Abstract
Arabidopsis APETALA2 (AP2) encodes a member of the AP2/EREBP (ethylene responsive element binding protein) class of transcription factors and is involved in the specification of floral organ identity, establishment of floral meristem identity, suppression of floral meristem indeterminancy, and development of the ovule and seed coat. Here, we show that loss-of-function ap2 mutations cause an increase in seed mass relative to that of wild-type seeds. Analysis of an allelic series of ap2 mutations showed that increases in seed mass corresponded with the severity of defects in flower structure, indicating that AP2 activity directly influences seed mass. Experiments with male-sterile plants and deflowered wild-type plants showed that reduced fertility of ap2 mutant plants due to abnormal flower structure accounted for only part of the increase in seed mass caused by strong ap2 mutant alleles. Reciprocal cross experiments showed that AP2 acts maternally to control seed mass. The maternal effect of AP2 on seed mass involves the regulation of both embryo cell number and cell size. We show further that ap2 mutations cause changes in the ratio of hexose to sucrose during seed development, opening the possibility that AP2 may control seed mass through its effects on sugar metabolism. Together, these results identify a role for AP2 in controlling seed mass.
Keywords: ap2, Arabidopsis, seed size, sugar metabolism
Seeds of higher plants consist of three major components, each with a different genotype. The embryo that develops into the vegetative plant is diploid with a zygotic complement of genomes contributed by its parents. The endosperm, a tissue system that serves a nutritive role for the developing embryo and/or germinating seedling, is triploid with two and one genome equivalents, respectively, contributed by the maternal and paternal parent. By contrast, the seed coat that surrounds the embryo and endosperm is strictly of maternal origin. Growth and development of the embryo, endosperm, and seed coat must be coordinated to produce the mature seed. Although seeds have been studied extensively, many aspects of seed development are not well understood, including the mechanisms that underlie seed size or mass.
A critical factor in determining plant fitness is seed mass. Seed mass is negatively correlated with the number of seeds produced and positively correlated with seedling survival (1–5). Small-seeded plants are considered to be efficient colonizers because they produce large numbers of seeds, whereas seedlings of large-seeded plants are thought to more effectively withstand resource restrictions and abiotic stresses. Moreover, seed mass can vary intraspecifically in response to environmental cues, although little is known of the specific regulatory processes involved.
Several factors that influence seed mass have been identified. For example, quantitative trait loci (QTL) that influence seed mass have been mapped in a number of crop plants (6–12). An analysis of genetic factors affecting Arabidopsis seed mass used a segregating population from a cross between small-seeded and large-seeded ecotypes to show that both maternal and nonmaternal QTL affect seed mass, implicating both the maternal and zygotic genomes in processes that determine the sizes of seeds (13). Although the specific genes associated with these QTL have not been identified, other studies implicate enzymes involved in sugar metabolism as candidates for factors that influence seed mass (14). A series of studies with fava bean suggest that the relative accumulation of hexose and sucrose during seed development plays roles in determining seed mass through changes in seed cell number and cell size (14–16).
Seed mass is also influenced by parent-of-origin effects (17, 18). Interploidy crosses between diploid and polyploid plants produce offspring with a maternal or paternal genomic excess. Seeds from Arabidopsis plants with a paternal genomic excess are larger than diploid seeds resulting from self-fertilization, whereas smaller seeds are obtained with a maternal genomic excess. The parental conflict theory (19, 20), proposed to account for these parent-of-origin effects, posits that the maternal plant will attempt to allocate resources equally among its progeny, whereas the paternal plant will try to maximize channeling of maternal resources to its progeny. Imprinting of maternal and paternal alleles expressed in the endosperm has been implicated to function in the control of seed mass (reviewed in refs. 21–23).
In this study, we focus on the role of Arabidopsis APETALA2 (AP2) in controlling seed mass. AP2 encodes the founding member of a family of plant-specific transcription factors that contains an AP2/EREBP (ethylene responsive element binding protein) domain, a conserved region of ≈60 aa involved in DNA binding (24–26). This transcription factor is involved in the specification of flower organ identity, establishment of flower meristem identity, and suppression of flower meristem indeterminancy (25, 27–31). AP2 is also required for ovule and seed coat development (24, 32, 33). Although the most conspicuous function of AP2 is in flower development, its transcripts are not restricted to flowers but are detected in leaves, stems, and seedlings also (24, 34), opening the possibility of a more global function for AP2. Here, we identify a previously undescribed function for AP2 by showing that loss-of-function ap2 mutations cause increases in seed mass.
Materials and Methods
Plant Materials, Growth Conditions, and Flowering Time Analysis. ap2-5, ap2-6, ap2-7, ap2-11, and the male-sterile mutant, CS4002, are in the Columbia (Col) ecotype. All lines except ap2-11 were obtained from the Arabidopis Biological Resources Stock Center (Ohio State University, Columbus). ap2-11 was isolated from a population of plants mutagenized with T-DNA (35). Plants were grown at 22°C under continuous light. Flowering time was assayed by counting the number of rosette and cauline leaves when the primary inflorescence was 10 cm high.
Protein Analysis. Fifteen mature dried seeds from wild-type plants or ap2 mutants were homogenized with 30 μl of extraction buffer (36) by using a microglass pestle and mortar (Kontes, Vineland, NJ). After centrifugation, 15 μl of each extract was used for SDS/PAGE (37). Protein content in 5 μl of each extract was determined by using the Bio-Rad RC/DC protein assay kit with BSA as the standard.
Microscopy. Mature dried seeds were imbibed for 1 h and dissected under the microscope to isolate mature embryos. Embryos were incubated in a buffer (50 mM sodium phosphate, pH 7.0/10 mM EDTA/1% Triton X-100/1% DMSO) at 37°C for 12 h, fixed with FAA (10% formalin/5% acetic acid/45% ethanol/0.01% Triton X-100) for 45 min, and rehydrated through an ethanol series. Embryos were then treated for 1 h in Hoyer's solution [chloral hydrate/water/glycerol (3:0.8:0.4)]. Observations were made with an Olympus compound microscope using differential interference contrast (DIC) optics. Cleared embryos were photographed, and organ and cell sizes were measured by using nih image analysis software (http://rsb.info.nih.gov/nih-image/index.html).
Sugar Analysis. Soluble sugars were extracted with 80% ethanol (38) from developing seeds (≈90–120) from three wild-type or ap2 mutant siliques. Extracts were evaporated in vacuo and dissolved in water at 1 μl per seed. d-Glucose, d-fructose, and sucrose levels in the aqueous extracts were determined enzymatically by using a kit from Boehringer Mannheim/R-Biopharm per the manufacturer's specifications.
Results
ap2 Mutants Set Large Seed. As part of a screen to identify genes that prevent the delay in flowering time that occurs when plants are grown on high concentrations of sucrose (34), we identified a line with floral defects similar to those of an ap2 mutant (27, 28). Sepals were transformed into carpeloid organs, petals failed to develop in the second whorl, and stamen number was reduced. Analyses showed that the mutation resulted from an 11-bp deletion in the AP2 gene (bases + 724 to + 734 relative to the transcription start site, GenBank accession no. U12546; ref. 24) in the region encoding the first AP2 domain of the protein. Thus, this mutation, designated ap2-11, is likely to be a null allele of AP2 analogous to the ap2-10 allele that is disrupted in the second AP2 domain (24). We verified that ap2-11, and another mutation, ap2-7, induced early flowering in Arabidopsis. As shown in Table 1, ap2 mutants had fewer rosette and total leaves at the time of flowering than did wild type. This early flowering defect accounted for the ability of the mutation to counteract the effect of sugars on flowering time.
Table 1. ap2 mutants are early flowering.
| Leaf no. at time of flowering*
|
|||
|---|---|---|---|
| Genotype | n | Rosette leaves | Total leaves |
| Wild type | 31 | 14.4 ± 1.2 | 18.4 ± 1.7 |
| ap2-7 | 27 | 11.8 ± 1.0 | 14.2 ± 1.2 |
| ap2-11 | 28 | 12.8 ± 1.0 | 15.1 ± 1.3 |
Means ± SD are shown. Differences between values for wild type and each ap2 mutant are significant at the 0.05 level
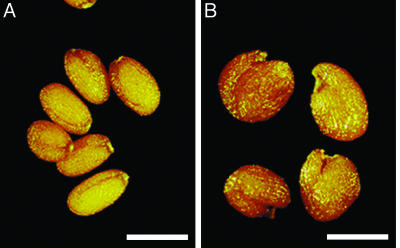
Because altered sugar metabolism has been shown to affect seed size (15), we examined the seeds of self-pollinated homozygous ap2-11 plants. We discovered that ap2-11 mutant seeds were larger than wild-type seeds, even after extensive backcrossing of the line with wild type. Fig. 1 shows that ap2-11 dried mature seeds were larger and more variable in shape than oblong-formed wild-type seeds.
Fig. 1.
ap2 mutants produce large seeds. Shown are mature dried seeds from wild type (A) and the ap2 mutant, ap2-11 (B). (Bars: 500 μm.)
To determine whether there was a direct link between AP2 activity and seed size, we analyzed quantitatively an allelic series of ap2 mutants in the Col ecotype that varied in their defects on flower morphology (27). Specifically, ap2-5, the weakest allele tested, had mutant flowers with six stamens, the same number present in wild-type flowers. ap2-6 and ap2-7 represent progressively stronger mutant alleles whose flowers contain three to four and one to three stamens, respectively. The morphology of ap2-11 flowers was most similar to that of ap2-7 and ap2-10.
We measured the weights of seeds from an allelic series of ap2 mutants and also determined the numbers of flowers per inflorescence, siliques elongated, and seeds per silique on the ap2 mutant plants. The latter set of parameters has been implicated to be associated with total seed yield (13). We limited our measurements to flowers and seeds produced on the primary inflorescence to control variability. Table 2 shows that, with the possible exception of the weak ap2-5 allele, ap2 mutants produced seeds that were heavier than those from wild-type plants. Specifically, ap2-6, ap2-7, and ap2-11 seeds were 1.8, 1.9, and 1.9 times heavier, respectively, than wild-type seeds. Similar results were obtained for seeds formed on secondary inflorescences (data not shown). Therefore, the effect of each mutant ap2 allele on seed weight correlated with defects induced on flower morphology. That is, mutants with the most defective flowers produced the largest seeds, suggesting that loss of AP2 activity is responsible for the observed effect on seed mass.
Table 2. ap2 mutants produce large seeds.
| Genotype | Seed weight* | Total seed weight, mg | Flower no. | Elongated silique no. | Seed weight per silique, mg | Seeds per silique |
|---|---|---|---|---|---|---|
| Wild type | 2.1 ± 0.1A | 62 ± 1.9 | 53 ± 2.3 | 53 ± 2.3 | 1.2 ± 0.03 | 56 ± 1.7 |
| ap2-5 | 2.3 ± 0.1B | 45 ± 2.7 | 44 ± 3.7 | 40 ± 2.7 | 1.1 ± 0.03 | 49 ± 1.3 |
| ap2-6 | 3.8 ± 0.2C | 17 ± 7.7 | 57 ± 5.0 | 21 ± 8.4 | 0.80 ± 0.29 | 23 ± 8.1 |
| ap2-7 | 4.0 ± 0.3C,D | 3.0 ± 2.3 | 53 ± 5.2 | 5.2 ± 2.8 | 0.60 ± 0.27 | 16 ± 7.4 |
| ap2-11 | 4.0 ± 0.1D | 4.5 ± 1.8 | 51 ± 1.0 | 7.3 ± 1.9 | 0.63 ± 0.28 | 16 ± 7.1 |
All values pertain to the primary inflorescence. Plants were grown concurrently under identical conditions. Similar results were obtained in an independent experiment that was performed in a different season of the year. Means ± SD are shown.
Weight of seeds produced on primary inflorescence is given in mg per 100 seeds. Seed weight values that differ at the 0.05 significance level are labeled with different letters. Seed weights for ap2-6 and ap2-7 and values for ap2-7 and ap2-11 are not significantly different
ap2 mutations also induced other defects in reproductive development. Although flower number on the primary inflorescences of mutant plants was similar to that of wild type, fertility was negatively affected by the ap2 mutations. For example, ap2 mutants produced fewer elongated siliques on the primary inflorescence compared with wild type. Pistils fail to elongate into siliques when ovules within the pistil have not been fertilized to a significant extent (39). Consistent with this result, average seed number per silique was lower in ap2 mutants as compared with that of wild type. The reduced fertility in ap2 mutants is most likely a consequence of defects in flower morphology. We conclude that reduction in AP2 activity causes plants to produce seeds whose mass is larger than that of wild type. However, the ap2 mutants produce fewer seeds than wild type because of defects in fertility caused by the mutation.
Effect of the ap2 Mutation on Seed Mass Is Not Solely Due to Its Effect on Fertility. Given the reduced fertility of plants with strong mutant alleles of AP2 (Table 2), we asked whether the large seed mass phenotype could result simply from allocation of extra resources to the few seeds produced. We hand pollinated three flowers at identical positions on secondary inflorescences of wild-type plants, two strong ap2 mutants (ap2-6 and ap2-7), and a male-sterile mutant (CS4002 in Col ecotype). For this set of experiments, flowers were pollinated with pollen of the same genotype, with the exception of male-sterile plants for which wild-type pollen was used. Manual pollination ensured that all siliques contained similar numbers of seeds.
As shown in Table 3, average seed weight from male-sterile maternal plants (2.8 mg per 100 seeds) was higher than that from wild-type maternal plants (2.3 mg), confirming that seed mass increased under conditions of reduced fertility. A similar result was obtained by removing all flowers on a wild-type plant with the exception of three flowers on each secondary inflorescence. The weight of seeds in the few siliques produced from these deflowered plants (2.7 ± 0.1 mg per 100 seeds) was similar to that obtained from pollinated male-sterile mutants. By contrast, seeds from ap2-6 and ap2-7 mutants were heavier (3.3 mg and 3.8 mg, respectively) than those from male-sterile mutants and deflowered wild-type plants, even though seed number per silique was comparable. Together, these results suggest that the effect of strong ap2 mutations on seed mass is not primarily due to its effect on fertility.
Table 3. AP2 acts maternally to control seed mass.
| Parent
|
||||
|---|---|---|---|---|
| Female | Male | Seed weight* | Seed no. per silique | Seed weight per silique, mg |
| Wild type | Wild type | 2.3 ± 0.1A | 58 ± 1.6 | 1.4 ± 0.02 |
| ap2-6 | 2.3 ± 0.1A | 58 ± 2.7 | 1.4 ± 0.1 | |
| ap2-7 | 2.2 ± 0.1A | 58 ± 4.6 | 1.3 ± 0.1 | |
| Male sterile CS4002 | Wild type | 2.8 ± 0.1B | 45 ± 6.9 | 1.3 ± 0.2 |
| ap2-7 | 2.8 ± 0.1B | 46 ± 6.3 | 1.3 ± 0.2 | |
| ap2-6 | Wild type | 3.3 ± 0.1C | 51 ± 7.0 | 1.7 ± 0.2 |
| ap2-6 | 3.3 ± 0.1C | 60 ± 2.7 | 2.0 ± 0.1 | |
|
ap2-7
|
Wild type | 3.8 ± 0.2D | 53 ± 5.4 | 2.0 ± 0.2 |
| ap2-7 | 3.8 ± 0.1D | 51 ± 4.4 | 1.9 ± 0.2 | |
Reciprocal crosses between wild-type plants and homozygous ap2 mutants were performed on secondary inflorescences. Plants were grown together in the same conditions. Three flowers at identical positions (11th to 13th flowers for wild type or male-sterile mutant, 10th to 15th flowers in ap2 mutants) were manually pollinated. Two to four inflorescences per plant and four to five plants were used for calculations. Similar results were obtained in an independent experiment that was performed in a different season of the year. Means ± SD are shown.
Weight is given as mg per 100 seeds. Seed weight values that differ at the 0.01 significance level are labeled with different letters
AP2 Acts Maternally to Influence Seed Mass. To obtain clues about the genetic control of seed mass, we asked whether AP2 acts maternally or zygotically. Table 3 shows the results of reciprocalcross experiments between two ap2 mutants, ap2-6 and ap2-7, and wild-type plants. The effect of the ap2 mutation on seed mass was observed only when maternal plants were homozygous for the mutation. Maternal ap2 mutants produced seeds of comparable weight regardless of the genotype used as the pollen donor, and these seeds were consistently heavier than those from maternal wild-type plants. ap2 mutant and wild-type pollen produced seeds whose weight was comparable to that of wildtype maternal plants. This result suggests that ap2 is a maternal-effect mutation that affects seed mass.
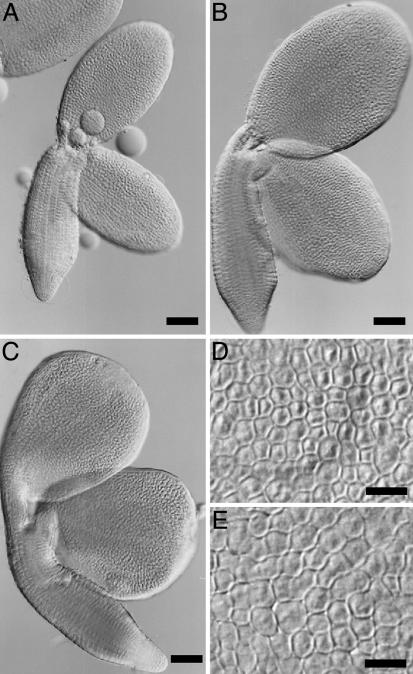
AP2 Controls Embryo Cell Number and Size. To determine whether the ap2-induced effects on seed mass reflected an increase in embryo size, we isolated and visualized mature embryos from wild-type and ap2-7 mutant seeds. ap2 mature embryos (Fig. 2 B and C) were significantly larger than those of wild type (Fig. 2 A). In addition, many mutant embryos had irregularly shaped cotyledons. Imaging experiments showed that the average area of an ap2 mutant cotyledon was 1.8 times larger than that of wild type (Table 4). The embryonic axis of mature ap2 mutant embryos was also bigger than that of wild type.
Fig. 2.
ap2 mutation causes an increase in both embryo cell size and number. (A–C) Mature embryos from wild type (A) and ap2-7 mutants (B and C) were dissected from mature dried seeds, cleared, and photographed. The embryo in C has an irregular shape. (D and E) Epidermal cell layer from the central region of cotyledons from wild-type (D) and ap2-7 mutant (E) embryos. [Bars: 100 μm (A–C) and 20 μm (D and E).]
Table 4. AP2 affects both cotyledon cell size and cell number.
| Genotype | Area per cotyledon, mm2 | Area per cell, μm2 |
|---|---|---|
| Wild type | 0.10 ± 0.01 (90) | 86 ± 4.2 (20) |
| ap2-7 | 0.18 ± 0.03 (106) | 112 ± 12 (21) |
| Ratio | 1.8 | 1.3 |
Entire cotyledons from mature dried embryos or their central regions were photographed. Imaging analysis software was used to calculate areas. Means ± SD are shown. The number of cotyledons analyzed is given in parentheses. Area differences between wild type and ap2-7 mutants are significant at the 0.001 level.
Organ size is determined by both cell number and size. We analyzed epidermal cells in the central regions of wild-type and ap2 mutant cotyledons to learn which parameter is affected by AP2 to control seed mass. Fig. 2 D and E shows that cells from ap2 mutant embryos were larger than those of wild type. The average area of an ap2 epidermal cell was 1.3 times larger than that of a corresponding wild-type cell (Table 4). Given differences in the areas of ap2 mutant and wild-type cotyledons (1.8 times) and cells (1.3 times), we conclude that the ap2 mutation must affect cotyledon cell number, with mutants having ≈1.4 times more cells than wild type (1.8/1.3 = 1.4). Thus, AP2 affects both embryo cell size and number.
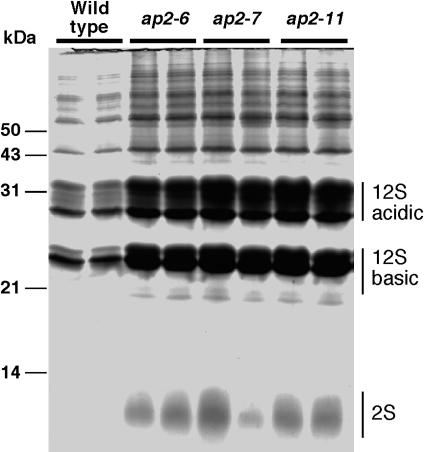
Because the increase in embryo cell size during the maturation phase results in large part from the accumulation of storage reserves, we analyzed protein levels in wild-type and ap2 mutant seeds, including the 12S and 2S storage proteins. As shown in Fig. 3, fractionation of protein extracts from identical numbers of wild-type and ap2 mutant seeds by SDS/PAGE revealed differences in protein levels. Moreover, the levels of all proteins, including the two major storage proteins in Arabidopsis, 12S and 2S, were uniformly higher in ap2 mutant seeds, suggesting that the overall proportion of individual proteins was not affected by the mutation. Measurement of protein levels in mature seeds, shown in Table 5, confirmed that ap2 mutant seeds accumulated more protein than did wild-type seeds.
Fig. 3.
ap2 mutant seeds contain more protein than do wild-type seeds. Protein extracts from an equal number of wild-type, ap2-6, ap2-7, and ap2-11 seeds were fractionated on a 12% SDS/polyacrylamide gel and stained. Although ap2 mutant seeds possess more proteins, the relative proportion of 12S and 2S storage proteins and other proteins are not affected by the mutations. Molecular mass markers are shown to the left of the gel.
Table 5. ap2 mutant seeds have higher protein levels than do wild-type seeds.
| Genotype | μg of protein per seed* | Ratio† |
|---|---|---|
| Wild type | 3.1 ± 0.1A | — |
| ap2-5 | 4.0 ± 0.4B | 1.3 |
| ap2-6 | 7.8 ± 0.2C | 2.5 |
| ap2-7 | 8.1 ± 0.2C | 2.6 |
| ap2-11 | 8.4 ± 0.7C | 2.7 |
Means ± SD are shown. Values that differ at the 0.05 significance level are labeled with different letters
Ratio of values from ap2 mutants to wild type
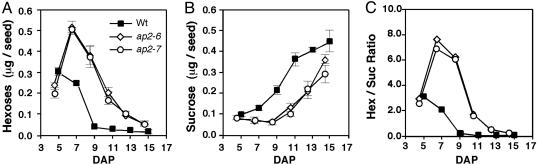
AP2 Affects Sugar Metabolism. ap2-11 was originally identified as a mutation that overcomes the negative effects of sugars on flowering time, opening the possibility that AP2 may function in the control of sugar metabolism and/or signaling. We examined this possibility by measuring glucose and fructose (hexose) and sucrose levels in wild-type and ap2-6 and ap2-7 seeds during development. As shown in Fig. 4 A and B, hexose was at its highest level early in wild-type seed development and declined late in seed development. Sucrose levels display the opposite accumulation pattern, becoming the predominant soluble sugar late in seed development. This pattern of soluble sugar accumulation mirrors results obtained by others (38). By contrast, accumulation of soluble sugars, particularly hexose, was altered significantly in ap2 mutants. Relative to wild type, hexose accumulated to a higher level in ap2 mutants and declined at a slower rate (Fig. 4A), whereas sucrose accumulation in ap2 mutants proceeded at a slower rate (Fig. 4B). As a result, the ratio of hexose to sucrose levels (Fig. 4C) reached a higher value and decreased at a slower rate in ap2 mutants relative to wild type. Together, these results suggest a role for AP2 in controlling soluble sugar accumulation during seed development.
Fig. 4.
Hexose and sucrose levels are altered during seed development in ap2 mutants. Time courses of hexose (glucose and fructose; A) and sucrose (B) accumulation and hexose/sucrose levels (C) in developing seeds from wild type and ap2-6 and ap2-7 mutants are shown. Results represent the average of three to four determinations with standard errors. Wild-type and ap2 mutant flowers were hand pollinated, and developing seeds were harvested at the indicated days after pollination (DAP).
Discussion
AP2 Activity Controls Seed Mass. AP2 was identified originally as a homeotic gene required for the specification of floral organ identity and was shown to encode an AP2/EREBP transcription factor (24, 25). Here, we show that loss-of-function ap2 mutants produce large seeds. Our studies using an allelic series of mutants provide strong support that AP2 functions directly in controlling seed mass (Table 2). A recent study provided additional support for this conclusion by showing that transgene suppression of AP2 activity in Arabidopsis produced defective flowers and large seeds (40). Thus, the extent of AP2 gene activity, based on its well defined role in flower development, plays a role in determining seed mass.
ap2 mutations negatively affect plant fertility, primarily through disruption of flower structure that limits the efficiency of self-pollination in mutant plants. Given that limited availability of resources in the maternal plant appears to account for the negative correlation between seed number and seed mass (4), ap2 mutants are expected to produce seeds larger than wild-type plants with normal fertility. We confirmed that reduced fertility did affect seed mass and could account for ≈33% of the increase in average seed weight observed in a strong ap2 mutant (Table 3). However, the remaining, much larger effect of AP2 on seed mass must be attributed to factors unrelated to fertility. The early flowering of plants with strong ap2 mutant alleles (Table 1) provides additional support for the conclusion that the effects of the ap2 mutation on seed mass is not solely due to its effects on fertility. Leaf number is closely related to the availability of reproductive resources in the maternal plant (13). ap2 mutants produced fewer leaves than wild type and, presumably, have fewer resources available. Yet, ap2 mutants gave rise to significantly larger seeds.
Maternal Effect of AP2 on Seed Mass. Experiments in which wildtype and ap2 mutant genotypes were crossed reciprocally (Table 3) provided an important clue about the mechanisms controlling seed mass by showing that AP2 acts through the maternal genome. Consistent with this finding are analyses of reciprocal crosses in several crop species showing that seed mass is often influenced by the genotype of the maternal plant (41). One interpretation of our result is that AP2 activity in maternal tissues, potentially including the seed coat, is more important than its activity in the embryo and endosperm in controlling seed mass. Alternatively, because the maternal plant contributes two genome equivalents to the triploid endosperm, seed mass may be affected by AP2 gene dosage in the endosperm. A QTL analysis of small- and large-seeded Arabidopsis ecotypes suggests that maternal tissues are important in determining seed mass (13). A number of QTL affecting seed mass colocalized with QTL for maternal traits such as seed number, fruit size, and plant resources, suggesting that they operate through maternal tissues. The gene identities of these QTL have not been determined, although AP2 does appear to colocalize to a QTL affecting seed mass at the bottom of chromosome IV (13). Conversely, a recent study used genetic experiments to conclude that AP2 acts through both the endosperm and maternal tissues to affect seed size (40).
The endosperm has been implicated to serve as the site of parent-of-origin effects on seed mass through the imprinting of genes thought to be involved in enhancing or suppressing endosperm size and, therefore, seed and embryo mass (21–23). Although little is known of the mechanisms mediating parent-of-origin effects on seed mass, no current evidence points to an involvement of AP2. For example, mutations in three genes, MEA, FIE, and FIS2, each induce endosperm phenotypes that mimic paternal genomic excess (39, 42–44). However, these mutations are inherited differently than AP2 is in that ap2 is a maternal sporophytic mutation, whereas mea, fie, and fis2, are female gametophytic mutations. Thus, AP2 does not act equivalently with these other genes, although they could participate in a common pathway. Two other genes, IKU1 and IKU2, have been proposed to mediate the effects of maternal and paternal dosage on seed size, because mutations in either gene affect many aspects of endosperm development (45). However, unlike AP2, both iku mutations are sporophytic recessive and not maternal-effect mutations. Thus, the IKU genes are also likely to operate differently than does AP2.
How does AP2 act maternally to control seed mass? Our experiments suggest that AP2 acts, in part, through its effects on both embryo cell number and size (Fig. 2 and Table 4). Others have shown that changes in cell number and size underlie ecotype variations in Arabidopsis seed size (13). Similar to our findings, this study of natural variation concluded that cell number differences are controlled by maternal factors. However, by contrast to our results with AP2, ecotype differences in cell size were attributed to nonmaternal allelic variation. Our finding that AP2 acts maternally to affect embryo cell size may indicate that it operates through a mechanism that was not uncovered by studies of natural ecotype variations.
The seed coat has been implicated to influence seed mass. For example, sporophytic mutations in cereals that affect development of a specialized seed coat tissue responsible for nutrient transfer to the endosperm cause defects in seed development and size (46–48). ap2 mutants have defective seed coats in that they lack epidermal plateaus and mucilage, their outer integument cells are larger than those of wild type and are irregular in shape, and mutant seeds are hypersensitive to bleach (24, 49). Although it is not clear whether these defects are directly associated with AP2 effects on seed mass, they emphasize that the maternal component of the seed is defective in ap2 mutants.
AP2, Seed Mass, and Soluble Sugar Metabolism. Our findings that the ap2 mutation acts maternally, potentially through the seed coat, to control seed mass (Table 3) and affects soluble sugar metabolism during seed development (Fig. 4) provide a potential explanation for the increase in embryo cell number in ap2 mutant seeds (Table 4). During seed development, modulation of hexose and sucrose levels has been implicated to control cellular activities (reviewed in ref. 15). Specifically, a high ratio of hexose to sucrose is correlated strongly with mitotic activity during the early morphogenesis phase of seed development, whereas a higher proportion of sucrose to hexose is associated with cell expansion and seed filling during the late maturation phase (14–16). For example, immature fava bean embryos cultured with high concentrations of hexose continue to undergo cell divisions whereas those cultured with high sucrose concentration enter the maturation phase (15). Thus, cell division during the morphogenesis phase and seed filling during the maturation phase appear to correlate with changes in hexose and sucrose levels.
ap2 mutations induced changes in the levels of hexose and sucrose in developing seeds. These changes resulted in hexose/sucrose ratios that reached a higher maximal level in ap2 mutant seeds compared with wild-type seeds and that remained high for a longer period of seed development in ap2 mutants than they did in wild type (Fig. 4). For example, hexose/sucrose ratios remained high at 9 and 11 DAP in ap2 mutants but was very low during the same period of wild-type seed development. We speculate that the altered hexose/sucrose ratios during ap2 mutant seed development may promote an extended period of cell division that could account for the increase in embryo cell number observed in ap2 mutants.
AP2 is a transcription factor. Thus, it is likely to affect seed mass by regulating the expression of other genes. Given the dramatic changes in hexose levels in ap2 mutants, potential targets of AP2 activity are enzymes involved in sugar metabolism, such as cell-wall-bound invertases. Sucrose is transported from photosynthetic organs to the seed coat, a maternally derived structure, where it is metabolized differently early and late in embryogenesis (14, 50–52). During the early phase of favabean seed development, cell-wall-bound invertases localized in thin-walled parenchyma, the innermost seed coat tissue, hydrolyze sucrose. Hexoses formed by the reaction move apoplastically and are transported into embryo cells. Cell-wall-bound invertase activity decreases during the late phase of seed development, and sucrose is transported directly into embryo cells where it is hydrolyzed by sucrose synthase. Cell-wall-bound invertase activity has been correlated with seed cell number both in fava bean cultivars that exhibit variations in seed size and in the maize miniature1 mutant that is defective in the enzyme (15, 53). Thus, AP2 may affect seed mass by controlling cell-wall-bound invertase activity.
We note that two other members of the AP2/EREBP transcription factor family have roles that appear to intersect with those of AP2. Mutations in the Arabidopsis WRINKLED1 gene lead to defects in carbohydrate metabolism and reductions in seed mass and oil content (38, 54). Arabidopsis AINTEGUMENTA gene is thought to play a role in controlling cell proliferation (55, 56). Ectopic expression of AINTEGUMENTA caused plant organs, including seeds, to increase in size in both transgenic Arabidopsis and tobacco (57). The increase in seed size in transgenic plants appears to result from an increase in cell number during seed development. Thus, other AP2/EREBP proteins may function in a pathway similar to that of AP2.
Acknowledgments
We thank Diane Jofuku and Jack Okamuro at Ceres Inc. for sharing results before publication and for their useful and supportive comments, and we thank Hirokazu Tsukaya at National Institute for Basic Biology for his helpful comments. The receipt of valuable plant materials from the Arabidopsis Biological Resource Center at Ohio State University is acknowledged. This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan to M.O. and from the Japan Society for the Promotion of Science to K.N., and by a fellowship from the Yamada Science Foundation to M.O.
Abbreviations: AP2, APETALA2; Col, Columbia; DAP, days after pollination; EREBP, ethylene responsive element binding protein; QTL, quantitative trait loci.
References
- 1.Coomes, D. A. & Grubb, P. J. (2003) Trends Ecol. Evol. 18, 283-291. [Google Scholar]
- 2.Silvertown, J. (1989) Trends Ecol. Evol. 4, 24-26. [DOI] [PubMed] [Google Scholar]
- 3.Stanton, M. L. (1984) Ecology 65, 1105-1112. [Google Scholar]
- 4.Venable, D. L. (1992) Am. Nat. 140, 287-304. [Google Scholar]
- 5.Westoby, M., Jurado, E. & Leishman, M. (1992) Trends Ecol. Evol. 7, 368-372. [DOI] [PubMed] [Google Scholar]
- 6.Grandillo, S. & Tanksley, S. D. (1996) Theor. Appl. Genet. 92, 935-951. [DOI] [PubMed] [Google Scholar]
- 7.Lu, C., Shen, L., Tan, Z., Xu, Y., He, P., Chen, Y. & Zhu, L. (1997) Theor. Appl. Genet. 94, 145-150. [DOI] [PubMed] [Google Scholar]
- 8.Maughan, P. J., Saghai Maroof, M. A. & Buss, G. R. (1996) Theor. Appl. Genet. 93, 574-579. [DOI] [PubMed] [Google Scholar]
- 9.Mian, M. A. R., Bailey, M. A., Tamulonis, J. P., Shipe, E. R., Carter, T. E., Parrott, W. A., Ashley, D. A., Hussey, R. S. & Boerma, H. R. (1996) Theor. Appl. Genet. 93, 1011-1016. [DOI] [PubMed] [Google Scholar]
- 10.Paterson, A. H., Lin, Y.-R., Li, Z., Schertz, K. F., Doebley, J. F., Pinson, S. R. M., Liu, S.-C., Stansel, J. W. & Irvine, J. E. (1995) Science 269, 1714-1718. [DOI] [PubMed] [Google Scholar]
- 11.Rami, J. F., Dufour, P., Trouche, G., Fliedel, G., Mestres, C., Davrieux, F., Blanchard, P. & Hamon, P. (1998) Theor. Appl. Genet. 97, 605-616. [Google Scholar]
- 12.Timmerman-Vaughan, G. M., McCallum, J. A., Frew, T. J., Weeden, N. F. & Russel, A. C. (1996) Theor. Appl. Genet. 93, 431-439. [DOI] [PubMed] [Google Scholar]
- 13.Alonso-Blanco, C., Blankestijn-De Vries, H., Hanhart, C. J. & Koornneef, M. (1999) Proc. Natl. Acad. Sci. USA 96, 4710-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber, H., Borisjuk, L. & Wobus, U. (1997) Trends Plant Sci. 2, 169-174. [Google Scholar]
- 15.Weber, H., Borisjuk, L. & Wobus, U. (1996) Plant J. 10, 823-834. [Google Scholar]
- 16.Weber, H., Heim, U., Golombek, S., Borisjuk, L., Manteuffel, R. & Wobus, U. (1998) Plant J. 16, 163-172. [DOI] [PubMed] [Google Scholar]
- 17.Scott, R. J., Spielman, M., Bailey, J. & Dickinson, H. G. (1998) Development (Cambridge, U.K.) 125, 3329-3341. [DOI] [PubMed] [Google Scholar]
- 18.Lin, B. Y. (1984) Genetics 107, 103-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haig, D. & Westoby, M. (1991) Philos. Trans. R. Soc. London B 333, 1-14. [Google Scholar]
- 20.Haig, D. & Westoby, M. (1989) Am. Nat. 134, 147-155. [Google Scholar]
- 21.Gehring, M., Choi, Y. & Fischer, R. L. (2004) Plant Cell 16, S203-S213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger, F. (2003) Curr. Opin. Plant Biol. 6, 42-50. [DOI] [PubMed] [Google Scholar]
- 23.Lohe, A. R. & Chaudhury, A. (2002) Curr. Opin. Plant Biol. 5, 19-25. [DOI] [PubMed] [Google Scholar]
- 24.Jofuku, K. D., Denboer, B. G. W., Vanmontagu, M. & Okamuro, J. K. (1994) Plant Cell 6, 1211-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamuro, J. K., den Boer, B. & Jofuku, K. D. (1993) Plant Cell 5, 1183-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riechmann, J. L. & Meyerowitz, E. M. (1998) Biol. Chem. 379, 633-646. [DOI] [PubMed] [Google Scholar]
- 27.Kunst, L., Klenz, J. E., Martinez-Zapater, J. & Haughn, G. W. (1989) Plant Cell 1, 1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowman, J. L., Smyth, D. R. & Meyerowitz, E. M. (1991) Development (Cambridge, U.K.) 112, 1-20. [DOI] [PubMed] [Google Scholar]
- 29.Irish, V. F. & Sussex, I. M. (1990) Plant Cell 2, 741-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowman, J. L., Alvarez, J., Weigel, D., Meyerowitz, E. M. & Smyth, D. R. (1993) Development (Cambridge, U.K.) 119, 721-743. [Google Scholar]
- 31.Schultz, E. A. & Haughn, G. W. (1993) Development (Cambridge, U.K.) 119, 745-765. [Google Scholar]
- 32.Modrusan, Z., Reiser, L., Feldmann, K. A., Fischer, R. L. & Haughn, G. W. (1994) Plant Cell 6, 333-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leon-Kloosterziel, K. M., Keijzer, C. J. & Koornneef, M. (1994) Plant Cell 6, 385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohto, M., Onai, K., Furukawa, Y., Aoki, E., Araki, T. & Nakamura, K. (2001) Plant Physiol. 127, 252-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki, T., Inagaki, S., Nakajima, S., Akashi, T., Ohto, M., Kobayashi, M., Seki, M., Shinozaki, K., Kato, T., Tabata, S., et al. (2004) Plant J. 38, 673-684. [DOI] [PubMed] [Google Scholar]
- 36.Naito, S., Dube, P. H. & Beachy, R. N. (1988) Plant. Mol. Biol. 11, 109-123. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli, U. K. (1970) Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 38.Focks, N. & Benning, C. (1998) Plant Physiol. 118, 91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohad, N., Margossian, L., Hsu, Y.-C., Williams, C., Repetti, P. & Fischer, R. L. (1996) Proc. Natl. Acad. Sci. USA 93, 5319-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jofuku, K. D., Omidyar, P. K., Gee, Z. & Okamuro, J. K. (2005) Proc. Natl. Acad. Sci. USA 102, 3117-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roach, D. A. & Wulff, R. D. (1987) Annu. Rev. Ecol. Syst. 18, 209-235. [Google Scholar]
- 42.Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M. A. & Gagliano, W. B. (1998) Science 280, 446-450. [DOI] [PubMed] [Google Scholar]
- 43.Chaudhury, A. M., Ming, L., Miller, C., Craig, S., Dennis, E. S. & Peacock, W. J. (1997) Proc. Natl. Acad. Sci. USA 94, 4223-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiyosue, T., Ohad, N., Yadegari, R., Hannon, M., Dinneny, J., Wells, D., Katz, A., Margossian, L., Harada, J. J., Goldberg, R. B. & Fischer, R. L. (1999) Proc. Natl. Acad. Sci. USA 96, 4186-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia, D., Saingery, V., Chambrier, P., Mayer, U., Jurgens, G. & Berger, F. (2003) Plant Physiol. 131, 1661-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maitz, M., Santandrea, G., Zhang, Z., Lal, S., Hannah, L. C., Salamini, F. & Thompson, R. D. (2000) Plant J. 23, 29-42. [DOI] [PubMed] [Google Scholar]
- 47.Cheng, W.-H., Taliercio, E. W. & Chourey, P. S. (1996) Plant Cell 8, 971-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Felker, F. C., Peterson, D. M. & Nelson, O. E. (1985) Am. J. Bot. 72, 248-256. [Google Scholar]
- 49.Windsor, J. B., Symonds, V. V., Mendenhall, J. & Lloyd, A. M. (2000) Plant J. 22, 483-493. [DOI] [PubMed] [Google Scholar]
- 50.Weber, H., Buchner, P., Borisjuk, L. & Wobus, U. (1996) Plant J. 9, 841-850. [DOI] [PubMed] [Google Scholar]
- 51.Weber, H., Borisjuk, L., Heim, U., Sauer, N. & Wobus, U. (1997) Plant Cell 9, 895-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber, H., Borisjuk, L., Heim, U., Buchner, P. & Wobus, U. (1995) Plant Cell 7, 1835-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vilhar, B., Kladnik, A., Blejec, A., Chourey, P. S. & Dermastia, M. (2002) Plant Physiol. 129, 23-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cernac, A. & Benning, C. (2004) Plant J. 40, 575-585. [DOI] [PubMed] [Google Scholar]
- 55.Elliott, R. C., Betzner, A. S., Huttner, E., Oakes, M. P., Tucker, W. Q. J., Gerentes, D., Perez, P. & Smyth, D. R. (1996) Plant Cell 8, 155-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klucher, K. M., Chow, H., Reiser, L. & Fischer, R. L. (1996) Plant Cell 8, 137-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizukami, Y. & Fischer, R. L. (2000) Proc. Natl. Acad. Sci. USA 97, 942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]