kit (Qiagen). RT-PCR of the TRIM5α SPRY domain was performed with the primers shown in Table 2, by using the SuperScript One-Step kit (Invitrogen). PCR products were sequenced directly.
Sequence Analysis. DNA sequences were aligned by using clustal x (13), with hand alignment of small indels based on amino acid sequence. Maximum likelihood analysis was performed with codeml in the paml 3.14 software package (14). Global synonymous changes per site (dS)/replacement changes per site (dN) ratios for the tree (Fig. 1) were calculated by a free-ratio model, which allows dN/dS to vary along different branches. We tested whether dN/dS values were >1 in two lineages, those leading to gibbon and human, by using two methods as described in ref. 15 (see Appendix 1, which is published as supporting information on the PNAS web site). Briefly, likelihoods were compared when the lineage was fixed at dN/dS = 1 relative to when the lineage was allowed to have a dN/dS >1 (16). In the second method, we calculated the dN/dS ratios relative to the reconstructed ancestor and tested for significant deviations from dN/dS = 1 (16).
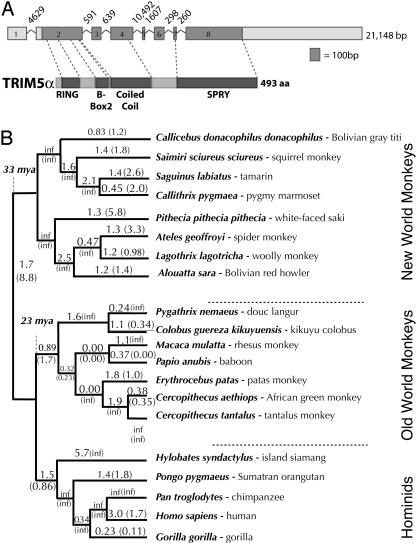
Fig. 1.
Ancient and variable positive selection has shaped TRIM5α evolution. (A) TRIM5α is the longest of six reported transcripts of the eight-exon human TRIM5 gene. It encodes a 493-aa protein consisting of a RING finger, a B-Box2, and a coiled-coil domain (signature domains of the TRIM family; ref. 24), as well as an α-isoform-specific SPRY domain. (B) TRIM5α was sequenced from a panel of primates representing 33 million years of evolutionary divergence. Values of dN/dS along each branch were calculated by using the free-ratio model of paml for either the whole gene or for the SPRY domain alone (in parentheses), as shown on a cladogram of the accepted primate phylogeny (38). A dN/dS value of >1 suggests that positive selection has acted along that lineage. inf refers to cases where dS = 0.
To detect selection in TRIM5α, multiple alignments were fitted to either the F3 × 4 or F61 models of codon frequencies. Likelihood ratio tests of the data were performed by using different sets of site-specific (NS sites) models as follows: M0 (one-ratio) to M3 (discrete); M1 (two-state, neutral, dN/dS > 1 disallowed) to M2 (selection, similar to model 1 but dN/dS > 1 allowed); and M7 (fit to a beta distribution, dN/dS > 1 disallowed) to M8 (similar to model 7 but dN/dS > 1 allowed). In all cases, permitting sites to evolve under positive selection gave a much better fit to the data (Table 1). These analyses also identified certain amino acid residues with high posterior probabilities (>0.95) of having evolved under positive selection (17, 18) (Fig. 2; see also Appendix 1).
Table 1. Positive selection in the TRIM5α gene.
| Data set | -2(ln λ) | df | P value | dN/dS | Proportion of sites, % |
|---|---|---|---|---|---|
| All primates | |||||
| M0 vs. M3 (k = 3) | 217.94 | 4 | P < 0.0001 | 2.4, 7.2 | 33, 4 |
| M1 vs. M2 | 109.88 | 2 | P < 0.0001 | 4.5 | 16 |
| M7 vs. M8 | 109.50 | 2 | P < 0.0001 | 4.2 | 18 |
| Hominids + OWMs | |||||
| M0 vs. M3 (k = 3) | 57.42 | 4 | P < 0.0001 | 1.8, 17 | 57, 2 |
| M1 vs. M2 | 39.16 | 2 | P < 0.0001 | 10 | 4 |
| M7 vs. M8 | 40.21 | 2 | P < 0.0001 | 10 | 4 |
We used likelihood ratio tests to determine whether any codon positions were associated with dN/dS significantly > 1 and hence possibly subject to positive Darwinian selection. Neutral models (M0, M1, and M7) were compared to selection models (M2 and M8), which allow a proportion of codons for which dN/dS exceeds 1, or models for heterogeneity of dN/dS among sites (M3). As indicated by the P values, all analyses find very strong evidence for the selection model. Note that in M3, two classes of sites are permitted to have dN/dS > 1. Also indicated are the proportion of codons that were found to have dN/dS > 1, with the associated dN/dS values shown. Analyses using the f61 model of codon frequencies are shown, but similar results were obtained by using the f3×4 frequency model (full paml results are available in Appendix 1).
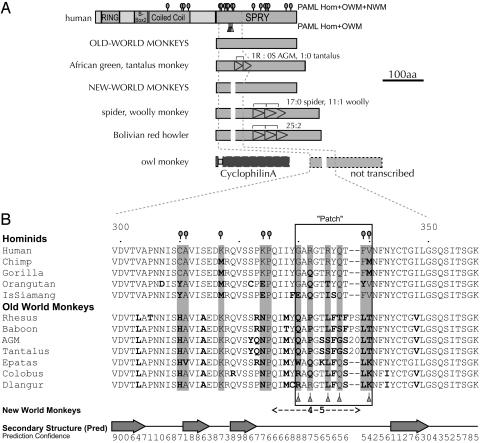
Fig. 2.
The SPRY domain is a hot spot for insertions/deletions and positive selection. (A) Codons highlighted by paml as being subject to positive selection with high posterior probabilities (P > 0.95) in an analysis of all primates are indicated as circles on stems above the TRIM5α protein schematic. In a more limited analysis of just the hominids and Old World monkeys (OWMs), only five residues in the entire protein were identified as evolving under positive selection (triangles on stems below the TRIM5α schematic). Representative SPRY domains from OWMs and New World monkeys are schematized, along with notable exceptions that have internal duplications (indicated by tandem arrowheads). These duplications appear to have accumulated more replacement (R) changes than synonymous (S) changes (R:S ratios indicated above). In owl monkeys, a CyclophilinA insertion (TRIM-Cyp) occurred between exons 7 and 8 (26), and it is believed that the 8th exon of TRIM5 containing the SPRY domain is not transcribed. (B) Codons identified as being under positive selection are indicated in gray background and by using the same symbols as in A. Changes relative to human are indicated in bold. NWMs have a large deletion in the area of the patch (see Data Set 1, which is published as supporting information on the PNAS web site). Secondary structure predictions and confidence values (0, low; 9,high) were made by using the psipred server (19). Arrows indicate β-strands, and lines indicate coils. According to this prediction, most of the TRIM5α protein is predicted to be comprised of α-helices, whereas the entire SPRY domain is predicted with high confidence to be β-strands and coils with no α-helices.
Secondary structure predictions and associated confidence values for the entire human TRIM5α protein were made by using the psipred (19) Protein Structure Prediction Server (http://bioinf.cs.ucl.ac.uk/psipred).
Viral Infection Assays. CRFK (feline renal fibroblast) cells were obtained from the American Type Culture Collection and grown in DMEM/10% FCS. The human and rhesus TRIM5α alleles with a hemagglutinin epitope tag (4) in the LPCX retroviral vector were obtained from the National Institutes of Health AIDS Reagent Program (donated by Joseph Sodroski). Site-directed mutagenesis of the rhesus or human genes was performed by the QuikChange kit (Stratagene) in a single step (oligonucleotide sequences available on request). Retroviral vectors were produced in 293T cells, and supernatants were used to infect CRFK cells. Twenty-four hours after infection, 3 mg/ml puromycin was added to the medium to select for transduced cells to obtain pools of cells that stably expressed exogenous TRIM5α genes. Expression was verified by Western blotting using a hemagglutinin antibody (Babco, Richmond, CA).
Single-cycle assays for HIV-1 were performed essentially as described in ref. 20. HIV-1 provirus was deleted for env, and GFP or luciferase was inserted into the nef region (20). SIVag-mTAN virus (21) was a kind gift of Ned Landau (The Salk Institute, San Diego). Virus was produced in 293T cells by cotransfection of the provirus with the VSV-G gene, titrated on cells without restriction factors, and frozen. Infection of CRFK cells with HIV-1 (GFP) was performed in 24-well plates with 3 × 104 cells per well by using 5-fold dilutions of virus that had been titrated previously to give between 1% and 80% infection. Two days after infection, the cells were fixed with 2% paraformaldehyde and analyzed by flow cytometry for GFP expression. Luciferase assays were performed with virus stocks that had been titrated previously to be in the linear range of the assay (between 10,000 and 1 million relative light units). Infections were performed in triplicate in 24-well plates, assayed with the luciferase assay kit (Promega) in a 96-well plate, and read on a luminometer.
Results
We sequenced the TRIM5α gene (≈1,482 bp of coding sequence) from 17 primate genomes that represent 33 million years of evolution (Fig. 1). Comparison of the rates of nonsynonymous (that alter the encoded amino acid) and synonymous DNA changes between species can be used to assess the types of selective pressures that have acted on a gene (22). For most protein-coding genes, dS exceeds dN because amino acid replacements are generally detrimental to protein function and therefore are culled out of the population (purifying selection). We find that many branches of the primate phylogeny, including internal branches, show evidence for TRIM5α evolution under positive selection (defined as dN/dS > 1.0; Fig. 1B). Thus, TRIM5α has been subject to positive selection for at least 33 million years. This selection has been strikingly variable because dN/dS ratios along different branches are significantly different from each other (P < 0.02; Appendix 1). In contrast, the positive selection on APOBEC3G was found to be more constant (11, 12) because ω values along each branch did not differ significantly from each other (P > 0.75; Appendix 1). Positive selection of TRIM5α is especially strong in the hominid clade, with the highest whole-gene dN/dS values of 5.7 and 3.0 found in the lineages leading to island siamang (gibbon) and human, respectively. We tested for the presence of positive selection in the gibbon and human branches by both comparative two-ratio likelihood tests by using paml (15) and Monte-Carlo simulation using k-estimator (16) (see Materials and Methods and Appendix 1). The average dS in TRIM5α is not unusually low; it is 0.084 between hominids and Old World monkeys (OWMs) and 0.153 between hominids and New World monkeys (NWMs), compared with previous estimates of 0.08 and 0.15, respectively, for substitution rates in various intronic and noncoding regions of primate genomes (23). Thus, we can rule out the possibility that selection has led to deflated dS values in TRIM5α, resulting in artificially high dN/dS ratios.
TRIM5 is a member of the large tripartite motif family in primate genomes, characterized by having RING finger, B-box, and coiled-coil domains (24). The α isoform of TRIM5 has an additional SPRY protein domain (Fig. 1 A), which is found in many proteins including those in the Ig superfamily (25). This SPRY domain has been shown previously to be essential for the restriction of HIV-1 (4). Although little is known about SPRY function, we found that this domain had undergone the most intense positive selection. This finding is evidenced by the high dN/dS values obtained in an analysis of this domain alone (in parenthesis on cladogram, Fig. 1B), including a striking dN/dS of 8.8 in the branch separating NWM from OWM and hominids (see also Fig. 4, which is published as supporting information on the PNAS web site). In addition to the strong positive selection, the SPRY domain has undergone an unusual number of insertions and deletions (Fig. 2 A). A small deletion has occurred in the lineage leading to the NWMs, whereas there have been two distinct instances of internal duplications, one in African green monkeys and close relatives and a different triplication in the lineage leading to spider, woolly, and howler monkeys. The African green monkeys and tantalus duplication has been verified in TRIM5α transcripts (GenBank accession nos. AY625003 and AY593973). We confirmed the woolly and spider monkey triplications by sequencing RT-PCR products (data not shown). When the duplicated and triplicated sequences within a single gene were aligned to each other, they showed an inflated number of nonsynonymous to synonymous changes (e.g., 25 replacement:2 synonymous changes for howler monkey; Fig. 2 A). Thus, internal duplications in the SPRY domain followed by positive selection predict that these sequences are functionally important for the ability of TRIM5α to restrict different viruses. Finally, in owl monkey, the entire SPRY domain of TRIM5α has been replaced by a retrotransposed CyclophilinA gene, now referred to as TRIM-Cyp (26). CyclophilinA has been reported to directly interact with viral capsid (27), and, because the SPRY domain can be functionally replaced by CyclophilinA, this finding suggests that the SPRY also might be a capsid-interacting domain.
We used a maximum-likelihood approach, by using the paml suite of programs (14), to determine whether specific codons of TRIM5α have been repeatedly subjected to positive selection in primates. Statistical results support the presence of positive selection with extraordinary confidence, regardless of whether we include or exclude NWMs in our analysis (Table 1). Analysis of the full data set identifies ≈18% of the codons as having evolved under positive selection with an average dN/dS ratio of >4 (Table 1, NSsites model 8 vs. 7). Of these, residues identified as being under positive selection with high confidence (>95% posterior probability) are illustrated on a schematic of the TRIM5α protein (circles on stems, Fig. 2) and fall in either the coiled-coil or SPRY domains. Some TRIM proteins have been shown to homomultimerize through their coiled-coil domains (24), but our results also suggest that the coiled-coil domain of TRIM5α may additionally participate in host defense. A secondary structure prediction of the SPRY domain suggests that the positively selected residues and insertions/deletions fall exclusively in predicted coils (Fig. 2B), which could represent specific interaction surfaces.
Because there are NWM-specific deletions in TRIM5α, excluding NWMs allows the opportunity to analyze all residues in hominids and OWMs, a more focused look at just the last 23 million years of primate phylogeny. Remarkably, the analysis of hominids and OWMs identified only five residues in the entire protein as being under positive selection with high confidence (triangles on stems, Fig. 2A). All five residues fall within an 11- to 13-aa segment of the SPRY domain (Fig. 2B), which we will refer to as the SPRY patch. Such tight clusters of positive selection are predicted to be points of physical contact between two proteins locked in genetic conflict. Similar paml analyses have successfully highlighted the known binding surface between ZP3, an egg-receptor protein, and sperm (28). Additionally, the positive selection in the major histocompatibility complex (MHC) proteins is confined to small segments of the protein known to constitute the antigen-recognition site (29). In this case, we had no a priori knowledge of the TRIM5α interaction interface crucial for viral restriction, so we tested our computational prediction that the SPRY patch identifies such a domain. Because TRIM5α was originally identified because of the fact that it confers resistance to HIV-1 infection in rhesus, but not in human, cells (4) we used this species-specific example of TRIM5α restriction to investigate the functional importance of the patch.
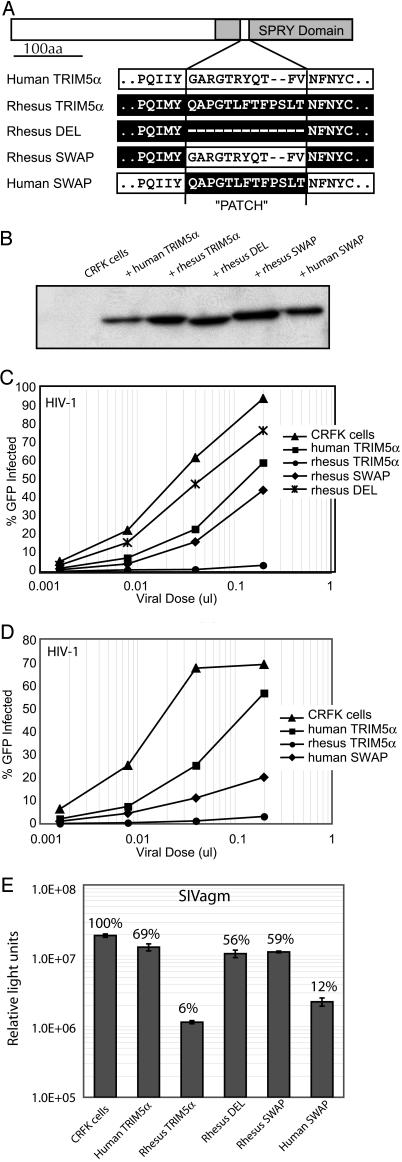
We constructed chimeric proteins between human and rhesus TRIM5α that either differed only in the 11- to 13-aa patch region or had the patch region deleted altogether (Fig. 3A). CRFK cells (feline cells that have no known retroviral restriction) were stably transduced to express these proteins at approximately similar levels (Fig. 3B) and were tested for restriction of increasing titer of a recombinant HIV-1 virus that expresses GFP (20). As has been previously reported (4), human TRIM5α showed a weak restriction of HIV-1, whereas rhesus TRIM5α almost completely blocked infection (Fig. 3C). When the patch is deleted from rhesus TRIM5α (rhesus DEL), it loses restriction not only against HIV-1 (Fig. 3C) but also against SIV from African green monkeys and N-tropic murine leukemia virus (Fig. 3E and data not shown). Thus, the patch we have identified based on positive selection may define a protein surface that is generally necessary for broad retroviral restriction.
Fig. 3.
The positively selected patch is critical for the species-specific retroviral restriction by TRIM5α. (A) Retroviral vectors (LPCX) containing hemagglutinin-tagged TRIM5α alleles from human and rhesus genomes were modified to delete the rhesus SPRY patch (Rhesus DEL), swap the human patch into rhesus TRIM5α (Rhesus SWAP), or swap the rhesus patch into the human TRIM5α (Human SWAP). The sequences in the region of the swaps are shown. (B) A Western blot (using a hemagglutinin antibody) shows stable CRFK lines expressing each construct at roughly equivalent levels. (C) Variants of the rhesus TRIM5α protein were assessed for their ability to restrict HIV-1. Single-cycle assays for HIV-1 infectivity were carried out in CRFK (feline renal fibroblasts) cells expressing exogenous TRIM5α genes. Experiments were performed by using 5-fold dilutions of an HIV-1 provirus that was deleted for env and where GFP was inserted into the nef region (20). HIV-positive cells were detected by flow cytometry for GFP expression. (D) A variant of the human TRIM5α (human SWAP) was tested in single-cycle assays for HIV-1 infectivity as in C. (E) CRFK cells that express the TRIM5α alleles were challenged with SIVagm virus, which expresses luciferase in place of the nef gene. Preliminary experiments were performed to establish a dose of virus within the linear range of infection (data not shown). All infections were performed in triplicate, and luciferase activity was measured 2 days after infection. The relative light units are shown on the y-axis, and the percent infection (compared with the CRFK control) is shown above the bar. SD of the data also are shown.
We next asked whether the patch is responsible for the species specificity of TRIM5α by substituting the human patch into rhesus TRIM5α and vice versa. Remarkably, we found that substitution of the human patch into the rhesus TRIM5α (rhesus SWAP) reduces its restrictive capability to a level that is close to that of human TRIM5α (Fig. 3C). Thus, the patch we have defined by using positive selection identifies a region necessary for species-specific restriction to HIV-1. We then tested the reciprocal chimera, human TRIM5α with the rhesus TRIM5α patch (human SWAP, Fig. 3D). The Human SWAP was significantly more restrictive against HIV-1 than human TRIM5α. Despite this gain-of-function effect, swapping the patch alone was not completely sufficient to recapitulate rhesus TRIM5α-like restriction, suggesting that other minor determinants of restriction also may exist.
We wanted to know whether the patterns seen with these chimeric proteins are specific to HIV-1 or are generally responsible for defining broad species-specific ability to restrict lentiviruses. To address this question, we challenged the TRIM5α-expressing CRFK cell lines with SIVagm and found that the reversal of species-specific restriction is the same as was observed for HIV-1 (Fig. 3E). Thus, human TRIM5α does not usually restrict SIVagm, but can acquire the ability to restrict SIVagm when the 13-aa patch from rhesus TRIM5α is added (human SWAP in Fig. 3E). Conversely, rhesus TRIM5α loses the ability to restrict SIVagm when its patch is replaced by the 11-aa patch from human TRIM5α (rhesus SWAP in Fig. 3E). Thus, the patch we have identified through positive selection determines the species-specificity of TRIM5α for restricting at least two highly divergent lentiviruses.
Discussion
Positive selection is a beacon for domains involved in genetic conflict. Based on analysis of positively selected codons, we were able to predict an important protein domain (potentially an interaction surface) that we then verified in vivo. In addition, we have illustrated strong positive selection of the TRIM5α gene throughout primate evolution. The ancient and intense selective pressures that have shaped the evolution of APOBEC3G (11) and TRIM5α indicate that the innate immune system is intricately evolved, just as the adaptive immune system is known to be (30). The tight clustering of positively selected residues in TRIM5α is in stark contrast to our earlier findings in APOBEC3G, where the codons that were identified as having been repeatedly subject to positive selection were scattered throughout the length of the gene, with no significant clustering (11). There are two possible reasons for this difference. First, the TRIM5α protein has a nonredundant domain structure, whereas APOBEC3G has resulted from two sequential duplications of a single cytidine deaminase domain to result in four potentially redundant structural copies of this domain (31, 32). Second, TRIM5α and APOBEC3G play different roles in their respective conflicts. Although it is in the host genome's best interest that TRIM5α improve binding to its target, it is in the host's interest that APOBEC3G avoid interactions with viral antagonists such as Vif in the case of HIV-1. Therefore, unlike TRIM5α, APOBEC3G is under pressure to mutate whichever domain is the current viral target.
What drives the evolution of TRIM5α? The antiquity of the positive selection rules out primate lentiviruses like HIV-1 as being the sole, or even the major, cause because they are believed to be <1 million years old (33). In addition, TRIM5α from human and OWM has been shown to be active against murine leukemia virus (6–9), a gammaretrovirus that is closely related to human endogenous retroviruses (34) that have episodically invaded primate genomes and continue to be active in the human genome (35–37). This finding suggests that TRIM5α evolution may have been strongly influenced by distinct episodes of endogenous retrovirus infection and subsequent retrotransposition events (36). HIV and other primate lentiviruses are likely to be newcomers to this conflict, with the OWM TRIM5α restriction against HIV-1 just an evolutionary coincidence. Nevertheless, as we have shown, the positive selection has had a profound impact on the species-specific restriction of both HIV-1 and SIVagm. Thus, the evolutionary histories of both TRIM5 and APOBEC3G indicate ancient adaptation to endogenous retrovirus-like elements (11, 12), yet both restriction systems were discovered because of their incidental activity against HIV. These findings indicate that the cellular arsenal honed against endogenous retroviruses is large and mostly undiscovered and may strongly impact lentiviral restriction.
Supplementary Material
Acknowledgments
We thank Monica Rodriguez and the Fred Hutchinson Cancer Research Center flow cytometry facility for technical assistance. We thank the National Institutes of Health AIDS Reagent Program for TRIM5α alleles contributed by Joseph Sodroski; Ned Landau (Salk Institute) for SIV vectors; Jeremy Luban and David Sayah for communicating results and sequences before publication; and Coriell, FrozenZoo, and Trent Colbert for primate genomic DNA. We thank our colleagues Sue Biggins, Julie Kerns, Danielle Vermaak, and Janet Young for comments on the manuscript, and Jorja Henikoff, Trygve Bakken, and an anonymous reviewer for valuable suggestions and assistance with the paml implementation. This work was supported by start-up funds from the Fred Hutchinson Cancer Research Center (to H.S.M.) and National Institutes of Health Grants R37 AI30927 (to M.E.) and T32 CA 09657 (to S.L.S.). H.S.M. is an Alfred P. Sloan Research Fellow in Computational and Evolutionary Molecular Biology.
Abbreviations: dS, synonymous changes per site; dN, replacement changes per site; HIV-1, HIV type 1; NWM, New World monkey; OWM, Old World monkey.
Data deposition: All sequences reported in this paper have been deposited in the GenBank database (accession nos. AY843504–AY843520).
References
- 1.Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. (2002) Nature 418, 646–650. [DOI] [PubMed] [Google Scholar]
- 2.Wiegand, H. L., Doehle, B. P., Bogerd, H. P. & Cullen, B. R. (2004) EMBO J. 23, 2451–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng, Y.-H., Irwin, D., Kurosu, T., Tokunaga, K., Sata, T. & Peterlin, B. M. (2004) J. Virol. 78, 6073–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stremlau, M., Owens, C. M., Perron, M. J., Kiessling, M., Autissier, P. & Sodroski, J. (2004) Nature 427, 848–853. [DOI] [PubMed] [Google Scholar]
- 5.Xu, L., Yang, L., Moitra, P. K., Hashimoto, K., Rallabhandi, P., Kaul, S., Meroni, G., Jensen, J. P., Weissman, A. M. & D'Arpa, P. (2003) Exp. Cell Res. 288, 84–93. [DOI] [PubMed] [Google Scholar]
- 6.Yap, M. W., Nisole, S., Lynch, C. & Stoye, J. P. (2004) Proc. Natl. Acad. Sci. USA 101, 10786–10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatziioannou, T., Perez-Caballero, D., Yang, A., Cowan, S. & Bieniasz, P. D. (2004) Proc. Natl. Acad. Sci. USA 101, 10774–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keckesova, Z., Ylinen, L. M. & Towers, G. J. (2004) Proc. Natl. Acad. Sci. USA 101, 10780–10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perron, M. J., Stremlau, M., Song, B., Ulm, W., Mulligan, R. C. & Sodroski, J. (2004) Proc. Natl. Acad. Sci. USA 101, 11827–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Valen, L. (1973) Evol. Theory 1, 1–30. [Google Scholar]
- 11.Sawyer, S. L., Emerman, M. & Malik, H. S. (2004) PLoS Biol. 2, 1278–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, J. & Webb, D. M. (2004) Hum. Mol. Genet. 13, 1785–1791. [DOI] [PubMed] [Google Scholar]
- 13.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang, Z. (1997) Comput. Appl. Biosci. 13, 555–556. [DOI] [PubMed] [Google Scholar]
- 15.Yang, Z. (1998) Mol. Biol. Evol. 15, 568–573. [DOI] [PubMed] [Google Scholar]
- 16.Comeron, J. M. (1999) Bioinformatics 15, 763–764. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen, R. & Yang, Z. (1998) Genetics 148, 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang, Z. & Bielawski, J. P. (2000) Trends Ecol. Evol. 15, 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGuffin, L. J., Bryson, K. & Jones, D. T. (2000) Bioinformatics 16, 404–405. [DOI] [PubMed] [Google Scholar]
- 20.Yamashita, M. & Emerman, M. (2004) J. Virol. 78, 5670–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mariani, R., Chen, D., Schrofelbauer, B., Navarro, F., Konig, R., Bollman, B., Munk, C., Nymark-McMahon, H. & Landau, N. R. (2003) Cell 114, 21–31. [DOI] [PubMed] [Google Scholar]
- 22.Hurst, L. D. (2002) Trends Genet. 18, 486–487. [DOI] [PubMed] [Google Scholar]
- 23.Li, W.-H. (1997) Molecular Evolution (Sinauer, Sunderland, MA).
- 24.Reymond, A., Meroni, G., Fantozzi, A., Merla, G., Cairo, S., Luzi, L., Riganelli, D., Zanaria, E., Messali, S., Cainarca, S., et al. (2001) EMBO J. 20, 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry, J., Mather, I. H., McDermott, M. F. & Pontarotti, P. (1998) Mol. Biol. Evol. 15, 1696–1705. [DOI] [PubMed] [Google Scholar]
- 26.Sayah, D. M., Sokolskaja, E., Berthoux, L. & Luban, J. (2004) Nature 430, 569–573. [DOI] [PubMed] [Google Scholar]
- 27.Luban, J., Bossolt, K. L., Franke, E. K., Kalpana, G. V. & Goff, S. P. (1993) Cell 73, 1067–1078. [DOI] [PubMed] [Google Scholar]
- 28.Swanson, W. J., Yang, Z., Wolfner, M. F. & Aquadro, C. F. (2001) Proc. Natl. Acad. Sci. USA 98, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes, A. L. & Nei, M. (1988) Nature 335, 167–170. [DOI] [PubMed] [Google Scholar]
- 30.Holmes, E. C. (2004) PLoS Biol. 2, 1267–1269. [Google Scholar]
- 31.Jarmuz, A., Chester, A., Bayliss, J., Gisbourne, J., Dunham, I., Scott, J. & Navaratnam, N. (2002) Genomics 79, 285–296. [DOI] [PubMed] [Google Scholar]
- 32.Wedekind, J. E., Dance, G. S. C., Sowden, M. P. & Smith, H. C. (2003) Trends Genet. 19, 207–216. [DOI] [PubMed] [Google Scholar]
- 33.Sharp, P. M., Bailes, E., Robertson, D. L., Gao, F. & Hahn, B. H. (1999) Biol. Bull. 196, 338–342. [DOI] [PubMed] [Google Scholar]
- 34.Andersson, M. L., Lindeskog, M., Medstrand, P., Westley, B., May, F. & Blomberg, J. (1999) J. Gen. Virol. 80, 255–260. [DOI] [PubMed] [Google Scholar]
- 35.Berkhout, B., Jebbink, M. & Zsiros, J. (1999) J. Virol. 73, 2365–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smit, A. F. (1999) Curr. Opin. Genet. Dev. 9, 657–663. [DOI] [PubMed] [Google Scholar]
- 37.Belshaw, R., Pereira, V., Katzourakis, A., Talbot, G., Paces, J., Burt, A. & Tristem, M. (2004) Proc. Natl. Acad. Sci. USA 101, 4894–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purvis, A. (1995) Philos. Trans. Biol. Sci. 348, 405–421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.