Abstract
Soon after the discovery of transforming growth factor-β (TGF-β), seminal work in vertebrate and invertebrate models revealed the TGF-β family to be central regulators of tissue morphogenesis. Members of the TGF-β family direct some of the earliest cell-fate decisions in animal development, coordinate complex organogenesis, and contribute to tissue homeostasis in the adult. Here, we focus on the role of the TGF-β family in mammalian stem-cell biology and discuss its wide and varied activities both in the regulation of pluripotency and in cell-fate commitment.
TGF-β family members regulate pluripotency and cell-fate commitment in mammalian development. Their wide and varied functions are determined by their interactions with cell-type-specific signaling and transcription factors.
Stem cells are defined by their ability to divide continually to maintain the stem-cell pool and to provide progeny that differentiate into other cell types. These defining qualities are established by internal transcriptional programs that interact with the local environment, or niche, to both promote stem-cell maintenance and drive cell-fate determination (Voog and Jones 2010; Young 2011; Scadden 2014; Kfoury and Scadden 2015). Members of the TGF-β family of signaling ligands are key components of the stem-cell niche for both embryonic and somatic stem cells and orchestrate diverse responses in different types of stem cells. Here, we provide an overview of the general characteristics of embryonic versus somatic stem cells, briefly introduce key concepts in understanding the core TGF-β family Smad signaling pathway, and then delve into the function of TGF-β family signaling in various embryonic and somatic stem-cell systems.
EMBRYONIC AND SOMATIC STEM CELLS
After fertilization, the zygote divides to form the morula and then the blastocyst. The blastocyst consists of an outer layer of cells, called the trophoblast, an inner cavity of fluid, called the blastocele, and an internal cluster of cells, called the inner cell mass (ICM) (Rossant 2008). The embryo forms from the cells of the ICM, and these cells can be isolated and cultured ex vivo to give rise to embryonic stem cells (ESCs) (Fig. 1A). Naïve epiblast cells from within the ICM are the source of mouse (m) ESCs (Gardner and Brook 1997; Batlle-Morera et al. 2008). mESCs have not undergone X-inactivation and can contribute to all three germ layers (endoderm, mesoderm, and ectoderm) of chimeric mice when they are injected into blastocysts, which is the key functional test for pluripotency in mESCs (Bradley et al. 1984). mESCs maintain a normal karyotype and are defined by their ability to proliferate without differentiation (self-renewal) and their potential to give rise to every cell type in the body (pluripotency) (Evans and Kaufman 1981; Martin 1981; Thomson 1998). Cells can also be isolated from primed epiblast cells derived from the postimplantation blastocyst (Brons et al. 2007; Tesar et al. 2007). These cells, called EpiSCs (postimplantation epiblast-derived stem cells), express many key transcription factors that are characteristic of mESCs and can differentiate into all three germ layers in teratoma assays, where cells are injected into immunodeficient mice to allow spontaneous differentiation. However, EpiSCs show X inactivation and are not capable of producing chimeric mice when injected into the blastocyst. These last two qualities indicate that EpiSCs do not possess the full developmental potency of mESCs (Fig. 1B).
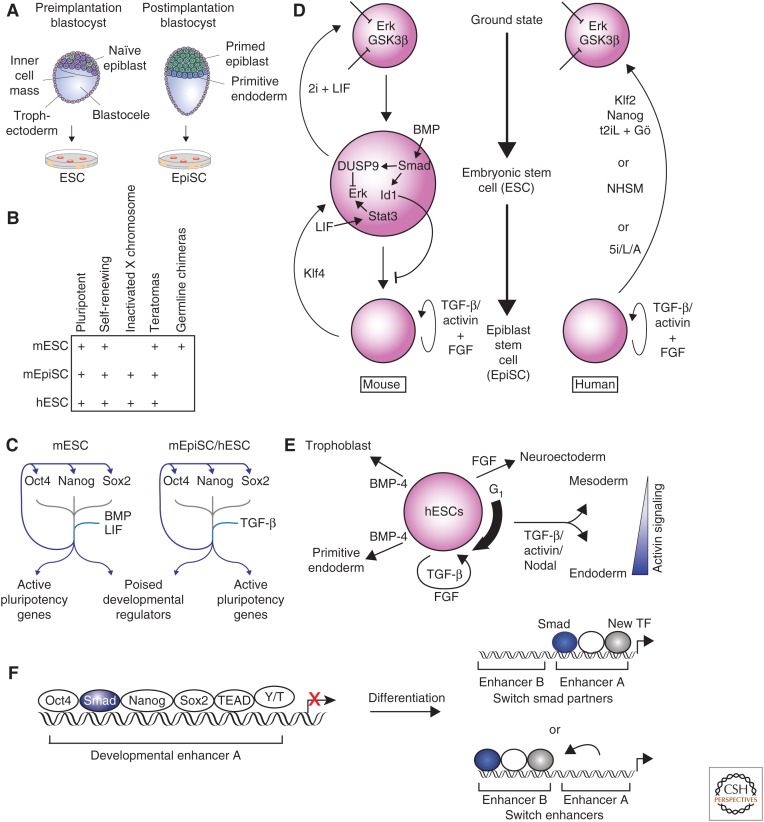
Figure 1.
Transforming growth factor β (TGF-β) family signaling in embryonic stem cells (ESCs). (A) Mouse embryonic stem cells (mESCs) are derived from naïve epiblast cells in the preimplantation blastocyst, whereas postimplantation epiblasts (EpiSCs) are derived from primed epiblast cells in the postimplantation blastocyst. (B) The defining characteristics of mESCs, mEpiSCs, and human (h) ESCs are shown. (C) Oct4 (Pou5f1), Nanog, and Sox2 are the key transcription factors that maintain ESC state. These factors co-occupy the genome at their own promoters, at the promoters of other key ESC genes, and at developmental regulators that are repressed but poised to be activated during differentiation. Bone morphogenetic protein (BMP) and leukemia inhibitory factor (LIF) reinforce this network in mESCs, and TGF-β family signaling reinforces this network in hESCs and EpiSCs. (D) Key pathways that interact with TGF-β family signaling to maintain mESC (left) and hESC (right) states are shown. BMP signaling is a key factor in the maintenance of mESC state, whereas TGF-β and/or activin maintain the mEpiSC and hESC states. (E) The role of TGF-β family signaling in hESCs differentiation is indicated. The thick arrow to the right of the hESC indicates that hESCs respond most efficiently to activin signaling with Smad2 and Smad3 activation during early G1 phase of cell cycle. Once cells differentiate into mesendoderm, the concentration of activin is a major determining factor in the differentiation toward mesoderm or endoderm. (F) Smad2 and/or Smad3 co-occupy the genome with the key transcription factors that maintain the ESC state (left). During differentiation into endoderm, the expression of the key ESC transcription factors is lost. Smad2 and/or Smad3 regulate induction of new genes by either continuing to occupy the same enhancer but with different combinations of transcription factors (top right) or by moving to new enhancers in association with new transcription factors (bottom right). Y/T, YAP and TAZ.
In ESCs, a remarkable pluripotent transcriptional circuitry poises the cells in a state that allows differentiation into all cell types while maintaining their ability to replicate as ESCs indefinitely. Oct4, Sox2, and Nanog were identified as the core factors of this transcription factor network that are critical to maintain the ESC state (Chen et al. 2008a; Orkin et al. 2008; MacArthur et al. 2009; Young 2011). These three transcription factors co-occupy DNA throughout the ESC genome to regulate their own expression, activate ESC genes, and repress developmental regulators (Fig. 1C) (Boyer et al. 2005). This network is modulated by interaction with additional transcription factors, signaling pathways, microRNAs (miRNAs), and chromatin regulators. Transcription factors, including Esrrb (Ivanova et al. 2006; Zhang et al. 2008; Martello et al. 2012), Sal4 (Wu et al. 2006; Zhang et al. 2006a), Tbx3 (Ivanova et al. 2006; Niwa et al. 2009), and Prdm14 (Chia et al. 2010) share binding sites with Oct4, Sox2, and Nanog, and are required to maintain the ESC state. This transcriptional network is further regulated by miRNAs (Marson et al. 2008; Viswanathan et al. 2008) and chromatin regulators, including the Polycomb group and SetDB1 (Boyer et al. 2006; Bilodeau et al. 2009). Finally, pluripotency is critically dependent on physical interactions between pluripotency factors and transcriptional mediators of key morphogen signaling pathways. Thus, signaling intermediaries of the Wnt, bone morphogenetic protein (BMP), and TGF-β pathways, as well as the Janus kinase and signal transducers and activators of transcription (JAK-STAT) pathway co-occupy the genome with Oct4, Sox2, and Nanog to promote the pluripotent state (Chen et al. 2008b; Cole et al. 2008; van den Berg et al. 2010; Mullen et al. 2011), but also play key roles in driving differentiation. This dichotomy defines a key feature of developmental systems, in which the environmental context plays a key role in defining the biological output in response to morphogen signaling.
During development, ESCs are present for a limited time, ultimately differentiating into cell types with increasingly restricted plasticity. Somatic stem cells are produced during differentiation and give rise to terminally differentiated cells that compose and carry out the specialized functions of distinct tissues. After tissues are formed, somatic stem cells remain in small numbers and can be called on to proliferate and differentiate to replace lost cells as a homeostatic mechanism and/or as a regenerative response to injury (Goodell et al. 2015). Both ESCs and somatic stem cells are capable of self-renewal and both are capable of differentiation. However, somatic stem cells give rise to a more restricted number of cell types compared with ESCs, which are pluripotent. Accordingly, in addition to being multipotent (differentiating into multiple cell types), somatic stem cells can be unipotent (differentiating into one known terminally differentiated cell type).
Both ESCs and somatic stem cells have the potential for use in human therapeutics, and each has different strengths and limitations. ESCs have the ability to differentiate into any adult tissue in the body, but could require longer differentiation protocols to reach terminal cells, and have oncogenic potential. Somatic stem cells have a more restricted repertoire, but, because they are more closely related to cells of specific tissues, they may require fewer steps to differentiate into terminal cell types. An additional difference is that somatic stem cells can be taken from adult patients and do not raise the same ethical concerns as production of ESCs from in vitro fertilization. Furthermore, tissues produced from somatic stem cells can be returned to the same patient without risk of immune rejection.
The importance of transcription factors to control cell identity was highlighted by the creation of induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka 2006). Transcription factors that were enriched in ESCs were ectopically expressed in different combinations in mouse embryonic fibroblasts (MEFs) to identify factors that could reprogram MEFs into ESCs. Four transcription factors were identified that were sufficient to drive reprogramming to a pluripotent state. These reprogrammed ESCs were referred to as iPSCs and were found to possess all the defining features of ESCs. This groundbreaking discovery was built on previous work that showed differentiated nuclei could be reprogrammed by transplantation into enucleated oocytes (Gurdon 1962) and cell identity can be altered with the forced expression of key transcription factors (Davis et al. 1987). These seminal studies reveal the cell state to be remarkably plastic. Although initial somatic cell reprogramming experiments were performed in mouse, where expression of Oct4, Sox2, Klf4, and c-Myc are sufficient for reprogramming to mouse iPSCs (Takahashi and Yamanaka 2006), subsequent generation of human iPSCs was achieved from human fibroblasts by expression of either Oct4, Sox2, Klf4, and c-Myc, or expression of Oct4, Nanog, Klf4, and c-Myc (Takahashi et al. 2007; Yu et al. 2007). Since their original description, multiple combinations of transcription factors, signaling molecules, and small molecules have been identified to drive this process. Furthermore, morphogen signaling, in particular the TGF-β family, plays key roles in reprogramming.
iPSCs are pluripotent, self-renew, and are very similar to ESCs in patterns of gene expression and chromatin structure (Chin et al. 2009; Deng et al. 2009; Guenther et al. 2010; Hawkins et al. 2010; Newman and Cooper 2010). The production of iPSCs from a patient’s somatic cells has already allowed the study of the effect of patient-specific genetic backgrounds on differentiation (Dimos et al. 2008; Park et al. 2008; Cherry and Daley 2012; Kiskinis et al. 2014). These cells also provide the potential to generate ESCs from a patient’s somatic cells, which could be differentiated into tissues for therapy without the risk of immune-mediated rejection. Genetic mutations could also be corrected in patient-derived iPSCs before differentiation into tissues so that the newly differentiated tissue would no longer contain disease-associated mutations. However, before the potential uses in regenerative medicine can be realized, it will first be necessary to address the safety concerns of implanting iPSCs or cells derived from iPSCs into patients.
TGF-β AND BMP SIGNALING
The TGF-β family is one of the largest families of secreted morphogens encoded in the mammalian genome (33 distinct genes) (Morikawa et al. 2016). The core Smad signaling pathway has been extensively reviewed (Massagué 2005), and therefore will only be briefly summarized here. TGF-β family members signal via transmembrane serine/threonine kinase receptors to form a unique signaling system in animals. There are 12 transmembrane kinase receptors encoded in mammalian genomes that are subdivided into five type II and 7 type I receptors. Signaling is initiated when TGF-β family ligands drive formation of a heterotetrameric complex of two type II and two type I receptors (Hinck 2012). This allows the type II receptor to transphosphorylate serine or threonine residues in the conserved “GS region” of the type I receptor, which in turn activates the Smad signaling pathway through direct carboxy-terminal phosphorylation of Smads. Smads are a family of unique transcriptional regulators that contain conserved Mad Homology 1 (MH1) and MH2 domains separated by a poorly conserved linker region. There are eight Smads in mammalian genomes, which are subdivided into three functional classes. The receptor-regulated R-Smads comprise Smads 1, 2, 3, 5, and 8 (which is also known as Smad9) and are activated when they dock with phosphorylated type I receptors, which then phosphorylate the last two serines in the R-Smad. This leads to dissociation of R-Smads from the receptor, interaction with the co-Smad, Smad4, and accumulation of R-Smad:Smad4 complexes in the nucleus. Importantly, TGF-β versus BMP-like pathway activation is provided through specific R-Smad MH2:type I receptor interactions. Thus, although not all combinations of type I receptors and R-Smads have been directly tested, evidence strongly suggests that the BMP type I receptors ALK1 (gene name, ACVRL1), ALK2 (gene name, ACVR1), BMPRIA/ALK3 (gene name, BMPRIA) and BMPRIB/ALK6 (gene name, BMPRIB) all preferentially activate R-Smads 1, 5, and 8, whereas the activin, TGF-β, and Nodal type I receptors ActRIB/ALK4 (gene name, ACVR1B), TβRI/ALK5 (gene name, TGFBR1) and ActRIC/ALK7 (gene name, ACVR1C) activate Smad2 and Smad3. A confusing aspect to TGF-β family function is often perceived by the ability of ligands to mix and match with different combinations of type II and type I receptors. However, because signaling to R-Smads is initiated by the phosphorylated type I receptor, the nature of the transcriptional response to ligand is dictated by the engaged type I receptor(s) in the heterotetrameric complex. Thus, Smad signaling can typically be grouped into one of two broad types, a BMP-like versus TGF-β-like response. Interestingly, in developmental models and in particular stem-cell models, these pathways often impose alternative biological outcomes, and, whereas many systems display ligand-dependent specificity in Smad activation in certain contexts, such as endothelial cells, TGF-β can also activate the BMP Smad pathway (Goumans et al. 2002). Finally, the third class of Smads, known as inhibitory Smads, are encoded by Smad6 and Smad7 and act as feedback inhibitors of Smad signaling. In the nucleus, the MH1 domain of almost all the R-Smads and Smad4 possess DNA-binding affinity (a splice isoform of Smad2 is the exception), although the MH1:DNA interaction is typically of low affinity and specificity. Thus, Smads rely on DNA-binding partners for recruitment to regulatory gene elements, where Smad complexes can stimulate or inhibit transcription via interaction with histone-modifying enzymes, such as the histone acetyl transferases CBP or p300, or histone deacetylases (HDACs), respectively, and SWI/SNF chromatin-remodeling complexes (Ross et al. 2006). The interaction of Smads with distinct DNA-binding partners that are expressed in a cell-type-specific manner thus provides an important mechanism underlying contextual responses to TGF-β family signaling.
EMBRYONIC STEM CELLS
Mouse Embryonic Stem Cells
Mouse ESCs can self-renew and are capable of differentiating into all three germ layers (Evans and Kaufman 1981; Martin 1981). The ESC state is maintained by the core circuitry of the transcription factors Oct4, Nanog, and Sox2, which physically interact and co-occupy the genome at many key genes including those encoding developmental regulators (Chen et al. 2008b; Marson et al. 2008). Among these targets are the genes encoding Oct4, Nanog, and Sox2 themselves, which help reinforce the transcriptional program to maintain the ESC state (Boyer et al. 2005). Among the genes bound by Oct4, Nanog, and Sox2 are many developmental regulators that are in the so-called “poised state,” which allows for rapid induction during differentiation while being repressed in ESCs (Bernstein et al. 2006).
mESCs were originally derived by culture on a layer of MEFs using media containing fetal calf serum (FCS) (Evans and Kaufman 1981; Martin 1981). The requirement for feeder cells, but not FCS, can be eliminated by culturing mESCs in the presence of leukemia inhibitory factor (LIF) (Smith et al. 1988; Williams et al. 1988). mESCs express the BMP receptors BMPRIA and BMPRII, and this knowledge led to the discovery that mESCs can be maintained in serum-free media supplemented only with LIF and BMP-2 or BMP-4 (Fig. 1D) (Ying et al. 2003). mESCs can be derived and maintained in media with FCS and LIF because FCS contains BMP (Ying et al. 2003). LIF signaling is mediated by activation of Stat3 (Niwa et al. 1998), and BMP signaling does not contribute to activation of Stat3 (Ying et al. 2003) or affect Stat3 transcription (Chen et al. 2008b). The requirement for BMP signaling in mESCs is due, in large part, to activation of Id1 and Id3, whereas BMP-induced mesoderm is blocked by physical interaction of Nanog and Smad1 (Suzuki et al. 2006). In the absence of BMP signaling, ectopic expression of either Id1 or Id3 is sufficient to maintain mESCs in culture (Ying et al. 2003). Chromatin immunoprecipitation and sequencing (ChIP-seq) analysis further revealed that Smad1 co-occupies the mESC genome at sites also occupied by Oct4, Nanog, Sox2, and Stat3 in normal mESC culture conditions with LIF and FCS (Chen et al. 2008b). Presumably this co-occupation of the genome is preserved in mESCs cultured with LIF and BMP alone, but this has not been tested. Importantly, while Oct4 binding is not dependent on Smad1, loss of Oct4 expression results in loss of Smad1 binding, suggesting that Smad1 requires Oct4 to stabilize binding to these sites (Chen et al. 2008b).
mESCs do not express the TGF-β type I receptor TβRI and thus do not activate Smad2 or Smad3 in response to TGF-β1 (Roelen et al. 1994). However, mESCs do activate Smad2 and 3 in response to activin and Nodal, likely via the ActRIB/ALK4 and ActRIC/ALK7 receptors (James et al. 2005). Studies using antibodies recognizing Smad3, or Smad2 and Smad3, show that Smad3 and, presumably, Smad2 co-occupy the genome with Oct4, Nanog, and Sox2 in mESCs (Mullen et al. 2011). Smad3 and Stat3 are also enriched at super-enhancers, which are large enhancer domains that regulate genes that determine cell identity (Hnisz et al. 2013; Whyte et al. 2013). Although Smad2 and/or Smad3 tend to co-occupy the genome with the master transcription factors in mESCs, lack of Smad2 or Smad3 phosphorylation results only in decreased proliferation, but does not affect pluripotency (James et al. 2005; Ogawa et al. 2006).
mESCs maintained on MEFs with FCS and LIF, or in the presence of LIF and BMP, possess all the qualities of ESCs, but remain a heterogeneous population and show variable expression of the transcription factor Nanog (Ying et al. 2008). mESC cultures also contain fibroblast growth factor 4 (FGF4), which activates the Erk MAPK pathway (Kunath et al. 2007). The requirement for BMP signaling can be bypassed with inhibitors of FGF and Erk MAPK signaling, suggesting that a major role of BMP signaling is to block the effects of Erk MAPK (Ying et al. 2008). BMP signaling activates expression of the dual specificity phosphatase DUSP9, dependent on Smad1 and 5, and thus acts to inhibit Erk MAPK activation, while induction of Id1 appears to act further downstream to block differentiation (Li et al. 2012). Furthermore, inhibition of glycogen synthase kinase 3β (GSK3β), which leads to activation of β-catenin, can be combined with FGF or Erk MAPK inhibition to replace both LIF and BMP (Wray et al. 2011; Yi et al. 2011). This insight has led to the identification of a naïve “ground state” of mESCs, which is independent of BMP and LIF signaling (Fig. 1D) (Ying et al. 2008).
Postimplantation epiblasts (EpiSCs) represent a later stage of development, but also express Oct4, Nanog, and Sox2, the key transcription factors of mESCs. These cells do not require LIF or BMP for maintenance and instead require FGF and activin signaling (Batlle-Morera et al. 2008; Vallier et al. 2009b,c). mESCs can be differentiated into EpiSCs by culture with activin and FGF, and this change is also associated with X-inactivation. The reverse differentiation does not occur, and culture of EpiSCs in LIF and BMP is not sufficient to cause EpiSCs to revert to mESCs (Guo et al. 2009). However, EpiSCs can be reprogrammed into mESCs, including reversal of X-inactivation, with ectopic expression of the transcription factor Klf4 (Guo et al. 2009). EpiSCs are distinct from ESCs, as they can form teratomas when injected into immune-deficient mice, but are not able to form germline chimeras when injected into mouse blastocysts (Fig. 1B) (Brons et al. 2007; Tesar et al. 2007).
Human Embryonic Stem Cells
Human (h) ESCs are most closely related developmentally to mouse EpiSCs, which are derived from the postimplantation epiblast (Brons et al. 2007; Tesar et al. 2007). hESCs are derived from the ICM (Thomson 1998), but culture conditions leading to the isolation of human pluripotent stem cells (hPSCs) select for cells that behave as EpiSCs. Thus, hESCs are often referred to as hPSCs to reflect this distinction. Indeed, hESCs and mouse EpiSCs respond to the TGF-β family in the same way, whereas mESCs have different requirements to maintain their identity. hESCs show X inactivation and are maintained in culture with FGF and activin or TGF-β (Amit et al. 2000; James et al. 2005; Hall et al. 2008; Shen et al. 2008; Silva et al. 2008). hESCs can form teratomas (Thomson 1998), which is a quality of both mESCs and EpiSCs, but evaluation of the ability of hESCs to generate chimeric mice from blastocyst injections are not performed for ethical reasons. The hESC state is maintained by expression of Oct4, Nanog, and Sox2. These transcription factors physically interact and co-occupy the genome at many key hESC genes, as well as those encoding developmental regulators (Fig. 1C, right) (Boyer et al. 2005). As also shown for mESCs, these transcription factors regulate their own expression, which reinforces the transcriptional program, and occupy genes poised for differentiation (Bernstein et al. 2006).
Initial culture conditions for hESCs required growing hESCs on MEFs in media containing FCS (Thomson 1998). Blocking TGF-β signaling in these conditions was found to cause differentiation (James et al. 2005), indicating that TGF-β signaling was required to maintain the hESC state. Removal of TGF-β signaling from the media or inhibition of Smad2 and Smad3 activation by the small molecule, TβRI kinase inhibitor SB431542, each resulted in differentiation of hESCs primarily down the neuroectoderm lineage (Vallier et al. 2009a). In addition to TGF-β signaling, FGF signaling is required to maintain hESCs in culture (Amit et al. 2000). Further analysis revealed that hESCs can be cultured without MEFs using an extracellular protein matrix composed primarily of laminin and collagen (Xu et al. 2001) in chemically defined media containing albumin, TGF-β1, FGF2, LiCl, γ-aminobutyric acid, and pipecolic acid (Beattie et al. 2005; Vallier 2005; Ludwig et al. 2006). This requirement was further refined using a basal media without albumin, which requires TGF-β1 or Nodal, FGF2, insulin, selenium, transferrin, and l-ascorbic acid (Chen et al. 2011).
The TGF-β/Smad pathway interacts with the master transcription factors that regulate hESC state and differentiation. Both Oct4 and Nanog can form a protein complex with Smad2 and likely Smad3 (Smad2/3) in hESCs (Vallier et al. 2009a; Beyer et al. 2013). Furthermore, ChIP-seq analysis performed using antibodies that recognize Smad3, or Smad2 and Smad3, revealed that these transcription factors tend to co-occupy the genome with Oct4, Nanog, and Sox2 in hESCs, including at the genes encoding Oct4, Nanog, and Sox2 (Brown et al. 2011; Mullen et al. 2011). Inhibiting Smad2 and Smad3 phosphorylation using SB431542 also leads to a reduction in Oct4 and Nanog expression, although Nanog expression is more sensitive to this loss of signaling (Vallier 2005; Greber et al. 2008; Xu et al. 2008).
Smad2 and Smad3 share ∼90% homology in amino acid sequence (Yagi et al. 1999), but have different roles in embryonic development. Smad2 deficiency results in embryonic lethality as a result of a failure to specify anterior visceral endoderm (AVE), which is a Nodal-dependent event (Waldrip et al. 1998; Weinstein et al. 1998), whereas Smad3 deficiency is not embryonic lethal and results in impaired immunity and increased incidence of colorectal cancers in mice (Zhu et al. 1998; Datto et al. 1999). In addition, Smad2 plays a more significant role in maintaining the hESC state than Smad3, as depletion of Smad2 results in increased hESC differentiation, whereas depletion of Smad3 has little effect on differentiation (Sakaki-Yumoto et al. 2013). The more significant role of Smad2 may result, in part, from a higher level of Smad2 binding at the gene encoding Nanog in hESCs and a decreased dependence of the interaction with Smad4 to facilitate DNA binding (Kim et al. 2011; Sakaki-Yumoto et al. 2013). These results are interesting, as Smad4 is particularly important in vivo to specify the anterior primitive streak during gastrulation (Chu et al. 2004), in which it functions with Smad2, and the Smad2 and Smad3 DNA-binding partner Foxh1, to drive mesendoderm specification. These studies highlight that, whereas Smad4 is often considered an obligate component of Smad signaling, it is dispensable for many biological responses to TGF-β and BMP (Sirard et al. 2000).
The concept of the naïve ground state in hESCs remains controversial. hESCs are more closely related to mouse EpiSCs than to mESCs. When mouse ESCs are cultured with inhibitors of Erk MAPK, inhibitors of GSK3β, and LIF, that is, in 2i + LIF medium, they can be maintained in a ground state characterized by stable gene expression and reduced population heterogeneity (Fig. 1D) (Ying et al. 2008). Initial attempts to generate ground-state ESCs from hESCs showed that culturing hESCs in 2i + LIF medium with ectopic expression of ground-state transcription factors led to ground-state qualities, but cells were not stable and required constitutive transgene expression (Hanna et al. 2010). Additional studies identified combinations of inhibitors that could promote qualities of mESCs (Chan et al. 2013; Gafni et al. 2013; Valamehr et al. 2014; Ware et al. 2014), but did not remove the requirement for FGF. Reprogramming of human fibroblasts into ESCs by expressing Oct4, Sox2, Klf4, c-Myc, and retinoic acid receptors produced human pluripotent cells with features of ground-state ESCs that could be maintained after removal of transgene expression (Wang et al. 2011). Subsequent studies showed that it is possible to induce ground-state characteristics by persistent ectopic expression of Klf2 and Nanog in hESCs (Takashima et al. 2014). The requirement for persistent ectopic expression of Klf2 and Nanog could be eliminated by addition of 2i plus LIF and Gö6983, a PKC inhibitor (Takashima et al. 2014). This cocktail was referred to as t2iL + Gö because the concentration of GSK3 inhibitor had to be titrated from standard 2i concentrations. The need for ectopic gene expression can be bypassed in hESCs grown on feeder cells and treated with a combination of four inhibitors plus LIF, TGF-β1, and FGF2, called naïve human stem-cell medium (NHSM) (Gafni et al. 2013) or five inhibitors plus LIF and activin (5i/L/A) (Fig. 1D) (Theunissen et al. 2014). Further analysis is needed to understand if these new conditions are sufficient to achieve bona fide ground state in human ESCs. Teratoma formation does not distinguish between hESCs and human pluripotent cells that are equivalent to mESCs or ground-state mESCs. Embryo chimera assays have been performed to distinguish between hESCs and earlier developmental states, but have not yet yielded consistent results (Gafni et al. 2013; Theunissen et al. 2014).
Induced Pluripotent Stem Cells
Although transcription factors are essential for reprogramming, signaling molecules contained in the media must also play a key role during the production of iPSCs and in their maintenance. Dissection of the mechanisms underlying reprogramming induced by Oct4, Sox2, Klf4, and c-Myc in mouse somatic cells using small molecule probes revealed that inhibition of TGF-β signaling enhances reprogramming efficiency (Maherali and Hochedlinger 2009). Further, molecular profiling defined three distinct transcriptional phases during reprogramming, including an essential early mesenchymal-to-epithelial transition (MET) initiation phase that is mediated by BMP signaling (Samavarchi-Tehrani et al. 2010) in cooperation with Klf4 (Chen et al. 2010; Li et al. 2010). This is followed by an intermediate maturation phase that represents a restriction point for acquisition of pluripotent competency. Finally, transition to the stabilized pluripotent state is associated with acquisition of the full pluripotency network and independence from transgene expression (Golipour et al. 2012; Polo et al. 2012). TGF-β is a potent inducer of the mesenchymal fate, thus providing one mechanism underlying TGF-β suppression of reprogramming (Li et al. 2010). More recent studies further reveal that c-Jun similarly provides a blockade to reprogramming by promoting the mesenchymal fate (Liu et al. 2015). Because the gene encoding c-Jun is also a TGF-β target gene (Pertovaara et al. 1989) and c-Jun, as a component of AP1, also interacts with Smad3 (Zhang et al. 1998), this may provide a pathway enforcing somatic identity in mesenchymal cells. As TGF-β signaling imposes a mesenchymal phenotype in a variety of cell types (Lamouille et al. 2014), these studies establish a key antagonistic interplay between TGF-β and BMP in the control of epithelial plasticity during somatic-cell reprogramming.
Reprogramming of mouse somatic cells in mESC media yields iPSCs that behave as mESCs (Takahashi and Yamanaka 2006); that is, BMP signaling also promotes their pluripotency (see above). Thus, reprogramming of mouse somatic cells to mESC is compatible with BMP signaling throughout the reprogramming process. In contrast, reprogramming of human somatic cells using the human homologs of Oct4, Sox2, Klf4, and c-Myc generates iPSCs that behave as hESCs (Takahashi et al. 2007; Yu et al. 2007); that is, TGF-β signaling promotes their pluripotency. However, epithelial character is a defining feature of hESCs and, as in mouse reprogramming, MET is important for conversion to the pluripotent state. For example, intermediary reprogrammed cells undergo MET in transition to stable human iPSCs (Teshigawara et al. 2016), and promotion of TGF-β signaling by Ezh2 during reprogramming promotes the mesenchymal phenotype and inhibits generation of iPSC (Rao et al. 2015). Collectively, these studies indicate that during the transition to transgene-independent hiPSCs, TGF-β-dependent epithelial-to-mesenchymal transition (EMT) pathways must be circumvented to allow TGF-β-dependent stabilization of the pluripotent state. One possibility is that key EMT transcription factors, such as Snai1 may be selectively directed to pluripotency regulatory target genes during reprogramming (Gingold et al. 2014; Unternaehrer et al. 2014). For example, the gene encoding Snai1 is a TGF-β target, and the Snai1 protein interacts with Nanog to paradoxically promote pluripotency-associated gene expression during mouse reprogramming (Gingold et al. 2014). How the biological output of cell-fate-determining pathways, such as those driven by TGF-β, are contextually modified to both block and promote pluripotency is an important area of investigation.
Embryonic Stem-Cell Differentiation
hESCs can be differentiated into the three germ layers as well as into trophoblast and primitive endoderm through modulation of TGF-β family signaling (Fig. 1E). TGF-β and FGF signaling together are required to maintain pluripotency (James et al. 2005), and loss of TGF-β signaling in the continued presence of FGF results in neuroectoderm differentiation (Smith et al. 2008; Vallier et al. 2009c). Loss of FGF signaling and continued activation of Smad2 and Smad3 in response to activin or Nodal are sufficient to direct hESCs to differentiate into mesendoderm (D’Amour et al. 2005), with increasing concentrations of activin promoting formation of definitive endoderm and lower concentrations favoring mesoderm (Gadue et al. 2006). Differentiation toward endoderm is also augmented by activation of Wnt and BMP signaling (Gadue et al. 2006; Teo et al. 2012), whereas signaling through BMP-4 in the absence of TGF-β or FGF promotes differentiation of extraembryonic trophoblast and primitive endoderm (Vallier et al. 2009c; Sakaki-Yumoto et al. 2013).
Many genes bound by master transcription factors and Smad2 and/or Smad3 in hESCs are repressed. These are often bivalent genes that have chromatin marks of both an activated (histone H3 lysine 4 trimethylation, H3K4me3) and repressed (histone H3 lysine 27 trimethylation, H3K27me3) state, and are poised for differentiation (Fig. 1C) (Bernstein et al. 2006). Several of these key developmental regulators are also bound by YAP and TAZ, two transcriptional regulators of the Hippo signaling pathway (Beyer et al. 2013). When the Hippo kinase pathway is activated by a variety of extrinsic cues, both YAP and TAZ are phosphorylated and localize to the cytoplasm. In contrast, when Hippo signaling is turned off, YAP and TAZ translocate to the nucleus where they interact with TEAD transcription factors (Varelas et al. 2008). YAP and TAZ also associate with Smad2 and/or Smad3, and together bind multiple pluripotency and developmental genes, likely in a complex with TEADs. These Hippo factors can recruit HDACs, which are associated with a repressed chromatin state and inhibition of gene expression (Beyer et al. 2013). Under pluripotency conditions, this complex predominantly suppresses the mesendoderm lineage. On endoderm differentiation, YAP, TAZ, and TEAD binding are lost at multiple genes for developmental regulators, whereas Smad2 and/or Smad3 remain bound through the DNA-binding, Smad partner, Foxh1. Foxh1 is expressed throughout the epiblast of pregastrulation mouse embryos (Weisberg et al. 1998) and in hESCs (Kim et al. 2011). In mESCs, Nodal signaling requires expression of Foxh1 to promote mesendoderm differentiation (Hoodless et al. 2001; Yamamoto et al. 2001), and studies in Xenopus show that FAST2 (homolog of Foxh1) is required for activin-mediated induction of Eomes expression (Ryan et al. 2000). Eomes is also a Smad2- and Smad3-binding partner that promotes endoderm differentiation together with Foxh1 (Kim et al. 2011; Teo et al. 2011; Beyer et al. 2013). Interestingly, TAZ and YAP can control nucleocytoplasmic shuttling of Smad2 and Smad3 (Varelas et al. 2008), and serve as sensors of mechanical force (Dupont et al. 2012). This control can couple TGF-β-Smad signaling to substrate rigidity, and, in the case of hESCs, provides for more efficient neural induction and motor neuron yield when cells are cultured on soft substrates (Sun et al. 2014).
During differentiation, Smad2 and Smad3 associate with new transcriptional partners, because the ESC master transcription factors are no longer expressed (Brown et al. 2011). Thus, instead of interacting with Oct4 and Nanog, Smad2 and/or Smad3 now co-occupy the genome with Foxh1 and Eomes at sites enriched for developmental regulators during endoderm differentiation (Brown et al. 2011; Kim et al. 2011). In some cases, association of Smad2 and/or Smad3 with new transcription factors occurs at enhancers that were not occupied in ESCs, such as at the gene encoding Eomes (Fig. 1F). At other genes, such as Mixl1, Smad2 and/or Smad3 remain at the same enhancers during endoderm differentiation but now associate with different combinations of transcription factors (Brown et al. 2011; Kim et al. 2011; Beyer et al. 2013). Studies of mESC differentiation have also found that Smad2 and/or Smad3 interact with Trim33/TIF1γ to establish open chromatin through removal of HP1γ from compacted chromatin. The loss of HP1γ results in more accessible DNA to allow heteromeric complexes with Smad4 to form at new enhancers (Xi et al. 2011). Another group, however, has proposed that Trim33/TIF1γ ubiquitylates Smad4, leading to Smad4 degradation and inhibition of endoderm differentiation (Morsut et al. 2010). Although these studies appear in contradiction, it is possible that selective loss of Smad4 may alter the nature of the biological output to TGF-β signaling, particularly given the specific role that Smad4 plays in endoderm specification during gastrulation (Chu et al. 2004).
TGF-β signaling regulates expression of protein-coding genes to control differentiation, but noncoding RNAs are also targeted by TGF-β signaling. Long noncoding RNAs (lncRNAs) are a class of RNAs that are polyadenylated and have the same structure as messenger RNAs (mRNAs) (Guttman et al. 2010) but tend to be retained in the nucleus with biological activity as RNAs (Rinn and Chang 2012). Increasing numbers of these lncRNAs have been described to play significant roles in development and differentiation (Marahrens et al. 1997; Klattenhoff et al. 2013; Sauvageau et al. 2013; Herriges et al. 2014; Jiang et al. 2015), and lncRNAs have shown a diverse range of functions, including regulation of chromatin structure (Rinn et al. 2007), recruitment of transcription complexes (Wang et al. 2012), and modulation of mRNA translation and stability (Gong and Maquat 2012; Kretz et al. 2014). More than 1300 lncRNAs are induced after 2 days of treatment with activin to differentiate hESCs toward endoderm (Sigova et al. 2013). In addition, the lncRNA DEANR1 was identified after 4 days of endoderm differentiation (Jiang et al. 2015). Depletion of DEANR1 results in reduced FOXA2 expression along with reduced expression of many other genes repressed with depletion of FOXA2 mRNA. Immunoprecipitation analysis using an antibody recognizing Smad2 and Smad3 shows that DEANR1 is associated with Smad2 and/or Smad3, and that Smad2 and/or Smad3 binding to the gene encoding Foxa2 is reduced in DEANR1-deficient cells. DEANR1 is located about 2.4 kb downstream from FOXA2, and these results suggest that DEANR1 helps recruit Smad2 and/or Smad3 to the gene encoding Foxa2 during endoderm differentiation (Jiang et al. 2015). The extent of lncRNA involvement in TGF-β family signaling is not yet known, but with further investigation it is likely that many additional lncRNAs will be identified that regulate differentiation either as direct transcriptional targets of TGF-β signaling or through interactions with Smad proteins.
Smad activity during ESC differentiation is also affected by the site of Smad phosphorylation. Cyclin D expression is regulated by the cell cycle (Neganova et al. 2009), and increasing cyclin D results in increased activity of CDK4 and CDK6 (Matsushime et al. 1992; Ewen et al. 1993; Kato et al. 1993), which phosphorylate Smad2 and Smad3 in their linker region, blocking nuclear localization (Matsuura et al. 2004; Pauklin and Vallier 2013). In early G1, cyclin D expression and the activity of CDK4 and CDK6 are low, allowing TGF-β signaling to phosphorylate the carboxy-terminal region of Smad2 and Smad3. Thus, in early G1, hESCs are most receptive to TGF-β signaling and induction of endoderm differentiation. As cells progress into late G1 and through the rest of the cell cycle, cyclin D expression and CDK4 and CDK6 activity increase, resulting in phosphorylation of the Smad2 and Smad3 linker regions, which blocks nuclear localization and inhibits transmission of the TGF-β signal. hESCs at later stages in the cell cycle are therefore less responsive to TGF-β signaling and have more tendency to differentiate toward neuroectoderm (Fig. 1E) (Pauklin and Vallier 2013). The linker region of Smads is also the target for input from additional signaling pathways. In particular, Erk MAPK and GSK3 converge on conserved sites in the linker that in turn promote ubiquitin-dependent degradation of Smad1 (Fuentealba et al. 2007) and Smad2 (Alarcon et al. 2009), thus providing cross talk with both receptor tyrosine kinase pathways and Wnt signaling.
PRIMORDIAL GERM CELLS
Germ cells are critical for reproduction and propagation of the species and arise from primordial germ cells (PGCs), which produce spermatozoa and oocytes. PGCs are distinctive stem-cell pools that differentiate from the epiblast during gastrulation (Lawson and Hage 1994). PGCs are found in the posterior streak region, where they are exposed to BMP-4 and BMP-8b ligands that are expressed in the extra-embryonic ectoderm adjacent to the epiblast (Lawson et al. 1999; Ying et al. 2000). BMP is critical for the specification and expansion of PGCs, and mice deficient in BMP ligands, receptors, or Smad mediators display significantly reduced PGCs (Lawson et al. 1999; Ying et al. 2000; Chang and Matzuk 2001; Tremblay et al. 2001; Hayashi et al. 2002; Arnold et al. 2006). The visceral endoderm also contributes to PGC development. BMP-2 and the type I BMP receptor, ALK2, are both expressed primarily in the visceral endoderm in the early embryo, and loss of either factor leads to a defect in PGC differentiation (Ying and Zhao 2001; de Sousa Lopes et al. 2004). Furthermore, genetic engineering of Smad1 alleles reveals that specification of PGCs is dependent not only on activation of Smad1 by the BMP receptor, but also on MAPK inputs to the Smad linker region (Aubin 2004). In vitro cultures of PGCs are also dependent on BMPs, which, together with a cocktail of other factors, support PGC proliferation in the absence of fibroblast feeders (Farini et al. 2005). Analysis of ex vivo epiblast cultures shows that BMP-4, but not BMP-8b, is required for activation of the genes encoding Blimp1/Prdm1 and Prdm14 (Ohinata et al. 2009), two key transcriptional regulators of the PGC lineage (Yamaji et al. 2008). Prdm1 and Prdm14 induction is also dependent on Wnt-mediated activation of the mesoderm factor T (Aramaki et al. 2013). Mesoderm induction is typically accompanied by an EMT. The role of mesoderm factors creates somewhat of a conundrum, as specification of PGCs depends critically on epithelial identity and E-cadherin-mediated cell–cell interactions (Okamura et al. 2003), which must be maintained as PGCs follow an extensive migratory path to the gonads. The role of BMP signaling may thus include enforcement of epithelial identity in the PGC niche, which could mirror its roll in somatic-cell reprogramming.
NEURAL STEM CELLS
The adult brain contains two populations of neural stem cells (NSCs). These stem cells are located in the subgranular zone (SGZ) of the hippocampal dentate gyrus and the subventricular zone (SVZ) lining the lateral ventricles (Fig. 2) (Kriegstein and Alvarez-Buylla 2009; Mu et al. 2010). NSCs from both the SGZ and SVZ have the potential to differentiate into neurons, astrocytes, and oligodendrocytes in vitro. Neural differentiation in the SGZ produces glutaminergic neurons, whereas neural differentiation in the SVZ gives rise to GABAergic and dopaminergic interneurons innervating the olfactory bulb (Kriegstein and Alvarez-Buylla 2009; Mu et al. 2010). BMP signaling plays an essential role in regulating the multipotency of NSCs in both the SGZ and SVZ, but differences have evolved in how the two stem-cell populations use BMP signaling. In midbrain development, signaling through the canonical TGF-β pathway inhibits Wnt-induced proliferation and expansion of neuroepithelial cells, the NSCs of early brain development (Falk et al. 2008). However, in the adult brain, TGF-β signaling has no detectable effect on NSCs and, instead, is required at later stages of neurogenesis (He et al. 2014).
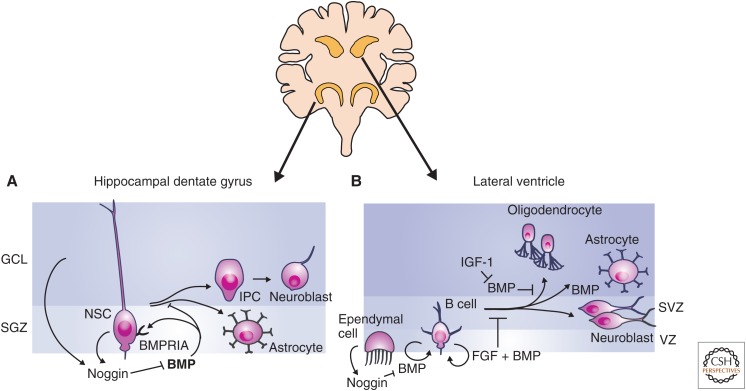
Figure 2.
Transforming growth factor β (TGF-β) family signaling in neural stem cells. Adult neural stem cells (NSCs) are primarily located in the subgranular zone (SGZ) of the hippocampal dentate gyrus and the subventricular zone (SVZ) lining the lateral ventricles. (A) Bone morphogenetic protein (BMP) signaling promotes the maintenance of the NSC state in the SGZ, and signaling through BMPRIA inhibits NSC proliferation. NSCs of the SGZ produce the BMP inhibitor Noggin. Loss of BMP signaling through repression of BMPRIA or inhibition of BMP signaling results in the formation of intermediate progenitor cells (IPCs), which differentiate into neuroblasts and astrocytes in vivo. BMP is drawn in bold in the dentate gyrus to indicate increased levels of BMP expression relative to SVZ. (B) In the SVZ, NSCs are called B cells and are maintained as slowly cycling NSCs by BMP and fibroblast growth factor (FGF) signaling. Noggin is produced by ependymal cells and inhibits of BMP signaling. BMP signaling promotes maintenance of the NSC state, but once cells begin to proliferate and differentiate, BMP signaling can also promote astrocyte differentiation. GCL, Granular cell layer.
NSCs from the SGZ of the dentate gyrus are characterized by slow cycling and expression of glial fibrillary acidic protein (GFAP), nestin, Sox2, and astrocyte-specific glutamate transporter (GLAST) (Shibata et al. 1997; Seri et al. 2001; Fukuda et al. 2003; Suh et al. 2007). NSCs can differentiate into intermediate progenitor cells (IPCs, also referred to as transit-amplifying or neural progenitor cells), which then give rise to neurons, astrocytes, and oligodendrocytes, or can directly give rise to terminal neural lineages (Fig. 2A) (Haubensak et al. 2004; Miyata 2004; Noctor et al. 2004). IPCs also express Sox2 and nestin, but are distinguished from NSCs by their more rapid proliferation, nonradial morphology, absence of GFAP expression, and lack of response to BMP signaling (Mira et al. 2010).
BMP signaling is a key factor controlling NSC maintenance. NSCs express the BMPRIA receptor and respond to BMP signaling with activation of Smad1, 5, and/or 8 (Mira et al. 2010; Sun et al. 2011). In addition, loss of BMPRIA or Smad4 expression results in an initial increase in proliferation of NSCs and neurogenesis (Mira et al. 2010), all suggesting that BMP signaling is responsible for maintaining NSCs in a slowly cycling, undifferentiated state. The BMP signal occurs primarily through the canonical BMP signaling pathway with activation of Smad1, 5, and/or 8, and not through the noncanonical Erk MAPK pathway (Sun et al. 2011). By slowing proliferation, BMP signaling may also preserve the ability to produce neurons later in life, as NSCs have limited potential for proliferation and lose the ability to proliferate with age (Hattiangady and Shetty 2008; Aizawa et al. 2011). Although loss of BMP signaling does result in an initial expansion in neural differentiation and NSC proliferation, longer-term BMP signaling blockade results in loss of both IPCs and immature neurons (Mira et al. 2010).
Inhibition of BMP signaling is a key point of control in regulating NSC differentiation. The BMP inhibitor Noggin (Balemans and Van Hul 2002) is expressed by cells of the dentate gyrus (Fan et al. 2003) and by NSCs themselves (Guo et al. 2011), showing both paracrine and autocrine inhibition of BMP signaling. The RNA-binding protein-FXR2 is expressed in NSCs where it binds Noggin mRNA and increases the rate of degradation. Thus, in NSCs of the SGV, FXR2 expression inhibits Noggin at the posttranscriptional level to promote NSC maintenance (Guo et al. 2011). Insulin-like growth factors (IGFs) affect oligodendrocyte differentiation (Masters et al. 1991), and the effect of IGF1 on neural differentiation is a result, at least in part, of induction of Noggin and Smad6 expression to inhibit BMP signaling (Hsieh 2004).
BMP signaling is also a regulator in the SVZ where NSCs are referred to as B cells and are located in the walls of the lateral ventricles (Fig. 2B). B cells express BMP-2, -4, and -7 (Lim et al. 2000; Peretto et al. 2002; Bonaguidi et al. 2008). The BMP inhibitor Noggin is not expressed in B cells or IPCs in the SVZ, and is instead expressed by the adjacent ependymal cells (Lim et al. 2000; Guo et al. 2011). As a result, FXR2 expression in B cells does not regulate Noggin production in the lateral ventricles, and loss of FXR2 does not affect B cells or IPCs in the SVZ (Guo et al. 2011). B cells also have a greater potential for expansion in vitro than NSCs of the SGZ (Seaberg and van der Kooy 2002; Bull and Bartlett 2005), but this defect can be rescued by inhibition of BMP signaling (Lim et al. 2000; Bonaguidi et al. 2008). Inhibition of BMP signaling has less effect on proliferation of B cells compared with NSCs of the SGZ, and this may reflect lower levels of BMP expression in B-cell cultures (Bonaguidi et al. 2008). BMP signaling helps maintain the B-cell state, but once differentiation is initiated, BMP signaling tends to promote astrocyte differentiation while suppressing neural and oligodendrocyte fates, and Noggin tends to have the opposite effect. BMP signaling also promotes neuroblast survival once cells have committed to the neural fate (Bond et al. 2012).
BMP signaling acts in concert with other signaling pathways including FGF signaling. FGF2 is required to maintain the NSC state in culture in conjunction with BMP (Sun et al. 2011). IGF1 promotes oligodendrocyte differentiation and induces expression of the BMP signaling inhibitors, Smad6 and Noggin. The addition of FGF2 and IGF1 to NSC cultures inhibits induction of Smad6 and Noggin gene expression and disrupts oligodendrocyte differentiation (Hsieh 2004). In addition, FGF2 signaling activates the Erk MAPK pathway, leading to phosphorylation of the Smad1 linker. Following phosphorylation of its linker regions, Smad1 is retained in the cytoplasm despite carboxy-terminal phosphorylation in response to BMP signaling (Pera 2003; Bilican et al. 2008). BMP signaling in B cells is also regulated by cyclin-dependent kinase inhibitors. p21CIP1 interacts with E2F transcription factors to inhibit expression of BMP-2, and loss of p21CIP1 expression results in increased cell proliferation, increased BMP-2 expression, and astrocyte differentiation (Kippin et al. 2005; Porlan et al. 2013).
HAIR FOLLICLE STEM CELLS
Hair follicle stem cells (HFSCs) are a slow-cycling population of cells that reside at the site of attachment of the arrector pili muscle, in a region called the bulge (Cotsarelis et al. 1990). Hair growth occurs in cycles. Telogen is the quiescent phase, anagen is the growth phase, and catagen is the follicle regression phase. In telogen (Fig. 3A), the bulge cells are located at a short distance from the dermal papilla (DP), which provides many of the signals to begin hair growth. The HFSCs in the closest proximity to the DP are referred to as hair germ (HG) cells. During anagen, the HFSCs receive signals to proliferate and differentiate into cells of the matrix, which produce the hair follicle and follicle-supporting cells. The expanding cell population pushes the DP further from the HFSCs, reducing the proliferative signal received by the HFSCs (Fig. 3B). In catagen, apoptosis leads to loss of the matrix cells and the DP moves back in close proximity to the HG cells.
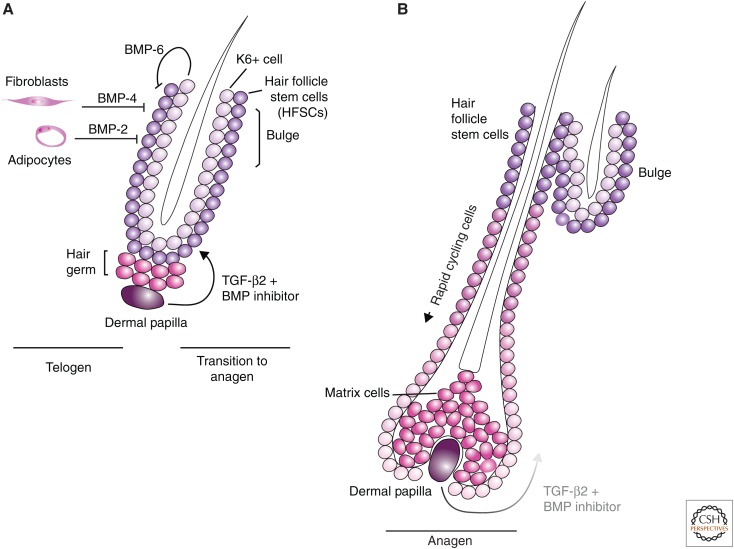
Figure 3.
Transforming growth factor β (TGF-β) family signaling in hair follicle stem cells. (A) Telogen represents the quiescent state of hair follicle development. Hair follicle stem cells (HFSCs) are located in the bulge. Cells including dermal fibroblasts and adipocytes and K6+ cells secrete bone morphogenetic protein (BMP) molecules, which maintain HFSCs in their slowly proliferating state. Expression of Noggin and TGF-β from the dermal papilla (DP) cells blocks BMP signaling and promotes proliferation of the HFSCs. The progenitor cells that differentiate from the HFSCs are responsible for growth of the hair follicle. (B) During anagen, the hair follicle grows, pushing the DP and the TGF-β2 produced by these cells further from the HFSCs in the bulge. At a certain point, the BMP signaling wins out over the TGF-β signal, resulting in HFSC quiescence and atrophy of the hair follicle.
HFSCs were originally identified in the bulge because of the retention of tritiated thymidine, which marked them as cycling more slowly than neighboring cells (Cotsarelis et al. 1990; Chen et al. 2008b). HFSCs are present as a single layer of cells adjacent to an inner layer of cells that stabilize the old hair (Hsu et al. 2011). HFSCs are identified by surface expression of CD34 and by expression of the transcription factors Tcf3, Sox9, Lhx2, and NFATc1, and of Lgr5 (Merrill et al. 2001; Vidal et al. 2005; Rhee et al. 2006; Horsley et al. 2008; Jaks et al. 2008).
BMP signaling promotes telogen, while inhibition of BMP signaling in coordination with induction of Wnt and FGF signaling induces hair growth. In telogen, the shaft is short and the base of the bulge cells (HG cells) is in contact with the DP. The follicle is maintained in telogen primarily by secretion of BMP by surrounding cell populations. BMP-2 is expressed by adipocytes and BMP-4 is expressed by dermal fibroblasts (Plikus et al. 2008). BMP-6 is produced by the keratin 6+ inner bulge cells, which also produce FGF18, to suppress proliferation of HFSCs (Hsu et al. 2011). BMP signaling leads to activation of Smad1, 5, and/or 8 through the BMPRIA receptor (Kulessa et al. 2000; Kobielak et al. 2003; Plikus et al. 2008). BMP activation is also associated with expression of active (nonphosphorylated) PTEN, which blocks PI3K-Akt signaling and thus decreases Wnt signaling because of stabilization of GSK3β (Kobielak et al. 2003; Zhang et al. 2006b). Thus, BMP signaling inhibits proliferation and antagonizes Wnt signaling to promote telogen.
The transition from telogen to anagen is marked by suppression of BMP signaling, which results in HFSC proliferation and differentiation. DP cells express the BMP inhibitor Noggin (Zimmerman et al. 1996) and, by late anagen, Noggin is expressed throughout the hair follicle (Botchkarev et al. 2001; Zhang et al. 2006b). Sostdc1 (also known as ectodin or USAG-1), another BMP inhibitor (Laurikkala et al. 2003; Ying et al. 2008; Wray et al. 2011; Yi et al. 2011) is also secreted by DP cells at increasing levels through telogen (Rendl et al. 2005; Greco et al. 2009). In addition, FGF7 and FGF10 are produced by DP cells, and induce increased HFSC proliferation (Hsu et al. 2011), in contrast to the effects of FGF18. Smad2 phosphorylation is detected in HFSCs just before the transition from telogen to anagen, and Smad2 is activated by TGF-β2 produced by DP cells but not by TGF-β1 or TGF-β3. The most pronounced effect of TGF-β2 signaling is observed in HG cells, which show a loss in Smad1, 5, and/or 8 activation and increased proliferation in response to TGF-β2 signaling. TGF-β2 signaling is dose-dependent; low doses promote proliferation, whereas high doses inhibit proliferation. This activity is mediated in significant part by transmembrane protein with epidermal growth factor–like and two follistatin-like domains 1 (Tmeff1, also known as Tomoregulin-1), which suppresses BMP signaling. Tmeff1 gene expression is activated by TGF-β2 signaling through the TGF-β type II receptor (TβRII), and the Tmeff1 promoter region is directly occupied by Smad2 and/or Smad3 (Oshimori and Fuchs 2012). With inhibition of BMP signaling, Wnt promotes cell proliferation and differentiation leading to follicle growth.
HEMATOPOIETIC STEM CELLS
Hematopoietic stem cells (HSCs) reside in the bone marrow in adult vertebrates and are the source of red blood cells, platelets, and white blood cells of both lymphoid and myeloid lineages (Wang and Wagers 2011). The HSC state is maintained by signals from the microenvironment or niche (Morrison and Scadden 2014), which is composed of cells, including mesenchymal stem/stromal cells (Dexter et al. 1977; Méndez-Ferrer et al. 2010), endothelial cells (Kiel et al. 2005), osteoblasts (Calvi et al. 2003; Zhang et al. 2003; Arai et al. 2004), and sympathetic nerve fibers (Yamazaki et al. 2011).
Under steady-state conditions, HSCs are maintained in a slowly proliferating state in which cells remain largely in G0 and divide every 1–2 months (Bradford et al. 1997; Cheshier et al. 1999; Sudo et al. 2000). Signals from stem-cell factor (SCF) (Ding et al. 2012), CXCL12 (Sugiyama et al. 2006) and TGF-β (Keller et al. 1988) are required for HSC maintenance (Fig. 4). Endothelial cells produce SCF, whereas mesenchymal stem cells (MSCs) produce both SCF and CXCL12 (Ding et al. 2012; Ding and Morrison 2013; Greenbaum et al. 2013). HSCs produce TGF-β, but it is in an inactive (latent) form (Yamazaki et al. 2009), and activated TGF-β must be provided by the niche. TGF-β signaling was originally reported to inhibit the proliferation of HSCs in vitro (Keller et al. 1988), and Smad2 and/or Smad3 were found to be phosphorylated in freshly isolated HSCs consistent with active TGF-β signaling (Yamazaki et al. 2009). The formation of lipid rafts occurs with the onset of proliferation and differentiation and is associated with activation of Akt, which leads to inactivation of the transcription factor, FoxO (Yamazaki et al. 2006), and this process is inhibited by TGF-β signaling (Yamazaki et al. 2009). Treatment of HSCs with TGF-β is also associated with maintenance of cytoplasmic localization of p57INK4B and cyclin D1 leading to inhibition of cell division (Yamazaki et al. 2009).
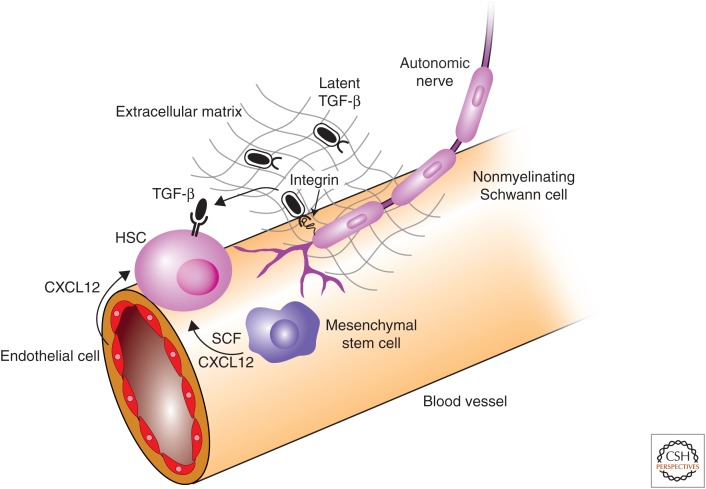
Figure 4.
Transforming growth factor β (TGF-β) family signaling in hematopoietic stem cells. The niche provides signaling support to maintain hematopoietic stem-cell (HSC) state. CXCL12 and stem-cell factor (SCF) are both required for HSC maintenance and are provided by mesenchymal stem cells (MSCs) and endothelial cells. Inactive (latent) TGF-β is produce by HSCs and other cells in the niche. Nonmyelinating Schwann cells express integrin αvβ8, which binds the latent TGF-β complex and recruits metalloproteinases that cleave latent TGF-β and release active TGF-β. TGF-β acts through TβRII on the surface of HCSs to inhibit proliferation and promote maintenance of HSC state.
Although TGF-β signaling inhibits proliferation of HSCs (Keller et al. 1988; Ottmann and Pelus 1988; Sitnicka et al. 1996), BMP promotes HSC specification and expansion during gastrulation in vertebrates and is required for in vitro HSC maintenance and proliferation (Bhatia et al. 1999; Kang et al. 2004). BMPs specify the HSC lineage in cooperation with Wnt signaling and the induction of Cdx and Hox gene expression and consequent transcriptional networks (Lengerke et al. 2008). The emergence of the HSC population during gastrulation also requires the TGF-β family accessory receptor, endoglin, which marks all cells of hematopoietic fate and binds both TGF-β and BMP receptor complexes (Barbara et al. 1999; Borges et al. 2012). Hematopoietic and endothelial progenitor cells are unusual in that they display strong TGF-β-dependent activation of Smad1 and show elevated expression of endoglin (Oh et al. 2000; Borges et al. 2012), which modulates the magnitude of TGF-β-dependent Smad1 versus Smad2 activation (Pece-Barbara et al. 2005). This may provide a mechanism to balance BMP- versus TGF-β-mediated Smad activation within the HSC niche and provides a key example of how cellular context directs biological outputs in response to TGF-β family signaling (Pece-Barbara et al. 2005).
Cross talk between TGF-β and BMP signaling has made mechanistic interpretation of genetic data challenging, and modulation of individual components of the TGF-β signaling pathway initially provided conflicting results. HSCs deficient in TβRI showed a defect in proliferation in vitro but not in vivo (Larsson 2003). This receptor was later found to be expressed at low levels in HSCs (Utsugisawa et al. 2006; Yamazaki et al. 2009), making the results more difficult to interpret. Overexpression of Smad7, an inhibitory Smad that antagonizes TGF-β family–induced Smad signaling, results in an expanded HSC pool in vivo while inhibiting HSC proliferation in vitro (Blank et al. 2006). Targeted deletion of Smad4 in HSCs, which would be predicted to disrupt TGF-β family signaling, caused decreased self-renewal in competition with wild-type HSCs in vivo and revealed no defect in proliferation in vitro (Karlsson et al. 2007).
Subsequent studies provide a clearer picture of the role of TGF-β signaling in HSCs. TβRII is highly expressed in HSCs, and HSCs deficient for Tgfbr2 show decreased Smad2 and/or Smad3 phosphorylation and an increased propensity to enter the cell cycle both in vitro and in vivo (Yamazaki et al. 2011). HSCs are defined by combinations of surface markers (Kiel et al. 2005) and have the ability to reconstitute all lineages of the blood, but they are not a homogenous population (Mercier et al. 2011). HSCs can be divided into myeloid-biased and lymphoid-biased HSCs. Each population is capable of reconstituting all blood lineages, but they show a tendency toward individual lineages (Challen et al. 2010). These subpopulations were found to respond differently to TGF-β signaling. Higher concentrations of TGF-β are associated with decreased proliferation of both myeloid and lymphoid HSCs, whereas lower concentrations of TGF-β signaling promote myeloid HSC proliferation and inhibit lymphoid HSC proliferation (Challen et al. 2010).
HSCs primarily secrete TGF-β1 as do other cell types in the niche (Yamazaki et al. 2009), but, in each case, TGF-β is secreted in a latent form and is not biologically active. Latent TGF-β forms a complex with TGF-β latency-associated protein (LAP) and is stored in the extracellular matrix (Annes et al. 2003). Nonmyelinating Schwann cells, which are associated with small autonomic axons adjacent to blood vessels in the bone marrow, were proposed to be a key regulator of TGF-β activity (Yamazaki et al. 2011). These cells express integrin αvβ8, which directs metalloproteinases to cleave latent TGF-β (Munger et al. 1999; Annes et al. 2003). Immunofluorescence analysis showed that nonmyelinating Schwann cells are located in close proximity to active TGF-β. In addition, loss of these specific Schwann cells after transection of sympathetic nerves results in a significant decrease in Smad2 and/or Smad3 phosphorylation in HSCs and increased proliferation (Yamazaki et al. 2011). The investigators propose that integrin αvβ8 binds latent TGF-β in the extracellular matrix leading to recruitment of metalloproteinases and cleavage of the latent TGF-β complex to release active TGF-β. Overall, these results present a complex picture in which the signal strength, differences in HSC subpopulations, and the niche all interact to determine the response to TGF-β signaling.
INTESTINAL STEM CELLS
Intestinal stem cells are located in the crypts of the small intestine and colon and provide a constant supply of intestinal epithelial cells that migrate to the villi and are then shed after several days (Fig. 5A) (Leblond and Stevens 1948; Creamer et al. 1961). Intestinal crypts contain two types of stem cells. Lgr5+ stem cells proliferate more rapidly and are responsible for the production of intestinal epithelial cells (Barker et al. 2007), while +4 stem cells are quiescent and are important for regeneration of the intestinal epithelium after injury (Sangiorgi and Capecchi 2008). The stem cells are interdigitated between Paneth cells, and Paneth cells provide many of the ligands in the niche to support the intestinal stem cells (Sato et al. 2011). Wnt ligands are a key signal promoting self-renewal and proliferation of intestinal stem cells and are expressed at higher levels in the crypts than the villi (Batlle et al. 2002; Gregorieff et al. 2005). In the presence of higher Wnt activity in the crypts, Notch signaling helps prevent differentiation of Lgr5+ cells by inhibiting differentiation into secretory cells. Wnt activity decreases as cells migrate into the villi, and, in the setting of reduced Wnt activity, Notch promotes differentiation of absorptive epithelial cells instead of secretory cells (Fre et al. 2005; van Es et al. 2005; Medema and Vermeulen 2011). The BMP gradient is opposite to that of Wnt (Haramis et al. 2004; Kosinski et al. 2007), and BMP-4 is detected in the intervillous mesenchyme at higher levels than in mesenchyme surrounding the crypts (Haramis et al. 2004; Kosinski et al. 2007). Noggin, Gremlin1 and 2, and Chordin-like 1 are BMP antagonists and are expressed at higher levels in the crypts or their underlying mesenchyme (He et al. 2004; Kosinski et al. 2007). Loss of BMP signaling in intestinal epithelium either through loss of BMPRIA expression, ectopic expression of Noggin, or elevated Gremlin1 expression results in an expansion of crypts suggesting that BMP signaling represses crypt formation and inhibits expansion of the stem-cell niche (Haramis et al. 2004; He et al. 2004; Davis et al. 2015). Treatment of crypt cells with Noggin leads to phosphorylation of PTEN and Akt and is associated with translocation of β-catenin to the nucleus, suggesting that BMP signaling may directly inhibit Wnt signaling in crypts (He et al. 2004). Furthermore, human mutations in the genes encoding Smad4 (Houlston et al. 1998) or BMPRIA (Zhou et al. 2001) are associated with juvenile polyposis coli, whereas hereditary mixed polyposis syndrome is associated with duplication of GREM1, the gene encoding Gremlin1 (Davis et al. 2015). These genetic disorders associated with loss of BMP activity lead to adenoma formation and colorectal cancer that arises from expansion of the intestinal stem-cell compartment. Understanding the signaling requirements for Lgr5+ intestinal stem cells has now led to efficient in vitro maintenance and expansion of intestinal stem cells with R-spondin1 (activation of Wnt signaling), Noggin (inhibition of BMP signaling), Notch, and EGF (Sato et al. 2009).
Figure 5.
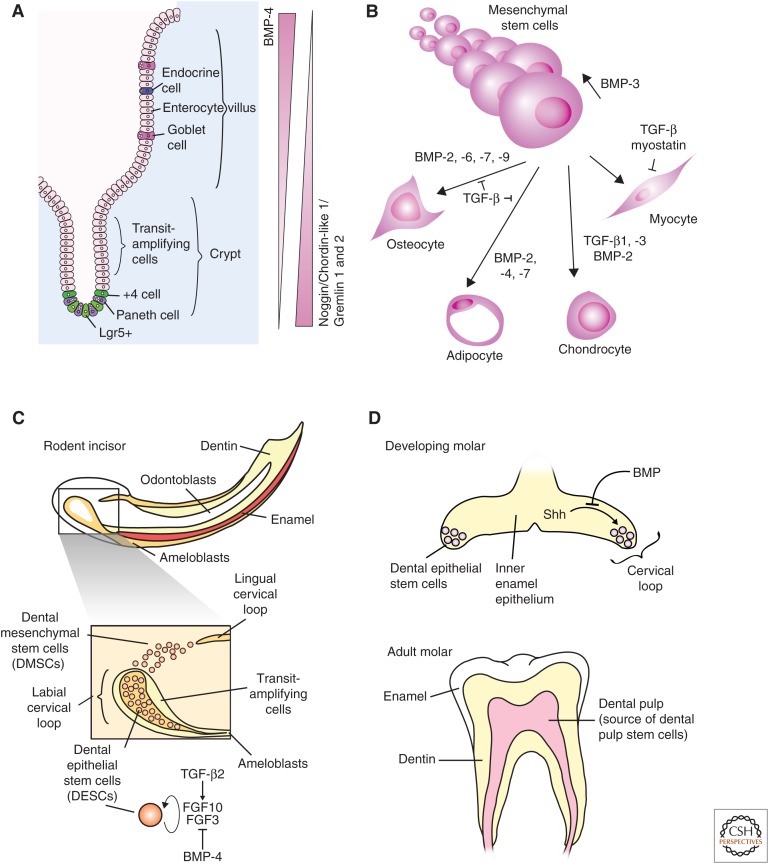
Transforming growth factor β (TGF-β) family signaling in intestinal, mesenchymal, and dental stem cells. (A) Bone morphogenic protein 4 (BMP-4) signaling promotes differentiation of intestinal stem cells, whereas inhibition of BMP signaling by Noggin, Gremlin 1 or 2, and Chordin-like 1 promote maintenance of the intestinal stem-cell fate. (B) Mesenchymal stem-cell (MSC) maintenance and differentiation is regulated by BMP and TGF-β signaling. BMP-3 signaling promotes MSC proliferation and the indicated BMPs promote osteocyte and adipocyte differentiation. TGF-β1 and -β3 and BMP-2 coordinate to promote chondrocyte differentiation, and both myostatin and TGF-β inhibit myoblast differentiation. (C) Rodent incisors continue to grow throughout life, and growth is maintained by dental epithelial stem cells (DESCs) and dental mesenchymal stem cells (DMSCs). DESCs differentiate into ameloblasts to produce enamel, and DMSCs differentiate into odontoblasts to produce dentin. TGF-β2 increases and BMP-4 represses expression of fibroblast growth factor 3 (FGF3) and FGF10. FGF3 and FGF10 inhibit differentiation and promote proliferation of DESCs. (D) Dental stem cells are present in the developing molar and are lost in the adult molar. Shh promotes maintenance of the DESCs in the developing molar until the signal is blocked by BMP (top). The adult molar does not contain DESCs, but the dental pulp is a source of MSCs (bottom).
MESENCHYMAL STEM CELLS
Adult MSCs are present in many adult tissues including the bone marrow, adipose tissue, muscle, and dental pulp. They can self-renew and have the potential to differentiate into multiple lineages including bone, cartilage, fat, tendon, and muscle (Friedenstein et al. 1968; Beresford et al. 1992; Pittenger et al. 1999). These cells are of clinical interest as they are readily accessible from adult patients and provide a source of connective tissues for wound repair. MSCs are defined as mononuclear cells that can attach to plastic, proliferate in culture, and differentiate into mesenchymal lineages (Prockop 1997). This definition encompasses a heterogeneous population of cells that can differ in gene-expression patterns and differentiation potential depending on the tissue of origin. MSCs were originally derived from the bone marrow (BMSCs), and BMSCs are often the point of comparison for other MSC populations. BMSCs have been differentiated into bone, cartilage, fat, tendon, and muscle cells (Friedenstein et al. 1968; Beresford et al. 1992; Pittenger et al. 1999), and were also found to be required to maintain HSCs in culture (Dexter et al. 1977). MSCs derived from tendons are called tendon-derived stem cells (TDSCs). TDSCs can be differentiated into osteocytes, chondrocytes, and adipocytes in vitro and can produce bone, cartilage, fat, and tendon-like tissue when injected in vivo (Bi et al. 2007). When compared with BMDCs, TDSCs express higher levels of BMPRIA, BMPRIB, and BMPRII and produce more osteocytes in response to BMP-2 (Rui et al. 2011). The dental papilla is a source of dental pulp stem cells (Smith et al. 1995; Chai et al. 2000). These stem cells are capable of differentiating into odontoblasts and producing dentin-like structures. When compared with BMSCs, they were initially found to be deficient in the production of osteocytes and adipocytes (Gronthos et al. 2000), but were later found to be capable of adipocyte differentiation under different conditions (Gronthos et al. 2002). MSCs were also recently characterized from rodent incisors. These MSCs are capable of differentiation into adipocytes, chondrocytes, and produce calcified tissue (Zhao et al. 2014). MSCs isolated from articular chondrocytes have the potential to produce chondrocytes, osteocytes, and adipocytes, but there are limited comparisons to BMSCs (Barbero et al. 2003; Tallheden et al. 2003). MSCs isolated from adipose tissue can differentiate into bone, cartilage, fat, and muscle. They share many, but not all surface markers with BMSCs and, unlike BMSCs, lipid-derived MSCs do not express detectable BMP-2 (Zuk et al. 2001, 2002).
MSC differentiation has been studied using primary MSC cultures as well as multipotent mesenchymal cell lines (Fig. 5B). C3H10T1/2 cells (Reznikoff et al. 1973) are mesenchymal cells derived from mouse embryos, and C2C12 cells (Yaffe and Saxel 1977) are multipotent myoblast cells. Both of these lines were used in many early studies that showed the differentiation potential of MSCs (Taylor and Jones 1979; Davis et al. 1987). Experiments using C3H10T1/2 cells established the significance of BMP in MSC differentiation with the discovery that BMP-2 and BMP-4 primarily promote differentiation of MSCs into osteocytes but also induce development of adipocytes and chondrocytes (Ahrens et al. 1993). BMP-2, -4, -6, -7, and -9 have all been shown to promote differentiation toward osteocytes using C3H10T1/2 and C2C12 lines (Luu et al. 2007). This process is more efficient in mouse MSCs compared with human MSCs, possibly as a result of variations in Noggin expression (Osyczka et al. 2004). In BMSCs, BMP-2 signaling leads to induction of expression of the key osteocyte transcription factor, Runx2 (also known as Cbfa1) (Komori et al. 1997; Otto et al. 1997). Although Runx2 turns out to be an indirect target of BMP signaling, Runx2 does physically interact with Smad1 and Smad5 during differentiation (Lee et al. 2000).
TGF-β signaling also induces Runx2 expression (Lee et al. 2000). However, the overall effect of TGF-β signaling on C2C12 cells is to inhibit osteocyte differentiation, and both TGF-β1 and TGF-β3 promote chondrocyte differentiation of BMSCs (Johnstone et al. 1998; Mackay et al. 1998; Pittenger et al. 1999). The TGF-β-dependent inhibition of osteocyte differentiation is mediated at least in part by the interaction between Smad3 and Runx2, which leads to recruitment of HDAC4 or 5 to sites bound by the Smad3–Runx2 complex, including the gene encoding osteocalcin (Kang et al. 2005). Inhibition of the TβRI kinase activity with a small molecule results in a loss of chondrocyte differentiation, while inhibition of BMP type I receptor function disrupts differentiation of BMSCs to chondroblasts, but does not inhibit progression of chondroblasts to chondrocytes (Hellingman et al. 2011).
BMP-2, -4, and -7 signaling promote adipocyte differentiation through activation of PPARγ expression, the key transcription factor regulating adipogenesis (Cristancho and Lazar 2011), whereas concomitantly inhibiting myocyte differentiation (Lee et al. 2000; Jin et al. 2006). BMP signaling induces nuclear localization of the transcription factor Schnurri, which can interact with Smad1, Smad4, and C/EBPα to induce expression of PPARγ (Jin et al. 2006). BMP-mediated activation of p38 MAPK also promotes adipogenesis by increasing PPARγ activity (Hata et al. 2003; Huang et al. 2009), whereas TGF-β antagonizes adipogenesis in vitro through Smad3-mediated inhibition of C/EBP activity (Choy and Derynck 2003).
There are two different classes of adipocytes. White adipocytes produce white fat that stores triglycerides. Brown adipocytes produce brown fat that is involved with energy expenditures, and promotion of brown fat over white fat may help reduce health problems related to obesity (Gesta et al. 2007). Pretreatment of C3H10T1/2 cells with BMP-7 for 3 days before adipocyte differentiation led to induction of C/EBPδ expression before differentiation and increased production of brown adipocytes compared with white adipocytes, including induction of expression of UCP1, a key protein that mediates energy expenditure (Tseng et al. 2008). Injection of BMP-7-treated C3H10T1/2 cells into immunodeficient mice also led to production of predominantly brown fat over white fat, and Bmp7−/− mice showed decreased production of brown fat (Tseng et al. 2008).
TGF-β and myostatin both inhibit muscle differentiation. Myogenic differentiation of C3H10T1/2 and C2C12 cells is inhibited by TGF-β signaling through phosphorylated Smad3 that can interact with and inhibit the function of MyoD1 in myogenesis. This process is dependent on Smad3 and not Smad2 (Liu et al. 2001). In vivo, myostatin is a key negative regulator of muscle differentiation, as genetically engineered mice bred for myostatin mutations and humans with naturally occurring mutations, all display notable increases in lean muscle mass (Allen et al. 2011). In C3H10T1/2 cells, myostatin binds to the type II receptor, ActRIIB (gene name, ACVR2B) in combination with ActRIB/ALK4, and TβRI leading to phosphorylation of Smad2 and Smad3. This interaction blocks BMP-7 signaling by competing for ActRIIB leading to inhibition of adipocyte differentiation, but does not inhibit BMP-2 signaling (Rebbapragada et al. 2003).
TGF-β and BMP signaling molecules are major factors directing MSC differentiation. In addition to those described above, BMP-3 signaling promotes MSC proliferation through activation of Smad2 phosphorylation (Stewart et al. 2010). It remains unclear how the TGF-β family directs such diverse responses in MSC differentiation, but this process likely involves coordination with other signaling pathways including Wnt-β-catenin signaling (Ross et al. 2000), the strength of TGF-β family signaling, and heterogeneity in response to TGF-β family signaling (Chen et al. 1998).
DENTAL STEM CELLS
Different populations of stem cells are present in rodent incisors, which show continued growth in adults and in molars, which do not continue to grow. Rodent incisors contain two stem-cell populations. Dental epithelial stem cells (DESCs) are present in the cervical loop and differentiate into ameloblasts to produce enamel, whereas dental mesenchymal stem cells (DMSCs) are located adjacent to the cervical loop and differentiate into odontoblasts to produce dentin (Fig. 5C) (Harada et al. 1999; Seidel et al. 2010; Zhao et al. 2014). These two stem-cell populations work together to maintain incisor growth. Conditional deletion of Smad4 also promotes expansion of DESCs in rodent incisors (Li et al. 2015) where both DESCs and DMSCs are expressed in the adult. In contrast, conditional loss of TβRI expression blocks TGF-β signaling and leads to a loss of DESCs and a defect in proliferation of transient-amplifying cells produced by DESCs. FGF10 is required for maintenance of DESCs (Harada et al. 1999, 2002), and loss of TβRI expression can be rescued with exogenous FGF10, suggesting that FGF10 acts downstream from TGF-β signaling (Zhao et al. 2011). Conditional deletion of Tgfb2 in the dental mesenchyme is associated with increased DESC differentiation, malformation of the incisors, decreased expression of FGF3 and FGF10, and increased expression of Wnt5. The effects of Tgfb2 deletion could be partially rescued by inhibition of Wnt signaling, suggesting that mesenchymal TGF-β2 production controls Wnt and FGF10 expression to regulate DESC maintenance and differentiation (Yang et al. 2014). The phenotype observed with loss of Smad4 expression is likely a result of loss of BMP signaling, as dental mesenchyme produces BMP-4, which inhibits FGF3 expression (Wang et al. 2007), and suggests that Smad4 may play a larger role in mediating BMP signaling in this niche compared with TGF-β signaling. DMSCs make up only ∼5% of mesenchymal tissue adjacent to the cervical loop (Zhao et al. 2014), and their differentiation is regulated by sonic hedgehog (Shh), which is produced by the neurovascular bundle. It is not yet clear whether DMSCs play a role in production of FGF3, FGF10, or Wnt proteins in response to TGF-β signaling in the incisor mesenchyme.
In adult molars, the cervical loop and DESCs are lost during development, which limits the growth potential of the teeth (Fig. 5D) (Tummers 2003). Conditional deletion of Smad4 or Bmpr1a results in persistence of the cervical loop in molars, and this finding was associated with expansion of DESCs as defined by expression of Sox2 (Li et al. 2015). This response was not observed with conditional deletion of Tgfb2. Deletion of Smad4 in dental epithelial cells was also associated with increased expression of Shh in the niche and the maintenance of Gli1 (GLI family zinc finger protein) positive cells. Gli1 expression is induced by Shh signaling, and Sox2 expression, which marks DESCs, persists in Gli1-positive cells (Li et al. 2015). These findings suggest that BMP signaling is responsible for loss of DESCs during molar maturation and acts by reducing Shh expression in the niche of the immature cervical loop.
CONCLUSIONS
The TGF-β family is a key regulator of stem-cell state and differentiation from the earliest stages of development to homeostasis of the adult organism. TGF-β and BMP signaling each play diverse roles in controlling both embryonic and adult stem-cell fate. These roles are determined by the interaction with the unique patterns of transcription factors and signaling receptors that define each cell type in combination with additional signals from their niches. Further understanding of how the TGF-β family regulates stem-cell state will help unlock the full potential of stem cells in regenerative medicine.
ACKNOWLEDGMENTS
We thank Amar Sahay, David Sykes, Kaveh Daneshvar, and Chan Zhou for helpful discussion and Tom DiCesare for assistance with graphics. This work is supported by National Institutes of Health (NIH) Grants DK090122 and DK104009 (A.C.M.), by the Massachusetts General Hospital Executive Committee on Research (A.C.M.), and by the Canadian Institutes of Health Research Grant FDN-143252 (J.L.W.).
Footnotes
Editors: Rik Derynck and Kohei Miyazono
Additional Perspectives on The Biology of the TGF-β Family available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Ahrens M, Ankenbauer T, Schröder D, Hollnagel A, Mayer H, Gross G. 1993. Expression of human bone morphogenetic proteins-2 or -4 in murine mesenchymal progenitor C3H10T1/2 cells induces differentiation into distinct mesenchymal cell lineages. DNA Cell Biol 12: 871–880. [DOI] [PubMed] [Google Scholar]
- Aizawa K, Ageyama N, Terao K, Hisatsune T. 2011. Primate-specific alterations in neural stem/progenitor cells in the aged hippocampus. Neurobiol Aging 32: 140–150. [DOI] [PubMed] [Google Scholar]
- Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. 2009. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell 139: 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen DL, Hittlel DS, McPherron AC. 2011. Expression and function of myostatin in obesity, diabetes, and exercise adaptation. Med Sci Sports Exerc 43: 1828–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, Itskovitz-Eldor J, Thomson JA. 2000. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol 227: 271–278. [DOI] [PubMed] [Google Scholar]
- Annes JP, Munger JS, Rifkin DB. 2003. Making sense of latent TGFβ activation. J Cell Sci 116: 217–224. [DOI] [PubMed] [Google Scholar]
- Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, Ito K, Koh GY, Suda T. 2004. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 118: 149–161. [DOI] [PubMed] [Google Scholar]
- Aramaki S, Hayashi K, Kurimoto K, Ohta H, Yabuta Y, Iwanari H, Mochizuki Y, Hamakubo T, Kato Y, Shirahige K, et al. 2013. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev Cell 27: 516–529. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Maretto S, Islam A, Bikoff EK, Robertson EJ. 2006. Dose-dependent Smad1, Smad5 and Smad8 signaling in the early mouse embryo. Dev Biol 296: 104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin J. 2004. In vivo convergence of BMP and MAPK signaling pathways: Impact of differential Smad1 phosphorylation on development and homeostasis. Genes Dev 18: 1482–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balemans W, Van Hul W. 2002. Extracellular regulation of BMP signaling in vertebrates: A cocktail of modulators. Dev Biol 250: 231–250. [PubMed] [Google Scholar]
- Barbara NP, Wrana JL, Letarte M. 1999. Endoglin is an accessory protein that interacts with the signaling receptor complex of multiple members of the transforming growth factor-β superfamily. J Biol Chem 274: 584–594. [DOI] [PubMed] [Google Scholar]
- Barbero A, Ploegert S, Heberer M, Martin I. 2003. Plasticity of clonal populations of dedifferentiated adult human articular chondrocytes. Arthritis Rheum 48: 1315–1325. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Batlle E, Henderson JT, Beghtel H, van den Born MMW, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, et al. 2002. β-Catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 111: 251–263. [DOI] [PubMed] [Google Scholar]
- Batlle-Morera L, Smith A, Nichols J. 2008. Parameters influencing derivation of embryonic stem cells from murine embryos. Genesis 46: 758–767. [DOI] [PubMed] [Google Scholar]
- Beattie GM, Lopez AD, Bucay N, Hinton A, Firpo MT, King CC, Hayek A. 2005. Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells 23: 489–495. [DOI] [PubMed] [Google Scholar]
- Beresford JN, Bennett JH, Devlin C, Leboy PS, Owen ME. 1992. Evidence for an inverse relationship between the differentiation of adipocytic and osteogenic cells in rat marrow stromal cell cultures. J Cell Sci 102: 341–351. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. [DOI] [PubMed] [Google Scholar]
- Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X, Wrana JL. 2013. Switch enhancers interpret TGF-β and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep 5: 1611–1624. [DOI] [PubMed] [Google Scholar]
- Bhatia M, Bonnet D, Wu D, Murdoch B, Wrana J, Gallacher L, Dick JE. 1999. Bone morphogenetic proteins regulate the developmental program of human hematopoietic stem cells. J Exp Med 189: 1139–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y, Ehirchiou D, Kilts TM, Inkson CA, Embree MC, Sonoyama W, Li L, Leet AI, Seo BM, Zhang L, et al. 2007. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med 13: 1219–1227. [DOI] [PubMed] [Google Scholar]
- Bilican B, Fiore-Heriche C, Compston A, Allen ND, Chandran S. 2008. Induction of Olig2+ precursors by FGF involves BMP signalling blockade at the Smad level. PLoS ONE 3: e2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. 2009. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev 23: 2484–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank U, Karlsson G, Moody JL, Utsugisawa T, Magnusson M, Singbrant S, Larsson J, Karlsson S. 2006. Smad7 promotes self-renewal of hematopoietic stem cells. Blood 108: 4246–4254. [DOI] [PubMed] [Google Scholar]
- Bonaguidi MA, Peng CY, McGuire T, Falciglia G, Gobeske KT, Czeisler C, Kessler JA. 2008. Noggin expands neural stem cells in the adult hippocampus. J Neurosci 28: 9194–9204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Bhalala OG, Kessler JA. 2012. The dynamic role of bone morphogenetic proteins in neural stem cell fate and maturation. Dev Neurobio 72: 1068–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges L, Iacovino M, Mayerhofer T, Koyano-Nakagawa N, Baik J, Garry DJ, Kyba M, Letarte M, Perlingeiro RCR. 2012. A critical role for endoglin in the emergence of blood during embryonic development. Blood 119: 5417–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev V, Botchkareva N, Nakamura M, Huber O, Funa K, Luaster R, Paus R, Gilchrest BA. 2001. Noggin is required for induction of the hair follicle growth phase in postnatal skin. FASEB J 15: 2205–2214. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nat Cell Biol 441: 349–353. [DOI] [PubMed] [Google Scholar]
- Bradford GB, Williams B, Rossi R, Bertoncello I. 1997. Quiescence, cycling, and turnover in the primitive hematopoietic stem cell compartment. Exp Hematol 25: 445–453. [PubMed] [Google Scholar]
- Bradley A, Evans M, Kaufman MH, Robertson E. 1984. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309: 255–256. [DOI] [PubMed] [Google Scholar]
- Brons IGM, Smithers LE, Trotter MWB, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195. [DOI] [PubMed] [Google Scholar]
- Brown S, Teo A, Pauklin S, Hannan N, Cho CHH, Lim B, Vardy L, Dunn NR, Trotter M, Pedersen R, et al. 2011. Activin/Nodal signaling controls divergent transcriptional networks in human embryonic stem cells and in endoderm progenitors. Stem Cells 29: 1176–1185. [DOI] [PubMed] [Google Scholar]
- Bull ND, Bartlett PF. 2005. The adult mouse hippocampal progenitor is neurogenic but not a stem cell. J Neurosci 25: 10815–10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, et al. 2003. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425: 841–846. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. 2000. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127: 1671–1679. [DOI] [PubMed] [Google Scholar]
- Challen GA, Boles NC, Chambers SM, Goodell MA. 2010. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-β1. Stem Cell 6: 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YS, Göke J, Ng JH, Lu X, Gonzales KAU, Tan CP, Tng WQ, Hong ZZ, Lim YS, Ng HH. 2013. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Stem Cell 13: 663–675. [DOI] [PubMed] [Google Scholar]
- Chang H, Matzuk MM. 2001. Smad5 is required for mouse primordial germ cell development. Mech Dev 104: 61–67. [DOI] [PubMed] [Google Scholar]
- Chen D, Ji X, Harris MA, Feng JQ, Karsenty G, Celeste AJ, Rosen V, Mundy GR, Harris SE. 1998. Differential roles for bone morphogenetic protein (BMP) receptor type IB and IA in differentiation and specification of mesenchymal precursor cells to osteoblast and adipocyte lineages. J Cell Biol 142: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Vega VB, Ng HH. 2008a. Transcriptional regulatory networks in embryonic stem cells. Cold Spring Harb Symp Quant Biol 73: 203–209. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008b. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu J, Yang J, Chen Y, Chen J, Ni S, Song H, Zeng L, Ding K, Pei D. 2010. BMPs functionally replace Klf4 and support efficient reprogramming of mouse fibroblasts by Oct4 alone. Cell Res 21: 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, et al. 2011. Chemically defined conditions for human iPSC derivation and culture. Nat Meth 8: 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry ABC, Daley GQ. 2012. Reprogramming cellular identity for regenerative medicine. Cell 148: 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheshier SH, Morrison SJ, Liao X, Weissman IL. 1999. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc Natl Acad Sci 96: 3120–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, et al. 2010. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468: 316–320. [DOI] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. 2009. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Stem Cell 5: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy L, Derynck R. 2003. Transforming growth factor-β inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. J Biol Chem 278: 9609–9619. [DOI] [PubMed] [Google Scholar]
- Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ. 2004. Differential requirements for Smad4 in TGFβ-dependent patterning of the early mouse embryo. Development 131: 3501–3512. [DOI] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. 2008. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev 22: 746–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM. 1990. Label-retaining cells reside in the bulge area of pilosebaceous unit: Implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61: 1329–1337. [DOI] [PubMed] [Google Scholar]
- Creamer B, Shorter RG, Bamforth J. 1961. The turnover and shedding of epithelial cells. I: The turnover in the gastro-intestinal tract. Gut 2: 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho AG, Lazar MA. 2011. Forming functional fat: A growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol 12: 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. 2005. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 23: 1534–1541. [DOI] [PubMed] [Google Scholar]
- Datto MB, Frederick JP, Pan L, Borton AJ, Zhuang Y, Wang XF. 1999. Targeted disruption of Smad3 reveals an essential role in transforming growth factor β-mediated signal transduction. Mol Cell Biol 19: 2495–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000. [DOI] [PubMed] [Google Scholar]
- Davis H, Irshad S, Bansal M, Rafferty H, Boitsova T, Bardella C, Jaeger E, Lewis A, Freeman-Mills L, Giner FC, et al. 2015. Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat Med 21: 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. 2009. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol 27: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa Lopes SMC, Roelen BA, Monteiro RM, Emmens R, Lin HY, Li E, Lawson KA, Mummery CL. 2004. BMP signaling mediated by ALK2 in the visceral endoderm is necessary for the generation of primordial germ cells in the mouse embryo. Genes Dev 18: 1838–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter TM, Allen TD, Lajtha LG. 1977. Conditions controlling the proliferation of haemopoietic stem cells in vitro. J Cell Physiol 91: 335–344. [DOI] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, et al. 2008. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321: 1218–1221. [DOI] [PubMed] [Google Scholar]
- Ding L, Morrison SJ. 2013. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495: 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, Morrison SJ. 2012. Endothelial and perivascular cellsmaintain haematopoietic stem cells. Nature 481: 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. 2012. Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156. [DOI] [PubMed] [Google Scholar]
- Ewen ME, Sluss HK, Sherr CJ, Matsushime H, Kato J, Livingston DM. 1993. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell 73: 487–497. [DOI] [PubMed] [Google Scholar]
- Falk S, Wurdak H, Ittner LM, Ille F, Sumara G, Schmid MT, Draganova K, Lang KS, Paratore C, Leveen P, et al. 2008. Brain area-specific effect of TGF-β signaling on Wnt-dependent neural stem cell expansion. Cell Stem Cell 2: 472–483. [DOI] [PubMed] [Google Scholar]
- Fan X, Xu H, Cai W, Yang Z, Zhang J. 2003. Spatial and temporal patterns of expression of Noggin and BMP4 in embryonic and postnatal rat hippocampus. Brain Res Dev 146: 51–58. [DOI] [PubMed] [Google Scholar]
- Farini D, Scaldaferri ML, Iona S, La Sala G, De Felici M. 2005. Growth factors sustain primordial germ cell survival, proliferation and entering into meiosis in the absence of somatic cells. Dev Biol 285: 49–56. [DOI] [PubMed] [Google Scholar]
- Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. 2005. Notch signals control the fate of immature progenitor cells in the intestine. Nat Cell Biol 435: 964–968. [DOI] [PubMed] [Google Scholar]
- Friedenstein AJ, Petrakova KV, Kurolesova AI, Frolova GP. 1968. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation 6: 230–247. [PubMed] [Google Scholar]
- Fuentealba LC, Eivers E, Ikeda A, Hurtado C, Kuroda H, Pera EM, De Robertis EM. 2007. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell 131: 980–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Kato F, Tozuka Y, Yamaguchi M, Miyamoto Y, Hisatsune T. 2003. Two distinct subpopulations of nestin-positive cells in adult mouse dentate gyrus. J Neurosci 23: 9357–9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadue P, Huber TL, Paddison PJ, Keller GM. 2006. Wnt and TGF-β signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci 103: 16806–16811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. 2013. Derivation of novel human ground state naïve pluripotent stem cells. Nature 504: 282–286. [DOI] [PubMed] [Google Scholar]
- Gardner RL, Brook FA. 1997. Reflections on the biology of embryonic stem (ES) cells. Int J Dev Biol 41: 235–243. [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR. 2007. Developmental origin of fat: Tracking obesity to its source. Cell 131: 242–256. [DOI] [PubMed] [Google Scholar]
- Gingold JA, Fidalgo M, Guallar D, Lau Z, Sun Z, Zhou H, Faiola F, Huang X, Lee DF, Waghray A, et al. 2014. A genome-wide RNAi screen identifies opposing functions of Snai1 and Snai2 on the Nanog dependency in reprogramming. Mol Cell 56: 140–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, Trcka D, Wrana JL. 2012. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell 11: 769–782. [DOI] [PubMed] [Google Scholar]
- Gong C, Maquat LE. 2012. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell MA, Nguyen H, Shroyer N. 2015. Somatic stem cell heterogeneity: Diversity in the blood, skin and intestinal stem cell compartments. Nat Rev Mol Cell Biol 16: 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, Dijke ten P. 2002. Balancing the activation state of the endothelium via two distinct TGF-β type I receptors. EMBO J 21: 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B, Lehrach H, Adjaye J. 2008. Control of early fate decisions in human ES cells by distinct states of TGFβ pathway activity. Stem Cells Dev 17: 1065–1077. [DOI] [PubMed] [Google Scholar]
- Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, Cruz-Racelis de la J, Fuchs E. 2009. A two-step mechanism for stem cell activation during hair regeneration. Stem Cell 4: 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum A, Hsu YMS, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, Link DC. 2013. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 495: 227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Pinto D, Begthel H, Destrée O, Kielman M, Clevers H. 2005. Expression pattern of Wnt signaling components in the adult Intestine. Gastroenterology 129: 626–638. [DOI] [PubMed] [Google Scholar]
- Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. 2000. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci 97: 13625–13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronthos S, Brahim J, Li W, Fisher LW, Cherman N, Boyde A, DenBesten P, Robey PG, Shi S. 2002. Stem cell properties of human dental pulp stem cells. J Dent Res 81: 531–535. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. 2010. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Stem Cell 7: 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. 2009. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136: 1063–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Zhang L, Christopher DM, Teng ZQ, Fausett SR, Liu C, George OL, Klingensmith J, Jin P, Zhao X. 2011. RNA-binding protein FXR2 regulates adult hippocampal neurogenesis by reducing Noggin expression. Neuron 70: 924–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB. 1962. Adult frogs derived from the nuclei of single somatic cells. Dev Biol 4: 256–273. [DOI] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type–specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Byron M, Butler J, Becker KA, Nelson A, Amit M, Itskovitz-Eldor J, Stein J, Stein G, Ware C, et al. 2008. X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. J Cell Physiol 216: 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. 2010. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci 107: 9222–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. 1999. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol 147: 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Toyono T, Toyoshima K, Yamasaki M, Itoh N, Kato S, Sekine K, Ohuchi H. 2002. FGF10 maintains stem cell compartment in developing mouse incisors. Development 129: 1533–1541. [DOI] [PubMed] [Google Scholar]
- Haramis APG, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJA, Clevers H. 2004. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 303: 1684–1686. [DOI] [PubMed] [Google Scholar]
- Hata K, Nishimura R, Ikeda F, Yamashita K, Matsubara T, Nokubi T, Yoneda T. 2003. Differential roles of Smad1 and p38 kinase in regulation of peroxisome proliferator-activating receptor γ during bone morphogenetic protein 2–induced adipogenesis. Mol Biol Cell 14: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattiangady B, Shetty AK. 2008. Aging does not alter the number or phenotype of putative stem/progenitor cells in the neurogenic region of the hippocampus. Neurobiol Aging 29: 129–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. 2004. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: A major site of neurogenesis. Proc Natl Acad Sci 101: 3196–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Stem Cell 6: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Kobayashi T, Umino T, Goitsuka R, Matsui Y, Kitamura D. 2002. SMAD1 signaling is critical for initial commitment of germ cell lineage from mouse epiblast. Mech Dev 118: 99–109. [DOI] [PubMed] [Google Scholar]
- He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, et al. 2004. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-β-catenin signaling. Nat Genet 36: 1117–1121. [DOI] [PubMed] [Google Scholar]
- He Y, Zhang H, Yung A, Villeda SA, Jaeger PA, Olayiwola O, Fainberg N, Wyss-Coray T. 2014. ALK5-dependent TGF-β signaling is a major determinant of late-stage adult neurogenesis. Nature 17: 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellingman CA, Davidson ENB, Koevoet W, Vitters EL, van den Berg WB, van Osch GJVM, van der Kraan PM. 2011. Smad signaling determines chondrogenic differentiation of bone-marrow-derived mesenchymal stem cells: Inhibition of Smad1/5/8P prevents terminal differentiation and calcification. Tissue Eng 17: 1157–1167. [DOI] [PubMed] [Google Scholar]
- Herriges MJ, Swarr DT, Morley MP, Rathi KS, Peng T, Stewart KM, Morrisey EE. 2014. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev 28: 1363–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinck AP. 2012. Structural studies of the TGF-βs and their receptors—Insights into evolution of the TGF-β superfamily. FEBS Lett 586: 1860–1870. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoodless PA, Pye M, Chazaud C, Labbé E, Attisano L, Rossant J, Wrana JL. 2001. FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes Dev 15: 1257–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E. 2008. NFATc1 balances quiescence and proliferation of skin stem cells. Cell 132: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlston R, Bevan S, Williams A, Young J, Dunlop M, Rozen P, Eng C, Markie D, Woodford-Richens K, Rodriguez-Bigas MA, et al. 1998. Mutations in DPC4 (SMAD4) cause juvenile polyposis syndrome, but only account for a minority of cases. Hum Mol Genet 7: 1907–1912. [DOI] [PubMed] [Google Scholar]
- Hsieh J. 2004. IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J Cell Biol 164: 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Pasolli HA, Fuchs E. 2011. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Song T-J, Li X, Hu L, He Q, Liu M, Lane MD, Tang Q-Q. 2009. BMP signaling pathway is required for commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc Natl Acad Sci 106: 12670–12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. 2006. Dissecting self-renewal in stem cells with RNA interference. Nat Cell Biol 442: 533–538. [DOI] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgård R. 2008. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet 40: 1291–1299. [DOI] [PubMed] [Google Scholar]
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. 2005. TGFβ/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development 132: 1273–1282. [DOI] [PubMed] [Google Scholar]
- Jiang W, Liu Y, Liu R, Zhang K, Zhang Y. 2015. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep 11: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Takagi T, Kanesashi SN, Kurahashi T, Nomura T, Harada J, Ishii S. 2006. Schnurri-2 controls BMP-dependent adipogenesis via interaction with Smad proteins. Dev Cell 10: 461–471. [DOI] [PubMed] [Google Scholar]
- Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. 1998. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res 238: 265–272. [DOI] [PubMed] [Google Scholar]
- Kang Q, Sun MH, Cheng H, Peng Y, Montag AG, Deyrup AT, Jiang W, Luu HH, Luo J, Szatkowski JP, et al. 2004. Characterization of the distinct orthotopic bone-forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene Ther 11: 1312–1320. [DOI] [PubMed] [Google Scholar]
- Kang JS, Alliston T, Delston R, Derynck R. 2005. Repression of Runx2 function by TGF-β through recruitment of class II histone deacetylases by Smad3. EMBO J 24: 2543–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson G, Blank U, Moody JL, Ehinger M, Singbrant S, Deng CX, Karlsson S. 2007. Smad4 is critical for self-renewal of hematopoietic stem cells. J Exp Med 204: 467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. 1993. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev 7: 331–342. [DOI] [PubMed] [Google Scholar]
- Keller JR, Mantel C, Sing GK, Ellingsworth LR, Ruscetti SK, Ruscetti FW. 1988. Transforming growth factor β1 selectively regulates early murine hematopoietic progenitors and inhibits the growth of IL-3-dependent myeloid leukemia cell lines. J Exp Med 168: 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kfoury Y, Scadden DT. 2015. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell 16: 239–253. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121: 1109–1121. [DOI] [PubMed] [Google Scholar]
- Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills AE, Gupta R, Baker J. 2011. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev Biol 357: 492–504. [DOI] [PubMed] [Google Scholar]
- Kippin TE, Martens DJ, van der Kooy D. 2005. p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev 19: 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, Han S, Peng T, Thams S, Mikkilineni S, et al. 2014. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Stem Cell 14: 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. 2013. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152: 570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak K, Pasolli HA, Alonso L, Polak L, Fuchs E. 2003. Defining BMP functions in the hair follicle by conditional ablation of BMP receptor IA. J Cell Biol 163: 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89: 755–764. [DOI] [PubMed] [Google Scholar]
- Kosinski C, Li VSW, Chan ASY, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW, Yuen ST, et al. 2007. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci 104: 15418–15423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. 2014. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493: 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. 2009. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci 32: 149–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulessa H, Turk G, Hogan BL. 2000. Inhibition of BMP signaling affects growth and differentiation in the anagen hair follicle. EMBO J 19: 6664–6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. 2007. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134: 2895–2902. [DOI] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. 2014. Molecular mechanisms of epithelial–mesenchymal transition. Nature 15: 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J. 2003. TGF-β signaling-deficient hematopoietic stem cells have normal self-renewal and regenerative ability in vivo despite increased proliferative capacity in vitro. Blood 102: 3129–3135. [DOI] [PubMed] [Google Scholar]
- Laurikkala J, Kassai Y, Pakkasjärvi L, Thesleff I, Itoh N. 2003. Identification of a secreted BMP antagonist, ectodin, integrating BMP, FGF, and SHH signals from the tooth enamel knot. Dev Biol 264: 91–105. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Hage WJ. 1994. Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found Symp 182: 68–84; discussion 84–91. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, Hogan BL. 1999. BMP4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev 13: 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond CP, Stevens CE. 1948. The constant renewal of the intestinal epithelium in the albino rat. Anat Rec 100: 357–377. [DOI] [PubMed] [Google Scholar]
- Lee KS, Kim HJ, Li QL, Chi XZ, Ueta C, Komori T, Wozney JM, Kim EG, Choi JY, Ryoo HM, et al. 2000. Runx2 is a common target of transforming growth factor β1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol 20: 8783–8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengerke C, Schmitt S, Bowman TV, Jang IH, Maouche-Chretien L, McKinney-Freeman S, Davidson AJ, Hammerschmidt M, Rentzsch F, Green JBA, et al. 2008. BMP and Wnt specify hematopoietic fate by activation of the Cdx–Hox pathway. Cell Stem Cell 2: 72–82. [DOI] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. 2010. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7: 51–63. [DOI] [PubMed] [Google Scholar]
- Li Z, Fei T, Zhang J, Zhu G, Wang L, Lu D, Chi X, Teng Y, Hou N, Yang X, et al. 2012. BMP4 signaling acts via dual-specificity phosphatase 9 to control ERK activity in mouse embryonic stem cells. Stem Cell 10: 171–182. [DOI] [PubMed] [Google Scholar]
- Li J, Feng J, Liu Y, Ho TV, Grimes W, Ho HA, Park S, Wang S, Chai Y. 2015. BMP-SHH signaling network controls epithelial stem cell fate via regulation of its niche in the developing tooth. Dev Cell 33: 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Tramontin AD, Trevejo JM, Herrera DG, García-Verdugo JM, Alvarez-Buylla A. 2000. Noggin antagonizes BMP signaling to create a niche for adult neurogenesis. Neuron 28: 713–726. [DOI] [PubMed] [Google Scholar]
- Liu D, Black BL, Derynck R. 2001. TGF-β inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev 15: 2950–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Han Q, Peng T, Peng M, Wei B, Li D, Wang X, Yu S, Yang J, Cao S, et al. 2015. The oncogene c-Jun impedes somatic cell reprogramming. Nat Cell Biol 17: 856–867. [DOI] [PubMed] [Google Scholar]
- Ludwig TE, Levenstein ME, Jones JM, Berggren WT, Mitchen ER, Frane JL, Crandall LJ, Daigh CA, Conard KR, Piekarczyk MS, et al. 2006. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol 24: 185–187. [DOI] [PubMed] [Google Scholar]
- Luu HH, Song WX, Luo X, Manning D, Luo J, Deng ZL, Sharff KA, Montag AG, Haydon RC, He TC. 2007. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J Orthop Res 25: 665–677. [DOI] [PubMed] [Google Scholar]
- MacArthur BD, Ma’ayan A, Lemischka IR. 2009. Systems biology of stem cell fate and cellular reprogramming. Nat Rev Mol Cell Biol 10: 672–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay A, Beck S, Murphy JM, Barry F, Chichester C, Pittenger M. 1998. Chondrogenic differentiation of cultured human mesenchymal stem cells from marrow. Tissue Eng 4: 415–428. [DOI] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K. 2009. TGFβ signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol 19: 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 11: 156–166. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Sugimoto T, Diamanti E, Joshi A, Hannah R, Ohtsuka S, Göttgens B, Niwa H, Smith A. 2012. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Stem Cell 11: 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci 78: 7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. 2005. Smad transcription factors. Genes Dev 19: 2783–2810. [DOI] [PubMed] [Google Scholar]
- Masters BA, Werner H, Roberts CT, LeRoith D, Raizada MK. 1991. Insulin-like growth factor I (IGF-I) receptors and IGF-I action in oligodendrocytes from rat brains. Reg Pept 33: 117–131. [DOI] [PubMed] [Google Scholar]
- Matsushime H, Ewen ME, Strom DK, Kato JY, Hanks SK, Roussel MF, Sherr CJ. 1992. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell 71: 323–334. [DOI] [PubMed] [Google Scholar]
- Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. 2004. Cylcin-dependent kinases regulate the antiproliferative function of Smads. Nature 430: 223–226. [DOI] [PubMed] [Google Scholar]
- Medema JP, Vermeulen L. 2011. Microenvironmental regulation of stem cells in intestinal homeostasis and cancer. Nature 474: 318–326. [DOI] [PubMed] [Google Scholar]
- Méndez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, MacArthur BD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, Frenette PS. 2010. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466: 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier FE, Ragu C, Scadden DT. 2011. The bone marrow at the crossroads of blood and immunity. Nat Rev Immunol 12: 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill BJ, Gat U, DasGupta R, Fuchs E. 2001. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev 15: 1688–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira H, Andreu Z, Suh H, Lie DC, Jessberger S, Consiglio A, Emeterio JS, Hortigüela R, Marqués-Torrejón MÁ, Nakashima K, et al. 2010. Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Stem Cell 7: 78–89. [DOI] [PubMed] [Google Scholar]
- Miyata T. 2004. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development 131: 3133–3145. [DOI] [PubMed] [Google Scholar]
- *.Morikawa M, Derynck R, Miyazono K. 2016. TGF-β and the TGF-β family: Context-dependent roles in cell and tissue physiology. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Scadden DT. 2014. The bone marrow niche for haematopoietic stem cells. Nature 505: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsut L, Yan KP, Enzo E, Aragona M, Soligo SM, Wendling O, Mark M, Khetchoumian K, Bressan G, Chambon P, et al. 2010. Negative control of Smad activity by ectodermin/Tif1 patterns the mammalian embryo. Development 137: 2571–2578. [DOI] [PubMed] [Google Scholar]
- Mu Y, Lee SW, Gage FH. 2010. Signaling in adult neurogenesis. Curr Opin Neurobiol 20: 416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Lovén J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. 2011. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147: 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J, Pittet JF, Kaminski N, Garat C, Matthay MA, et al. 1999. The integrin αv β6 binds and activates latent TGF β1: A mechanism for regulating pulmonary inflammation and fibrosis. Cell 96: 319–328. [DOI] [PubMed] [Google Scholar]
- Neganova I, Zhang X, Atkinson S, Lako M. 2009. Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene 28: 20–30. [DOI] [PubMed] [Google Scholar]
- Newman AM, Cooper JB. 2010. Lab-specific gene expression signatures in pluripotent stem cells. Stem Cell 7: 258–262. [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev 12: 2048–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. 2009. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460: 118–122. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. 2004. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 7: 136–144. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Saito A, Matsui H, Suzuki H, Ohtsuka S, Shimosato D, Morishita Y, Watabe T, Niwa H, Miyazono K. 2006. Activin–Nodal signaling is involved in propagation of mouse embryonic stem cells. J Cell Sci 120: 55–65. [DOI] [PubMed] [Google Scholar]
- Oh SP, Seki T, Goss KA, Imamura T, Yi Y, Donahoe PK, Li L, Miyazono K, Dijke ten P, Kim S, et al. 2000. Activin receptor-like kinase 1 modulates transforming growth factor-β1 signaling in the regulation of angiogenesis. Proc Natl Acad Sci 97: 2626–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M. 2009. A signaling principle for the specification of the germ cell lineage in mice. Cell 137: 571–584. [DOI] [PubMed] [Google Scholar]
- Okamura D, Kimura T, Nakano T, Matsui Y. 2003. Cadherin-mediated cell interaction regulates germ cell determination in mice. Development 130: 6423–6430. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Wang J, Kim J, Chu J, Rao S, Theunissen TW, Shen X, Levasseur DN. 2008. The transcriptional network controlling pluripotency in ES cells. Cold Spring Harb Symp Quant Biol 73: 195–202. [DOI] [PubMed] [Google Scholar]
- Oshimori N, Fuchs E. 2012. Paracrine TGF-β signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Stem Cell 10: 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osyczka AM, Diefenderfer DL, Bhargave G, Leboy PS. 2004. Different effects of BMP-2 on marrow stromal cells from human and rat bone. Cells Tissues Organs 176: 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann OG, Pelus LM. 1988. Differential proliferative effects of transforming growth factor-β on human hematopoietic progenitor cells. J Immunol 140: 2661–2665. [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89: 765–771. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. 2008. Disease-specific induced pluripotent stem cells. Cell 134: 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S, Vallier L. 2013. The cell-cycle state of stem cells determines cell fate propensity. Cell 155: 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece-Barbara N, Vera S, Kathirkamathamby K, Liebner S, Di Guglielmo GM, Dejana E, Wrana JL, Letarte M. 2005. Endoglin null endothelial cells proliferate faster and are more responsive to transforming growth factor β1 with higher affinity receptors and an activated Alk1 pathway. J Biol Chem 280: 27800–27808. [DOI] [PubMed] [Google Scholar]
- Pera EM. 2003. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev 15: 3023–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretto P, Cummings D, Modena C, Behrens M, Venkatraman G, Fasolo A, Margolis FL. 2002. BMP mRNA and protein expression in the developing mouse olfactory system. J Comp Neurol 451: 267–278. [DOI] [PubMed] [Google Scholar]
- Pertovaara L, Sistonen L, Bos TJ, Vogt PK, Keski-Oja J, Alitalo K. 1989. Enhanced jun gene expression is an early genomic response to transforming growth factor β stimulation. Mol Cell Biol 9: 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. 1999. Multilineage potential of adult human mesenchymal stem cells. Science 284: 143–147. [DOI] [PubMed] [Google Scholar]
- Plikus MV, Mayer JA, la Cruz de D, Baker RE, Maini PK, Maxson R, Chuong CM. 2008. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature 451: 340–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. 2012. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151: 1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porlan E, Morante-Redolat JM, Marqués-Torrejón MÁ, Andreu-Agulló C, Carneiro C, Gómez-Ibarlucea E, Soto A, Vidal A, Ferrón SR, Fariñas I. 2013. Transcriptional repression of Bmp2 by p21. Nature 16: 1567–1575. [DOI] [PubMed] [Google Scholar]
- Prockop DJ. 1997. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science 276: 71–74. [DOI] [PubMed] [Google Scholar]
- Rao RA, Dhele N, Cheemadan S, Ketkar A, Jayandharan GR, Palakodeti D, Rampalli S. 2015. Ezh2-mediated H3K27me3 activity facilitates somatic transition during human pluripotent reprogramming. Sci Rep 5: 8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada A, Benchabane H, Wrana JL, Celeste AJ, Attisano L. 2003. Myostatin signals through a transforming growth factor β-like signaling pathway to block adipogenesis. Mol Cell Biol 23: 7230–7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendl M, Lewis L, Fuchs E. 2005. Molecular dissection of mesenchymal–epithelial interactions in the hair follicle. PLoS Biol 3: e331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznikoff CA, Brankow DW, Heidelberger C. 1973. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res 33: 3231–3238. [PubMed] [Google Scholar]
- Rhee H, Polak L, Fuchs E. 2006. Lhx2 maintains stem cell character in hair follicles. Science 312: 1946–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelen BA, Lin HY, Knezević V, Freund E, Mummery CL. 1994. Expression of TGF-βs and their receptors during implantation and organogenesis of the mouse embryo. Dev Biol 166: 716–728. [DOI] [PubMed] [Google Scholar]
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. 2000. Inhibition of adipogenesis by Wnt signaling. Science 289: 950–953. [DOI] [PubMed] [Google Scholar]
- Ross S, Cheung E, Petrakis TG, Howell M, Kraus WL, Hill CS. 2006. Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription. EMBO J 25: 4490–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J. 2008. Stem cells and early lineage development. Cell 132: 527–531. [DOI] [PubMed] [Google Scholar]
- Rui YF, Lui PPY, Lee YW, Chan KM. 2011. Higher BMP receptor expression and BMP-2-induced osteogenic differentiation in tendon-derived stem cells compared with bone-marrow-derived mesenchymal stem cells. Int Orthop 36: 1099–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Garrett N, Bourillot P, Stennard F, Gurdon JB. 2000. The Xenopus eomesodermin promoter and its concentration-dependent response to activin. Mech Dev 94: 133–146. [DOI] [PubMed] [Google Scholar]
- Sakaki-Yumoto M, Liu J, Ramalho-Santos M, Yoshida N, Derynck R. 2013. Smad2 is essential for maintenance of the human and mouse primed pluripotent stem cell state. J Biol Chem 288: 18546–18560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. 2010. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Stem Cell 7: 64–77. [DOI] [PubMed] [Google Scholar]
- Sangiorgi E, Capecchi MR. 2008. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40: 915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. 2009. Single Lgr5 stem cells build crypt. Nature 459: 262–265. [DOI] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. 2011. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469: 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. 2013. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2: e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden DT. 2014. Nice neighborhood: Emerging concepts of the stem cell niche. Cell 157: 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaberg RM, van der Kooy D. 2002. Adult rodent neurogenic regions: The ventricular subependyma contains neural stem cells, but the dentate gyrus contains restricted progenitors. J Neurosci 22: 1784–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, Cao T, Carano RAD, Curran T, Schober M, et al. 2010. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development 137: 3753–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seri B, García-Verdugo JM, McEwen BS, Alvarez-Buylla A. 2001. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J Neurosci 21: 7153–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD, Fan G. 2008. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci 105: 4709–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Yamada K, Watanabe M, Ikenaka K, Wada K, Tanaka K, Inoue Y. 1997. Glutamate transporter GLAST is expressed in the radial glia-astrocyte lineage of developing mouse spinal cord. J Neurosci 17: 9212–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, et al. 2013. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci 110: 2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva SS, Rowntree RK, Mekhoubad S, Lee JT. 2008. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci 105: 4820–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirard C, Kim S, Mirtsos C, Tadich P, Hoodless PA, Itie A, Maxson R, Wrana JL, Mak TW. 2000. Targeted disruption in murine cells reveals variable requirement for Smad4 in transforming growth factor β-related signaling. J Biol Chem 275: 2063–2070. [DOI] [PubMed] [Google Scholar]
- Sitnicka E, Ruscetti FW, Priestley GV, Wolf NS, Bartelmez SH. 1996. Transforming growth factor β1 directly and reversibly inhibits the initial cell divisions of long-term repopulating hematopoietic stem cells. Blood 88: 82–88. [PubMed] [Google Scholar]
- Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. 1988. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336: 688–690. [DOI] [PubMed] [Google Scholar]
- Smith AJ, Tobias RS, Cassidy N, Bégue-Kirn C, Ruch JV, Lesot H. 1995. Influence of substrate nature and immobilization of implanted dentin matrix components during induction of reparative dentinogenesis. Connect Tissue Res 32: 291–296. [DOI] [PubMed] [Google Scholar]
- Smith JR, Vallier L, Lupo G, Alexander M, Harris WA, Pedersen RA. 2008. Inhibition of activin/Nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev Biol 313: 107–117. [DOI] [PubMed] [Google Scholar]
- Stewart A, Guan H, Yang K. 2010. BMP-3 promotes mesenchymal stem cell proliferation through the TGF-β/activin signaling pathway. J Cell Physiol 223: 658–66. [DOI] [PubMed] [Google Scholar]
- Sudo K, Ema H, Morita Y, Nakauchi H. 2000. Age-associated characteristics of murine hematopoietic stem cells. J Exp Med 192: 1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Kohara H, Noda M, Nagasawa T. 2006. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25: 977–988. [DOI] [PubMed] [Google Scholar]
- Suh H, Consiglio A, Ray J, Sawai T, D’Amour KA, Gage FH. 2007. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Stem Cell 1: 515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Hu J, Zhou L, Pollard SM, Smith A. 2011. Interplay between FGF2 and BMP controls the self-renewal, dormancy and differentiation of rat neural stem cells. J Cell Sci 124: 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Yong KMA, Villa-Diaz LG, Zhang X, Chen W, Philson R, Weng S, Xu H, Krebsbach PH, Fu J. 2014. Hippo/YAP-mediated rigidity-dependent motor neuron differentiation of human pluripotent stem cells. Nat Mater 13: 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Raya A, Kawakami Y, Morita M, Matsui T, Nakashima K, Gage FH, Rodríguez-Esteban C, Izpisúa Belmonte JC. 2006. Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc Natl Acad Sci 103: 10294–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. [DOI] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, et al. 2014. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158: 1254–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallheden T, Dennis JE, Lennon DP, Sjögren-Jansson E, Caplan AI, Lindahl A. 2003. Phenotypic plasticity of human articular chondrocytes. J Bone Joint Surg Am 85A: 93–100. [DOI] [PubMed] [Google Scholar]
- Taylor SM, Jones PA. 1979. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell 17: 771–779. [DOI] [PubMed] [Google Scholar]
- Teo AKK, Arnold SJ, Trotter MWB, Brown S, Ang LT, Chng Z, Robertson EJ, Dunn NR, Vallier L. 2011. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev 25: 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo AKK, Ali Y, Wong KY, Chipperfield H, Sadasivam A, Poobalan Y, Tan EK, Wang ST, Abraham S, Tsuneyoshi N, et al. 2012. Activin and BMP4 synergistically promote formation of definitive endoderm in human embryonic stem cells. Stem Cells 30: 631–642. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RDG 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448: 196–199. [DOI] [PubMed] [Google Scholar]
- Teshigawara R, Hirano K, Nagata S, Ainscough J, Tada T. 2016. OCT4 activity during conversion of human intermediately reprogrammed stem cells to iPSCs through mesenchymal–epithelial transition. Development 143: 15–23. [DOI] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. 2014. Systematic identification of culture conditions for induction and maintenance of naïve human pluripotency. Cell Stem Cell 15: 471–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147. [DOI] [PubMed] [Google Scholar]
- Tremblay KD, Dunn NR, Robertson EJ. 2001. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development 128: 3609–3621. [DOI] [PubMed] [Google Scholar]
- Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, et al. 2008. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 454: 1000–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummers M. 2003. Root or crown: A developmental choice orchestrated by the differential regulation of the epithelial stem cell niche in the tooth of two rodent species. Development 130: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Unternaehrer JJ, Zhao R, Kim K, Cesana M, Powers JT, Ratanasirintrawoot S, Onder T, Shibue T, Weinberg RA, Daley GQ. 2014. The epithelial–mesenchymal transition factor SNAIL paradoxically enhances reprogramming. Stem Cell Rep 3: 691–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsugisawa T, Moody JL, Aspling M, Nilsson E, Carlsson L, Karlsson S. 2006. A road map toward defining the role of Smad signaling in hematopoietic stem cells. Stem Cells 24: 1128–1136. [DOI] [PubMed] [Google Scholar]
- Valamehr B, Robinson M, Abujarour R, Rezner B, Vranceanu F, Le T, Medcalf A, Lee TT, Fitch M, Robbins D, et al. 2014. Platform for induction and maintenance of transgene-free hiPSCs resembling ground state pluripotent stem cells. Stem Cell Rep 2: 366–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L. 2005. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci 118: 4495–4509. [DOI] [PubMed] [Google Scholar]
- Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MWB, Cho CHH, Martinez A, Rugg-Gunn P, et al. 2009a. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development 136: 1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L, Touboul T, Brown S, Cho C, Bilican B, Alexander M, Cedervall J, Chandran S, Hrlund-Richter L, Weber A, et al. 2009b. Signaling pathways controlling pluripotency and early cell fate decisions of human induced pluripotent stem cells. Stem Cells 27: 2655–2666. [DOI] [PubMed] [Google Scholar]
- Vallier L, Touboul T, Chng Z, Brimpari M, Hannan N, Millan E, Smithers LE, Trotter M, Rugg-Gunn P, Weber A, et al. 2009c. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS ONE 4: e6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg DLC, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. 2010. An Oct4-centered protein interaction network in embryonic stem cells. Stem Cell 6: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, et al. 2005. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nat Cell Biol 435: 959–963. [DOI] [PubMed] [Google Scholar]
- Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. 2008. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol 10: 837–848. [DOI] [PubMed] [Google Scholar]
- Vidal VPI, Chaboissier MC, Lützkendorf S, Cotsarelis G, Mill P, Hui CC, Ortonne N, Ortonne JP, Schedl A. 2005. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr Biol 15: 1340–1351. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. 2008. Selective blockade of microRNA processing by Lin28. Science 320: 97–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voog J, Jones DL. 2010. Stem cells and the niche: A dynamic duo. Stem Cell 6: 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrip WR, Bikoff EK, Hoodless PA, Wrana JL, Robertson EJ. 1998. Smad2 signaling in extraembryonic tissues determines anterior–posterior polarity of the early mouse embryo. Cell 92: 797–808. [DOI] [PubMed] [Google Scholar]
- Wang LD, Wagers AJ. 2011. Dynamic niches in the origination and differentiation of haematopoietic stem cells. Nat Rev Mol Cell Biol 12: 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, Maas RL, Chuong CM, Schimmang T, Thesleff I. 2007. An integrated gene regulatory network controls stem cell proliferation in teeth. Plos Biol 5: e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang J, Liu H, Lu D, Chen X, Zenonos Z, Campos LS, Rad R, Guo G, Zhang S, et al. 2011. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor γ and liver receptor homolog 1. Proc Natl Acad Sci 108: 18283–18288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. 2012. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, Jimenez-Caliani AJ, Deng X, Cavanaugh C, Cook S, et al. 2014. Derivation of naïve human embryonic stem cells. Proc Natl Acad Sci 111: 4484–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein M, Yang X, Li C, Xu X, Gotay J, Deng CX. 1998. Failure of egg cylinder elongation and mesoderm induction in mouse embryos lacking the tumor suppressor Smad2. Proc Natl Acad Sci 95: 9378–9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg E, Winnier GE, Chen X, Farnsworth CL, Hogan BL, Whitman M. 1998. A mouse homologue of FAST-1 transduces TGF β superfamily signals and is expressed during early embryogenesis. Mech Dev 79: 17–27. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and Mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. 1988. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336: 684–687. [DOI] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A. 2011. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol 13: 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Chen X, Zhang J, Loh YH, Low TY, Zhang W, Zhang W, Sze SK, Lim B, Ng HH. 2006. Sall4 interacts with Nanog and co-occupies nanog genomic sites in embryonic stem cells. J Biol Chem 281: 24090–24094. [DOI] [PubMed] [Google Scholar]
- Xi Q, Wang Z, Zaromytidou AI, Zhang XHF, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, et al. 2011. A poised chromatin platform for TGF-β access to master regulators. Cell 147: 1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Inokuma M, Denham J, Golds K, Kundu P, Gold J, Carpenter MK. 2001. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol 19: 971–974. [DOI] [PubMed] [Google Scholar]
- Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, et al. 2008. NANOG Is a direct target of TGFβ/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell 3: 196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe D, Saxel O. 1977. A myogenic cell line with altered serum requirements for differentiation. Differentiation 7: 159–166. [DOI] [PubMed] [Google Scholar]
- Yagi K, Goto D, Hamamoto T, Takenoshita S, Kato M, Miyazono K. 1999. Alternatively spliced variant of Smad2 lacking exon 3: Comparison with wild-type Smad2 and Smad3. J Biol Chem 274: 703–709. [DOI] [PubMed] [Google Scholar]
- Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M. 2008. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet 40: 1016–1022. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Meno C, Sakai Y, Shiratori H, Mochida K, Ikawa Y, Saijoh Y, Hamada H. 2001. The transcription factor FoxH1 (FAST) mediates Nodal signaling during anterior-posterior patterning and node formation in the mouse. Genes Dev 15: 1242–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Iwama A, Takayanagi SI, Morita Y, Eto K, Ema H, Nakauchi H. 2006. Cytokine signals modulated via lipid rafts mimic niche signals and induce hibernation in hematopoietic stem cells. EMBO J 25: 3515–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Iwama A, Takayanagi SI, Eto K, Ema H, Nakauchi H. 2009. TGF-β as a candidate bone marrow niche signal to induce hematopoietic stem cell hibernation. Blood 113: 1250–1256. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Ema H, Karlsson G, Yamaguchi T, Miyoshi H, Shioda S, Taketo MM, Karlsson S, Iwama A, Nakauchi H. 2011. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell 147: 1146–1158. [DOI] [PubMed] [Google Scholar]
- Yang G, Zhou J, Teng Y, Xie J, Lin J, Guo X, Gao Y, He M, Yang X, Wang S. 2014. Mesenchymal TGF-β signaling orchestrates dental epithelial stem cell homeostasis through Wnt signaling. Stem Cells 32: 2939–2948. [DOI] [PubMed] [Google Scholar]
- Yi F, Pereira L, Hoffman JA, Shy BR, Yuen CM, Liu DR, Merrill BJ. 2011. Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat Cell Biol 13: 762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y, Zhao GQ. 2001. Cooperation of endoderm-derived BMP2 and extraembryonic ectoderm-derived BMP4 in primordial germ cell generation in the mouse. Dev Biol 232: 484–492. [DOI] [PubMed] [Google Scholar]
- Ying Y, Liu XM, Marble A, Lawson KA, Zhao GQ. 2000. Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol Endocrinol 14: 1053–1063. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. 2003. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115: 281–292. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. 2011. Control of the embryonic stem cell state. Cell 144: 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feng XH, Derynck R. 1998. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature 394: 909–913. [DOI] [PubMed] [Google Scholar]
- Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, Ross J, Haug J, Johnson T, Feng JQ, et al. 2003. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425: 836–841. [DOI] [PubMed] [Google Scholar]
- Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y, Yang J, Ma Y, Chai L, et al. 2006a. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol 8: 1114–1123. [DOI] [PubMed] [Google Scholar]
- Zhang J, He XC, Tong WG, Johnson T, Wiedemann LM, Mishina Y, Feng JQ, Li L. 2006b. Bone morphogenetic protein signaling inhibits hair follicle anagen induction by restricting epithelial stem/progenitor cell activation and expansion. Stem Cells 24: 2826–2839. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang J, Wang T, Esteban MA, Pei D. 2008. Esrrb activates Oct4 transcription and sustains self-renewal and pluripotency in embryonic stem cells. J Biol Chem 283: 35825–35833. [DOI] [PubMed] [Google Scholar]
- Zhao H, Li S, Han D, Kaartinen V, Chai Y. 2011. Alk5-mediated transforming growth factor β signaling acts upstream of fibroblast growth factor 10 to regulate the proliferation and maintenance of dental epithelial stem cells. Mol Cell Biol 31: 2079–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Feng J, Seidel K, Shi S, Klein O, Sharpe P, Chai Y. 2014. Secretion of Shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor. Cell Stem Cell 14: 160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XP, Woodford-Richens K, Lehtonen R, Kurose K, Aldred M, Hampel H, Launonen V, Virta S, Pilarski R, Salovaara R, et al. 2001. Germline mutations in BMPR1A/ALK3 cause a subset of cases of juvenile polyposis syndrome and of Cowden and Bannayan–Riley–Ruvalcaba syndromes. Am J Hum Genet 69: 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Richardson JA, Parada LF, Graff JM. 1998. Smad3 mutant mice develop metastatic colorectal cancer. Cell 94: 703–714. [DOI] [PubMed] [Google Scholar]
- Zimmerman LB, De Jesús-Escobar JM, Harland RM. 1996. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell 86: 599–606. [DOI] [PubMed] [Google Scholar]
- Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH. 2001. Multilineage cells from human adipose tissue: Implications for cell-based therapies. Tissue Eng 7: 211–228. [DOI] [PubMed] [Google Scholar]
- Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, Alfonso ZC, Fraser JK, Benhaim P, Hedrick MH. 2002. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell 13: 4279–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]