Abstract
Contemporary climate change is characterized both by increasing mean temperature and increasing climate variability such as heat waves, storms, and floods. How populations and communities cope with such climatic extremes is a question central to contemporary ecology and biodiversity conservation. Previous work has shown that species diversity can affect ecosystem functioning and resilience. Here, we show that genotypic diversity can replace the role of species diversity in a species-poor coastal ecosystem, and it may buffer against extreme climatic events. In a manipulative field experiment, increasing the genotypic diversity of the cosmopolitan seagrass Zostera marina enhanced biomass production, plant density, and faunal abundance, despite near-lethal water temperatures due to extreme warming across Europe. Net biodiversity effects were explained by genotypic complementarity rather than by selection of particularly robust genotypes. Positive effects on invertebrate fauna suggest that genetic diversity has second-order effects reaching higher trophic levels. Our results highlight the importance of maintaining genetic as well as species diversity to enhance ecosystem resilience in a world of increasing uncertainty.
Keywords: global change, ecosystem functioning, ecological resilience, seagrass
Concerns about the accelerating loss of biodiversity have motivated an influential research program on the consequences of biodiversity loss at the species and functional group level (summarized in refs. 1–4). Although there are some important exceptions, most studies have found that in primary producers in particular, local species richness is positively correlated with a number of important ecosystem properties, such as productivity and resilience (5–8). Here we ask whether these results can be generalized to the genetic level, which represents the most fundamental aspect of biodiversity.
We hypothesized that genetic diversity may functionally replace the role of species diversity in species-poor aquatic macrophyte stands. The term species poor as it is used here refers to the low number of structuring primary producers. Seagrass meadows are a prominent example because they are often composed of only one or two species (9). Despite low species richness, seagrasses form the structural foundation of productive marine communities worldwide (10, 11), providing essential services such as nutrient cycling, habitat for fish and invertebrate species, and coastal erosion control (10, 12). In contrast to their species richness, their genotypic diversity can be very high and differs dramatically across all spatial scales (13, 14). This fact prompts the question of the ecological significance of such variation in diversity.
In a manipulative field experiment, we studied the role of genotypic diversity for ecosystem functioning in a coastal ecosystem dominated by eelgrass Zostera marina, the most abundant seagrass species of the northern hemisphere (15). Our experiment coincided with a period of an unprecedented heat wave that hit Europe in 2003 (16, 17). According to historical climate records, the return time for this extreme warming event was predicted to be >10,000 years (17). Nevertheless, it can been viewed as precursor for increasing climatic variability predicted for the coming decades (17, 18), and as such it provided an unparalleled opportunity to assess the short-term ecological response of a coastal community to rapid global warming. Because anthropogenic alterations of the world's climate are already resulting in range shifts and species extinctions (19–21), an understanding of factors that enhance resistance and resilience (22) of populations and communities in the face of climatic extremes is fundamental for biodiversity conservation and environmental management.
Methods
Study Species. Eelgrass (Z. marina L.), a monoecious marine flowering plant or seagrass, is the dominant macrophyte species of shallow sedimentary shorelines in the northern hemisphere (15). Like all seagrasses (and many land plants), eelgrass reproduces predominantly vegetatively (23). In the study area, the southwestern Baltic Sea, 2-year observations of genetic diversity in permanent plots of 1 m × 1m(n = 24) indicated that only 7% of the yearly shoot turnover was due to sexual reproduction (seedlings) and 93% was due to vegetative (clonal) growth (T.B.H.R., unpublished data). The genetic diversity of eelgrass has two clearly defined components: the genomic diversity in a sample of genotypes (24) and the number of genotypes (clones) per area (25). In contrast to previous experiments (26), both components can now be precisely separated by using high-resolution molecular markers (27). Here, we use DNA micro-satellites that are assumed to be selectively neutral to identify clones (genotypes) for a replicated manipulation of genotypic (clonal) diversity.
Study Site and Experimental Design. We tested the hypothesis that higher genotypic diversity increases ecosystem functioning in a field experiment that manipulated the number of eelgrass genotypes (clones) in plots situated at a shallow protected estuary of the southwestern Baltic Sea (Maasholm, Germany, 54°41′N, 10°12′E). The experiment was conducted by using SCUBA in a water depth of 1.6–1.8 m. At our high-latitude study site, growth of macrophytes is highly seasonal and confined to May–September. Thus, our experimental period from May 16 through Sep. 26, 2003, covered almost the complete growing season. To identify donor genotypes, we set up a marked grid of 10 m × 9.3 m in the vicinity (<20 m distance) of the experimental site in mid-April 2003. A leaf tip (3–5 cm) of candidate donor plants was sampled at 33.3-cm intervals (i.e., 31 × 29 = 899 grid points) and genotyped by using microsatellite markers (see below). This method allowed us to construct a detailed clonal map (see Fig. 4, which is published as supporting information on the PNAS web site). After selection of the appropriate subareas within the grid (Fig. 4), naturally occurring clusters of eelgrass leaf shoots (ramets) were harvested without breaking the rhizome connections and were reassembled into desired mixtures without leaving the ambient water. Shoots of all experimental units were planted within 24 h and secured in the sediment with wire staples.
The experimental layout consisted of 12 blocks of 1 × 1 m, 2 m distant from each other, which were placed into clearings within a large uninterrupted eelgrass meadow. Each block was separated into four subplots of 0.25 m2 that were assigned two one-genotype treatments (monocultures), and one three-genotype treatment and one six-genotype treatment (diversity treatments). Each subplot (experimental unit) received 18 eelgrass ramets (see Fig. 5, which is published as supporting information on the PNAS web site). Selection of genotype combinations and the allocation of specific combinations to plots and subplots were completely randomized. The chosen levels of genotypic diversity matched the range encountered in the field [range: 1–6 genotypes per 0.25 m2; mean (1 SD) = 2.8 (1.6) genotypes; T.B.H.R., unpublished data).
Genetic Methods. We assigned leaf shoots to clones by using the multilocus genotype of nine polymorphic DNA microsatellites previously developed for Z. marina (GenBank accession nos. AJ009898, AJ009900, AJ009904, and AJ249303–AJ249307). Genotyping was performed by using standard protocols (27, 28). Error probabilities of not detecting a unique genotype (29) were low (P < 0.001). Among the 126 genotypes identified, we selected six of the largest eight clones to obtain sufficient clonal replicates. Upon termination of the experiment, all leaf shoots (n = 2,262) were retyped to determine the final contribution of each genotype in the mixtures. As predicted by an exhaustive sampling before setting up the sampling grid, 18% of genotypes were “contaminations” from smaller clones that were hidden within the sampling grid. Six of 48 experimental units had more than 30% nontarget genotypes and were therefore excluded from an analysis of overyield and complementarity/selection effects (see below). Among the six genotypes selected, we found no significant effect of individual heterozygosity on clone area in the field (r2 = 0.19, P = 0.39), nor on their yield in the experiment (r2 = 0.01, P = 0.88).
Experimental Conditions and Response Variables. During the experiment, water temperatures were logged continuously at the pier of the Leibniz Institute for Marine Science in Kiel fjord, 45 km south of Maasholm. To reconstruct the temperature time series at the experimental site, which is shallower than Kiel fjord, we compared 42 days of temperature data from the experimental site with Kiel fjord records. Two separate linear regression models before and after the peak temperatures (i.e., warming and cooling) were used to predict Maasholm temperature data from Kiel fjord recordings (r2 = 0.93 and 0.77, respectively, both P < 0.0001).
Plots were inspected every 3 weeks for the intrusion of foreign genotypes. Shoot densities were counted every 3–6 weeks. Most plots (42/48) showed a net increase in leaf shoot density during the experimental period. Sediment cores (10 ml, 10 cm depth) were sampled in triplicate in each plot on Sep. 14, 2003. Upon arrival in the laboratory within 12 h, porewater was obtained by centrifugation (3,000 × g). The concentration of ammonium as the limiting macronutrient (30) was determined by using an autoanalyzer according to standard protocols. At the termination of the experiment, all plants were excavated, measured, dried to weight constancy, and weighed to the nearest 0.1 mg. We sampled epifauna in mesh bags (0.5-mm mesh size diameter). Samples were preserved in formalin (4%). Epifauna was analyzed in a one-genotype treatment and the six-genotype treatment of each block (i.e., n = 12). Taxonomic determination and counting took place under magnification (×6 to ×12). Amphipods were not determined to species.
Data Analysis. Data on eelgrass biomass (above plus belowground), shoot number, and pore water ammonium concentration were examined for a biodiversity effect by using a general linear model that incorporated genotypic diversity (continuous predictor) and block. In these analyses only, a single one-genotype replicate per block was dropped at random to achieve homogeneity of variances and a balanced design (that is, n = 12 for all three diversity levels). Reported results obtained with the intended treatment levels (i.e., one, three, and six genotypes) are conservative. When considering realized genotype numbers that included nonfocal clones, statistical models generally became more significant with respect to the factor genotypic diversity. The rate of recovery from climate perturbation was estimated by calculating a linear regression of shoot density (ln transformed) against time for each experimental unit separately, and the sampling date with the lowest abundance of eelgrass (i.e., peak of perturbation) was used as a starting point. The obtained regression slopes were then tested for differences among diversity as above.
We calculated net biodiversity effects (ΔY) according to ref. 31, which was adapted for genotypic data. Replicates of monocultures (n = 4) were randomly assigned to their respective polycultures. ΔY was tested against zero with two-sided t tests. Linear regression was used to test for an increasing effect size from three to six genotypes. Subsequently, ΔY values were partitioned into complementarity and selection, and were tested for statistical significance as above. We were able to consider only the six target genotypes in this analysis.
Epifaunal abundance and diversity were compared between one-genotype and six-genotype treatments by using linear regression (including blocking factor). We first performed a multivariate analysis of variance with the six most abundant species to test for an overall effect on faunal richness. Univariate tests were then performed for total abundance, and for three functional groups (grazers, filter feeders, and detritivores). All data were transformed as appropriate (usually square root or log10) to meet assumptions of the analysis (homogeneity of variances/normality).
Results
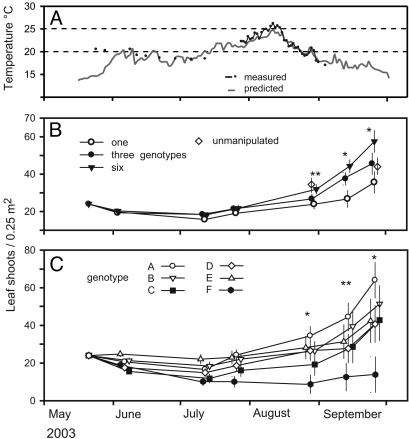
At the study site, water temperatures frequently attained levels above which Z. marina ceases growing (ca. 20°C; refs. 26 and 32), or even starts to die off (ca. 25°C; ref. 33; Fig. 1A). Accordingly, heat-related mortality reduced shoot numbers in the experiment by ≈50% from May through August (Fig. 1 B and C). This result contrasts markedly with several independent experiments that used a similar planting technique during normal climatic conditions; these experiments consistently showed low mortality and a 3-fold increase in shoot density from May through August (30, 34), making it unlikely that transplant shock was responsible for this decline (but see ref. 35). During the first major warming event at the beginning of June, unusual signs of heat-stress-related mortality were already present (discolored meristems, root necrosis, and leaf loss). In a subsample of nonmanipulated plants, half (10/20) had apparently died from heat stress during the warmest period (beginning of August). In nearby meadows, these losses amounted to a 48–52% decline in leaf shoot density and a 56% reduction in shoot height compared with data from 2001, 2002, and 2004 (n = 10–12, t test, all P < 0.0001), years for which average summer temperatures were recorded. The frequency of plants that showed discolored meristems was highly correlated with the number of leaf shoots in experimental plots during peak perturbation, further supporting a role for heat stress for plant mortality (response shoot density on Aug. 28, 2004; r2 = 0.52, P = 0.0013). In the experiment, shoot densities increased markedly only when water temperatures dropped below 20°C in mid-August. By then, the diverse experimental plots had attained natural shoot densities observed in nearby untouched meadows (Fig. 1B; t test, P = 0.58). After mid-August, three- and six-genotype treatments diverged markedly from the monocultures and showed faster recovery of shoot density (Fig. 1B). Higher recovery rates were statistically supported when we used the slopes of plotwise temporal regressions of shoot density (ln transformed) vs. time as response variable. Slopes were significantly higher with increasing genotypic diversity, suggesting faster recovery and possibly, higher ecological resilience [linear model, including blocking factor, F(1,23) = 8.2, P = 0.009].
Fig. 1.
Water temperature and eelgrass growth. (A) Water temperatures from direct measurements at the experimental site, and predicted vales from a 45 km distant measurement point. The critical temperatures above which Z. marina ceases growing, or starts dying off, are depicted by dashed lines. (B and C) Comparison of mean leaf shoot density (±1 SE) among one-, three-, and six-genotype treatments, and among the six target genotypes, A–F, as monocultures (n = 4) is shown. B also gives natural shoot densities at the experimental site. Data points are set off for clarity. Asterisks indicate results of general linear models including blocking factor, initial density as covariate, and genotypic diversity as continuous predictor at each date. *, P < 0.05; **, P < 0.01.
Our design allowed a comparative analysis of all clones because they were planted in replicated one-genotype treatments. The genotypes A–F showed very different responses to the climate event [one-way ANOVA; response variable final shoot density, F(5,18) = 4.04, P = 0.01]. Although genotype A increased almost 3-fold over the experimental period, genotype F never recovered from a 50% loss in shoot density during the heat wave [Fig. 1C; linear contrast, F(1,18) = 14.1, P = 0.001].
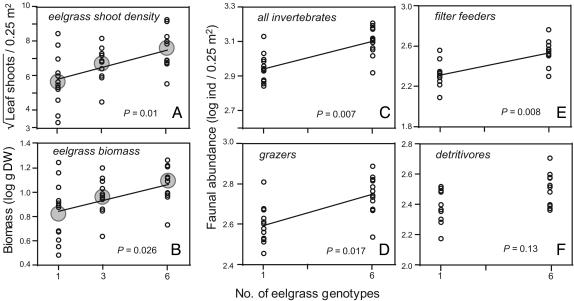
When the experiment was sampled at the end of the growth period, there was a positive correlation between genotypic diversity and both shoot number and dry biomass (Fig. 2 A and B). The yield of experimental plots slightly exceeded the highest yields observed in monocultures whereas the variances were progressively reduced [F test, variance one-genotype vs. six-genotype treatment; biomass F(24,12) = 3.78, P = 0.01; shoot number F(24,12) = 2.74, P = 0.035]. Such variance reduction may be due either to a lack of combinatorial replication at the highest diversity level (six genotypes), or to niche complementarity, i.e., a true diversity effect (36). Positive diversity effects were supported in an analysis of net biodiversity effects (ΔY, or overyield) that compared the final shoot number or biomass expected from the one-genotype plots with the observed yield (31). Significant ΔY values were present for both variables, shoot density and biomass (t tests against null-expectation ΔY = 0, P = 0.045 and 0.007 for biomass and shoot density, respectively). There was, however, no additional increase in ΔY between mixtures of three and six genotypes, i.e., the slopes of the regressions were not significant. On average, six-genotype treatments had 14.9 more shoots (+34%) and 2.59 g of more dry mass (+26%) than that predicted from monocultures.
Fig. 2.
Effects of increasing genotypic diversity on eelgrass (Z. marina) and associated fauna. Shoot number, square-root transformed (A), and dry biomass, log10 transformed (B), in eelgrass (Z. marina) plots as functions of their genotypic diversity. (C–F) Comparisons of the abundance of associated invertebrates (abundance log10 transformed). Regression models take spatial heterogeneity (blocking factor) into account. Shaded circles in A and B indicate treatment means. DW, dry weight.
A separation of genotypic diversity effects into complementarity and selection based on the comparison of diversity-treatments with replicated one-genotype treatments (31) revealed significant complementarity for biomass and shoot number, whereas selection effects were significantly negative for both variables (Table 1). Complementarity may indicate increased facilitation (37, 38), or niche differentiation among different genotypes (31). We therefore checked whether porewater nutrients were used more effectively in polycultures. However, sediment ammonium, the limiting macronutrient throughout the study area (30), did not decrease under increasing diversity. Rather, it was positively correlated with shoot number (linear model including blocking factor, r2 = 0.31, P = 0.007). This correlation implies additional nitrogen accumulation in the rhizosphere when shoot density was high.
Table 1. Partitioning of the net effects of genotypic diversity into complementarity and selection.
| Selection
|
Complementarity
|
|||
|---|---|---|---|---|
| Value | P (t test) | Value | P (t test) | |
| Biomass (g DW) | -3.17 | 0.011 | 4.123 | 0.018 |
| Leaf shoots/0.25 m2 | -6.47 | 0.009 | 12.14 | 0.016 |
Selection and complementarity values are pooled over three- and six- genotype treatments. Slopes of linear regressions between three- and six- genotype treatments were never significant (all P > 0.2). Only target genotypes entered this analysis. DW, dry weight.
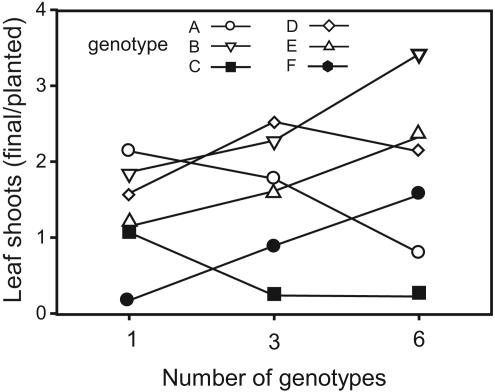
Although marked differences in monoculture yields among the six clones A–F (Fig. 1C) would suggest dominance of the best genotypes in the mixtures, selection effects were negative. This result demonstrates that effects of genotypic diversity were not due to greater chances of obtaining mixtures with more productive genotypes in diverse plots (the sampling effect; ref. 39). Negative selection effects are interesting and may imply either that genotypes performing well in monocultures performed proportionally worse in mixtures, or that they can be indicative of positive interactions (facilitation) enhancing particularly weak genotypes (37). A closer analysis revealed that both explanations apply. Selection effects occurred because plant mortality was particularly reduced in three genotypes that were weak in monoculture (genotypes D–F; Figs. 1C and 3), and because the best monoculture genotype (A) performed only about average in three- and six-genotype treatments (Fig. 3).
Fig. 3.
Rate of increase in leaf shoots (final/planted) of eelgrass genotypes A–F in monocultures and in three- and six-genotype treatments.
Second-order effects of increasing genotypic diversity were a higher abundance, but not diversity, of several epifaunal species or taxonomic groups that are closely associated with eelgrass (Fig. 2 C–F). A multivariate analysis with the six most abundant species showed a significant increase in faunal abundance in the six-genotype treatments compared with monocultures [repeated-measures ANOVA on log10-transformed abundances, F(1,10) = 7.67, P = 0.019]. We then divided all species into three functional groups: epiphyte grazers on eelgrass leaves, filter feeders, and detritivores. Filter feeders, mostly juvenile bivalves (Mytilus edulis, Mya truncata, and Cerastoderma edule), responded most favorably to increases in genotypic diversity, followed by grazers, including snails (Rissoa membranacea, Rissoa inconspicua, Hydrobia stagnorum, Bittium reticulatum, and Littorina saxatilis) and isopods (Idotea baltica and Idotea chelipes). In contrast, detritivorous crustaceans (Jassa spp. and Corophium spp.) showed no significant increase (Fig. 2F). Whereas the former two functional groups are directly dependent on a dense and productive leaf canopy, the latter group depends on plant detritus that may also be transported laterally into the experimental plots. Invertebrate diversity (as Simpson's index) showed no significant response to increasing genotypic diversity (P = 0.6; data not shown).
We performed an overyield analysis for faunal abundances similar to the eelgrass data and found that there were on average 72 more bivalve individuals (+22%) in six-genotype compared with one-genotype plots than expected from their abundance in monocultures (t test, P = 0.052). Overall, however, the increase in shoot densities and leaf area in the six-genotype treatments were the main factors responsible for higher abundance of epifauna in the plots. When we standardized the abundance of species groups on a leaf shoot basis, none of the comparisons among one- and six-genotype treatments remained significant.
Discussion
Some of the planet's most productive plant communities, such as salt marshes, reed stands (40), kelp beds (41), or seagrass meadows (9), consist of one or few dominant species. This observation is seemingly at odds with previous results that identified species diversity and functional diversity as key variables to explain high productivity and stability (4, 42). Our results reconcile this apparent contradiction and generalize from previous work on the role of species and functional group diversity to the genetic level, the most fundamental aspect of biodiversity. We have found that in the face of an extreme heat wave, genotypic diversity plays a role analogous to species and functional group diversity by increasing the rate of recovery after perturbation (7, 37, 43). Whether or not this is related to resilience, the rapidity with which the organism returns to the preperturbation state (22), or a higher productivity of diverse mixtures must remain an open question because our experimental units had not attained natural densities when the heat wave hit soon after planting.
Because the 2003 heat wave did not constitute an experimental treatment, it is difficult to predict whether positive effects of genotypic diversity would have been present without disturbance as well. Other recent work supports a role for genotypic diversity, particularly under disturbance (35). Whereas that study found only transient effects after intense bird grazing, we find strong effects that persist over at least one growth period.
Our study on genetic diversity tests the combined effects of complementarity and selection for net biodiversity effects (31). Complementarity effects were strongly positive, indicating facilitation and/or increased niche complementarity among genotypes. Surprisingly, we found that overall selection effects were negative, apparently because some genotypes that are strong in monoculture are weak performers in mixtures (Fig. 3). Had genotype A, in particular, just performed as well in mixture as in monoculture, net biodiversity effects would have been even stronger.
Research on the effects of global change has focused thus far on large-scale range shifts or evolutionary responses to shifting mean temperatures (20, 21). In contrast, knowledge of the effects of extreme events (44) is rare (but see ref. 45), although such events are predicted to increase in the coming decades (17, 18). For many species this leaves little time for range shifts (46) or local adaptation (47), highlighting the importance of short-term responses of local communities and their structuring species to increasing climate stress. This applies particularly to sessile species that cannot rapidly shift their distributional range, such as all plants.
Biodiversity experiments should ideally be replicated at multiple levels, including diversity, composition, and multiple sites (48). Our experiment was performed in an area of particularly high genetic and clonal diversity (14), with alternating low and high clonal diversities occurring side by side within the same meadow (Fig. 4). It will be interesting to conduct similar experiments at sites of lower clonal diversities to assess the robustness of our results. In our experiment, we opted for proper replication of a relatively small selection of genotypes to disentangle the contribution of complementarity and selection to overall biodiversity effects. As a consequence, the number of unique genotype combinations was relatively low, increasing the risk that some effects observed were due to specific interactions among genotypes (the composition effect; refs. 48 and 49). Compositional effects were identified in terrestrial grasslands particularly among members of different functional guilds, for example nitrogen fixing and nonfixing plants (49). A priori, such effects seem difficult to envisage among a sample of genotypes, given that individuals of the same species are much more alike. Notwithstanding, our data support a net biodiversity effect for the variable shoot density when all putative compositional effects are removed, i.e., when replicates of genotype combinations are averaged in an overyield analysis (paired t test, expected vs. observed yield, n = 7, P = 0.019).
Strong effects of genotypic diversity on fauna associated with eelgrass were surprising because the spacing of the experimental units was relatively close, permitting migration of crustaceans and gastropods between experimental units. In this respect, reported effects of eelgrass genotypic diversity on faunal abundance should be regarded as conservative estimates. Herbivorous gastropods and juvenile bivalves constitute important components of the food web in seagrass ecosystems as they crop epiphytic and planktonic algae, respectively, and may enhance the persistence of seagrass beds even in the face of eutrophication (50, 51). Larger, mobile consumers such as fish and crabs also use seagrass habitat as feeding and breeding habitat (12). Hence, the mediation of herbivore abundance by genotypic diversity of a primary producer may constitute a positive feedback loop (52) that links consumer dynamics with resilience and persistence of ecologically important seagrass beds (51). The epifaunal species enhanced in the experiment are consumed by a number of fish species several of which are commercially important (see Table 2, which is published as supporting information on the PNAS web site). We speculate that genotypic diversity of seagrass could have far-reaching effects across several trophic levels, including grazers, their predators, and human fisheries.
In conclusion, our study shows that climatic extremes can have immediate effects on coastal communities, and that genetic diversity may enhance recovery after such perturbations. Although we focus here on short-term processes within one generation of a long-lived clonal plant, evolutionary responses of seagrass populations are likely given the striking differences in clonal performance. Ongoing losses of genetic diversity sensu latu through habitat fragmentation (53) or genetic erosion (54) may not only endanger the evolutionary potential of populations (55). In addition, they may have immediate consequences for associated communities, ecosystem functioning, and ecosystem services provided by primary producers. These results have implications for biodiversity conservation and environmental management, which so far has focused on the maintenance of species diversity in particularly species-rich ecosystems (56). Our results suggest that conserving genetic and genotypic diversity in species-poor ecosystems that have no redundancy at the species level may be just as important for strengthening the resilience of dependent communities in the face of global change and increasing climatic extremes.
Supplementary Material
Acknowledgments
We thank W. Lampert for continual encouragement and support; A. Bockelmann, M. V. Ruggiero, S. Carstensen, S. Liedtke, D. Albrecht, T. Sonntag, R. Neuhaus, M. Dahl, G. Corno, I. Dankert, and A. Hasselmeyer for technical or field assistance; F. Nevoigt and N. Langhanki for meteorological data; H. Hillebrand for statistical advice; H. Lotze, A. Bockelmann, and J. L. Olsen for comments; and the Institute of Marine Geology (GEOMAR) for logistical support. T.B.H.R. and B.W. were supported by individual grants from the Deutsche Forschungsgemeinschaft (Grants Re 1108/3 and -4 and Wo 818/1-2).
References
- 1.Wardle, D. A. (2002) Communities and Ecosystems: Linking the Aboveground and Belowground Components (Princeton Univ. Press, Princeton).
- 2.Tilman, D. (1999) Ecology 80, 1455–1474. [Google Scholar]
- 3.Kinzig, A. P., Pacala, S. W. & Tilman, D. (2002) The Functional Consequences of Biodiversity (Princeton Univ. Press, Princeton).
- 4.Loreau, M., Naeem, S. & Inchausti, P. (2002) Biodiversity and Ecosystem Functioning: Synthesis and Perspective (Oxford Univ. Press, Oxford).
- 5.Hector, A., Schmid, B., Beierkuhnlein, C., Caldeira, M. C., Diemer, M., Dimitrakopoulos, P. G., Finn, J. A., Freitas, H., Giller, P. S., Good, J., et al. (1999) Science 286, 1123–1127. [DOI] [PubMed] [Google Scholar]
- 6.Engelhardt, K. A. M. & Ritchie, M. E. (2001) Nature 411, 687–689. [DOI] [PubMed] [Google Scholar]
- 7.Tilman, D., Wedin, D. & Knops, J. (1996) Nature 379, 718–720. [Google Scholar]
- 8.Loreau, M., Naeem, S., Inchausti, P., Bengtsson, J., Grime, J. P., Hector, A., Hooper, D. U., Huston, M. A., Raffaelli, D., Schmid, B., et al. (2001) Science 294, 804–808. [DOI] [PubMed] [Google Scholar]
- 9.Hemminga, M. A. & Duarte, C. M. (2000) Seagrass Ecology (Cambridge Univ. Press, Cambridge, U.K.).
- 10.Duarte, C. M. (2002) Environ. Conserv. 29, 192–206. [Google Scholar]
- 11.Williams, S. L. & Heck, K., Jr. (2001) in Marine Community Ecology, eds. Bertness, M. D., Gaines, S. D. & Hay, M. E. (Sinaur, Sunderland, MA), pp. 317–338.
- 12.Micheli, F. & Peterson, C. H. (1999) Conserv. Biol. 13, 869–881. [Google Scholar]
- 13.Reusch, T. B. H., Stam, W. T. & Olsen, J. L. (2000) Mol. Ecol. 9, 127–140. [DOI] [PubMed] [Google Scholar]
- 14.Olsen, J. L., Stam, W. T., Coyer, J. A., Reusch, T. B. H., Billingham, M., Boström, C., Calvert, E., Christie, H., Granger, S., La Lumiere, R., et al. (2004) Mol. Ecol. 13, 1923–1941. [DOI] [PubMed] [Google Scholar]
- 15.den Hartog, C. (1970) Verh. K. Ned. Akad. Wet. Afd. Natuurkd. II 59, 1–275. [Google Scholar]
- 16.Luterbacher, J., Dietrich, D., Xoplaki, E., Grosjean, M. & Wanner, H. (2004) Science 303, 1499–1503. [DOI] [PubMed] [Google Scholar]
- 17.Schär, C., Vidale, P. L., Lüthi, D., Frei, C., Häberli, C., Liniger, M. A. & Appenzeller, C. (2004) Nature 427, 332–335. [DOI] [PubMed] [Google Scholar]
- 18.Palmer, T. N. & Räisänen, J. (2002) Nature 415, 512–514. [DOI] [PubMed] [Google Scholar]
- 19.Hughes, L. (2000) Trends Ecol. Evol. 15, 56–61. [DOI] [PubMed] [Google Scholar]
- 20.Thomas, C. D., Cameron, A., Green, R. E., Bakkenes, M., Beaumont, L. J., Collingham, Y. C., Erasmus, B. F. N., de Siqueira, M. F., Graininger, A., Hannah, L., et al. (2004) Nature 427, 145–148. [DOI] [PubMed] [Google Scholar]
- 21.Parmesan, C., Ryrholm, N., Stefanescu, C., Hill, J. K., Thomas, C. D., Descimon, H., Huntely, B., Kaila, L., Kullberg, J., Tammaru, T., et al. (1999) Nature 399, 579–583. [Google Scholar]
- 22.Pimm, S. L. (1984) Nature 307, 321–326. [Google Scholar]
- 23.Tomlinson, P. B. (1974) Aquaculture 4, 107–130. [Google Scholar]
- 24.Hämmerli, A. & Reusch, T. B. H. (2003) Mol. Ecol. 12, 619–629. [DOI] [PubMed] [Google Scholar]
- 25.Widen, B., Cronberg, N. & Widen, M. (1994) Folia Geobot. Phytotaxon. 29, 245–263. [Google Scholar]
- 26.Williams, S. L. (2001) Ecol. Appl. 11, 1472–1488. [Google Scholar]
- 27.Reusch, T. B. H. (2000) Mol. Ecol. 9, 371–373. [DOI] [PubMed] [Google Scholar]
- 28.Reusch, T. B. H. (2002) Limnol. Oceanogr. 47, 78–85. [Google Scholar]
- 29.Parks, J. C. & Werth, C. R. (1993) Am. J. Bot. 80, 537–544. [DOI] [PubMed] [Google Scholar]
- 30.Worm, B. & Reusch, T. B. H. (2000) Mar. Ecol. Prog. Ser. 200, 159–166. [Google Scholar]
- 31.Loreau, M. & Hector, A. (2001) Nature 412, 72–76. [DOI] [PubMed] [Google Scholar]
- 32.Rasmussen, E. (1973) Ophelia 11, 1–507. [Google Scholar]
- 33.Greve, T. M., Borum, J. & Pedersen, O. (2003) Limnol. Oceanogr. 48, 210–216. [Google Scholar]
- 34.Reusch, T. B. H. & Williams, S. L. (1998) Oecologia 113, 428–441. [DOI] [PubMed] [Google Scholar]
- 35.Hughes, A. R. & Stachowicz, J. J. (2004) Proc. Natl. Acad. Sci. USA 101, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huston, M. A. & McBride, A. C. (2002) in Biodiversity and Ecosystem Functioning, eds. Loreau, M., Naeem, S. & Inchausti, P. (Oxford Univ. Press, New York), pp. 47–60.
- 37.Mulder, C. P. H., Uliassi, D. D. & Doak, D. F. (2001) Proc. Natl. Acad. Sci. USA 98, 6704–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cardinale, B. J., Palmer, M. A. & Collins, S. L. (2002) Nature 415, 426–429. [DOI] [PubMed] [Google Scholar]
- 39.Huston, M. A. (1997) Oecologia 110, 449–460. [DOI] [PubMed] [Google Scholar]
- 40.Chambers, R. M., Meyerson, L. A. & Saltonstall, K. (1999) Aquat. Bot. 64, 261–273. [Google Scholar]
- 41.Paine, R. T. (2002) Science 296, 736–739. [DOI] [PubMed] [Google Scholar]
- 42.Giller, P. S., Hillebrand, H., Berninger, U. G., Gessner, M. O., Hawkins, S., Inchausti, P., Inglis, C., Leslie, H., Malmqvist, B., Monaghan, M. T., et al. (2004) Oikos 104, 423–436. [Google Scholar]
- 43.Kennedy, T. A., Naeem, S., Howe, K. M., Knops, J. M. H., Tilman, D. & Reich, P. (2002) Nature 417, 636–638. [DOI] [PubMed] [Google Scholar]
- 44.Gaines, S. D. & Denny, M. W. (1993) Ecology 74, 1677–1692. [Google Scholar]
- 45.Holmgren, M., Scheffer, M., Ezcurra, E., Gutierrez, J. R. & Mohren, G. M. J. (2001) Trends Ecol. Evol. 16, 89–94. [DOI] [PubMed] [Google Scholar]
- 46.Barry, J. P., Baxter, C. H., Sagarin, R. D. & Gilman, S. E. (1995) Science 267, 672–675. [DOI] [PubMed] [Google Scholar]
- 47.Etterson, J. R. & Shaw, R. G. (2001) Science 294, 151–154. [DOI] [PubMed] [Google Scholar]
- 48.Schmid, B., Hector, A., Huston, M. A., Inchausti, P., Nijs, I., Leadley, P. W. & Tilman, D. (2002) in Biodiversity and Ecosystem Functioning: Synthesis and Perspective, eds. Loreau, M., Naeem, S. & Inchausti, P. (Oxford Univ. Press, New York), pp. 61–78.
- 49.Tilman, D., Knops, J., Wedin, D., Reich, P., Ritchie, M. & Siemann, E. (1997) Science 277, 1300–1302. [Google Scholar]
- 50.Neckles, H. A., Wetzel, R. L. & Orth, R. J. (1993) Oecologia 93, 285–295. [DOI] [PubMed] [Google Scholar]
- 51.Duffy, J. E., Richardson, J. P. & Canuel, E. A. (2003) Ecol. Lett. 6, 637–645. [Google Scholar]
- 52.Worm, B. & Duffy, J. E. (2003) Trends Ecol. Evol. 18, 628–632. [Google Scholar]
- 53.Olesen, J. M. & Jain, S. K. (1994) in Conservation Genetics, eds. Loeschke, V., Tomiuk, J. & Jain, S. K. (Birkhäuser, Basel), pp. 417–426.
- 54.Avise, J. C. & Hamrick, J. L. (1996) Conservation Genetics: Case Histories from Nature (Chapman & Hall, New York).
- 55.Myers, N. & Knoll, A. H. (2001) Proc. Natl. Acad. Sci. USA 98, 5389–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myers, N., Mittermeier, R. A., Mittermeier, C. G., da Fonseca, G. A. B. & Kent, J. (2001) Nature 403, 853–858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.