Abstract
Guanine nucleotide-exchange proteins activate ADP-ribosylation factors by accelerating the replacement of bound GDP with GTP. Mammalian brefeldin A-inhibited guanine nucleotide-exchange proteins, BIG1 and BIG2, are important activators of ADP-ribosylation factors for vesicular trafficking. To identify proteins that interact with BIG2, we used cDNA constructs encoding BIG2 sequences in a yeast two-hybrid screen of a human heart library. Clone p2-5-3, encoding a form of human exocyst protein Exo70, interacted with BIG2 amino acids 1–643 and 1–832, but not 644–832, which was confirmed by coimmunoprecipitation of in vitro-translated BIG2 N-terminal segments and 2-5-3. By immunofluorescence microscopy, endogenous BIG2 and Exo70 in HepG2 cells were visualized at Golgi membranes and apparently at the microtubule-organizing center (MTOC). Both were identified in purified centrosomes. Immunoreactive Exo70 and BIG2 partially or completely overlapped with γ-tubulin at the MTOC in cells inspected by confocal microscopy. In cells incubated with brefeldin A, most of the BIG2, Exo70, and trans-Golgi protein p230 were widely dispersed from their perinuclear concentrations, but small amounts always remained, apparently at the MTOC. After disruption of microtubules with nocodazole, BIG2 and Exo70 were widely distributed in cells and remained only partially colocalized with p230, BIG2 more so than Exo70. We conclude that in HepG2 cells BIG2 and Exo70 interact in trans-Golgi network and centrosomes, as well as in exocyst structures or complexes that move along microtubules to the plasma membrane, consistent with a functional association in both early and late stages of vesicular trafficking.
Keywords: microtubule-organizing center, vesicular trafficking, centrosome, Golgi, ADP-ribosylation factor
Guanine nucleotide-exchange proteins (GEPs) for ADP-ribosylation factors (ARFs) are ubiquitously expressed proteins comprising multiple domains including the Sec7 domain that is responsible for ARF activation and its inhibition by brefeldin A (BFA) (1). These proteins accelerate the replacement of bound GDP on inactive ARF with GTP to generate active ARF. GEPs are grouped according to size, structure, and biochemical properties. Two mammalian BFA-inhibited GEPs, BIG1 and BIG2, were purified together in a macromolecular complex (2, 3). They are 74% identical in amino acid sequence, with 90% identity in their Sec7 domains (2). BIG1 and BIG2 are found in multiple intracellular compartments. They appear to colocalize at the trans-Golgi network (4, 5), but move independently among intracellular compartments, e.g., after serum starvation of HepG2 cells, BIG1, but not BIG2, is present in nuclear structures (6). BIG2 mutations were related to an autosomal recessive periventricular heterotopia with microcephaly, a congenital human developmental disorder (7). No disease involving BIG1 has been reported.
Most early studies of mammalian ARFs were focused on ARF1 (class I), which is critical in initiating the formation of COP1- and clathrin-coated vesicles (8), although understanding of class III ARF6, which is of major importance in the regulation of plasma membrane and endosomal membrane events, as well as actin cytoskeleton dynamics, is growing rapidly (9). Protein trafficking intermediates in vesicular or tubular/saccular forms move along microtubule or actin tracks with the aid of motor proteins. The minus ends of microtubules are anchored in the microtubule-organizing center (MTOC) with plus ends extended to the cell surface. In polarized cells, plus end-directed motors, kinesins, deliver cargo to the apical or basolateral cell membranes and minus end-directed motors, cytoplasmic dyneins, move vesicles toward centriolar regions (10). The MTOC, or centrosome, is a unique structure with a centriole that is surrounded by pericentriolar molecules. Golgi structures are formed around the centrosome, and their integrity depends on microtubules (10). The structural organization of Golgi, MTOC, and microtubules provides efficiency in sorting, recycling, and translocation of proteins. In exocytic pathways, the exocyst complex, which comprises eight proteins (Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84) important in vesicle transport and tethering, is crucial for cell growth, polarity, and communication (11, 12). However, relationships among cytoskeletal structures and the formation, translocation, and fusion of vesicles remain ill-defined.
We had earlier identified proteins that interact with BIG1 or BIG2 by using yeast two-hybrid screens. BIG2 was described as an A kinase-anchoring protein based on its interaction with regulatory subunits of protein kinase A (13), and BIG1 interacted with FK506-binding protein 13 (14). Charych et al. (15) reported interaction of BIG2 with β-subunits of the GABAA receptor. Here, we describe an interaction of the N-terminal region of the BIG2 molecule with Exo70, a protein of the exocyst complex required for exocytosis. BIG2 and Exo70, which copurified with polymerized tubulin, were partially colocalized in Golgi membranes and the MTOC. These studies are consistent with a functional association of key proteins involved in early and late events in vesicular trafficking.
Materials and Methods
Antibodies. Polyclonal antibodies against Exo70 (antibody 2536) and BIG2 (antibody 779) were raised in rabbits and affinity-purified with the immunizing antigen by BioSource International (Camarillo, CA) or our laboratory. Peptides used as immunogens were acetyl-CTKNPEKYIKYGVEQ-amide and acetyl-CGEHARSDSGKVSTENGD-amide for human Exo70 and BIG2, respectively, with the N-terminal cysteine added to facilitate coupling to keyhole limpet hemocyanin. The antibody against Exo70 is directed against sequence near the C terminus, which is encoded in p2-5-3, the Exo70 cDNA used to prepare our GFP-Exo70 construct, and full-length human Exo70 (see Fig. 8, which is published as supporting information on the PNAS web site). The BIG1 antibody has been described (4). Polyclonal antibodies against c-myc and hemagglutinin (HA) were purchased from Santa Cruz Biotechnology. Mouse mAbs against GM130 and p230 were purchased from BD Biosciences, mouse mAbs against α-tubulin and γ-tubulin were purchased from Sigma, and mouse mAbs against Sec6 were purchased from Calbiochem.
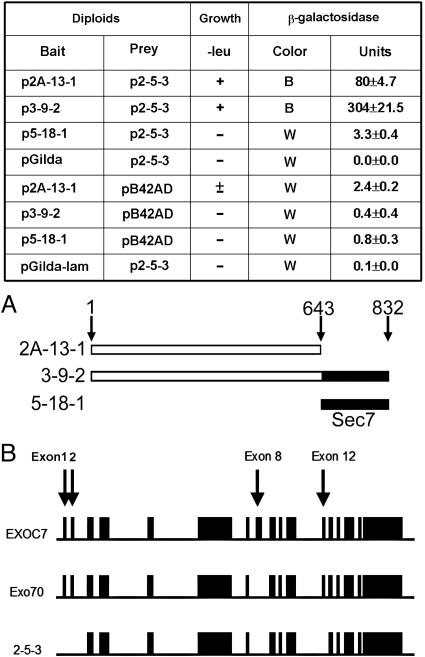
Yeast Two-Hybrid Experiments. Use of the MATCHMAKER LexA yeast two-hybrid system (Clontech) has been described (13). Briefly, three constructs (Fig. 1A) encoding BIG2 amino acids 1–643, 1–832, and 644–832 (Sec7 domain) in pGilda plasmid (p2A-13-1, p3-9-2, and p5-18-1, respectively) were used as bait to screen a human heart cDNA library. Positive colonies were confirmed by yeast mating experiments and recovered in KC8 Escherichia coli. Purified positive AD/library plasmids were sequenced by using a 373 DNA sequenator (Applied Biosystems).
Fig. 1.
Interaction between BIG2 and Exo70-related gene products in yeast two-hybrid experiments. (A) Three cDNAs encoding proteins corresponding to BIG2 amino acids 1–643 (2A-13-1), 1–832 (3-9-2), and 644–832 (5-18-1) were used as bait in yeast two-hybrid screen of a human heart cDNA library. In yeast mating experiments, two N-terminal segments of BIG2 (amino acids 1–643 and 1–832) but not the Sec7 domain (amino acids 644–832), interacted with 2-5-3. True positives exhibited growth on –leu plates and β-galactosidase activity (mean ± SD, n = 3). B, blue; W, white. pGilda and pB42AD are empty vectors only. pGilda-lam is a negative control with an unrelated protein. (B) Gene structures of full-length human Exo70 (EXOC7, Q9UPT5, GI: 26393663), the 2-5-3 cDNA retrieved in the yeast two-hybrid screen, and Exo70 clone (BC011045, GI: 15029668, IMAGE: 4215228) that was used for our overexpression experiment. Exons 1, 2, 8, 12, and the initial four codons of exon 3 are not present in p2-5-3 and exon 8 is not present in GFP-Exo70. Comparison of amino acid sequences of 2-5-3, Exo70, and EXOC7 is in Fig. 8.
Protein Synthesis in Reticulocyte Lysate. As described (13), the p2-5-3 insert was excised with EcoRI and XhoI from the pB42AD fusion plasmid and inserted into plasmid pGADT7 (Clontech) to add an N-terminal HA tag. Bait inserts encoding BIG2 (amino acids 1–643) and BIG2 (amino acids 1–832), excised with EcoRI and SalI, were ligated into plasmid pG-BKT7 (Clontech) to add an N-terminal c-myc tag. Procedures for coimmunoprecipitation of 35S-labeled proteins are in Supporting Text, which is published as supporting information on the PNAS web site.
Polymerization of Tubulin with Taxol from HepG2 Cells. The protocol of Vallee (16) was used. This and Western blotting procedures are described in Supporting Text.
Purification of Centrosomes. The protocol of Moudjou and Bornes (17) was used. See Supporting Text for details.
GFP-Exo70 Construct and Transfection. A GFP tag was added to the N terminus of a human Exo70 clone from Open Biosystems, Huntsville, AL (IMAGE: 4215228) with the Invitrogen Gateway system. Comparison of sequences of this clone and p2-5-3 with human gene sequence (EXOC7, Q9UPT5, GI: 26393663) reveals that p2-5-3 lacks exons 1, 2, 8, 12, and part of 3 and Exo70 lacks exon 8 as shown diagrammatically in Fig. 1B. Transfection of HepG2 cells with GFP-Exo70 was performed with Nucleofector T-28 protocol (2 × 106 HepG2 cells per 4 μg of GFP-Exo70/100 μl of Nucleofector solution V) (Amaxa Biosystems, Cologne, Germany). Transfected cells were incubated overnight at 37°C with 5% CO2.
Immunofluorescence Microscopy. HepG2 cells (1 × 105) were incubated on four-well, collagen I-coated culture slides (Becton Dickinson) overnight before experiments. After incubation (37°C, 1 h) without or with BFA (10 μg/ml) or nocodazole (10 μg/ml), as indicated, cells were washed with Dulbecco's PBS (DPBS) containing CaCl2 and MgCl2 (1 mM each) and fixed in 4% paraformaldehyde (15 min, room temperature). Cells were permeabilized with 0.1% Triton X-100 in DPBS for 4 min, incubated for 1.5 h in DPBS containing 10% goat serum and 3% skim milk, and reacted with anti-BIG2 (3.5 μg/ml), anti-Exo70 (13 μg/ml), anti-GM130 (1:100), anti-p230 (1:100), and anti-α-tubulin (1:500) for 2 h followed by secondary antibodies (fluorescein- or Texas red-labeled anti-rabbit or anti-mouse IgG) for 1 h at room temperature. Slides were mounted and counter-stained with DAPI (Vectashield with DAPI, Vector Laboratories). For γ-tubulin staining (1:1,000), cells were fixed in 100% methanol at –20°C for 8 min (not permeabilized). For visualization of both endogenous Exo70 and BIG2, Exo70 was stained as described above, and anti-BIG2 antibodies were labeled with Alexa Fluor 488 (Molecular Probes) according to the manufacturer's instructions.
Results
Interaction of BIG2 with Exo70 in a Yeast Two-Hybrid System. BIG2 constructs encoding amino acids 1–643 (2A-13-1) and 1–832 (3-9-2) were used as bait to screen a human heart cDNA library, which yielded clone p2-5-3. In yeast mating experiments, p2-5-3 did not interact with the Sec7 domain (5-18-1), which corresponds to amino acids 644–832 of BIG2, but interactions with 2A-13-1 and 3-9-2 were confirmed (Fig. 1). An interaction of BIG2 (amino acids 1–643 and 1–832) with 2-5-3 protein was also shown by coimmunoprecipitation of in vitro-translated, epitope-tagged proteins synthesized in a reticulocyte lysate system with anti-myc or anti-HA antibodies (Fig. 2A). Protein 2-5-3 was identified as a variant of human Exo70 (Fig. 1B), which is an exocyst protein required for exocytosis. Comparison of amino acid sequences predicted by nucleotide sequences of p2-5-3 with human Exo70 gene sequence indicates that exon 1, which encodes an N-terminal coiled-coil domain, exon 2, exon 8, exon 12, and part of exon 3 are missing in 2-5-3 (Figs. 1B and 8).
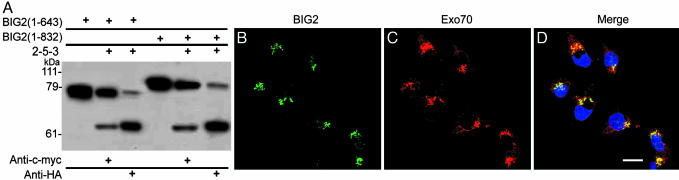
Fig. 2.
Coimmunoprecipitation of in vitro-translated BIG2 and 2-5-3 and colocalizatin of endogenous BIG2 and Exo70. (A) Coimmunoprecipitation of in vitro-translated BIG2 (amino acids 1–643 and 1–832) and 2-5-3. Samples (5 μl) of [35S]methionine-labeled proteins as indicated were mixed and precipitated with anti-c-myc (myc-BIG2) or anti-HA (HA-2-5-3) antibodies. Samples (10 μl) of each BIG2 protein without immunoprecipitation are also shown. Blots are representative of two experiments. (B–D) Localization of endogenous BIG2 and Exo70 in HepG2 cells. Both BIG2 (green, B) and Exo70 (red, C) are seen partially colocalized in the perinuclear region (D). Nuclei were counterstained with DAPI. (Scale bar: 20 μm.)
Colocalization of Endogenous BIG2 and Exo70 in HepG2 Cells. We used confocal immunofluorescence microscopy to visualize endogenous BIG2 and Exo70 proteins in HepG2 cells. The proteins were seen in a punctuate, perinuclear distribution where they appeared to be partially colocalized (Fig. 2 B–D). Exo70 immunoreactivity was, however, much more widely scattered throughout the cytoplasm and occasionally observed also at the plasma membrane (data not shown). BIG2 appeared to be colocalized in part, with cis-Golgi marker GM130 (Fig. 9, which is published as supporting information on the PNAS web site) and trans-Golgi marker p230 (see Fig. 5). Exo70 immunoreactivity likewise partially coincided with that of p230 and GM130 (see Figs. 6 and 9). These observations suggested that BIG2 and Exo70 might interact in some perinuclear membranes that could be at least partially in the trans-Golgi network.
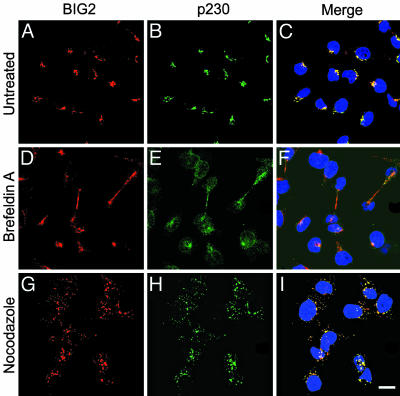
Fig. 5.
Effects of BFA and nocodazole on BIG2 distribution in HepG2 cells. (A–C) No treatment. (D–F) Treatment with BFA, 10 μg/ml, 1 h. (G–I) Treatment with nocodazole, 10 μg/ml, 1 h. Before treatment, BIG2 (red) and p230 (trans-Golgi network marker, green) were concentrated in perinuclear structures, but scattered widely after incubation with BFA, with colocalization apparently in the perinuclear region and slender intercellular connections perhaps during cytokinesis. Incubation with nocodazole dispersed BIG2 and p230 with seeming colocalization in numerous small cytoplasmic structures. (Scale bar: 20 μm.)
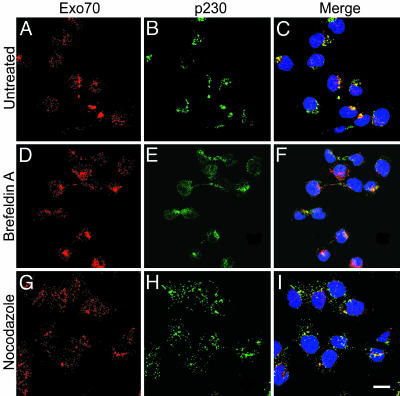
Fig. 6.
Effects of BFA and nocodazole on Exo70 distribution in HepG2 cells. (A–C) No treatment. (D–F) Treatment with BFA, 10 μg/ml, 1 h. (G–I) Treatment with nocodazole, 10 μg/ml, 1 h. Exo70 (red) was partially colocalized with p230 (green) in perinuclear regions and also in fine punctuate distribution in cytoplasm and nuclei. After incubation with BFA, Exo70 was largely dispersed from concentration with p230 in perinuclear regions, but the fine punctuate staining persisted. In cells incubated with nocodazole, Exo70 and p230 immunoreactivity appeared still partially coincident in widely distributed structures. (Scale bar: 20 μm.)
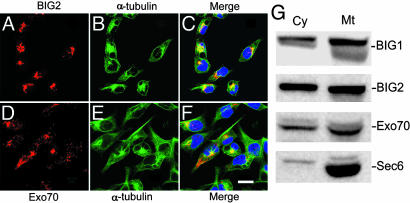
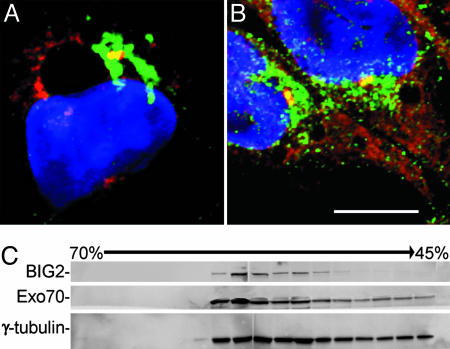
Interaction of BIG2 and Exo70 at the MTOC. Because the colocalization of BIG2 and Exo70 in a discrete region closely associated with Golgi structures appeared consistent with the position of a MTOC, we used antibodies against α-tubulin to mark microtubules and γ-tubulin to identify centrosomes. As seen in Fig. 3, immunoreactive Exo70 was seen along α-tubulin-positive linear structures, more concentrated near the nucleus in most cells. BIG2 was not seen clearly in the cell periphery, but appeared to coincide with more central microtubule structures. Western blot analyses of proteins in preparations of microtubules purified by taxol polymerization demonstrated the presence of BIG2 (and BIG1) as well as Exo70 and Sec6, which is another component of the exocyst complex (Fig. 3G). Immunoreactive BIG2 (Fig. 4A) and Exo70 (Fig. 4B) partially or completely overlapped with the γ-tubulin at the MTOC inspected by confocal microscopy. In those fractions from sucrose-density gradient purification of centrosomes that contained the largest amounts of γ-tubulin, both BIG2 and Exo70 were seen on Western blots (Fig. 4C). All data are consistent with the presence of BIG2 and Exo70 in structures or protein complexes associated with microtubules and present in the MTOC.
Fig. 3.
Association of BIG2 and Exo70 with microtubules. Both BIG2 (red, A and C) and Exo70 (red, D and F) appeared concentrated with α-tubulin (green, B, C, E, and F) in a perinuclear region, with only Exo70 clearly seen along more peripheral microtubules. (Scale bar: 20 μm.) (G) Proteins in taxol-polymerized microtubules from HepG2 cells separated by SDS/PAGE and reacted with antibodies against BIG1, BIG2, Exo70, and Sec6. Cy, cytosol (37 μg per lane, 0.02% of total); Mt, microtubule (10.5 μg per lane, 4.2% of total).
Fig. 4.
Association of BIG2 and Exo70 with centrosomes in HepG2 cells. Both BIG2 (green, A) and Exo70 (green, B) were partially localized with γ-tubulin (red) at centrosome. (Scale bar: 10 μm.) (C) Immunoreactive BIG2, Exo70, and γ-tubulin in fractions from sucrose density-gradient purification of centrosomes.
Effects of BFA and Nocodazole on Localization of BIG2 and Exo70. After incubation of HepG2 cells with BFA for 1 h, Golgi structure was disrupted as evidenced by dispersion of the trans-Golgi network marker p230 from its concentration in the perinuclear region of untreated cells (Figs. 5 and 6). BIG2 was also widely dispersed, but what appeared to be almost complete colocalization in concentrated perinuclear structures before BFA was only partial in the peripheral regions and in slender intercellular connections (Fig. 5). Exo70, which was visible in other structures in addition to Golgi at baseline, was also largely dispersed from Golgi-like concentrations (Fig. 6). After BFA incubation, small amounts of BIG2 and Exo70 remained with γ-tubulin-stained centrosomes (data not shown). In cells incubated with nocodazole, BIG2 (Fig. 5), Exo70 (Fig. 6), and p230 were distributed widely in punctuate concentrations throughout the cytoplasm. BIG2 more frequently overlapped with p230 (Fig. 5) than did Exo70 (Fig. 6). Partial colocalization of BIG2 and Exo70 after nocodazole treatment appeared consistent with the presence of both BIG2 and Exo70 in structures that interacted with microtubules (Fig. 10, which is published as supporting information on the PNAS web site).
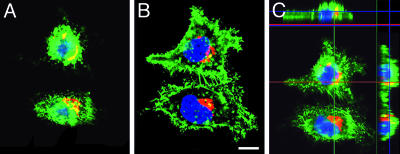
GFP-Exo70 Overexpression in HepG2 Cells. The distribution of GFP-Exo70 overexpressed in HepG2 cells was very different from that of endogenous Exo70. GFP-Exo70 was seen throughout the cytoplasm (not nucleus) and in the multiple filopodia-like plasma membrane protrusions (Fig. 7). In cells overexpressing GFP-Exo70, the perinuclear distribution of BIG2 was quite similar to that in nontransfected cells. GFP-Exo70 and BIG2 appeared to be colocalized in a limited region in the upper part of the cells (Fig. 7 A and C).
Fig. 7.
Overexpression of GFP-Exo70 in HepG2 cells. (A and B) Overexpressed GFP-Exo70 is shown throughout the cells (except nuclei) that had filopodia-like protrusions. Extent of apparent colocalization of GFP-Exo70 (green) and BIG2 (red) was greater in an apical (A) than basal (B) focal plane. (Scale bar: 10 μm.) (C) Reconstructed image from a stack of 20 sections with 0.88-μm intervals shows areas of colocalization of BIG2 and GFP-Exo70. Center X-Y, above X-Z, and right Y-Z planes are at the indicated positions (red/green lines).
Discussion
The p2-5-3 cDNA retrieved from a human heart library in a yeast two-hybrid screen using a construct encoding the N-terminal 832 aa of BIG2 as bait appears to encode a part of human Exo70. Direct interaction of two N-terminal polypeptide fragments of BIG2 (residues 1–643 and 1–832) with p2-5-3 was confirmed by yeast mating experiments and coimmunoprecipitation of the in vitro-synthesized proteins. Because Exo70 is an exocyst component, an association of BIG2 with exocyst formation and/or function was suspected. Absence of the ARF-activating Sec7 domain from the BIG2 (amino acids 1–643) molecule that interacted with the protein product 2-5-3 seems to indicate that the relationship between BIG2 and exocyst perhaps does not depend on ARF activation. Based on a comparison of the predicted protein sequences of full-length Exo70 (gene EXOC7) and p2-5-3 (Figs. 1 and 8), the N-terminal coiled-coil domain encoded in exon 1 of Exo70, which is missing in p2-5-3, is not required for its association with BIG2.
Although the interaction sites of Exo70 and BIG2 have not been defined, it appears that the Sec7 domain plus the C-terminal 954 aa of BIG2 and Exo70 sequence encoded in exons 1, 2, 8, 12, and part of 3, which are missing in 2-5-3, are not critical. Exon 8 of human Exo70 is also not present in rat Exo70 cDNA (GenBank accession no. NP-073182, GI: 12083643), but the rest of the predicted amino acid sequences are almost identical. Because we have not characterized the endogenous Exo70 protein (or proteins) in HepG2 cells, it is not known which structural elements encoded in exons missing from 2-5-3 may be involved in the observations reported. Our data support the conclusion that the Exo70 mRNA is alternatively spliced, and based on Western blot analyses (data not shown), multiple Exo70 proteins are synthesized.
The exocyst complex, which has been demonstrated in plasma membrane and the perinuclear region (trans-Golgi network and recycling endosomes) of mammalian cells comprises Exo70 and seven other proteins (18, 19). Structure and function of the exocyst complex were first studied and the components were named in Saccharomyces cerevisiae. Identification of the eight mammalian orthologues was completed when Matern et al. (20) reported that human Sec3 is the counterpart of yeast Sec3p, although it lacks the yeast Rho-binding sequence and does not apparently use an interaction with Rho to mark secretory sites in the plasma membrane. Their data (20) emphasized the importance of Exo70 for specification of exocyst localization at the mammalian cell plasma membrane, which could be relevant to the actions of BIG2 in association with the exocyst complex.
In HepG2 cells, BIG2 appeared colocalized with both p230 and GM130, markers for trans-Golgi and cis-Golgi structures, respectively, whereas Exo70 partially overlapped but did not coincide with either of them. Small amounts of BIG2 and Exo70 colocalized in the perinuclear region. Zhao et al. (5) reported that overexpressed BIG2 did not colocalize with the cis-Golgi marker p58 or GBF1, a BFA-resistant GEF that was present in cis-Golgi membranes. Shinotsuka et al. (21) concluded from studies of overexpressed BIG2 in HeLa cells and rat hepatocyes that BIG2 did not function in BFA inhibition of vesicular trafficking at the cis-Golgi, but acted rather in the trans-Golgi network. Colocalization of endogenous BIG2 and Exo70 in this area could be consistent with their joint function in pathways involving exocytotic vesicle formation and release.
The functions of BIG2 and Exo70 at the MTOC may be related, but not limited, to vesicle formation and translocation. Centrosomes play a key role in microtubule dynamics and in mitosis, although without them chromosome segregation can still occur (22). For cells in interphase, centrosomes and microtubules are important in overall shape, polarity, and motility. Proteomic analysis identified many molecules in centrosomes with multiple potential functions not yet understood (23). In addition to having roles in protein trafficking, BIG2 and Exo70 may participate in microtubule nucleation and mitosis. Early in mitosis, ARF1 is inactivated and peripheral proteins are released from Golgi membranes before their fragmentation and dispersion (24). ARF1 inactivation is also necessary for the subsequent chromosome segregation and cytokinesis (24). There is no information regarding an involvement of BIG2 in these processes. Exo70 was found to induce depolymerization of microtubules in vitro (25), but similar action in mitotic cells has apparently not been reported.
The treatment of HepG2 cells with BFA or nocodazole provided further evidence of a relationship of BIG2 and Exo70. Both BIG2 and Exo70 dispersed along with trans-Golgi protein p230 when BFA disrupted Golgi structure. Small amounts of both BIG2 and Exo70 remained, however, associated with γ-tubulin, in a perinuclear structure identified as the MTOC. BIG2 was also seen (Fig. 5) along with p230 in slender, elongated connections between cells, perhaps representing partition of Golgi elements in the final stages of cell division. After addition of BFA to cells, specific proteins are released from Golgi structures at different rates before the membranes with their intrinsic proteins are dispersed, and sensitivity to BFA concentration differs among cells. This may be why Vega and Hsu (26) did not observe loss of Exo70 from Golgi structures in PC12 cells incubated for 30 min with BFA (5 μg/ml), whereas we saw the effects after 1 h with 10 μg/ml. Disruption of microtubules with nocodazole dispersed BIG2, Exo70, and Golgi markers, with BIG2 and Exo70 remaining partially colocalized. All observations were consistent with an association of BIG2 and Exo70 in the MTOC and microtubule-associated exocytotic vesicles.
Overexpressed and endogenous Exo70 were distributed quite differently in HepG2 cells. GFP-Exo70, unlike the endogenous protein, was present throughout the cytoplasm and in the striking filopodia-like plasma membrane protrusions that were not apparent in the absence of Exo70 overexpression. They appear identical to the multiple protrusions that were described on polarized normal rat kidney cells overexpressing GFP-Exo70 (25). Matern et al. (20) had overexpressed both GFP-Exo70 and Exo70-GFP and analogous pairs of four other GFP-tagged exocyst subunits in polarized Madin–Darby canine kidney cells. They reported that only Exo70-GFP appeared confined to the lateral plasma membrane, although it was not localized like the endogenous protein, and it failed to generate tight intercellular contacts. Wang et al. (25) found that overexpression of GFP-Exo70 (but not GFP-Exo84) resulted in the disruption of microtubule architecture in the area of cytoplasm under regions of plasma membrane protrusions, and microtubule polymerization in vitro was inhibited by recombinant Exo70. The interaction of BIG2 with Exo70 in these processes suggests a mechanism for the congenital defects described in two families with mutations in the BIG2 gene (7).
The MTOC is the site near the nucleus at which clustered minus ends of microtubules anchored on rings of γ-tubulin project from a pair of centrioles in the centrosome (22). Microscopically, we found both endogenous Exo70 and BIG2 apparently associated with γ-tubulin at the MTOC. Although Exo70 and BIG2 appeared concentrated there, they were also seen along microtubules, which were identified by reaction with antibodies against α-tubulin. In microtubules prepared by polymerization of tubulin with taxol, BIG2 (and BIG1) as well as exocyst proteins Exo70 and Sec6 were identified. Wang et al. (25) found exocyst subunits in polymerized microtubules from normal rat kidney cells and noted that Exo70, along with other exocyst subunits, was present in commercial preparations of microtubule-associated protein-rich tubulin. Earlier reports of a relationship between Exo70 and microtubules include the detailed studies of Vega and Hsu (26), who demonstrated the presence of an exocyst complex at the MTOC in undifferentiated PC12 cells. In differentiated cells, it was associated with microtubules and concentrated at the growth cone. Their evidence led the authors to conclude that the exocyst complex may have a critical function at the intersection of cell signal transduction pathways and the cytoskeleton network (microtubules and septins) in targeting vesicles to the appropriate plasma membrane site for exocytosis (26). The partial colocalization of Exo70 with BIG2 may be especially relevant in this regard.
The role of BIG2 in BFA-inhibited activation of class I ARFs is only incompletely understood and cannot with present information be related mechanistically to the function of microtubules and Exo70. It can be said that the interactions between 2-5-3 and BIG2 described here did not depend on, and were not apparently affected by, the Sec7 domain part of BIG2 molecule that is responsible for its acceleration of GTP binding to class I ARFs and is critical for BFA-sensitive vesicular transport in the Golgi network. BIG2 (amino acids 1–643), the region N-terminal to the Sec7 domain does contain three short sequences termed A kinase-anchoring protein domains, which in vitro studies showed to have different specificities for association with each of the four cAMP-dependent protein kinase regulatory subunits (13). RIα, however, is the only one for which interaction was demonstrated in the human heart cDNA two-hybrid screen (13). A potential function for BIG2 in the integration/communication among cAMP signaling, vesicular trafficking, and cell cycle events is clearly of considerable interest. The association of BIG2 with Exo70 is a link between early and late events in vesicular trafficking pathways.
Supplementary Material
Acknowledgments
We thank Dr. Christian Combs and Daniela Malide (National Heart, Lung, and Blood Institute Confocal Microscopy Core Facility) and Michael W. Spencer (National Heart, Lung, and Blood Institute Pathology Core Facility) for invaluable help and Dr. Vincent C. Manganiello for important discussions and manuscript review.
Abbreviations: ARF, ADP-ribosylation factor; BFA, brefeldin A; BIG, BFA-inhibited guanine nucleotide-exchange protein; HA, hemagglutinin; MTOC, microtubule-organizing center.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY869728).
References
- 1.Jackson, C. L. & Casanova, J. E. (2000) Trends Cell Biol. 10, 60–67. [DOI] [PubMed] [Google Scholar]
- 2.Togawa, A., Morinaga, N., Ogasawara, M., Moss, J. & Vaughan, M. (1999) J. Biol. Chem. 274, 12308–12315. [DOI] [PubMed] [Google Scholar]
- 3.Morinaga, N., Moss, J. & Vaughan, M. (1997) Proc. Natl. Acad. Sci. USA 94, 12926–12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamaji, R., Adamik, R., Takeda, K., Togawa, A., Pacheco-Rodriguez, G., Ferrans, V. J., Moss, J. & Vaughan, M. (2000) Proc. Natl. Acad. Sci. USA 97, 2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao, X., Lasell, T. K. & Melançon, P. (2002) Mol. Biol. Cell 13, 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padilla, P. I., Pacheco-Rodriguez, G., Moss, J. & Vaughan, M. (2004) Proc. Natl. Acad. Sci. USA 101, 2752–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheen, V. L., Ganesh, V. S., Topcu, M., Sebire, G., Bodell, A., Hill, R. S., Grant, P. E., Shugart, Y. Y., Imitola, J., Khoury, S. J., et al. (2004) Nat. Genet. 36, 69–76. [DOI] [PubMed] [Google Scholar]
- 8.Kirchhausen, T. (2000) Nat. Rev. Mol. Cell. Biol. 1, 187–198. [DOI] [PubMed] [Google Scholar]
- 9.Donaldson, J. G. (2003) J. Biol. Chem. 278, 41573–41576. [DOI] [PubMed] [Google Scholar]
- 10.Thyberg, J. & Moskalewski, S. (1999) Exp. Cell Res. 246, 263–279. [DOI] [PubMed] [Google Scholar]
- 11.Whyte, J. R. & Munro, S. (2002) J. Cell Sci. 115, 2627–2637. [DOI] [PubMed] [Google Scholar]
- 12.Hsu, S. C., TerBush, D., Abraham, M. & Guo, W. (2004) Int. Rev. Cytol. 233, 243–265. [DOI] [PubMed] [Google Scholar]
- 13.Li, H., Adamik, R., Pacheco-Rodriguez, G., Moss, J. & Vaughan, M. (2003) Proc. Natl. Acad. Sci. USA 100, 1627–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padilla, P. I., Chang, M. J., Pacheco-Rodriguez, G., Adamik, R., Moss, J. & Vaughan, M. (2003) Proc. Natl. Acad. Sci. USA 100, 2322–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charych, E. I., Yu, W., Miralles, C. P., Serwanski, D. R., Li, X., Rubio, M. & De Blas, A. L. (2004) J. Neurochem. 90, 173–189. [DOI] [PubMed] [Google Scholar]
- 16.Vallee, R. B. (1982) J. Cell Biol. 92, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moudjou, M. & Bornens, M. (1998) in Cell Biology: A Laboratory Handbook, ed. Celis, J. (Academic, San Diego), pp. 111–119.
- 18.Folsch, H., Pypaert, M., Maday, S., Pelletier, L. & Mellman, I. (2003) J. Cell Biol. 163, 351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeaman, C., Grindstaff, K. K., Wright, J. R. & Nelson, W. J. (2001) J. Cell Biol. 155, 593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matern, H. T., Yeaman, C., Nelson, W. J. & Scheller, R. H. (2001) Proc. Natl. Acad. Sci. USA 98, 9648–9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinotsuka, C., Yoshida, Y., Kawamoto, K., Takatsu, H. & Nakayama, K. (2002) J. Biol. Chem. 277, 9468–9473. [DOI] [PubMed] [Google Scholar]
- 22.Rieder, C. L., Faruki, S. & Khodjakov, A. (2001) Trends Cell Biol. 11, 413–419. [DOI] [PubMed] [Google Scholar]
- 23.Andersen, J. S., Wilkinson, C. J., Mayor, T., Mortensen, P., Nigg, E. A. & Mann, M. (2003) Nature 426, 570–574. [DOI] [PubMed] [Google Scholar]
- 24.Altan-Bonnet, N., Phair, R. D., Polishchuk, R. S., Weigert, R. & Lippincott-Schwartz, J. (2003) Proc. Natl. Acad. Sci. USA 100, 13314–13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, S., Liu, Y., Adamson, C. L., Valdez, G., Guo, W. & Hsu, S. C. (2004) J. Biol. Chem. 279, 35958–35966. [DOI] [PubMed] [Google Scholar]
- 26.Vega, I. E. & Hsu, S. C. (2001) J. Neurosci. 21, 3839–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.