Abstract
Huntington's disease is a late-onset neurodegenerative disease caused by a CAG trinucleotide repeat in the gene encoding the huntingtin protein. Despite its well-defined genetic origin, the molecular and cellular mechanisms underlying the disease are unclear and complex. Here, we review some of the currently known functions of the wild-type huntingtin protein and discuss the deleterious effects that arise from the expansion of the CAG repeats, which are translated into an abnormally long polyglutamine tract. Finally, we outline some of the therapeutic strategies that are currently being pursued to slow down the disease.
The genetic basis of Huntington’s disease is well defined. However, the deleterious effects that arise from the abnormal protein are complex, and effective therapeutic interventions have yet to be identified.
INTRODUCTION TO HUNTINGTON’S DISEASE: GENETICS AND PATHOLOGY
Huntington’s disease (HD) is an autosomal dominant condition characterized by movement disorders and cognitive decline. Typically, the motor defects include chorea and loss of coordination. Psychiatric symptoms, such as depression, psychosis, and obsessive–compulsive disorder, are also common in HD and are particularly distressing for patients (Rosenblatt 2007). The prevalence of the mutation is four to ten cases per 100,000 in populations of Western European origin.
HD is characterized by a general shrinkage of the brain and degeneration of the striatum (caudate nucleus and putamen), with specific loss of efferent medium spiny neurons (MSNs) (Reiner et al. 1988). Although the striatum appears to be the most affected region of the brain, a regionally specific thinning of the cortical ribbon was found in patients with HD (Rosas et al. 2002). Such loss of cortical mass is an early event in the pathology of HD and proceeds from posterior to anterior cortical regions with disease progression. This regionally selective cortical degeneration may explain the heterogeneity of clinical expression in HD. Additional features are often present in HD patients, such as weight loss, skeletal-muscle wasting, and cardiac failure (Arenas et al. 1998; Aziz et al. 2008). Although generally less investigated than neurological signs, these additional signs might be due to the ubiquitous expression of mutant huntingtin (the toxic protein that causes HD).
HD is due to mutations in the HTT gene encoding huntingtin, a ubiquitously expressed protein of 350 kDa (Huntington's Disease Collaborative Research Group 1993). Huntingtin contains a polyglutamine tract encoded by uninterrupted CAG trinucleotide repeats in the first exon of HTT. Wild-type alleles contain up to 35 CAG repeats, whereas HD patients carry expansions of 36 or more repeats (Rubinsztein et al. 1996). Although complete penetrance of HD is observed for CAG sizes of ≥42, only a proportion of those with a CAG repeat length of 36–41 shows signs or symptoms of HD within a normal life span (Rubinsztein et al. 1996; Brinkman et al. 1997). For a review of polyglutamine-containing proteins and their role in neurodegenerative disease, see Pearce and Kopito (2017).
There is a strong inverse correlation between the number of CAG repeats and the age of onset of symptoms: larger CAG repeat expansions are generally associated with earlier ages of onset (Andrew et al. 1993). However, the CAG repeat number only partially explains 65%–71% of the variance in the age of onset, which also appears to be influenced by additional environmental and genetic factors, like modifier genes (Rosenblatt et al. 2001). Moreover, monozygotic twins have been reported to show different clinical symptoms, suggesting that epigenetic factors or tissue-specific variation in CAG repeats, because of somatic instability, may influence the disease (Georgiou et al. 1999).
HD is also characterized by the phenomenon of anticipation, where the age of onset tends to decrease in successive generations. This decrease is due to the unstable nature of the CAG repeats that tend to increase in size, particularly when passed through the male germline (Trottier et al. 1994). Although germline instability can explain the phenomenon of anticipation, somatic instability has been proposed as a mechanism underlying the tissue specificity of the disease.
To study the pathophysiology of HD, several mouse models have been generated. For an exhaustive description of those models, see reviews by Menalled and Chesselet (2002) and Lee et al. (2013).
WILD-TYPE HUNTINGTIN: STRUCTURE AND FUNCTIONS
Huntingtin is an ∼350 kDa protein containing the polyglutamine sequence at the NH2 terminus and multiple consensus sequences called HEAT (huntingtin, elongation factor 3, protein phosphatase 2A, and TOR1 [target of rapamycin 1]) repeats that are important for protein–protein interactions. HEAT motifs have a helix–turn–helix structure that is tightly packed to form a superhelix hydrophobic core that resists dissociation after proteolytic cleavage (Li et al. 2006). These motifs are often present in proteins involved in intracellular trafficking, such as clathrin adaptors and COPI (coat protein complex I) coatomer (Neuwald and Hirano 2000), and are possibly responsible for the scaffolding role of Huntingtin in the formation of protein complexes (Takano and Gusella 2002).
Huntingtin is a cytoplasmic protein with partial nuclear localization. Recently, its nuclear localization sequence (NLS) has been described in the NH2 terminus of the protein (Desmond et al. 2012). It spans between amino acids 174 and 207 and interacts with karyopherin β2, a protein that mediates nuclear import of proteins. This NLS comprises three consensus components: a basic-charged sequence, a downstream-conserved arginine, and a proline-tyrosine sequence.
Huntingtin also contains a nuclear export sequence in the COOH terminus (Xia et al. 2003). Moreover, the N-terminal sequence of huntingtin interacts with Tpr, a nuclear pore protein that is involved in nuclear export. Polyglutamine expansions decrease this interaction and increase the nuclear accumulation of huntingtin (Cornett et al. 2005).
Huntingtin is widely expressed in humans and rodents, with highest levels in the neurons of the central nervous system, where it appears to localize predominantly in the cytoplasm and be associated to vesicle membranes (DiFiglia et al. 1995). In particular, huntingtin is enriched in scattered striatal large neurons and in all corticostriatal neurons (Fusco et al. 1999).
PHYSIOLOGICAL FUNCTIONS OF HUNTINGTIN
Since the discovery of HTT as the gene responsible for HD, efforts have been made to elucidate the function of wild-type huntingtin, and several roles have been described so far. Here, we summarize the most studied ones.
Huntingtin Is Necessary for Embryonic Development
Huntingtin is required for early embryonic development, as knockout mice show embryonic lethality around day 8.5, before the emergence of the nervous system (Nasir et al. 1995; Zeitlin et al. 1995). Moreover, recent studies reveal that huntingtin plays a crucial role in neurogenesis. In fact, huntingtin was shown to be required for the maintenance of the lineage potential of primitive neuronal stem cells during the process of neural induction (Nguyen et al. 2013). Furthermore, huntingtin has a crucial role in neurulation controlling homotypic interactions between neuroepithelial cells (Lo Sardo et al. 2012). This function is executed by inhibiting both the activity of the metalloprotease ADAM10 and N-cadherin cleavage. This observation was also made in vivo, as defects in neural tube morphogenesis that were observed in huntingtin-knockdown zebra fish embryos could be rescued after treatment with GI254023X, an ADAM10 inhibitor.
Huntingtin Acts as a Protein Scaffold
Wild-type huntingtin is a well-characterized scaffolding protein. It interacts with β-tubulin and binds to microtubules (Hoffner et al. 2002). It also interacts with the dynein/dynactin complex (Caviston et al. 2007), regulating several intracellular trafficking processes. Recently, huntingtin has been shown to localize to spindle poles during mitosis, controlling spindle orientation in mouse neuronal cells (Godin et al. 2010). In the absence of huntingtin, dynein/dynactin and NuMA were dispersed around the spindle poles. Therefore, huntingtin possibly functions as a scaffold molecule that orchestrates the assembly of the dynein/dynactin complex.
Huntingtin as a Transcriptional Regulator
The nuclear localization confers huntingtin a role in transcriptional regulation (Kegel et al. 2002). Although numerous transcription factors are known to interact with mutant huntingtin, less is known about the interactions with the wild-type protein. A well-known target of huntingtin-mediated transcriptional regulation is the gene encoding brain-derived neurotrophic factor (BDNF) (Zuccato et al. 2003). In the cytoplasm, wild-type huntingtin sequesters and inhibits the activity of REST/NRSF (repressor element-1 transcription factor/neuron restrictive silencer factor), a transcription factor that negatively regulates BDNF transcription. Recently, it has been shown that huntingtin interacts with methyl-CpG-binding protein 2 in mouse and cellular models of HD. This interaction may also modulate the huntingtin-mediated expression of BDNF (McFarland et al. 2014).
Huntingtin in the Synapse
A new emerging role of huntingtin is in synaptic connectivity. Huntingtin is associated with synaptic vesicles in the presynaptic terminal (DiFiglia et al. 1995), as well as in the postsynaptic density (Marcora and Kennedy 2010), where it is associated with the scaffolding protein PSD95 (Sun et al. 2001). For many years, the role of huntingtin in this compartment was obscure. A recent study showed that huntingtin is required for a correct formation of cortical and striatal excitatory synapses (McKinstry et al. 2014). In particular, when huntingtin was silenced in developing mouse cortex, an increase in excitatory synapse formation in the cortex and striatum was observed at P21, followed by gliosis.
MECHANISMS OF PATHOGENESIS IN HD
Despite the well-known genetic origin of HD, the number and variety of molecular alterations reported in HD is broad and not completely understood. Although it is known that toxicity in HD arises from a gain of function of the mutant protein, given that expression of an expanded polyglutamine is toxic itself, a contribution of a loss of function of the wild-type protein cannot be discarded because deletion or inactivation of wild-type huntingtin also leads to neurodegeneration (O'Kusky et al. 1999; Dragatsis et al. 2000).
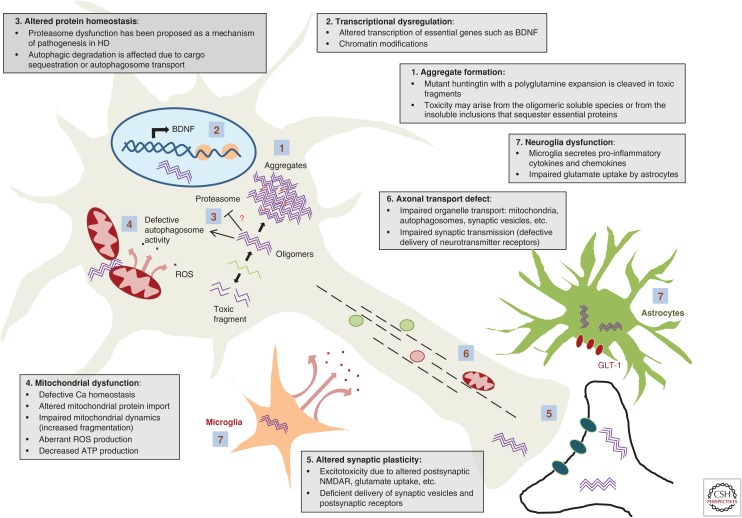
We outline here some of the mechanisms of pathogenesis described to date and focus in particular on those that are related to potential targets for therapy (Fig. 1).
Figure 1.
Schematic of selected mechanisms of pathogenesis in Huntington’s disease (HD). BDNF, Brain-derived neurotrophic factor; ROS, reactive oxygen species; NMDAR, N-methyl-D-aspartate receptor.
Mutant Huntingtin Aggregation: Is It Protective or Deleterious?
The hallmark of HD, and common to other polyglutamine disorders, is the presence of aggregates in the brain. These were initially considered crucial in HD pathology. Similar to other polyglutamine-containing proteins, mutant huntingtin aggregation proceeds by nucleated growth polymerization (Perutz and Windle 2001), leading to polyglutamine strands forming a β-sheet held together by hydrogen bonds (Perutz et al. 1994), which results in an amyloid structure (McGowan et al. 2000; Chen et al. 2002).
HD aggregates, initially found in the nucleus (DiFiglia et al. 1997; Becher et al. 1998) and later also in the cytoplasm and neuronal processes in the brain of HD patients (Gutekunst et al. 1999), are composed mainly of the expanded mutant huntingtin but also of many other proteins including ubiquitin (DiFiglia et al. 1997; Becher et al. 1998), proteasome subunits and chaperones (Cummings et al. 1998; Warrick et al. 1999), transcription factors (Huang et al. 1998; Steffan et al. 2000), or even the wild-type form of huntingtin (Kazantsev et al. 1999; Busch et al. 2003). Hence, the idea of a deleterious effect as a consequence of the loss of functional proteins sequestered into these aggregates was quite appealing. Another argument on behalf of their pathogenicity is that the number of polyglutamines correlates with both the rate of aggregation and the onset of the disease (Becher et al. 1998; Martindale et al. 1998; Perutz and Windle 2001), which suggests a direct link between aggregation and cell toxicity (Hackam et al. 1998).
Contrary to this intuitive hypothesis, there are also arguments supporting the ideas that aggregation does not correlate with toxicity and that aggregates might be just coincidental or even protective in HD (Saudou et al. 1998; Kim et al. 1999) and other polyglutamine disorders (Klement et al. 1998; Cummings et al. 1999). Single living neuron studies inferred an inverse correlation between the presence of aggregates and cell death (Arrasate et al. 2004) and suggested a protective role for these inclusions by sequestering toxic soluble species. In this line, the toxicity of the different mutant huntingtin species is currently a matter of debate: monomeric huntingtin forms soluble oligomers that precede fibrils and inclusions (Poirier et al. 2002; Mukai et al. 2005; Legleiter et al. 2010), and many reports point at these oligomers as the toxic species (Takahashi et al. 2008; Lajoie and Snapp 2010, 2013). Understanding which are the genuine harmful species is crucial to designing therapeutic strategies.
The aggregates in adult-onset HD are typically cytoplasmic, whereas those in juvenile-onset disease were proposed to be more frequent in the nucleus (DiFiglia et al. 1997; Becher et al. 1998). A recent study has suggested that some aggregates that appear to be nuclear may indeed be perinuclear (i.e., cytoplasmic). These perinuclear aggregates have been proposed to be the toxic species, which appear to cause cell death by abnormally activating the cell cycle (Liu et al. 2015). This study may resolve some of the controversy about the roles of aggregates in HD, as it appeared that the truly nuclear aggregates were relatively benign, compared with the perinuclear aggregates, and that diffuse mutant huntingtin does not impact cell death. However, the conclusions are still somewhat at odds with previous studies that suggested the mutant huntingtin was most toxic in its nonaggregated state (Arrasate et al. 2004).
Molecular chaperones promote efficient folding and prevent aggregation (Hartl et al. 2011), and increased levels of HSP40, HSP70, or HSP100 inhibit polyglutamine-induced protein aggregation and prevent its toxicity (Carmichael et al. 2000; Jana et al. 2000; Krobitsch and Lindquist 2000). A genomic screen recently identified that inhibition of glutaminyl-peptide cyclotransferase (QPCT), using small interference RNAs or small molecule inhibitors, leads to increased levels of the small heat shock protein (HSP) αB-crystallin, and consequently reduces aggregation and toxicity in several models of HD (Jimenez-Sanchez et al. 2015).
Huntingtin Is Cleaved in Toxic Fragments
Accumulation of pathogenic N-terminal fragments of huntingtin is characteristic in HD (Davies et al. 1997; Kim et al. 1999; Mende-Mueller et al. 2001). These fragments come from diverse origins, including proteolysis by caspases (Wellington et al. 1998; Kim et al. 2001; Hermel et al. 2004; Graham et al. 2006), calpains (Bizat et al. 2003; Kim et al. 2003; Gafni et al. 2004), and other proteases. In addition, alternative mechanisms might contribute, such as the aberrant splicing of the first exon of huntingtin protein (Sathasivam et al. 2013).
Although both wild-type and expanded huntingtin get cleaved, the presence of mutant fragments correlates with increased toxicity, which might be because of their higher propensity to form nuclear versus cytoplasmic less-toxic aggregates (Hackam et al. 1998; Lunkes and Mandel 1998; Kim et al. 1999; Lunkes et al. 2002). Also, the nature of these fragments may vary between tissues, which might contribute to differences in cell susceptibility (Mende-Mueller et al. 2001; Toneff et al. 2002; Wellington et al. 2002). Hence, inhibiting the formation of these fragments has been pursued as a therapeutic strategy, which could be achieved also indirectly—for example, by modifying the susceptibility of cleavage by phosphorylation of huntingtin by Cdk5 (Luo et al. 2005), or phosphorylation of a domain that impairs calpain cleavage (Schilling et al. 2006).
Mutant Huntingtin Disrupts Transcription
Transcriptional dysregulation has long been considered a major pathogenic mechanism in HD. DNA microarray studies have revealed that expression profiles of a number of genes are profoundly altered in HD (Luthi-Carter et al. 2000; Sipione et al. 2002). The activation domains of many transcription factors are composed of glutamine-rich regions, suggesting that they may interfere with expanded polyglutamines. Indeed, mutant huntingtin interacts with regulators of transcription, such as p53, cAMP response element-binding (CREB) protein, and CREB-binding protein (CBP), involved in cell proliferation and survival (Steffan et al. 2000; Nucifora et al. 2001; Sugars et al. 2004); PGC-1α (peroxisome proliferator-activating receptor-γ coactivator-1 α), which is necessary for energy metabolism (Cui et al. 2006; Chaturvedi et al. 2010); Sp1 and its coactivator TAFII130, which affects transcription of genes such as D2 dopamine receptor (Dunah et al. 2002; Zhai et al. 2005); and cystathionine γ-lyase, the biosynthetic enzyme for cysteine (Paul et al. 2014); among many others.
The increased susceptibility of the striatum in HD has been attributed to a reduction in the levels of BDNF, a pro-survival factor produced cortically to promote survival of striatal neurons. Impairment in transcription (Zuccato et al. 2001) or in axonal transport of BDNF (Gauthier et al. 2004) or its receptor TrkB (Liot et al. 2013) are all mechanisms that have been proposed to contribute to this deficit. In addition, corticostriatal synaptic defects in mouse models have been recently attributed to defects in BDNF signaling, rather than reduced BDNF levels, through an impact on postsynaptic p75 neurotrophin receptor (Plotkin et al. 2014), which along with TrkB binds to BDNF and is also implicated in HD (Brito et al. 2013; Jiang et al. 2013; Simmons et al. 2013).
Alterations in Gene Expression beyond Transcription: Epigenetics and Noncoding RNAs
Gene expression dysregulation in HD might also arise from variations in the epigenetic landscape, as well as in the regulation of noncoding RNAs. A first hint of deregulation of histone modification in HD came from the study of CBP, a transcriptional coactivator with histone acetyltransferase (HAC) functions. Expanded polyglutamines can bind to the HAC domain of CBP as well as other HACs, which disrupts their histone acetylation activity. Likewise, histone deacetylase (HDAC) inhibitors prevent neurodegeneration in cells, Drosophila, or mouse models of HD (McCampbell et al. 2001; Steffan et al. 2001; Ferrante et al. 2003; Hockly et al. 2003). More recently, genetic inhibition of HDAC4 has been shown to restore neurological dysfunction and extend life span in HD mouse models independently of its HDAC function but owing to reduced aggregate formation through decreased interaction between expanded polyglutamines and its glutamine-rich domain (Mielcarek et al. 2013).
In an effort to determine chromatin structural modifications in the genes downregulated in HD, a genome-wide approach identified a specific H3K4me3 pattern, a mark of active chromatin and transcription initiation, which correlated with transcriptional dysregulation in the R6/2 HD mouse and human HD brain (Vashishtha et al. 2013). Along similar lines, DNA methylation in promoter regions, which results in gene repression or silencing, was changed in a significant fraction of the genes altered in HD (Ng et al. 2013), although how mutant huntingtin triggers DNA methylation is currently unknown.
Gene expression is also influenced by noncoding RNAs. In HD human brain, miRNA deregulation has been reported (Johnson et al. 2008; Packer et al. 2008; Martí et al. 2010). Moreover, huntingtin has been found in RNA structures such as P bodies (Savas et al. 2008), stress granules (Ratovitski et al. 2012), or dendritic RNA granules (Savas et al. 2010), where it could influence protein expression at a posttranscriptional level. In Drosophila models of the related polyglutamine disease spinocerebellar ataxia type 3, expression of an untranslated CAG triplet expansion was sufficient to confer toxicity (Li et al. 2008). RNA toxicity mechanisms include aberrant protein–RNA interactions and sequestration of proteins, but also the hairpin secondary structure formed by CAG RNAs resemble double-stranded RNA (dsRNA) structures that are substrates for Dicer (Handa et al. 2003), cleaving them into shorter repeats that silence specific genes (Krol et al. 2007). Cleaved RNAs from CAG-expanded huntingtin may also become neurotoxic through Ago2-mediated gene silencing of CTG-containing genes (Bañez-Coronel et al. 2012).
Impairment of Protein Degradation Systems: Ubiquitin–Proteasome System and Autophagy
Two major degradation pathways exist to degrade intracellular proteins: the ubiquitin–proteasome system (UPS), which efficiently degrades wild-type huntingtin, and the autophagy–lysosome system, which seems to be important in degrading the expanded mutant forms (Ravikumar et al. 2002; Shibata et al. 2006).
Although most efforts have focused on finding strategies to upregulate these systems to reduce the levels of mutant protein, the influence that mutant huntingtin has in UPS and autophagy has also been a matter of research.
Early reports described an impairment in proteasome activity as a consequence of the expression of polyglutamine-expanded huntingtin (Bence et al. 2001; Jana et al. 2001; Verhoef et al. 2002; Bennett et al. 2005), a phenomenon that might be explained by either the sequestration of components of the UPS into inclusions (Davies et al. 1997; DiFiglia et al. 1997; Waelter et al. 2001) or the interaction between the proteasome and aggregation-resistant forms of huntingtin (Holmberg et al. 2004; Venkatraman et al. 2004).
Conversely, some groups did not observe deficits in UPS activity in HD (Bett et al. 2009; Maynard et al. 2009; Schipper-Krom et al. 2014a). Studies in single neurons and in mouse models have addressed this contradiction by revealing that an initial UPS impairment is followed by its normalization coinciding with the appearance of inclusions, suggesting an adaptive mechanism (Mitra et al. 2009; Ortega et al. 2010). More recently, it was shown that proteasomes can completely degrade expanded polyglutamines (Juenemann et al. 2013), which, together with the observation that proteasomes can be dynamically recruited to inclusions without affecting their activity (Schipper-Krom et al. 2014b), favor a competent UPS in HD.
Although an increased number of autophagosomes was described in HD models (Kegel et al. 2000), autophagosome formation is not affected by either mutant or wild-type huntingtin (Zheng et al. 2010). A closer look to the autophagic machinery revealed that, although formed, HD autophagosomes cannot optimally sequester substrates (Martinez-Vicente et al. 2010). This observation might be explained by the recently hypothesized role of wild-type huntingtin as a protein scaffold to recruit the autophagy machinery in selective autophagy, although the consequences of the triplet expansion in this scaffold function have not been addressed (Ochaba et al. 2014). Additionally, it has been proposed that HD autophagosomes have impaired axonal transport (Wong and Holzbaur 2014), which leads to inefficient autophagosome–lysosome fusion and decreased degradation of autophagosome content (Ravikumar et al. 2005; Jahreiss et al. 2008).
Altered Synaptic Plasticity and Neuronal Homeostasis in HD
Neuronal and synaptic abnormalities are early pathological events in HD (Usdin et al. 1999; Cummings et al. 2006; Milnerwood et al. 2006). Neuronal homeostasis might be compromised not only by decreased transcription of essential genes in neurotransmission and signaling but also by defects in the delivery of proteins and organelles along their axons. Pathogenic huntingtin inhibits fast axonal transport of organelles (Li et al. 2001; Gunawardena et al. 2003; Szebenyi et al. 2003; Lee et al. 2004; Trushina et al. 2004), a phenomenon that has been explained by aggregates blocking axons (Li et al. 2001; Lee et al. 2004), aggregate sequestration of motor proteins (Gunawardena et al. 2003; Trushina et al. 2004), or loss of function of wild-type huntingin (Gunawardena et al. 2003; Trushina et al. 2004). Huntingtin facilitates vesicle trafficking by serving as a scaffold between cargoes, microtubules, and motor proteins such as dyneins or kinesins (Caviston et al. 2007; Colin et al. 2008), an interaction mediated through huntingtin-associated protein 1 (HAP1), which appears to be disrupted in disease (Gauthier et al. 2004; McGuire et al. 2006). Polyglutamine-expanded huntingtin may also have an indirect effect by enhancing JNK3 phosphorylation of kinesin heavy chain, which disrupts its binding to microtubules in cellular and animal models of HD, thereby perturbing fast axonal transport (Morfini et al. 2009).
Axonal transport is required to correct delivery to neuronal membranes to ensure synaptic transmission. In HD, a failed delivery of receptors, such as GABA(A) (γ-aminobutyric acid type A) or AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) receptors, inhibits synaptic excitability. HAP1 is the scaffold linking these receptors to the kinesin motor KIF5, and this interaction is interrupted by mutant huntingtin (Twelvetrees et al. 2010; Mandal et al. 2011; Yuen et al. 2012). Mutant huntingtin also inhibits cortical transport and release of BDNF (Gauthier et al. 2004), or the retrograde transport in the striatum of its receptor TrkB (Liot et al. 2013), necessary to promote survival signals in the cell body.
MSNs in the striatum experience the most prominent degeneration in HD. The observation that MSNs were selectively affected by glutamatergic signals (Coyle and Schwarcz 1976; McGeer and McGeer 1976; Beal et al. 1986) leads to the hypothesis that striatal neurons in HD could be harmed by excessive neurotransmission, mainly through glutamate stimulation of NMDA receptors, resulting in neuronal cell death via a process termed excitotoxicity.
Alterations in the levels of the different subunits of postsynaptic NMDAR in the striatum could explain their aberrant activity in HD (Cepeda et al. 2001; Ali and Levine 2006; Benn et al. 2007; Fan et al. 2007), which may predispose striatal neurons to excitotoxic damage (Laforet et al. 2001; Zeron et al. 2002). In addition, mutations in HD might also affect trafficking of NMDAR in striatal neurons (Fan et al. 2007; Marco et al. 2013). But also, the balance between synaptic (pro-survival) and extrasynaptic (detrimental) NMDAR activity is altered in HD (Okamoto et al. 2009; Milnerwood et al. 2010). Excitotoxicity might result from increased glutamate release or from impaired uptake and clearance, as downregulation of GLT1 glial glutamate transporter has been observed in HD (Liévens et al. 2001; Shin et al. 2005; Estrada-Sánchez et al. 2009). Therapeutic agents targeting excitotoxicity may act directly on NMDAR, such as memantine (Okamoto et al. 2009; Milnerwood et al. 2010), or modulate levels of excitatory neurotransmitters, such as 3-hydroxikynurenine and quinolinic acid, both metabolites of the kynurenine pathway, the major tryptophan degradative pathway, which is perturbed in HD (Giorgini et al. 2005; Guidetti et al. 2006; Campesan et al. 2011; Zwilling et al. 2011).
Mitochondrial Dysfunction in HD
Altered mitochondrial function resulting in defects in ATP production, Ca++ buffering capacity, and apoptosis is associated with neurodegeneration in HD (Sawa et al. 1999; Panov et al. 2002). Some evidence suggests that mutant huntingtin can interact with the outer mitochondrial membrane, resulting in mitochondrial calcium abnormalities (Panov et al. 2002; Choo et al. 2004). Mutant huntingtin also interferes with normal organellar axonal transport and can therefore reduce transport of mitochondria to synapses, as well as ATP production (Orr et al. 2008; Song et al. 2011; Shirendeb et al. 2012).
Decreased transcription of mitochondrial genes may also contribute to mitochondrial defects, such as repression of PGC-1α, a nuclear coactivator that regulates the expression of genes that mediate mitochondrial biogenesis and respiration (Cui et al. 2006), or depletion of the enzyme necessary for synthesizing cysteine, which maintains mitochondrial homeostasis (Paul et al. 2014). Also, transport of proteins into mitochondria could be defective because huntingtin interacts and inhibits TIM23, a component of the inner mitochondrial membrane transport complex, and this defect may contribute to respiratory dysfunction and neuronal cell death (Yano et al. 2014).
Mitochondria are dynamic organelles that undergo fusion–fission cycles in response to stimuli and metabolic demands. Fragmentation leads to caspase activation and apoptosis, and, therefore, inhibiting mitochondria fission delays cell death (Youle and Karbowski 2005). Expanded huntingtin interferes with mitochondrial dynamics and interacts with a central regulator of protein fission, dynamin-related protein 1 (Drp-1), increasing its enzymatic activity and mitochondrial fragmentation. Conversely, overexpression of a negative form of Drp-1 or fusion-promoting enzymes inhibits mutant huntingtin-induced mitochondrial fragmentation and toxicity (Wang et al. 2009; Song et al. 2011; Shirendeb et al. 2012), and selective Drp-1 inhibitors have proved beneficial to slow down disease progression in several HD models (Guo et al. 2013).
A consequence of mitochondrial malfunction is the aberrant production of reactive oxygen species (ROS), which in turn causes more damage to mitochondria. Postmortem brain of HD patients and experimental models of HD show evidence of oxidative damage (Perluigi et al. 2005; Stoy et al. 2005; Sorolla et al. 2008). Therefore, antioxidants are currently being tested to ameliorate levels of ROS and to help in mitochondrial dysfunction.
Cell-to-Cell Transmission of Aggregates
Emerging evidence suggests that prion-like transmission from cell to cell of proteins like tau or α-synuclein spreads these disease-associated proteins to different brain regions (Lee et al. 2010; Guo and Lee 2014). This “infectious” property has also been associated with polyglutamine proteins, where internalization of exogenous polyglutamine aggregates serves as seeds for nucleating aggregation of cytoplasmic soluble polyglutamines (Ren et al. 2009). This first report suggested that aggregates are internalized from the extracellular space, but, more recently, cell-to-cell transfer of aggregates through tunneling nanotubes, actin-rich membrane bridges that connect cells and mediate the transfer of cytoplasmic content (Rustom et al. 2004) such as prions (Gousset et al. 2009), were suggested as an alternative route (Costanzo et al. 2013).
In vivo studies have shed some light on this hypothesis. Human embryonic stem cell (hESC)-derived neurons that were integrated into corticostriatal organotypic brain slices of an R6/2 mouse or injected into the cortex acquired mutant huntingtin aggregates after 2 or 4 weeks, respectively, which correlated with alterations in neuron integrity. Furthermore, corticostriatal co-cultures revealed that mutant huntingtin spread from R6/2 cortex to wild-type MSNs in the striatum but not in the opposite direction (Pecho-Vrieseling et al. 2014), suggesting that propagation occurred in a pre- to postsynaptic path, which was confirmed by using inhibitors of synaptic vesicle fusion (Pecho-Vrieseling et al. 2014).
Astrocyte and Microglial Dysfunction in HD
Although huntingtin aggregates are more prominent in neurons than in non-neuronal glial cells (Shin et al. 2005), probably because of the lack of cell division in neurons or to a less efficient protein homeostasis system (Tydlacka et al. 2008), glial cells also contribute to disease in HD, and reactive gliosis is observed in many HD mouse models (Reddy et al. 1998; Lin et al. 2001; Yu et al. 2003) and in postmortem brains of HD patients (Myers et al. 1991; Sapp et al. 2001).
Astrocytes are the major type of glia. They provide support to neurons and enable uptake of extracellular glutamate, preventing excitotoxicity. In an effort to discern the role of astrocytes in HD pathology, an N-terminal huntingtin with 160Q was selectively expressed in astrocytes. Despite no obvious degeneration of glia or neurons, these mice developed late-onset neurological symptoms, which correlated with reduced levels of the GLT-1 glutamate transporter (Bradford et al. 2009). When mutant huntingtin was expressed in both astrocytes and neurons, it worsened the phenotype relative to neuronal-only expression, confirming the contribution of astroglia to disease (Bradford et al. 2010).
Additional defects might contribute to pathology, such as impaired secretion of the chemokine CCL5 (Chou et al. 2008) or BDNF (Wang et al. 2012) from HD astrocytes. Also, striatal astrocytes from R6/2 and Q175 HD mouse models showed reduced levels of Kir4.1 K+ channels, which lead to increased extracellular K+ and neuronal excitability, whereas viral delivery of Kir4.1 attenuated the R6/2 mouse phenotype (Tong et al. 2014).
Growing evidence implicates neuroinflammation in neurodegeneration. In HD, increased secretion of proinflammatory cytokines and chemokines has been reported in late but also in early presymptomatic gene carriers (Tai et al. 2007; Björkqvist et al. 2008; Wild et al. 2011), suggesting that neuroinflammation is not only a reactive process but also an active player in disease progression.
Huntingtin is expressed in immune cells, resulting in cell-autonomous microglial activation and secretion of proinflammatory cytokines, as a consequence of elevated transcription of myeloid lineage-determining factors PU.1 and C/EBPs (CCAAT/enhancer-binding proteins) (Crotti et al. 2014). In the peripheral immune system, mutant huntingtin also impacts inflammatory responses through inhibition of NF-κB signaling (Träger et al. 2014). Signaling through CB2 cannabinoid receptors might also explain inflammation in HD (Palazuelos et al. 2009; Bouchard et al. 2012). In addition, both microglia and peripheral cells expressing mutant huntingtin showed reduced migration in response to chemotactic signals (Kwan et al. 2012b). Moreover, bone-marrow transplantation with wild-type cells restored the levels of cytokines and chemokines and partially suppressed pathology in HD mouse models (Kwan et al. 2012a).
Interplay between Mutant Huntingtin and Other Aggregate-Prone Proteins
Mutant huntingtin is also associated with other brain pathologies. α-Synuclein, the component of Lewy bodies in Parkinson’s disease, is found close to huntingtin aggregates (Charles et al. 2000), and excess α-synuclein expression is associated with increased mutant huntingtin aggregation (Furlong et al. 2000; Herrera and Outeiro 2012). In agreement with these findings, when α-synuclein was knocked out in an R6/1 mouse, the number of inclusions was reduced, and the disease progression attenuated (Tomás-Zapico et al. 2012). Overexpression of α-synuclein has a negative effect on autophagy (Winslow et al. 2010) and worsens the disease phenotype in R6/1 and N171-82Q mouse models (Corrochano et al. 2012). Conversely, its depletion was beneficial and correlated with an increase in autophagy in these mice, which explains the crosstalk between these two diseases (Corrochano et al. 2012). Because mutant huntingtin is an autophagy substrate, these observations are likely a major contributor to the crosstalk between α-synuclein and huntingtin aggregation, as opposed to any obvious cross-seeding.
An imbalance in the levels of tau isoforms containing either three or four microtubule binding repeats (3R or 4R) with an increased 4R/3R ratio is sufficient to cause neurodegeneration. This relation is increased in HD mice as a consequence of splicing defects and enhances HD pathology (Fernández-Nogales et al. 2014). Tau phosphorylation is also affected by HD mouse models, which might have consequences in HD progression (Blum et al. 2014).
THERAPY FOR HD
HD as a Tractable Therapeutic Problem
Although HD is rare, it does receive a great deal of research attention. One reason is that HD has some features that make it more likely to be a tractable problem than other neurodegenerative conditions. First, the autosomal dominant nature of the condition means that the diagnosis is almost definitive and can be made before death. Thus, it is possible to accurately model and study the disease in vitro and in vivo. Perhaps more important, one can be sure that patients are suffering from a reasonably homogeneous condition. In other dementing illnesses, the diagnosis is seldom definitive, and postmortem analysis often shows a mix of pathologies. Second, the familial nature of the condition means that diagnosis can be made before symptom onset. Early diagnosis is a crucial advantage, as it means that therapy can begin before major neuronal loss, and by which point in the illness, it may be more difficult to slow progression and impossible to correct existing deficits. Finally, the Huntington’s community of patients, their families, and their doctors have a history of cooperation, which has made large-scale clinical trials possible. This collaboration is clear not only from trials that have taken place but also from long-term longitudinal studies of disease progression that have provided rich data sources to inform the design of future trials, particularly with regard to appropriate trial end points (Tabrizi et al. 2013).
Current Treatment for HD
There are no known disease-modifying drugs currently available for HD. Treatment is symptomatic only. Tetrabenazine is the only drug with a licensed indication for HD in the United Kingdom, where it is used to treat choreiform movements. Trials of cholinesterase inhibitors used to treat the cognitive problems seen in Alzheimer’s disease have been largely negative in HD (Cubo et al. 2006). Psychiatric symptoms, which are often the most troubling for patients, are often treated with standard drug treatments used in non-HD patients. For example, psychosis is treated with atypical antipsychotics and depression with selective serotonin uptake inhibitor (SSRI) or selective norepinephrine uptake inhibitor (SNRI) antidepressants (Phillips et al. 2008). With the exception of one open label trial with venlafaxine, these treatments are supported largely by case studies or small series (Holl et al. 2010). The current care of people with HD involves many paramedical disciplines, including speech and language therapy, physiotherapy, nursing, and social care.
Therapeutic Trials Based on Potential Pathogenic Mechanisms
Gene Silencing
Silencing the expression of the mutant huntingtin gene is attractive, as one might expect it to provide an effective treatment by dealing with the pathology at its source. Indeed, trials in rodents have found the approach to be efficacious in ameliorating symptoms and pathology using either RNA interference (Drouet et al. 2009; Stanek et al. 2014) or antisense oligonucleotides (Kordasiewicz et al. 2012). Although attractive, this approach has a number of potential difficulties. These include allele specificity, off-target effects, and delivery. Recent nonhuman primate trials have shown promising safety data, and a small safety trial of antisense oligonucleotides in HD patients was planned for 2015 (McBride et al. 2011; Grondin et al. 2012).
Antiapoptotics/Caspase Inhibition
The tetracycline antibiotic minocycline is a caspase inhibitor, although like many of the compounds described here has pleiotropic mechanisms of action, including antioxidant and cytokine-modulating properties. It was initially shown to prolong life expectancy in a mouse model of HD, although subsequent work in the same mouse model was not encouraging (Chen et al. 2000; Menalled et al. 2010). Nevertheless, following small safety trials, a larger trial of patients with an end point of change in total functional capacity compared with historical controls suggested no benefit of minocycline. This finding highlights the importance of careful reproduction of preclinical data before moving to clinical trials.
Transglutaminase Inhibition
The glutamine residues in huntingtin are cross-linked by transglutaminase. Transglutaminase inhibitors, such as cystamine, have produced promising results in mouse models of the disease (Dedeoglu et al. 2002; Karpuj et al. 2002). A safety and dose-finding study of a cystamine dimer, cysteamine, has been performed (Dubinsky and Gray 2006), and a larger trial involving 96 HD patients is underway.
Mitochondria, Oxidative Stress, and Excitotoxicity
Using antioxidants to decrease oxidative stress has been a putative therapeutic strategy for a number of neurodegenerative diseases. This approach is another example in which treatment of mouse models suggested benefit, in this case with the NMDA receptor antagonist remacemide and the antioxidant coenzyme Q10 (Ferrante et al. 2002). Unfortunately, these results were not recapitulated in large clinical trials of patients (Huntington Study Group 2001). More recently, in the largest proposed trial in HD to date, the 2CARE study of high-dose coenzyme Q10 was stopped early due to a combination of futility and safety concerns. Other clinical trials with other NMDA receptor antagonists have also been disappointing (Kremer et al. 1999), as have trials of creatine, a potential antioxidant with previous positive results in mice. Trials of creatine in symptomatic patients have been disappointing, but further trials have been undertaken in at-risk individuals with some more positive findings on imaging (Rosas et al. 2014).
Upregulating Autophagy
Autophagy upregulation using a variety of drugs has shown amelioration of the HD phenotype and pathology in cellular, fly, fish, and mice models, as reviewed by Hochfeld et al. (2013). Conversely, inhibition of autophagy has been shown to worsen phenotypes, including using antioxidants that are autophagy inhibitors, and this observation may provide an explanation for the relative failure of antioxidant-based strategies (Underwood et al. 2010). The repertoire of drugs that upregulate autophagy has been expanded and includes compounds, such as rilmenidine, that have a benign side-effect profile and long records of safe human use. A safety trial of rilmenidine is currently underway in HD patients in Cambridge, United Kingdom.
Transplantation
Once symptoms are manifest, it may be difficult to reverse deficits with drug treatment, as neurons have been lost. One approach to correcting this deficit is to transplant new neuronal tissue, a strategy that has shown some promise in animal models of the disease (Dunnett et al. 1998). Early human studies have shown potential for graft survival, although a recent long-term follow-up study was less encouraging, and mutant huntingtin was found in transplanted tissue (Barker et al. 2013; Cicchetti et al. 2014). The variable results in terms of graft survival, safety, and outcome may reflect differences in protocol and procedure, which may lead to more successful trials now that a proof of principle has been established.
Other Clinical Trials
Other large trials have been reported in HD. Latrepirdine (Dimebon) was originally synthesized as an antihistamine and showed some early promise as a treatment for Alzheimer’s disease. Results from the relatively large DIAMOND trial in HD patients suggested some improvement in cognition, but the subsequent larger HORIZON trial showed no benefit, and the company involved is no longer taking this compound forward as a potential therapy (Kieburtz et al. 2010). Other compounds that may help with symptoms but may not be disease modifying have also been studied in large-scale trials. Pridopidine is a drug that may have a beneficial effect on movement via its effect on dopamine signaling. A large recent trial did not reach statistical significance for the primary motor end point, but did suggest some promise in motor scores overall. Nondrug strategies are also being pursued; for example, recent trials of physical activity and rehabilitation in HD patients have been reported (Busse et al. 2013).
The increase in understanding the basic science of HD has yet to translate into an effective disease-modifying therapy. Feasibility of large-scale trials of various sorts has been demonstrated, and a wide variety of approaches are currently being pursued, which one hopes will start to bring benefits in the near future.
ACKNOWLEDGMENTS
We are grateful for funding from a Wellcome Trust Principal Research Fellowship (D.C.R.), a Wellcome Trust/MRC Strategic Grant on Neurodegeneration, the Alzheimer’s Disease Biomedical Research Unit, and Addenbrooke's Hospital.
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Diseases available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this subject collection.
- Ali NJ, Levine MS. 2006. Changes in expression of N-methyl-D-aspartate receptor subunits occur early in the R6/2 mouse model of Huntington’s disease. Dev Neurosci 28: 230–238. [DOI] [PubMed] [Google Scholar]
- Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman MA. 1993. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nat Genet 4: 398–403. [DOI] [PubMed] [Google Scholar]
- Arenas J, Campos Y, Ribacoba R, Martín MA, Rubio JC, Ablanedo P, Cabello A. 1998. Complex I defect in muscle from patients with Huntington’s disease. Ann Neurol 43: 397–400. [DOI] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431: 805–810. [DOI] [PubMed] [Google Scholar]
- Aziz NA, van der Burg JMM, Landwehrmeyer GB, Brundin P, Stijnen T, EHDI Study Group, Roos RAC. 2008. Weight loss in Huntington disease increases with higher CAG repeat number. Neurology 71: 1506–1513. [DOI] [PubMed] [Google Scholar]
- Bañez-Coronel M, Porta S, Kagerbauer B, Mateu-Huertas E, Pantano L, Ferrer I, Guzmán M, Estivill X, Martí E. 2012. A pathogenic mechanism in Huntington’s disease involves small CAG-repeated RNAs with neurotoxic activity. PLoS Genet 8: e1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker RA, Mason SL, Harrower TP, Swain RA, Ho AK, Sahakian BJ, Mathur R, Elneil S, Thornton S, Hurrelbrink C, et al. 2013. The long-term safety and efficacy of bilateral transplantation of human fetal striatal tissue in patients with mild to moderate Huntington’s disease. J Neurol Neurosurg Psychiatry 84: 657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF, Kowall NW, Ellison DW, Mazurek MF, Swartz KJ, Martin JB. 1986. Replication of the neurochemical characteristics of Huntington’s disease by quinolinic acid. Nature 321: 168–171. [DOI] [PubMed] [Google Scholar]
- Becher MW, Kotzuk JA, Sharp AH, Davies SW, Bates GP, Price DL, Ross CA. 1998. Intranuclear neuronal inclusions in Huntington’s disease and dentatorubral and pallidoluysian atrophy: Correlation between the density of inclusions and IT15 CAG triplet repeat length. Neurobiol Dis 4: 387–397. [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292: 1552–1555. [DOI] [PubMed] [Google Scholar]
- Benn CL, Slow EJ, Farrell LA, Graham R, Deng Y, Hayden MR, Cha JHJ. 2007. Glutamate receptor abnormalities in the YAC128 transgenic mouse model of Huntington's disease. Neuroscience 147: 354–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EJ, Bence NF, Jayakumar R, Kopito RR. 2005. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell 17: 351–365. [DOI] [PubMed] [Google Scholar]
- Bett JS, Cook C, Petrucelli L, Bates GP. 2009. The ubiquitin-proteasome reporter GFPu does not accumulate in neurons of the R6/2 transgenic mouse model of Huntington's disease. PLoS ONE 4: e5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizat N, Hermel JM, Boyer F, Jacquard C, Créminon C, Ouary S, Escartin C, Hantraye P, Kajewski S, Brouillet E. 2003. Calpain is a major cell death effector in selective striatal degeneration induced in vivo by 3-nitropropionate: implications for Huntington’s disease. J Neurosci 23: 5020–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkqvist M, Wild EJ, Thiele J, Silvestroni A, Andre R, Lahiri N, Raibon E, Lee RV, Benn CL, Soulet D, et al. 2008. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J Exp Med 205: 1869–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum D, Herrera F, Francelle L, Mendes T, Basquin M, Obriot H, Demeyer D, Sergeant N, Gerhardt E, Brouillet E, et al. 2014. Mutant huntingtin alters Tau phosphorylation and subcellular distribution. Hum Mol Genet 24: 76–85. [DOI] [PubMed] [Google Scholar]
- Bouchard J, Truong J, Bouchard K, Dunkelberger D, Desrayaud S, Moussaoui S, Tabrizi SJ, Stella N, Muchowski PJ. 2012. Cannabinoid receptor 2 signaling in peripheral immune cells modulates disease onset and severity in mouse models of Huntington's disease. J Neurosci 32: 18259–18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford J, Shin JY, Roberts M, Wang CE, Li XJ, Li S. 2009. Expression of mutant huntingtin in mouse brain astrocytes causes age-dependent neurological symptoms. Proc Natl Acad Sci 106: 22480–22485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford J, Shin JY, Roberts M, Wang CE, Sheng G, Li S, Li XJ. 2010. Mutant huntingtin in glial cells exacerbates neurological symptoms of Huntington disease mice. J Biol Chem 285: 10653–10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman RR, Mezei MM, Theilmann J, Almqvist E, Hayden MR. 1997. The likelihood of being affected with Huntington disease by a particular age, for a specific CAG size. Am J Hum Genet 60: 1202–1210. [PMC free article] [PubMed] [Google Scholar]
- Brito V, Puigdellívol M, Giralt A, del Toro D, Alberch J, Ginés S. 2013. Imbalance of p75(NTR)/TrkB protein expression in Huntington's disease: Implication for neuroprotective therapies. Cell Death Dis 4: e595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Engemann S, Lurz R, Okazawa H, Lehrach H, Wanker EE. 2003. Mutant huntingtin promotes the fibrillogenesis of wild-type huntingtin: A potential mechanism for loss of huntingtin function in Huntington's disease. J Biol Chem 278: 41452–41461. [DOI] [PubMed] [Google Scholar]
- Busse M, Quinn L, Debono K, Jones K, Collett J, Playle R, Kelly M, Simpson S, Backx K, Wasley D, et al. 2013. A randomized feasibility study of a 12-week community-based exercise program for people with Huntington's disease. J Neurol Phys Ther 37: 149–158. [DOI] [PubMed] [Google Scholar]
- Campesan S, Green EW, Breda C, Sathyasaikumar KV, Muchowski PJ, Schwarcz R, Kyriacou CP, Giorgini F. 2011. The kynurenine pathway modulates neurodegeneration in a Drosophila model of Huntington's disease. Curr Biol 21: 961–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael J, Chatellier J, Woolfson A, Milstein C, Fersht AR, Rubinsztein DC. 2000. Bacterial and yeast chaperones reduce both aggregate formation and cell death in mammalian cell models of Huntington’s disease. Proc Natl Acad Sci 97: 9701–9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston JP, Ross JL, Antony SM, Tokito M, Holzbaur ELF. 2007. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc Natl Acad Sci 104: 10045–10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepeda C, Ariano MA, Calvert CR, Flores-Hernández J, Chandler SH, Leavitt BR, Hayden MR, Levine MS. 2001. NMDA receptor function in mouse models of Huntington disease. J Neurosci Res 66: 525–539. [DOI] [PubMed] [Google Scholar]
- Charles V, Mezey E, Reddy PH, Dehejia A, Young TA, Polymeropoulos MH, Brownstein MJ, Tagle DA. 2000. α-Synuclein immunoreactivity of huntingtin polyglutamine aggregates in striatum and cortex of Huntington’s disease patients and transgenic mouse models. Neurosci Lett 289: 29–32. [DOI] [PubMed] [Google Scholar]
- Chaturvedi RK, Calingasan NY, Yang L, Hennessey T, Johri A, Beal MF. 2010. Impairment of PGC-1α expression, neuropathology and hepatic steatosis in a transgenic mouse model of Huntington’s disease following chronic energy deprivation. Hum Mol Genet 19: 3190–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Ona VO, Li M, Ferrante RJ, Fink KB, Zhu S, Bian J, Guo L, Farrell LA, Hersch SM, et al. 2000. Minocycline inhibits caspase-1 and caspase-3 expression and delays mortality in a transgenic mouse model of Huntington disease. Nat Med 6: 797–801. [DOI] [PubMed] [Google Scholar]
- Chen S, Berthelier V, Hamilton JB, O'Nuallain B, Wetzel R. 2002. Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry (Mosc) 41: 7391–7399. [DOI] [PubMed] [Google Scholar]
- Choo YS, Johnson GVW, MacDonald M, Detloff PJ, Lesort M. 2004. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet 13: 1407–1420. [DOI] [PubMed] [Google Scholar]
- Chou SY, Weng JY, Lai HL, Liao F, Sun SH, Tu PH, Dickson DW, Chern Y. 2008. Expanded-polyglutamine huntingtin protein suppresses the secretion and production of a chemokine (CCL5/RANTES) by astrocytes. J Neurosci 28: 3277–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti F, Lacroix S, Cisbani G, Vallières N, Saint-Pierre M, St-Amour I, Tolouei R, Skepper JN, Hauser RA, Mantovani D, et al. 2014. Mutant huntingtin is present in neuronal grafts in Huntington disease patients. Ann Neurol 76: 31–42. [DOI] [PubMed] [Google Scholar]
- Colin E, Zala D, Liot G, Rangone H, Borrell-Pagès M, Li XJ, Saudou F, Humbert S. 2008. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J 27: 2124–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornett J, Cao F, Wang CE, Ross CA, Bates GP, Li SH, Li XJ. 2005. Polyglutamine expansion of huntingtin impairs its nuclear export. Nat Genet 37: 198–204. [DOI] [PubMed] [Google Scholar]
- Corrochano S, Renna M, Carter S, Chrobot N, Kent R, Stewart M, Cooper J, Brown SDM, Rubinsztein DC, Acevedo-Arozena A. 2012. α-Synuclein levels modulate Huntington’s disease in mice. Hum Mol Genet 21: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Abounit S, Marzo L, Danckaert A, Chamoun Z, Roux P, Zurzolo C. 2013. Transfer of polyglutamine aggregates in neuronal cells occurs in tunneling nanotubes. J Cell Sci 126: 3678–3685. [DOI] [PubMed] [Google Scholar]
- Coyle JT, Schwarcz R. 1976. Lesion of striatal neurones with kainic acid provides a model for Huntington’s chorea. Nature 263: 244–246. [DOI] [PubMed] [Google Scholar]
- Crotti A, Benner C, Kerman BE, Gosselin D, Lagier-Tourenne C, Zuccato C, Cattaneo E, Gage FH, Cleveland DW, Glass CK. 2014. Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat Neurosci 17: 513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubo E, Shannon KM, Tracy D, Jaglin JA, Bernard BA, Wuu J, Leurgans SE. 2006. Effect of donepezil on motor and cognitive function in Huntington disease. Neurology 67: 1268–1271. [DOI] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. 2006. Transcriptional repression of PGC-1α by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell 127: 59–69. [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Mancini MA, Antalffy B, DeFranco DB, Orr HT, Zoghbi HY. 1998. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat Genet 19: 148–154. [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Reinstein E, Sun Y, Antalffy B, Jiang Y, Ciechanover A, Orr HT, Beaudet AL, Zoghbi HY. 1999. Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 mice. Neuron 24: 879–892. [DOI] [PubMed] [Google Scholar]
- Cummings DM, Milnerwood AJ, Dallérac GM, Waights V, Brown JY, Vatsavayai SC, Hirst MC, Murphy KPSJ, 2006. Aberrant cortical synaptic plasticity and dopaminergic dysfunction in a mouse model of Huntington’s disease. Hum Mol Genet 15: 2856–2868. [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. 1997. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90: 537–548. [DOI] [PubMed] [Google Scholar]
- Dedeoglu A, Kubilus JK, Jeitner TM, Matson SA, Bogdanov M, Kowall NW, Matson WR, Cooper AJL, Ratan RR, Beal MF, et al. 2002. Therapeutic effects of cystamine in a murine model of Huntington’s disease. J Neurosci 22: 8942–8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond CR, Atwal RS, Xia J, Truant R. 2012. Identification of a karyopherin β1/β2 proline-tyrosine nuclear localization signal in huntingtin protein. J Biol Chem 287: 39626–39633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, Young C, Martin E, Vonsattel JP, Carraway R, Reeves SA. 1995. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron 14: 1075–1081. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. 1997. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277: 1990–1993. [DOI] [PubMed] [Google Scholar]
- Dragatsis I, Levine MS, Zeitlin S. 2000. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet 26: 300–306. [DOI] [PubMed] [Google Scholar]
- Drouet V, Perrin V, Hassig R, Dufour N, Auregan G, Alves S, Bonvento G, Brouillet E, Luthi-Carter R, Hantraye P, et al. 2009. Sustained effects of nonallele-specific Huntingtin silencing. Ann Neurol 65: 276–285. [DOI] [PubMed] [Google Scholar]
- Dubinsky R, Gray C. 2006. CYTE-I-HD: Phase I dose finding and tolerability study of cysteamine (Cystagon) in Huntington's disease. Mov Disord 21: 530–533. [DOI] [PubMed] [Google Scholar]
- Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, Mouradian MM, Young AB, Tanese N, Krainc D. 2002. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science 296: 2238–2243. [DOI] [PubMed] [Google Scholar]
- Dunnett SB, Carter RJ, Watts C, Torres EM, Mahal A, Mangiarini L, Bates G, Morton AJ. 1998. Striatal transplantation in a transgenic mouse model of Huntington’s disease. Exp Neurol 154: 31–40. [DOI] [PubMed] [Google Scholar]
- Estrada-Sánchez AM, Montiel T, Segovia J, Massieu L. 2009. Glutamate toxicity in the striatum of the R6/2 Huntington’s disease transgenic mice is age-dependent and correlates with decreased levels of glutamate transporters. Neurobiol Dis 34: 78–86. [DOI] [PubMed] [Google Scholar]
- Fan MMY, Fernandes HB, Zhang LYJ, Hayden MR, Raymond LA. 2007. Altered NMDA receptor trafficking in a yeast artificial chromosome transgenic mouse model of Huntington's disease. J Neurosci 27: 3768–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Nogales M, Cabrera JR, Santos-Galindo M, Hoozemans JJM, Ferrer I, Rozemuller AJM, Hernández F, Avila J, Lucas JJ. 2014. Huntington’s disease is a four-repeat tauopathy with tau nuclear rods. Nat Med 20: 881–885. [DOI] [PubMed] [Google Scholar]
- Ferrante RJ, Andreassen OA, Dedeoglu A, Ferrante KL, Jenkins BG, Hersch SM, Beal MF. 2002. Therapeutic effects of coenzyme Q10 and remacemide in transgenic mouse models of Huntington's disease. J Neurosci 22: 1592–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante RJ, Kubilus JK, Lee J, Ryu H, Beesen A, Zucker B, Smith K, Kowall NW, Ratan RR, Luthi-Carter R, et al. 2003. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington's disease mice. J Neurosci 23: 9418–9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong RA, Narain Y, Rankin J, Wyttenbach A, Rubinsztein DC. 2000. α-Synuclein overexpression promotes aggregation of mutant huntingtin. Biochem J 346: 577–581. [PMC free article] [PubMed] [Google Scholar]
- Fusco FR, Chen Q, Lamoreaux WJ, Figueredo-Cardenas G, Jiao Y, Coffman JA, Surmeier DJ, Honig MG, Carlock LR, Reiner A. 1999. Cellular localization of huntingtin in striatal and cortical neurons in rats: Lack of correlation with neuronal vulnerability in Huntington's disease. J Neurosci 19: 1189–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni J, Hermel E, Young JE, Wellington CL, Hayden MR, Ellerby LM. 2004. Inhibition of calpain cleavage of huntingtin reduces toxicity: Accumulation of calpain/caspase fragments in the nucleus. J Biol Chem 279: 20211–20220. [DOI] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pagès M, Dompierre JP, Rangone H, Cordelières FP, De Mey J, MacDonald ME, Lessmann V, Humbert S, et al. 2004. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 118: 127–138. [DOI] [PubMed] [Google Scholar]
- Georgiou N, Bradshaw JL, Chiu E, Tudor A, O'Gorman L, Phillips JG. 1999. Differential clinical and motor control function in a pair of monozygotic twins with Huntington’s disease. Mov Disord 14: 320–325. [DOI] [PubMed] [Google Scholar]
- Giorgini F, Guidetti P, Nguyen Q, Bennett SC, Muchowski PJ. 2005. A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease. Nat Genet 37: 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin JD, Colombo K, Molina-Calavita M, Keryer G, Zala D, Charrin BC, Dietrich P, Volvert ML, Guillemot F, Dragatsis I, et al. 2010. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron 67: 392–406. [DOI] [PubMed] [Google Scholar]
- Gousset K, Schiff E, Langevin C, Marijanovic Z, Caputo A, Browman DT, Chenouard N, de Chaumont F, Martino A, Enninga J, et al. 2009. Prions hijack tunnelling nanotubes for intercellular spread. Nat Cell Biol 11: 328–336. [DOI] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, et al. 2006. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 125: 1179–1191. [DOI] [PubMed] [Google Scholar]
- Grondin R, Kaytor MD, Ai Y, Nelson PT, Thakker DR, Heisel J, Weatherspoon MR, Blum JL, Burright EN, Zhang Z, et al. 2012. Six-month partial suppression of Huntingtin is well tolerated in the adult rhesus striatum. Brain J Neurol 135: 1197–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidetti P, Bates GP, Graham RK, Hayden MR, Leavitt BR, MacDonald ME, Slow EJ, Wheeler VC, Woodman B, Schwarcz R. 2006. Elevated brain 3-hydroxykynurenine and quinolinate levels in Huntington disease mice. Neurobiol Dis 23: 190–197. [DOI] [PubMed] [Google Scholar]
- Gunawardena S, Her LS, Brusch RG, Laymon RA, Niesman IR, Gordesky-Gold B, Sintasath L, Bonini NM, Goldstein LSB. 2003. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron 40: 25–40. [DOI] [PubMed] [Google Scholar]
- Guo JL, Lee VMY. 2014. Cell-to-cell transmission of pathogenic proteins in neurodegenerative diseases. Nat Med 20: 130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Disatnik MH, Monbureau M, Shamloo M, Mochly-Rosen D, Qi X. 2013. Inhibition of mitochondrial fragmentation diminishes Huntington's disease-associated neurodegeneration. J Clin Invest 123: 5371–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, Jones R, Rye D, Ferrante RJ, Hersch SM, Li XJ. 1999. Nuclear and neuropil aggregates in Huntington's disease: Relationship to neuropathology. J Neurosci 19: 2522–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackam AS, Singaraja R, Wellington CL, Metzler M, McCutcheon K, Zhang T, Kalchman M, Hayden MR. 1998. The influence of huntingtin protein size on nuclear localization and cellular toxicity. J Cell Biol 141: 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa V, Saha T, Usdin K. 2003. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res 31: 6243–6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. 2011. Molecular chaperones in protein folding and proteostasis. Nature 475: 324–332. [DOI] [PubMed] [Google Scholar]
- Hermel E, Gafni J, Propp SS, Leavitt BR, Wellington CL, Young JE, Hackam AS, Logvinova AV, Peel AL, Chen SF, et al. 2004. Specific caspase interactions and amplification are involved in selective neuronal vulnerability in Huntington’s disease. Cell Death Differ 11: 424–438. [DOI] [PubMed] [Google Scholar]
- Herrera F, Outeiro TF. 2012. α-Synuclein modifies huntingtin aggregation in living cells. FEBS Lett 586: 7–12. [DOI] [PubMed] [Google Scholar]
- Hochfeld WE, Lee S, Rubinsztein DC. 2013. Therapeutic induction of autophagy to modulate neurodegenerative disease progression. Acta Pharmacol Sin 34: 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, Sathasivam K, Ghazi-Noori S, Mahal A, Lowden PAS, et al. 2003. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci 100: 2041–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffner G, Kahlem P, Djian P. 2002. Perinuclear localization of huntingtin as a consequence of its binding to microtubules through an interaction with β-tubulin: Relevance to Huntington's disease. J Cell Sci 115: 941–948. [DOI] [PubMed] [Google Scholar]
- Holl AK, Wilkinson L, Painold A, Holl EM, Bonelli RM. 2010. Combating depression in Huntington's disease: Effective antidepressive treatment with venlafaxine XR. Int Clin Psychopharmacol 25: 46–50. [DOI] [PubMed] [Google Scholar]
- Holmberg CI, Staniszewski KE, Mensah KN, Matouschek A, Morimoto RI. 2004. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J 23: 4307–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Faber PW, Persichetti F, Mittal V, Vonsattel JP, MacDonald ME, Gusella JF. 1998. Amyloid formation by mutant huntingtin: Threshold, progressivity and recruitment of normal polyglutamine proteins. Somat Cell Mol Genet 24: 217–233. [DOI] [PubMed] [Google Scholar]
- Huntington Study Group, 2001. A randomized, placebo-controlled trial of coenzyme Q10 and remacemide in Huntington's disease. Neurology 57: 397–404. [DOI] [PubMed] [Google Scholar]
- Huntington’s Disease Collaborative Research Group. 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72: 971–983. [DOI] [PubMed] [Google Scholar]
- Jahreiss L, Menzies FM, Rubinsztein DC. 2008. The itinerary of autophagosomes: From peripheral formation to kiss-and-run fusion with lysosomes. Traffic 9: 574–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana NR, Tanaka M, Wang Gh, Nukina N. 2000. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: Their role in suppression of aggregation and cellular toxicity. Hum Mol Genet 9: 2009–2018. [DOI] [PubMed] [Google Scholar]
- Jana NR, Zemskov EA, Wang Gh, Nukina N. 2001. Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum Mol Genet 10: 1049–1059. [DOI] [PubMed] [Google Scholar]
- Jiang M, Peng Q, Liu X, Jin J, Hou Z, Zhang J, Mori S, Ross CA, Ye K, Duan W. 2013. Small-molecule TrkB receptor agonists improve motor function and extend survival in a mouse model of Huntington’s disease. Hum Mol Genet 22: 2462–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Sanchez M, Lam W, Hannus M, Sönnichsen B, Imarisio S, Fleming A, Tarditi A, Menzies F, Ed Dami T, Xu C, et al. 2015. siRNA screen identifies QPCT as a druggable target for Huntington’s disease. Nat Chem Biol 11: 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. 2008. A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiol Dis 29: 438–445. [DOI] [PubMed] [Google Scholar]
- Juenemann K, Schipper-Krom S, Wiemhoefer A, Kloss A, Sanz Sanz A, Reits EAJ. 2013. Expanded polyglutamine-containing N-terminal huntingtin fragments are entirely degraded by mammalian proteasomes. J Biol Chem 288: 27068–27084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpuj MV, Becher MW, Springer JE, Chabas D, Youssef S, Pedotti R, Mitchell D, Steinman L. 2002. Prolonged survival and decreased abnormal movements in transgenic model of Huntington disease, with administration of the transglutaminase inhibitor cystamine. Nat Med 8: 143–149. [DOI] [PubMed] [Google Scholar]
- Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. 1999. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc Natl Acad Sci 96: 11404–11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel KB, Kim M, Sapp E, McIntyre C, Castaño JG, Aronin N, DiFiglia M. 2000. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci 20: 7268–7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel KB, Meloni AR, Yi Y, Kim YJ, Doyle E, Cuiffo BG, Sapp E, Wang Y, Qin ZH, Chen JD, et al. 2002. Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal binding protein, and represses transcription. J Biol Chem 277: 7466–7476. [DOI] [PubMed] [Google Scholar]
- Kieburtz K, McDermott MP, Voss TS, Corey-Bloom J, Deuel LM, Dorsey ER, Factor S, Geschwind MD, Hodgeman K, Kayson E, et al. 2010. A randomized, placebo-controlled trial of latrepirdine in Huntington disease. Arch Neurol 67: 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Lee HS, LaForet G, McIntyre C, Martin EJ, Chang P, Kim TW, Williams M, Reddy PH, Tagle D, et al. 1999. Mutant huntingtin expression in clonal striatal cells: Dissociation of inclusion formation and neuronal survival by caspase inhibition. J Neurosci 19: 964–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Yi Y, Sapp E, Wang Y, Cuiffo B, Kegel KB, Qin ZH, Aronin N, DiFiglia M. 2001. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington’s disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc Natl Acad Sci 98: 12784–12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Roh JK, Yoon BW, Kang L, Kim YJ, Aronin N, DiFiglia M. 2003. Huntingtin is degraded to small fragments by calpain after ischemic injury. Exp Neurol 183: 109–115. [DOI] [PubMed] [Google Scholar]
- Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, Clark HB, Zoghbi HY, Orr HT. 1998. Ataxin-1 nuclear localization and aggregation: Role in polyglutamine-induced disease in SCA1 transgenic mice. Cell 95: 41–53. [DOI] [PubMed] [Google Scholar]
- Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, Artates JW, Weiss A, Cheng SH, Shihabuddin LS, et al. 2012. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron 74: 1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer B, Clark CM, Almqvist EW, Raymond LA, Graf P, Jacova C, Mezei M, Hardy MA, Snow B, Martin W, et al. 1999. Influence of lamotrigine on progression of early Huntington disease: A randomized clinical trial. Neurology 53: 1000–1011. [DOI] [PubMed] [Google Scholar]
- Krobitsch S, Lindquist S. 2000. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci 97: 1589–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Fiszer A, Mykowska A, Sobczak K, de Mezer M, Krzyzosiak WJ. 2007. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol Cell 25: 575–586. [DOI] [PubMed] [Google Scholar]
- Kwan W, Magnusson A, Chou A, Adame A, Carson MJ, Kohsaka S, Masliah E, Möller T, Ransohoff R, Tabrizi SJ, et al. 2012a. Bone marrow transplantation confers modest benefits in mouse models of Huntington’s disease. J Neurosci 32: 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan W, Träger U, Davalos D, Chou A, Bouchard J, Andre R, Miller A, Weiss A, Giorgini F, Cheah C, et al. 2012b. Mutant huntingtin impairs immune cell migration in Huntington disease. J Clin Invest 122: 4737–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforet GA, Sapp E, Chase K, McIntyre C, Boyce FM, Campbell M, Cadigan BA, Warzecki L, Tagle DA, Reddy PH, et al. 2001. Changes in cortical and striatal neurons predict behavioral and electrophysiological abnormalities in a transgenic murine model of Huntington's disease. J Neurosci 21: 9112–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie P, Snapp EL. 2010. Formation and toxicity of soluble polyglutamine oligomers in living cells. PLoS ONE 5: e15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie P, Snapp EL. 2013. Detecting soluble polyQ oligomers and investigating their impact on living cells using split-GFP. Methods Mol Biol 1017: 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WCM, Yoshihara M, Littleton JT. 2004. Cytoplasmic aggregates trap polyglutamine-containing proteins and block axonal transport in a Drosophila model of Huntington's disease. Proc Natl Acad Sci 101: 3224–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Desplats P, Sigurdson C, Tsigelny I, Masliah E. 2010. Cell-to-cell transmission of non-prion protein aggregates. Nat Rev Neurol 6: 702–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CYD, Cantle JP, Yang XW. 2013. Genetic manipulations of mutant huntingtin in mice: New insights into Huntington’s disease pathogenesis. FEBS J 280: 4382–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legleiter J, Mitchell E, Lotz GP, Sapp E, Ng C, DiFiglia M, Thompson LM, Muchowski PJ, 2010. Mutant huntingtin fragments form oligomers in a polyglutamine length-dependent manner in vitro and in vivo. J Biol Chem 285: 14777–14790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li SH, Yu ZX, Shelbourne P, Li XJ. 2001. Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington’s disease mice. J Neurosci 21: 8473–8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Serpell LC, Carter WJ, Rubinsztein DC, Huntington JA. 2006. Expression and characterization of full-length human huntingtin, an elongated HEAT repeat protein. J Biol Chem 281: 15916–15922. [DOI] [PubMed] [Google Scholar]
- Li LB, Yu Z, Teng X, Bonini NM. 2008. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature 453: 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liévens JC, Woodman B, Mahal A, Spasic-Boscovic O, Samuel D, Kerkerian-Le Goff L, Bates GP. 2001. Impaired glutamate uptake in the R6 Huntington’s disease transgenic mice. Neurobiol Dis 8: 807–821. [DOI] [PubMed] [Google Scholar]
- Lin CH, Tallaksen-Greene S, Chien WM, Cearley JA, Jackson WS, Crouse AB, Ren S, Li XJ, Albin RL, Detloff PJ. 2001. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum Mol Genet 10: 137–144. [DOI] [PubMed] [Google Scholar]
- Liot G, Zala D, Pla P, Mottet G, Piel M, Saudou F. 2013. Mutant Huntingtin alters retrograde transport of TrkB receptors in striatal dendrites. J Neurosci 33: 6298–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KY, Shyu YC, Barbaro BA, Lin YT, Chern Y, Thompson LM, Shen CKJ, Marsh JL. 2015. Disruption of the nuclear membrane by perinuclear inclusions of mutant huntingtin causes cell-cycle re-entry and striatal cell death in mouse and cell models of Huntington’s disease. Hum Mol Genet 24: 1602–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Sardo V, Zuccato C, Gaudenzi G, Vitali B, Ramos C, Tartari M, Myre MA, Walker JA, Pistocchi A, Conti L, et al. 2012. An evolutionary recent neuroepithelial cell adhesion function of huntingtin implicates ADAM10-Ncadherin. Nat Neurosci 15: 713–721. [DOI] [PubMed] [Google Scholar]
- Lunkes A, Mandel JL. 1998. A cellular model that recapitulates major pathogenic steps of Huntington’s disease. Hum Mol Genet 7: 1355–1361. [DOI] [PubMed] [Google Scholar]
- Lunkes A, Lindenberg KS, Ben-Haïem L, Weber C, Devys D, Landwehrmeyer GB, Mandel JL, Trottier Y. 2002. Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol Cell 10: 259–269. [DOI] [PubMed] [Google Scholar]
- Luo S, Vacher C, Davies JE, Rubinsztein DC. 2005. Cdk5 phosphorylation of huntingtin reduces its cleavage by caspases: Implications for mutant huntingtin toxicity. J Cell Biol 169: 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthi-Carter R, Strand A, Peters NL, Solano SM, Hollingsworth ZR, Menon AS, Frey AS, Spektor BS, Penney EB, Schilling G, et al. 2000. Decreased expression of striatal signaling genes in a mouse model of Huntington’s disease. Hum Mol Genet 9: 1259–1271. [DOI] [PubMed] [Google Scholar]
- Mandal M, Wei J, Zhong P, Cheng J, Duffney LJ, Liu W, Yuen EY, Twelvetrees AE, Li S, Li XJ, et al. 2011. Impaired α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor trafficking and function by mutant huntingtin. J Biol Chem 286: 33719–33728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco S, Giralt A, Petrovic MM, Pouladi MA, Martínez-Turrillas R, Martínez-Hernández J, Kaltenbach LS, Torres-Peraza J, Graham RK, Watanabe M, et al. 2013. Suppressing aberrant GluN3A expression rescues synaptic and behavioral impairments in Huntington’s disease models. Nat Med 19: 1030–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcora E, Kennedy MB. 2010. The Huntington’s disease mutation impairs Huntingtin’s role in the transport of NF-κB from the synapse to the nucleus. Hum Mol Genet 19: 4373–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí E, Pantano L, Bañez-Coronel M, Llorens F, Miñones-Moyano E, Porta S, Sumoy L, Ferrer I, Estivill X. 2010. A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res 38: 7219–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D, Hackam A, Wieczorek A, Ellerby L, Wellington C, McCutcheon K, Singaraja R, Kazemi-Esfarjani P, Devon R, Kim SU, et al. 1998. Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat Genet 18: 150–154. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, et al. 2010. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci 13: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard CJ, Böttcher C, Ortega Z, Smith R, Florea BI, Díaz-Hernández M, Brundin P, Overkleeft HS, Li JY, Lucas JJ, et al. 2009. Accumulation of ubiquitin conjugates in a polyglutamine disease model occurs without global ubiquitin/proteasome system impairment. Proc Natl Acad Sci 106: 13986–13991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Pitzer MR, Boudreau RL, Dufour B, Hobbs T, Ojeda SR, Davidson BL. 2011. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington’s disease. Mol Ther 19: 2152–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCampbell A, Taye AA, Whitty L, Penney E, Steffan JS, Fischbeck KH. 2001. Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc Natl Acad Sci 98: 15179–15184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland KN, Huizenga MN, Darnell SB, Sangrey GR, Berezovska O, Cha JHJ, Outeiro TF, Sadri-Vakili G. 2014. MeCP2: A novel Huntingtin interactor. Hum Mol Genet 23: 1036–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeer EG, McGeer PL. 1976. Duplication of biochemical changes of Huntington’s chorea by intrastriatal injections of glutamic and kainic acids. Nature 263: 517–519. [DOI] [PubMed] [Google Scholar]
- McGowan DP, van Roon-Mom W, Holloway H, Bates GP, Mangiarini L, Cooper GJ, Faull RL, Snell RG. 2000. Amyloid-like inclusions in Huntington’s disease. Neuroscience 100: 677–680. [DOI] [PubMed] [Google Scholar]
- McGuire JR, Rong J, Li SH, Li XJ. 2006. Interaction of Huntingtin-associated protein-1 with kinesin light chain: Implications in intracellular trafficking in neurons. J Biol Chem 281: 3552–3559. [DOI] [PubMed] [Google Scholar]
- McKinstry SU, Karadeniz YB, Worthington AK, Hayrapetyan VY, Ozlu MI, Serafin-Molina K, Risher WC, Ustunkaya T, Dragatsis I, Zeitlin S, et al. 2014. Huntingtin is required for normal excitatory synapse development in cortical and striatal circuits. J Neurosci 34: 9455–9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menalled LB, Chesselet MF. 2002. Mouse models of Huntington’s disease. Trends Pharmacol Sci 23: 32–39. [DOI] [PubMed] [Google Scholar]
- Menalled LB, Patry M, Ragland N, Lowden PAS, Goodman J, Minnich J, Zahasky B, Park L, Leeds J, Howland D, et al. 2010. Comprehensive behavioral testing in the R6/2 mouse model of Huntington’s disease shows no benefit from CoQ10 or minocycline. PLoS ONE 5: e9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende-Mueller LM, Toneff T, Hwang SR, Chesselet MF, Hook VY. 2001. Tissue-specific proteolysis of Huntingtin (htt) in human brain: Evidence of enhanced levels of N- and C-terminal htt fragments in Huntington’s disease striatum. J Neurosci 21: 1830–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielcarek M, Landles C, Weiss A, Bradaia A, Seredenina T, Inuabasi L, Osborne GF, Wadel K, Touller C, Butler R, et al. 2013. HDAC4 reduction: A novel therapeutic strategy to target cytoplasmic huntingtin and ameliorate neurodegeneration. PLoS Biol 11: e1001717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milnerwood AJ, Cummings DM, Dallérac GM, Brown JY, Vatsavayai SC, Hirst MC, Rezaie P, Murphy KPSJ. 2006. Early development of aberrant synaptic plasticity in a mouse model of Huntington’s disease. Hum Mol Genet 15: 1690–1703. [DOI] [PubMed] [Google Scholar]
- Milnerwood AJ, Gladding CM, Pouladi MA, Kaufman AM, Hines RM, Boyd JD, Ko RWY, Vasuta OC, Graham RK, Hayden MR, et al. 2010. Early increase in extrasynaptic NMDA receptor signaling and expression contributes to phenotype onset in Huntington's disease mice. Neuron 65: 178–190. [DOI] [PubMed] [Google Scholar]
- Mitra S, Tsvetkov AS, Finkbeiner S. 2009. Single neuron ubiquitin-proteasome dynamics accompanying inclusion body formation in huntington disease. J Biol Chem 284: 4398–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfini GA, You YM, Pollema SL, Kaminska A, Liu K, Yoshioka K, Björkblom B, Coffey ET, Bagnato C, Han D, et al. 2009. Pathogenic huntingtin inhibits fast axonal transport by activating JNK3 and phosphorylating kinesin. Nat Neurosci 12: 864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai H, Isagawa T, Goyama E, Tanaka S, Bence NF, Tamura A, Ono Y, Kopito RR. 2005. Formation of morphologically similar globular aggregates from diverse aggregation-prone proteins in mammalian cells. Proc Natl Acad Sci 102: 10887–10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RH, Vonsattel JP, Paskevich PA, Kiely DK, Stevens TJ, Cupples LA, Richardson EP, Bird ED. 1991. Decreased neuronal and increased oligodendroglial densities in Huntington’s disease caudate nucleus. J Neuropathol Exp Neurol 50: 729–742. [DOI] [PubMed] [Google Scholar]
- Nasir J, Floresco SB, O'Kusky JR, Diewert VM, Richman JM, Zeisler J, Borowski A, Marth JD, Phillips AG, Hayden MR. 1995. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell 81: 811–823. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Hirano T. 2000. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res 10: 1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CW, Yildirim F, Yap YS, Dalin S, Matthews BJ, Velez PJ, Labadorf A, Housman DE, Fraenkel E. 2013. Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc Natl Acad Sci 110: 2354–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen GD, Gokhan S, Molero AE, Mehler MF. 2013. Selective roles of normal and mutant huntingtin in neural induction and early neurogenesis. PLoS ONE 8: e64368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucifora FC, Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson VL, et al. 2001. Interference by huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science 291: 2423–2428. [DOI] [PubMed] [Google Scholar]
- Ochaba J, Lukacsovich T, Csikos G, Zheng S, Margulis J, Salazar L, Mao K, Lau AL, Yeung SY, Humbert S, et al. 2014. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc Natl Acad Sci 111: 16889–16894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Pouladi MA, Talantova M, Yao D, Xia P, Ehrnhoefer DE, Zaidi R, Clemente A, Kaul M, Graham RK, et al. 2009. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat Med 15: 1407–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Kusky JR, Nasir J, Cicchetti F, Parent A, Hayden MR. 1999. Neuronal degeneration in the basal ganglia and loss of pallido-subthalamic synapses in mice with targeted disruption of the Huntington's disease gene. Brain Res 818: 468–479. [DOI] [PubMed] [Google Scholar]
- Orr AL, Li S, Wang CE, Li H, Wang J, Rong J, Xu X, Mastroberardino PG, Greenamyre JT, Li XJ. 2008. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci 28: 2783–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega Z, Díaz-Hernández M, Maynard CJ, Hernández F, Dantuma NP, Lucas JJ. 2010. Acute polyglutamine expression in inducible mouse model unravels ubiquitin/proteasome system impairment and permanent recovery attributable to aggregate formation. J Neurosci 30: 3675–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. 2008. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci 28: 14341–14346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazuelos J, Aguado T, Pazos MR, Julien B, Carrasco C, Resel E, Sagredo O, Benito C, Romero J, Azcoitia I, et al. 2009. Microglial CB2 cannabinoid receptors are neuroprotective in Huntington's disease excitotoxicity. Brain J Neurol 132: 3152–3164. [DOI] [PubMed] [Google Scholar]
- Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. 2002. Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat Neurosci 5: 731–736. [DOI] [PubMed] [Google Scholar]
- Paul BD, Sbodio JI, Xu R, Vandiver MS, Cha JY, Snowman AM, Snyder SH. 2014. Cystathionine γ-lyase deficiency mediates neurodegeneration in Huntington's disease. Nature 509: 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Pearce MMP, Kopito RR. 2017. Prion-like characteristics of polyglutamine-containing proteins. Cold Spring Harb Perspect Med 10.1101/cshperspect.a024257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecho-Vrieseling E, Rieker C, Fuchs S, Bleckmann D, Esposito MS, Botta P, Goldstein C, Bernhard M, Galimberti I, Müller M, et al. 2014. Transneuronal propagation of mutant huntingtin contributes to non-cell autonomous pathology in neurons. Nat Neurosci 17: 1064–1072. [DOI] [PubMed] [Google Scholar]
- Perluigi M, Poon HF, Maragos W, Pierce WM, Klein JB, Calabrese V, Cini C, De Marco C, Butterfield DA. 2005. Proteomic analysis of protein expression and oxidative modification in r6/2 transgenic mice: A model of Huntington disease. Mol Cell Proteomics 4: 1849–1861. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Windle AH. 2001. Cause of neural death in neurodegenerative diseases attributable to expansion of glutamine repeats. Nature 412: 143–144. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Johnson T, Suzuki M, Finch JT. 1994. Glutamine repeats as polar zippers: Their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci 91: 5355–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips W, Shannon KM, Barker RA. 2008. The current clinical management of Huntington’s disease. Mov Disord 23: 1491–1504. [DOI] [PubMed] [Google Scholar]
- Plotkin JL, Day M, Peterson JD, Xie Z, Kress GJ, Rafalovich I, Kondapalli J, Gertler TS, Flajolet M, Greengard P, et al. 2014. Impaired TrkB receptor signaling underlies corticostriatal dysfunction in Huntington's disease. Neuron 83: 178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier MA, Li H, Macosko J, Cai S, Amzel M, Ross CA. 2002. Huntingtin spheroids and protofibrils as precursors in polyglutamine fibrilization. J Biol Chem 277: 41032–41037. [DOI] [PubMed] [Google Scholar]
- Ratovitski T, Chighladze E, Arbez N, Boronina T, Herbrich S, Cole RN, Ross CA. 2012. Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell Cycle 11: 2006–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. 2002. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet 11: 1107–1117. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O'Kane CJ, Brown SDM, Rubinsztein DC. 2005. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet 37: 771–776. [DOI] [PubMed] [Google Scholar]
- Reddy PH, Williams M, Charles V, Garrett L, Pike-Buchanan L, Whetsell WO, Miller G, Tagle DA. 1998. Behavioural abnormalities and selective neuronal loss in HD transgenic mice expressing mutated full-length HD cDNA. Nat Genet 20: 198–202. [DOI] [PubMed] [Google Scholar]
- Reiner A, Albin RL, Anderson KD, D'Amato CJ, Penney JB, Young AB. 1988. Differential loss of striatal projection neurons in Huntington disease. Proc Natl Acad Sci 85: 5733–5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren PH, Lauckner JE, Kachirskaia I, Heuser JE, Melki R, Kopito RR. 2009. Cytoplasmic penetration and persistent infection of mammalian cells by polyglutamine aggregates. Nat Cell Biol 11: 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas HD, Liu AK, Hersch S, Glessner M, Ferrante RJ, Salat DH, van der Kouwe A, Jenkins BG, Dale AM, Fischl B. 2002. Regional and progressive thinning of the cortical ribbon in Huntington's disease. Neurology 58: 695–701. [DOI] [PubMed] [Google Scholar]
- Rosas HD, Doros G, Gevorkian S, Malarick K, Reuter M, Coutu JP, Triggs TD, Wilkens PJ, Matson W, Salat DH, et al. 2014. PRECREST: A phase II prevention and biomarker trial of creatine in at-risk Huntington disease. Neurology 82: 850–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt A. 2007. Neuropsychiatry of Huntington's disease. Dialogues Clin Neurosci 9: 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt A, Brinkman RR, Liang KY, Almqvist EW, Margolis RL, Huang CY, Sherr M, Franz ML, Abbott MH, Hayden MR, et al. 2001. Familial influence on age of onset among siblings with Huntington disease. Am J Med Genet 105: 399–403. [PubMed] [Google Scholar]
- Rubinsztein DC, Leggo J, Coles R, Almqvist E, Biancalana V, Cassiman JJ, Chotai K, Connarty M, Crauford D, Curtis A, et al. 1996. Phenotypic characterization of individuals with 30-40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36-39 repeats. Am J Hum Genet 59: 16–22. [PMC free article] [PubMed] [Google Scholar]
- Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. 2004. Nanotubular highways for intercellular organelle transport. Science 303: 1007–1010. [DOI] [PubMed] [Google Scholar]
- Sapp E, Kegel KB, Aronin N, Hashikawa T, Uchiyama Y, Tohyama K, Bhide PG, Vonsattel JP, DiFiglia M. 2001. Early and progressive accumulation of reactive microglia in the Huntington disease brain. J Neuropathol Exp Neurol 60: 161–172. [DOI] [PubMed] [Google Scholar]
- Sathasivam K, Neueder A, Gipson TA, Landles C, Benjamin AC, Bondulich MK, Smith DL, Faull RLM, Roos RAC, Howland D, et al. 2013. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci 110: 2366–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou F, Finkbeiner S, Devys D, Greenberg ME. 1998. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell 95: 55–66. [DOI] [PubMed] [Google Scholar]
- Savas JN, Makusky A, Ottosen S, Baillat D, Then F, Krainc D, Shiekhattar R, Markey SP, Tanese N. 2008. Huntington’s disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc Natl Acad Sci 105: 10820–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Ma B, Deinhardt K, Culver BP, Restituito S, Wu L, Belasco JG, Chao MV, Tanese N. 2010. A role for huntington disease protein in dendritic RNA granules. J Biol Chem 285: 13142–13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa A, Wiegand GW, Cooper J, Margolis RL, Sharp AH, Lawler JF, Greenamyre JT, Snyder SH, Ross CA. 1999. Increased apoptosis of Huntington disease lymphoblasts associated with repeat length-dependent mitochondrial depolarization. Nat Med 5: 1194–1198. [DOI] [PubMed] [Google Scholar]
- Schilling B, Gafni J, Torcassi C, Cong X, Row RH, LaFevre-Bernt MA, Cusack MP, Ratovitski T, Hirschhorn R, Ross CA, et al. 2006. Huntingtin phosphorylation sites mapped by mass spectrometry. Modulation of cleavage and toxicity. J Biol Chem 281: 23686–23697. [DOI] [PubMed] [Google Scholar]
- Schipper-Krom S, Juenemann K, Jansen AH, Wiemhoefer A, van den Nieuwendijk R, Smith DL, Hink MA, Bates GP, Overkleeft H, Ovaa H, et al. 2014a. Dynamic recruitment of active proteasomes into polyglutamine initiated inclusion bodies. FEBS Lett 588: 151–159. [DOI] [PubMed] [Google Scholar]
- Schipper-Krom S, Juenemann K, Jansen AH, Wiemhoefer A, van den Nieuwendijk R, Smith DL, Hink MA, Bates GP, Overkleeft H, Ovaa H, et al. 2014b. Dynamic recruitment of active proteasomes into polyglutamine initiated inclusion bodies. FEBS Lett 588: 151–159. [DOI] [PubMed] [Google Scholar]
- Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. 2006. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem 281: 14474–14485. [DOI] [PubMed] [Google Scholar]
- Shin JY, Fang ZH, Yu ZX, Wang CE, Li SH, Li XJ. 2005. Expression of mutant huntingtin in glial cells contributes to neuronal excitotoxicity. J Cell Biol 171: 1001–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirendeb UP, Calkins MJ, Manczak M, Anekonda V, Dufour B, McBride JL, Mao P, Reddy PH. 2012. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum Mol Genet 21: 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DA, Belichenko NP, Yang T, Condon C, Monbureau M, Shamloo M, Jing D, Massa SM, Longo FM. 2013. A small molecule TrkB ligand reduces motor impairment and neuropathology in R6/2 and BACHD mouse models of Huntington’s disease. J Neurosci 33: 18712–18727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipione S, Rigamonti D, Valenza M, Zuccato C, Conti L, Pritchard J, Kooperberg C, Olson JM, Cattaneo E. 2002. Early transcriptional profiles in huntingtin-inducible striatal cells by microarray analyses. Hum Mol Genet 11: 1953–1965. [DOI] [PubMed] [Google Scholar]
- Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, Poquiz P, Tjong J, Pouladi MA, Hayden MR, et al. 2011. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med 17: 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorolla MA, Reverter-Branchat G, Tamarit J, Ferrer I, Ros J, Cabiscol E. 2008. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic Biol Med 45: 667–678. [DOI] [PubMed] [Google Scholar]
- Stanek LM, Sardi SP, Mastis B, Richards AR, Treleaven CM, Taksir T, Misra K, Cheng SH, Shihabuddin LS. 2014. Silencing mutant huntingtin by adeno-associated virus-mediated RNA interference ameliorates disease manifestations in the YAC128 mouse model of Huntington’s disease. Hum Gene Ther 25: 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, Wanker EE, Bates GP, Housman DE, Thompson LM. 2000. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci 97: 6763–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, et al. 2001. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413: 739–743. [DOI] [PubMed] [Google Scholar]
- Stoy N, Mackay GM, Forrest CM, Christofides J, Egerton M, Stone TW, Darlington LG. 2005. Tryptophan metabolism and oxidative stress in patients with Huntington’s disease. J Neurochem 93: 611–623. [DOI] [PubMed] [Google Scholar]
- Sugars KL, Brown R, Cook LJ, Swartz J, Rubinsztein DC. 2004. Decreased cAMP response element-mediated transcription: An early event in exon 1 and full-length cell models of Huntington’s disease that contributes to polyglutamine pathogenesis. J Biol Chem 279: 4988–4999. [DOI] [PubMed] [Google Scholar]
- Sun Y, Savanenin A, Reddy PH, Liu YF. 2001. Polyglutamine-expanded huntingtin promotes sensitization of N-methyl-D-aspartate receptors via post-synaptic density 95. J Biol Chem 276: 24713–24718. [DOI] [PubMed] [Google Scholar]
- Szebenyi G, Morfini GA, Babcock A, Gould M, Selkoe K, Stenoien DL, Young M, Faber PW, MacDonald ME, McPhaul MJ, et al. 2003. Neuropathogenic forms of huntingtin and androgen receptor inhibit fast axonal transport. Neuron 40: 41–52. [DOI] [PubMed] [Google Scholar]
- Tabrizi SJ, Scahill RI, Owen G, Durr A, Leavitt BR, Roos RA, Borowsky B, Landwehrmeyer B, Frost C, Johnson H, et al. 2013. Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington's disease in the TRACK-HD study: Analysis of 36-month observational data. Lancet Neurol 12: 637–649. [DOI] [PubMed] [Google Scholar]
- Tai YF, Pavese N, Gerhard A, Tabrizi SJ, Barker RA, Brooks DJ, Piccini P. 2007. Microglial activation in presymptomatic Huntington’s disease gene carriers. Brain J Neurol 130: 1759–1766. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Kikuchi S, Katada S, Nagai Y, Nishizawa M, Onodera O. 2008. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum Mol Genet 17: 345–356. [DOI] [PubMed] [Google Scholar]
- Takano H, Gusella JF. 2002. The predominantly HEAT-like motif structure of huntingtin and its association and coincident nuclear entry with dorsal, an NF-kB/Rel/dorsal family transcription factor. BMC Neurosci 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomás-Zapico C, Díez-Zaera M, Ferrer I, Gómez-Ramos P, Morán MA, Miras-Portugal MT, Díaz-Hernández M, Lucas JJ. 2012. α-Synuclein accumulates in huntingtin inclusions but forms independent filaments and its deficiency attenuates early phenotype in a mouse model of Huntington's disease. Hum Mol Genet 21: 495–510. [DOI] [PubMed] [Google Scholar]
- Toneff T, Mende-Mueller L, Wu Y, Hwang SR, Bundey R, Thompson LM, Chesselet MF, Hook V. 2002. Comparison of huntingtin proteolytic fragments in human lymphoblast cell lines and human brain. J Neurochem 82: 84–92. [DOI] [PubMed] [Google Scholar]
- Tong X, Ao Y, Faas GC, Nwaobi SE, Xu J, Haustein MD, Anderson MA, Mody I, Olsen ML, Sofroniew MV, et al. 2014. Astrocyte Kir4.1 ion channel deficits contribute to neuronal dysfunction in Huntington’s disease model mice. Nat Neurosci 17: 694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Träger U, Andre R, Lahiri N, Magnusson-Lind A, Weiss A, Grueninger S, McKinnon C, Sirinathsinghji E, Kahlon S, Pfister EL, et al. 2014. HTT-lowering reverses Huntington’s disease immune dysfunction caused by NFκB pathway dysregulation. Brain J Neurol 137: 819–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier Y, Biancalana V, Mandel JL. 1994. Instability of CAG repeats in Huntington’s disease: Relation to parental transmission and age of onset. J Med Genet 31: 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trushina E, Dyer RB, Badger JD, Ure D, Eide L, Tran DD, Vrieze BT, Legendre-Guillemin V, McPherson PS, Mandavilli BS, et al. 2004. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol 24: 8195–8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twelvetrees AE, Yuen EY, Arancibia-Carcamo IL, MacAskill AF, Rostaing P, Lumb MJ, Humbert S, Triller A, Saudou F, Yan Z, et al. 2010. Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron 65: 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tydlacka S, Wang CE, Wang X, Li S, Li XJ. 2008. Differential activities of the ubiquitin-proteasome system in neurons versus glia may account for the preferential accumulation of misfolded proteins in neurons. J Neurosci 28: 13285–13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood BR, Imarisio S, Fleming A, Rose C, Krishna G, Heard P, Quick M, Korolchuk VI, Renna M, Sarkar S, et al. 2010. Antioxidants can inhibit basal autophagy and enhance neurodegeneration in models of polyglutamine disease. Hum Mol Genet 19: 3413–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin MT, Shelbourne PF, Myers RM, Madison DV. 1999. Impaired synaptic plasticity in mice carrying the Huntington's disease mutation. Hum Mol Genet 8: 839–846. [DOI] [PubMed] [Google Scholar]
- Vashishtha M, Ng CW, Yildirim F, Gipson TA, Kratter IH, Bodai L, Song W, Lau A, Labadorf A, Vogel-Ciernia A, et al. 2013. Targeting H3K4 trimethylation in Huntington disease. Proc Natl Acad Sci 110: E3027–E3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL. 2004. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell 14: 95–104. [DOI] [PubMed] [Google Scholar]
- Verhoef LGGC, Lindsten K, Masucci MG, Dantuma NP. 2002. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum Mol Genet 11: 2689–2700. [DOI] [PubMed] [Google Scholar]
- Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE. 2001. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell 12: 1393–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lim PJ, Karbowski M, Monteiro MJ. 2009. Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum Mol Genet 18: 737–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lin F, Wang J, Wu J, Han R, Zhu L, Difiglia M, Qin Z. 2012. Expression of mutant N-terminal huntingtin fragment (htt552-100Q) in astrocytes suppresses the secretion of BDNF. Brain Res 1449: 69–82. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. 1999. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet 23: 425–428. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Ellerby LM, Hackam AS, Margolis RL, Trifiro MA, Singaraja R, McCutcheon K, Salvesen GS, Propp SS, Bromm M, et al. 1998. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J Biol Chem 273: 9158–9167. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Ellerby LM, Gutekunst CA, Rogers D, Warby S, Graham RK, Loubser O, van Raamsdonk J, Singaraja R, Yang YZ, et al. 2002. Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington’s disease. J Neurosci 22: 7862–7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild E, Magnusson A, Lahiri N, Krus U, Orth M, Tabrizi SJ, Björkqvist M. 2011. Abnormal peripheral chemokine profile in Huntington's disease. PLoS Curr 3: RRN1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, Lichtenberg M, Menzies FM, Ravikumar B, Imarisio S, et al. 2010. α-Synuclein impairs macroautophagy: Implications for Parkinson’s disease. J Cell Biol 190: 1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, Holzbaur ELF. 2014. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci 34: 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Lee DH, Taylor J, Vandelft M, Truant R. 2003. Huntingtin contains a highly conserved nuclear export signal. Hum Mol Genet 12: 1393–1403. [DOI] [PubMed] [Google Scholar]
- Yano H, Baranov SV, Baranova OV, Kim J, Pan Y, Yablonska S, Carlisle DL, Ferrante RJ, Kim AH, Friedlander RM. 2014. Inhibition of mitochondrial protein import by mutant huntingtin. Nat Neurosci 17: 822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Karbowski M. 2005. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol 6: 657–663. [DOI] [PubMed] [Google Scholar]
- Yu ZX, Li SH, Evans J, Pillarisetti A, Li H, Li XJ. 2003. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington’s disease. J Neurosci 23: 2193–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen EY, Wei J, Zhong P, Yan Z. 2012. Disrupted GABAAR trafficking and synaptic inhibition in a mouse model of Huntington’s disease. Neurobiol Dis 46: 497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. 1995. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat Genet 11: 155–163. [DOI] [PubMed] [Google Scholar]
- Zeron MM, Hansson O, Chen N, Wellington CL, Leavitt BR, Brundin P, Hayden MR, Raymond LA. 2002. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington’s disease. Neuron 33: 849–860. [DOI] [PubMed] [Google Scholar]
- Zhai W, Jeong H, Cui L, Krainc D, Tjian R. 2005. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell 123: 1241–1253. [DOI] [PubMed] [Google Scholar]
- Zheng S, Clabough EBD, Sarkar S, Futter M, Rubinsztein DC, Zeitlin SO. 2010. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet 6: e1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, et al. 2001. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science 293: 493–498. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, et al. 2003. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet 35: 76–83. [DOI] [PubMed] [Google Scholar]
- Zwilling D, Huang SY, Sathyasaikumar KV, Notarangelo FM, Guidetti P, Wu HQ, Lee J, Truong J, Andrews-Zwilling Y, Hsieh EW, et al. 2011. Kynurenine 3-monooxygenase inhibition in blood ameliorates neurodegeneration. Cell 145: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]