Significance
Copper (Cu) is an essential trace metal nutrient in health and is increasingly recognized for its role in the control of infection. The pathogen Escherichia coli encounters host niches with mild to high acidity and elevated copper levels. Our study shows that this bacterium can alter its metabolism and harness the amino acid glutamine to suppress the effects of acid stress and copper toxicity. Given the abundance of glutamine in systemic circulation and its importance in the host immune system, our work provides a new insight into the ways in which bacterial pathogens can adapt and survive host-imposed antibacterial strategies.
Keywords: copper stress, glutamate biosynthesis, GOGAT, acid tolerance
Abstract
Copper (Cu) is a key antibacterial component of the host innate immune system and almost all bacterial species possess systems that defend against the toxic effects of excess Cu. The Cu tolerance system in Gram-negative bacteria is composed minimally of a Cu sensor (CueR) and a Cu export pump (CopA). The cueR and copA genes are encoded on the chromosome typically as a divergent but contiguous operon. In Escherichia coli, cueR and copA are separated by two additional genes, ybaS and ybaT, which confer glutamine (Gln)-dependent acid tolerance and contribute to the glutamate (Glu)-dependent acid resistance system in this organism. Here we show that Cu strongly inhibits growth of a ∆copA mutant strain in acidic cultures. We further demonstrate that Cu stress impairs the pathway for Glu biosynthesis via glutamate synthase, leading to decreased intracellular levels of Glu. Addition of exogenous Glu rescues the ∆copA mutant from Cu stress in acidic conditions. Gln is also protective but this relies on the activities of YbaS and YbaT. Notably, expression of both enzymes is up-regulated during Cu stress. These results demonstrate a link between Cu stress, acid stress, and Glu/Gln metabolism, establish a role for YbaS and YbaT in Cu tolerance, and suggest that subtle changes in core metabolic pathways may contribute to overcoming host-imposed copper toxicity.
The efflux of excess transition metal ions such as copper (Cu) is an important feature of bacterial physiology, particularly during the interactions between a bacterial pathogen and its host. Several lines of evidence have established a role for Cu as a host-derived antibacterial agent that contributes to nutritional immunity (1). In turn, the ability to export Cu from the bacterial cytoplasm is now recognized as a key determinant of bacterial virulence (2–5). At the biochemical level, the mechanisms of bacterial Cu export are well understood and, in Gram-negative species, are exemplified by the Cue/Cop regulon in Escherichia coli (6). This system consists of a Cu(I)-sensing transcriptional regulator (CueR), which controls expression of a transmembrane efflux pump (CopA) that exports Cu(I) from the cytoplasm to the periplasm, and a periplasmic cuprous oxidase (CueO) that converts Cu(I) to the less toxic Cu(II) form.
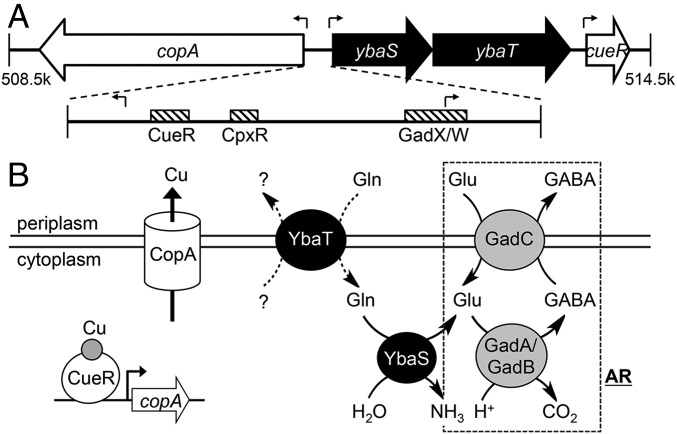
In the E. coli chromosome, copA and cueR are separated by an operon annotated as ybaST, which is encoded in the same orientation as cueR and is divergent from copA (Fig. 1A). ybaS encodes a glutaminase that catalyses the hydrolysis of l-glutamine (Gln) to generate l-glutamate (Glu) and ammonia (Fig. 1B). The glutaminase activity of YbaS confers Gln-dependent acid tolerance and contributes to the Glu-dependent system for acid resistance (AR) (7, 8). In this AR mechanism, Glu is converted to γ-aminobutyric acid (GABA) by two glutamate decarboxylases (GadA and GadB). This process consumes a proton and raises the cytoplasmic pH (9). The intracellular Glu pool can be replenished if an extracellular supply is present and this import occurs via the permease GadC (Fig. 1B). ybaT encodes an amino acid permease that may also contribute to Glu-dependent AR by supplying Gln to YbaS for hydrolysis (Fig. 1B), but its substrate specificity remains to be established.
Fig. 1.
Clustering of Cu tolerance and acid tolerance genes in E. coli. (A) Genomic context of copA and cueR in E. coli. Figure shows the approximate locations of genes on the reference genome (RefSeq NC_000913.3). Block arrows represent directions of ORFs. Transcription start sites are indicated by bent line arrows. Striped boxes represent binding sites for transcription factors (CueR, accttccagcaaggggaaggt; CpxR, gtaaaagtccgtaaa; and GadX/W, taaatcaggatgcctgaaaatcggcaccggggtg). (B) Biochemical function of CopA, CueR, YbaS, and YbaT. CueR is a Cu sensor, whereas CopA is a Cu efflux pump. Both constitute the central mechanism for Cu tolerance in E. coli. YbaS is a glutaminase, whereas YbaT is a putative Gln-importing permease. The dashed box shows components of the Glu-dependent acid resistance system (AR), namely the glutamate decarboxylases GadA and GadB, as well as the Glu/GABA-antiporter GadC. Both YbaS and YbaT are thought to support the function of this AR.
Here we examine the significance of the synteny of Cu tolerance and acid tolerance genes in relation to E. coli physiology. Using a ∆copA deletion mutant strain, we show a link between Cu stress, acid stress, and Glu/Gln metabolism, and establish a role for YbaS and YbaT in Cu tolerance. Our results suggest that subtle changes in bacterial metabolism may contribute to overcoming host-imposed copper toxicity during nutritional immunity.
Results
Organization of copA and cueR in E. coli.
The nucleotide sequence of the copA-ybaST-cueR locus from E. coli was used to query all complete bacterial genome sequences in the National Center for Biotechnology Information database (SI Appendix, SI Methods). The search results indicated that the synteny of copA, ybaST, and cueR is unique to Escherichia and Shigella genera (SI Appendix, Fig. S1). In the case of E. coli, this gene cluster is part of the core genome (10) and is found in 208/209 complete genomes (>95% sequence conservation), which include strains representative of environmental, commensal, and all pathogenic types.
The ybaST insertion is absent from other Enterobactericeae such as Salmonella and Klebsiella. In these organisms, copA is divergent from but contiguous with cueR (SI Appendix, Fig. S2), which is the canonical arrangement for a merR-like operon. One exception was Serratia marcescens, in which cueR and copA are separated by a cluster of genes encoding for the biosynthesis of the antibiotic prodigiosin (pigA-O). Expression of pig genes is repressed by Cu but the physiological relevance of this observation is unclear (11). In agreement with a previous report, ybaS is also present in nine additional genera from the Enterobacteriacae family, including Edwardsiella and Yersinia. However, consistent with its established function in acid tolerance, ybaS in these genomes is frequently encoded adjacent to gadA/B or gadC genes for AR (SI Appendix, Fig. S3).
Cu Stress in a ∆copA Mutant Is Enhanced During Growth in Acidic Conditions.
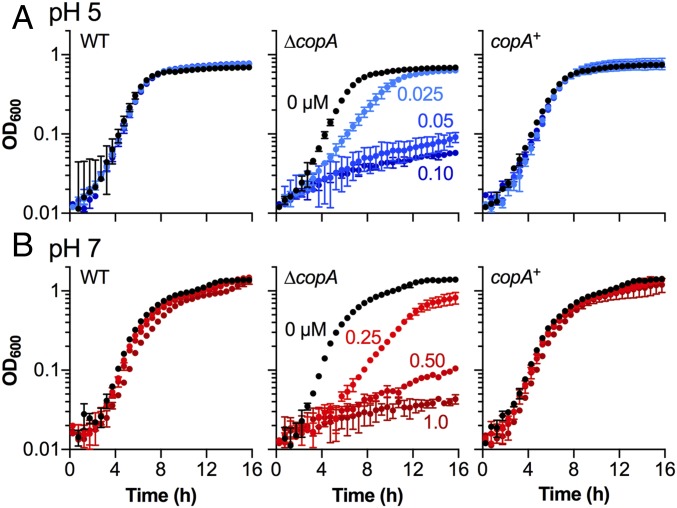
To determine whether there was a link between Cu and acid stress, we examined the inhibitory effects of added Cu on the growth of E. coli in minimal medium buffered at pH 5 and pH 7. Addition of up to 1.0 µM Cu did not impact growth of the wild type (WT) strain at either pH but it inhibited growth of the ∆copA mutant (Fig. 2). Notably, the amount of Cu required to completely suppress growth at pH 5 (0.1 µM, Fig. 2A) was less than the amount required at pH 7 (1.0 µM, Fig. 2B), suggesting that Cu stress in the ∆copA mutant was enhanced during growth in acidic conditions. The Cu-tolerant phenotype was restored upon expression of copA via plasmid-mediated complementation (Fig. 2). Identical results were obtained using the ∆copA mutants of other pathogenic and nonpathogenic strains of E. coli (SI Appendix, Fig. S4), indicating that the interplay between Cu stress and acid stress is a conserved feature of E. coli physiology.
Fig. 2.
Cu stress during growth under different pH conditions. Growth of E. coli EC958 WT, ∆copA mutant, and copA+ complemented mutant: (A) at pH 5 in the presence of 0–0.10 µM added Cu and (B) at pH 7 in the presence of 0–1.0 µM added Cu. Data were averaged from three independent experiments. Error bars represent ±SD.
Addition of Cu to 0.05 µM was sufficient to affect growth at pH 5 and not at pH 7 (SI Appendix, Fig. S4 and Fig. 2). However, this treatment led to a comparable rise in total intracellular Cu levels in midexponential ΔcopA cells during growth at both pH values as determined by inductively coupled plasma MS (SI Appendix, Fig. S5). Hence, the increase in Cu stress at pH 5 did not correlate with an increase in the amounts of trapped Cu. Nevertheless, Cu may become more bioavailable during growth at pH 5 due to protonation of thiols and amines, leading to a decrease in the Cu buffering capacity of the extracellular medium or intracellular milieu. To report for bioavailable Cu, we used a lacZ transcriptional reporter fused to the copA promoter (PcopA-lacZ) (12). The latter is activated by CueR, the primary Cu sensor in E. coli (Fig. 1B). In agreement with a previous observation (13), background PcopA-lacZ activity in midexponential ∆copA cells was higher than in WT and it did not increase further in response to added Cu (SI Appendix, Fig. S6A), suggesting that this mutant trapped trace amounts of Cu in its cytoplasm. Nevertheless, PcopA-lacZ activities in the ∆copA mutant were comparable, irrespective of the pH of the culture medium (SI Appendix, Fig. S6A). Similarly, the response of the PcopA-lacZ reporter to Cu was comparable in WT cultures grown at pH 5 and pH 7 (SI Appendix, Fig. S6B). Thus, the present evidence did not support an increased cellular level or bioavailability of Cu at pH 5 under the experimental conditions used here.
Exogenous Glu and Gln Protect the ∆copA Mutant from Cu Stress at pH 5.
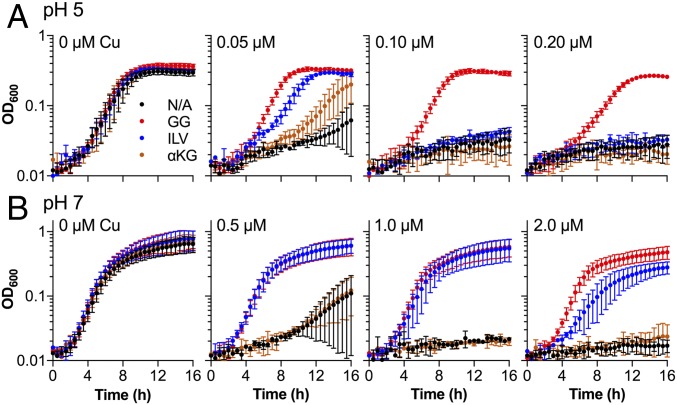
Given the established role of ybaS in Gln-dependent acid tolerance, its demonstrated ability to contribute to Glu-dependent AR, and its synteny with copA and cueR (Fig. 1), we hypothesized that Cu stress during growth in acidic conditions is linked to Glu and/or Gln utilization. Indeed, addition of exogenous Glu and Gln (0.5 mM total) rescued the growth of the ∆copA mutant in Cu-supplemented medium at pH 5 (Fig. 3A) and pH 7 (Fig. 3B). This protection was also observed for other pathogenic and nonpathogenic strains of E. coli (SI Appendix, Fig. S7). Compared with Gln, protection by Glu extended to higher Cu concentrations (SI Appendix, Fig. S8).
Fig. 3.
Protective effects of amino acids. E. coli UTI89 ∆copA mutant was cultured (A) at pH 5 in the presence of 0–0.2 µM added Cu or (B) at pH 7 in the presence of 0–2.0 µM added Cu. The medium was supplemented with water (black, N/A); Glu and Gln (red, GG); Ile, Leu, and Val (blue, ILV); or α-ketoglutarate (orange, α-KG). The total concentration of amino acids in each experiment was 0.5 mM. Data were averaged from three independent experiments. Error bars represent ±SD.
Asp and Asn were also protective at pH 5 (SI Appendix, Fig. S9A). In contrast, Arg, which is responsible for an alternative acid resistance system in E. coli (9), was not protective at pH 5 (SI Appendix, Fig. S9B). Cys and the Cys-containing tripeptide glutathione were strongly protective (SI Appendix, Fig. S9C), likely because these thiol-containing molecules are high-affinity chelators of Cu. We also tested branched-chain amino acids (BCAAs, i.e., Ile, Leu, and Val), which are known to rescue Cu-sensitive mutants of E. coli from Cu stress at pH 7 (13, 14). These amino acids rescued growth of the ∆copA mutant strongly at pH 7 (Fig. 3B) but only weakly at pH 5 (Fig. 3A), especially compared with Glu and/or Gln. These results indicated that Cu stress at pH 5 led to a high requirement for exogenous Glu and/or Gln but not BCAAs. Nevertheless, addition of Glu and/or Gln did not restore Cu tolerance of the ∆copA mutant to WT levels. This observation and the varying protective effects of each amino acid are discussed below.
Cu Stress Leads to Depletion in Intracellular Glu Concentrations.
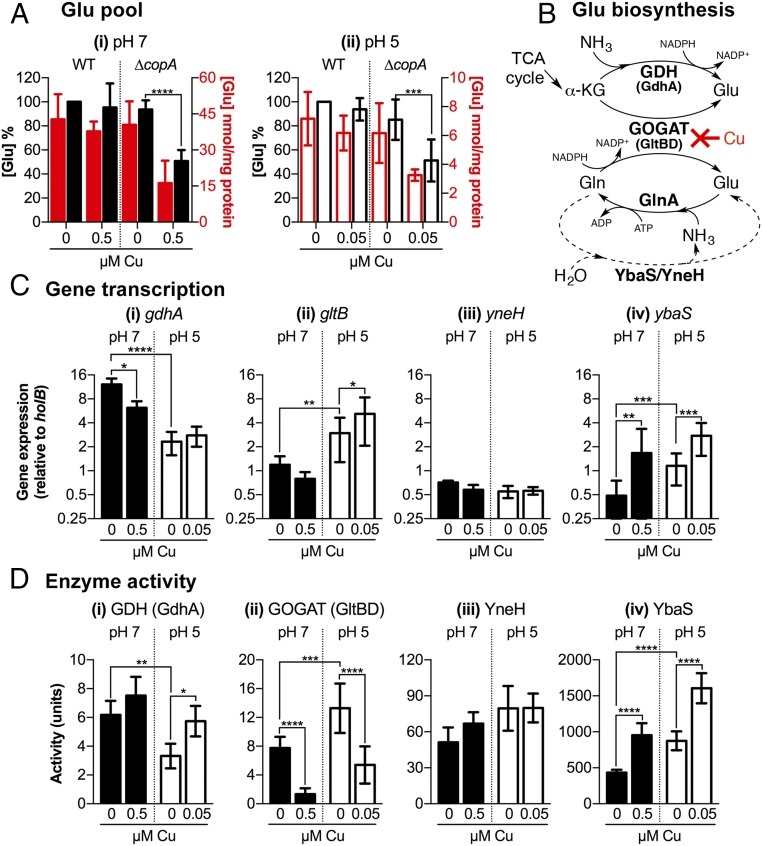
Glutamate is a key metabolite in E. coli, accounting for nearly 40% of all intracellular metabolites during exponential growth (15). The observed auxotrophy for Glu during Cu stress (Fig. 3 and SI Appendix, Fig. S8) indicated that the internal pool of this amino acid was depleted. To test this hypothesis, E. coli was cultured with or without added Cu to the midexponential phase and the concentrations of free Glu in harvested cells were measured. As anticipated, growth in Cu-supplemented medium diminished the Glu pools in the ∆copA mutant (Fig. 4A). This effect was observed at both pH 5 (Fig. 4 A, ii) and pH 7 (Fig. 4 A, i) but less Cu was required at pH 5, consistent with our earlier finding that less Cu was required to inhibit the ∆copA mutant at pH 5. Intracellular Glu levels in WT cells remained unaffected under these conditions (Fig. 4A).
Fig. 4.
Effects of Cu on glutamate biosynthesis in E. coli. (A) Intracellular Glu concentrations. E. coli UTI89 WT and ∆copA mutant were cultured at (i) pH 7 or (ii) pH 5 with or without added Cu as indicated. Intracellular concentrations of Glu were shown as absolute values (red columns) or as a percentage relative to untreated WT (black columns). Data were averaged from five independent biological replicates. (B) Glu biosynthesis pathways. De novo synthesis of Glu begins with α-KG from the TCA cycle. This process is catalyzed either by GDH using ammonia as the nitrogen donor (Top pathway) or by GOGAT using Gln as the nitrogen donor (Middle pathway). Glu can also be generated by the hydrolysis of Gln. This process is catalyzed by YbaS or YneH (Bottom pathway). Our data show that excess Cu inhibits biosynthesis at the GOGAT step. The figure also shows one route of Glu consumption via GS, which also generates Gln. (C) Expression of Glu biosynthesis genes. The ∆copA mutant was cultured at pH 7 (black columns) or pH 5 (white columns) with or without added Cu as indicated. Amounts of (i) gdhA, (ii) gltB, (iii) yneH, and (iv) ybaS transcripts relative to holB were measured by qPCR. Data were averaged from six independent biological replicates. (D) Activities of Glubiosynthesis enzymes. The ∆copA mutant was cultured at pH 7 (black columns) or pH 5 (white columns) with or without added Cu as indicated. Activities of (i) GDH, (ii) GOGAT, (iii) YneH, and (iv) YbaS were measured in cell-free lysis extracts. Data were averaged from six independent biological replicates. (A, C, and D) Error bars represent ±SD; ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05.
Our measurements further revealed that basal Glu levels in midexponential ∆copA cells cultured at pH 5 without any added Cu (∼9 nmol Glu per milligram of protein, Fig. 4 A, ii) were lower than in cells cultured at pH 7 (∼44 nmol Glu per milligram of protein, Fig. 4 A, i). This Glu-starved phenotype was likely associated with the consumption of this amino acid to maintain the internal pH during growth in mild acid, which could occur via the GadA/B decarboxylases (Fig. 1B) (16, 17). Hence, the observed importance of exogenous Glu but not BCAAs or other amino acids during Cu stress at pH 5 (Fig. 3A) may reflect the unique role for Glu for growth in acidic conditions.
Excess Cu Inhibits Glu Biosynthesis via Glutamate Synthase.
Glutamate in E. coli is synthesized from α-ketoglutarate (α-KG), an intermediate in the TCA cycle, via two pathways (Fig. 4B). Glutamate dehydrogenase (GDH or GdhA) generates Glu from the reductive amination of α-KG. In the alternative pathway, condensation of Glu with ammonia by glutamine synthetase (GS or GlnA) yields Gln. Subsequent reductive transamination of Gln with α-KG by glutamate synthase [GltBD or glutamine oxoglutarate aminotransferase (GOGAT)] generates two Glu molecules (a net gain of one). Together, GDH, GS, and GOGAT constitute the central pathway for nitrogen assimilation in E. coli. The GDH route is thought to be most efficient when ammonia is abundant, whereas GOGAT is important during ammonia limitation (18).
Addition of the Glu precursor α-KG failed to protect the ∆copA mutant from Cu stress at pH 5 and pH 7 (Fig. 3), implying that either GDH or GOGAT, or both, was inactive. Growth at pH 5 in the presence of added Cu did not reduce gdhA transcription (Fig. 4 C, i) or GDH activity (Fig. 4 D, i) in midexponential ∆copA cells. At pH 7, there was a decrease in gdhA transcription (Fig. 4 C, i) but there was no loss in GDH activity (Fig. 4 D, i). By contrast, growth in Cu-supplemented medium at pH 5 reduced the activity of GOGAT in midexponential ∆copA cells by ∼50% (Fig. 4 D, ii). GOGAT activity was also lost during growth in Cu-supplemented medium at pH 7 but the amount of Cu required to achieve this effect was again higher than at pH 5 (Fig. 4 D, ii). There was no change in the levels of gltB transcription in these Cu-treated cultures (Fig. 4 C, ii), suggesting that Cu exerted an inhibitory effect at the protein level. Both GOGAT and GDH remained active in WT cells during growth at either pH, with or without added Cu (SI Appendix, Fig. S10).
Cu-stressed ∆copA cultures displayed additional phenotypes consistent with GOGAT deficiency. There was a small but reproducible increase in GDH activity in ∆copA cells grown in Cu-rich medium at pH 5 (Fig. 4 D, i). Up-regulation of GDH has been shown to compensate for the decrease in Glu biosynthesis in ∆gltD and ∆gltB mutant strains of E. coli and Salmonella, respectively (19). Decreasing the amount of ammonia in the culture medium further exacerbated Cu stress in ∆copA cells at both pH 7 and pH 5 (SI Appendix, Fig. S11), consistent with GDH becoming more inefficient and increased reliance on GOGAT for making Glu (18) (Fig. 4B).
Mechanism of GOGAT Inhibition.
GOGAT contains two [4Fe–4S] clusters in the active site. The decrease in GOGAT activity during Cu stress at pH 5 and pH 7 corresponded with reductions in the activities of two additional [4Fe–4S]-containing enzymes, namely NADH dehydrogenase I (NUO) and succinate dehydrogenase (SDH) (SI Appendix, Fig. S12). These results suggested that Cu stress in our experimental conditions interfered with the synthesis, maturation, and incorporation of Fe–S clusters into enzymes via the Isc pathway as proposed previously (13, 14, 20). However, contrary to these prior reports, the amounts of Cu used in these experiments were not sufficient to induce expression of sufA and sufB, two genes that encode components of the Suf pathway for Fe–S cluster repair (SI Appendix, Fig. S13 A and B, discussed below).
Mature, solvent-accessible [4Fe–4S] clusters are also known to be destroyed by excess Cu (13, 21). To determine whether GOGAT was poisoned directly by Cu, the ∆copA mutant was cultured without any added Cu and, upon reaching midexponential phase, was exposed to added Cu (0–10 µM) for 30 min. Consistent with our hypothesis, this treatment had no effect on GDH activity but it reduced GOGAT activity in a dose-dependent manner (SI Appendix, Fig. S14), suggesting that at least one of its [4Fe–4S] clusters is solvent exposed. As a control, we confirmed that the activity of NUO remained unaffected (SI Appendix, Fig. S14), likely because its [4Fe–4S] clusters are protected by the protein scaffold. However, the doses of Cu that poisoned GOGAT (∼10 µM) were higher than doses that suppressed growth (<1 µM) (Fig. 2 and SI Appendix, Fig. S4). Hence, we propose that the loss of GOGAT during Cu stress was associated primarily with the loss of [4Fe–4S] cluster biosynthesis (also see SI Appendix, Discussion S1).
An Implication for the Loss of GOGAT at pH 5.
The varying protective effects of amino acids during Cu stress as described earlier can be reconciled with an impaired GOGAT activity. Exogenous Glu was protective (Fig. 3) likely because it bypassed GOGAT and supplied cells with the final reaction product (Fig. 4B). Similarly, Asp and Asn were protective (SI Appendix, Fig. S9A) because these amino acids could act as substrates for an alternative route to Glu. The three enzymes involved in this pathway, aspartate aminotransferase (AspC) and the two asparaginases (AnsA and AnsB) (SI Appendix, Fig. S9A), are presumably Cu tolerant because they do not require Fe–S clusters or other transition metal ions as cofactor.
As noted earlier, exogenous Glu did not restore Cu tolerance of the ∆copA mutant to WT levels, likely because multiple other Fe–S clusters were also inactivated (e.g., NUO and SDH). Whereas addition of Glu would alleviate the demand on GOGAT and upstream Fe–S enzymes that ultimately feed into the Glu pool (e.g., aconitase), it would not relieve the block on other defective enzymes such as the Fe–S dehydratases in the pathway for BCAA biosynthesis. Similarly, addition of BCAAs only partially rescued the ∆copA mutant from Cu stress (Fig. 3), likely because GOGAT and other Fe–S enzymes outside the BCAA biosynthesis pathway remained inactive.
It is important to highlight that at pH 5, the protective effect of BCAAs was diminished relative to Glu (Fig. 3A). This observation was consistent with the established requirement for Glu, but not BCAAs, for acid tolerance (Fig. 1B). The Km values for the two Glu decarboxylases in E. coli have been reported to range between 1 and 15 mM (22–24). Our estimate of the intracellular Glu pool at pH 5 was within this range (∼9 nmol/mg protein or ∼3 mM). Hence, we anticipate that the inability to synthesize Glu via GOGAT (and the corresponding decrease in the Glu pool) would impact the efficiency of acid tolerance under our experimental conditions.
Cu Stress Induces Expression of YbaS and YbaT.
Exogenous Gln was also protective, albeit to a lesser extent compared with Glu (SI Appendix, Fig. S8). This observation was counterintuitive, given that Gln is a cosubstrate for GOGAT (Fig. 4B). However, like Asp and Asn, Gln can be converted to Glu independently from GOGAT by two separate glutaminases, namely YbaS (glutaminase A) and YneH (glutaminase B) (Fig. 4B). Although Cu stress did not affect YneH activity in midexponential ∆copA cells (Fig. 4 D, iii), it increased the activity of YbaS (Fig. 4 D, iv). This increase was observed during growth at pH 5 and pH 7, although again the amount of Cu required to achieve this effect was less at pH 5 (Fig. 4 D, iv). The up-regulation in YbaS activity correlated with an increase in the levels of ybaS (Fig. 4 C, iv) and ybaT (SI Appendix, Fig. S13C) expression. In contrast, expression of yneH was not altered in response to Cu (Fig. 4 C, iii).
In the simplest model, induction of ybaST in response to Cu stress would occur in a CueR-dependent manner (Fig. 1). To test this hypothesis, we constructed a ∆copA∆cueR double mutant. This mutant was reproducibly more Cu sensitive than was the ∆copA parent strain at pH 5 (SI Appendix, Fig. S15 and SI Appendix, Discussion S2). Nevertheless, Cu treatment also increased the amounts of ybaST transcripts in the ∆copA∆cueR mutant compared with the untreated control (SI Appendix, Fig. S16), suggesting that CueR did not directly regulate expression of ybaST, at least under our experimental conditions.
Although the ybaS and ybaT genes for acid tolerance were induced during Cu stress, the copA gene for Cu tolerance did not appear to be up-regulated during acid stress. Basal expression levels of copA (SI Appendix, Fig. S17) and activities of the PcopA-lacZ fusion (SI Appendix, Fig. S6) in midexponential WT cells were comparable regardless of the growth pH. Thus, the evidence described in this work collectively points to an indirect and potentially complex relationship between Cu tolerance and acid tolerance systems.
The Protective Effect of Gln Is Suppressed in a ∆ybaST Mutant.
Our results led us to propose that YbaS and YbaT act as a compensatory pathway that offsets the loss in GOGAT and protects the intracellular Glu pool via hydrolysis of Gln and regeneration of Glu (Figs. 1B and 4B). The glutaminase activity of YbaS also produces ammonia, which contributes to overall acid tolerance (Fig. 1B) (7). This model predicts that the protective effect of exogenous Gln would be suppressed if YbaS and YbaT were inactive. To test this proposal, we mutated the entire ybaST operon in the ∆copA genetic background. The resulting ∆copA∆ybaST mutant strain was confirmed to display no measurable YbaS activity (SI Appendix, Fig. S18).
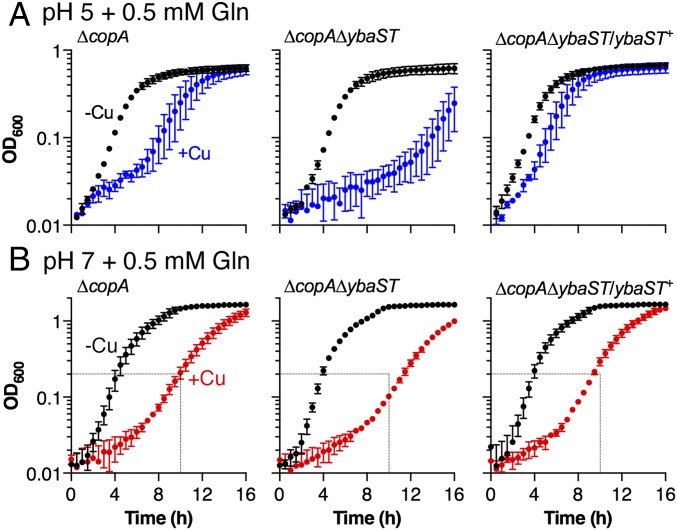
When cultured in the absence of added Gln, the ∆copA∆ybaST mutant displayed a Cu-sensitive phenotype that was comparable to the ∆copA parent strain (SI Appendix, Fig. S19). However, when the growth medium at pH 5 was supplemented with Gln (0.5 mM), the ∆copA∆ybaST double mutant was demonstrably more Cu sensitive than was the ∆copA parent strain, as evidenced by a prolonged lag phase in the presence of Cu (Fig. 5A). This observation indicated that the absence of functional ybaS and ybaT genes led to an increased sensitivity to Cu in the presence of Gln, or alternatively, a diminished protective effect of Gln during Cu stress. This phenotype was observed regardless of ammonia availability (SI Appendix, Fig. S20). Expression of the ybaST operon on a plasmid fully restored YbaS activity (SI Appendix, Fig. S18) and, subsequently, the protective effect of Gln (Fig. 5A). By contrast, deletion of ybaST had only a minor impact on Gln protection at pH 7, suggesting that this operon plays a less important role at pH 7 (Fig. 5B). These results confirmed that the protective effect of Gln against Cu stress during growth in acidic conditions required ybaS and ybaT, and they established a link between Cu stress, acid tolerance, and Gln utilization.
Fig. 5.
Effects of ∆ybaST mutation on Gln-dependent Cu tolerance. E. coli EC958 ∆copA mutant, ∆copA∆ybaST double mutant, and ∆copA∆ybaST/ybaST+ complemented mutant were cultured (A) at pH 5 in the presence of 0 µM (black, −Cu) or 0.05 µM (blue, +Cu) added Cu or (B) at pH 7 in the presence of 0 µM (black, −Cu) or 0.5 µM (red, +Cu) added Cu. The medium was supplemented with 0.5 mM Gln. The inoculum, used in each experiment was precultured in the same pH. Data were averaged from three independent experiments. Error bars represent ±SD.
Discussion
The antibacterial activity of Cu is an important component of the mammalian innate immune defense (1). In response, bacterial pathogens mount a survival strategy that relies on the efflux of excess Cu ions from their cytoplasm. The copA genes of clinically significant pathogens, including Mycobacterium tuberculosis (3), Streptococcus pneumoniae (2), and Klebsiella pneumoniae (4), have been identified as a virulence factor in animal models of infection. We have previously described the synergistic action of Cu ions with other antibacterial agents that may be derived from the host, such as nitric oxide (25) and hydrogen peroxide (26). Here we provide evidence that Cu ions and acid are also strong costressors.
Cu is a highly competitive metal that outcompetes weaker binding metals from sites in metalloproteins, leading to Cu intoxication in cells. Several proteins that contain Fe (particularly Fe–S clusters) (13, 14, 21, 26, 27), Zn (28), and Mn (29) have now been identified as targets of Cu poisoning. Because metalloproteins account for nearly half of all enzymes in cells (30), precisely which enzymes are mismetallated by Cu and the ensuing changes in bacterial physiology may vary, depending on the specific organism and experimental conditions. The latter do not always approximate the natural environment of the organism under investigation. In the case of E. coli, the unprecedented synteny of Cu tolerance genes with Gln-dependent acid tolerance genes in its chromosome (Fig. 1A) may provide an insight. Importantly, this genetic arrangement is conserved in E. coli, implying strong selection pressure.
E. coli resides primarily in the lower intestines of mammals. During its interaction with the animal host, this bacterium experiences mild and extreme fluctuations in external pH in the stomach, intestinal lumen, genitourinary tract, and phagolysosomes of epithelial and innate immune cells. Recent evidence suggests that E. coli also encounters elevated levels of Cu in at least some of these sites. Survival of a ∆copA mutant within murine macrophages was impaired compared with the WT (5). In uropathogenic E. coli, copA, as well as cueO, cusC, and cusF genes for Cu tolerance were highly expressed during human urinary tract infection, and this observation correlated with increased Cu concentrations in the urine of infected patients (31).
The combination of Cu and low pH poses a unique challenge to E. coli metabolism. Our in vitro work showed that excess Cu ions in the E. coli cytoplasm may impair acid tolerance by disrupting Glu biosynthesis via GOGAT. However, our data also suggested that E. coli may use alternative enzymes, namely YbaS and YbaT, to overcome this block in Glu synthesis if exogenous Gln is supplied. Intriguingly, Gln is the most abundant amino acid in systemic circulation. Approximately 600 µM of free Gln is present in human blood plasma and nearly a third of this supply is turned over by gastrointestinal mucosa epithelial cells (32, 33). Gln is also indispensable for the proliferation and antimicrobial activity of innate immune cells (34). It is plausible that to survive in vivo, E. coli can alter its metabolism to access host Gln stores using the mechanisms identified in this work. Notably, both ybaS and ybaT were also up-regulated along with copA during extraintestinal urinary tract infection in humans (31).
Other enteric bacteria, for example Salmonella and Klebsiella, share common colonization routes and niches with E. coli, and they are presumably also exposed to the combination of acid and Cu stress. Like E. coli, both organisms use GOGAT to synthesize Glu. However, neither relies on Glu for acid tolerance (35) and thus poisoning of the Glu biosynthesis pathway would not have the same impact on their survival. It is worth noting that in these organisms, copA and cueR are contiguous, and homologs for YbaS and YbaT are absent (SI Appendix, Fig. S2). The key targets of Cu stress in these organisms remain to be identified but for K. pneumoniae, [4Fe–4S] dehydratases in the pathway for branched-chain amino acid biosynthesis are major candidates, at least during colonization of the lung (4).
How is ybaST up-regulated by Cu? The intergenic region between copA and ybaST contains three regulatory sites (Fig. 1A): CueR, the primary Cu sensor in E. coli and regulator of copA; CpxR, which controls the global response to envelope stress; and GadX/W, which governs transcription of acid resistance genes including gadA/B and gadC. MerR-like regulators regulate transcription of divergent sequences in both directions (36) but CueR does not appear to also control ybaST (SI Appendix, Fig. S16). Meanwhile, the putative role of CpxR in ybaST transcription has not been experimentally tested (37). In contrast, regulation of ybaST by GadX and GadW is well established, and this occurs in a RpoS-dependent manner (38–40). Notably, RpoS and CpxR also contribute to the cellular adaptation to excess Cu (41, 42). Finally, our results link Cu stress to central nitrogen metabolism and thus additional control by the nitrogen response regulators, such as NtrB and NtrC, may be involved (18). These regulators sense intracellular Gln/Glu/α-KG ratios, which may shift during conditions of Cu stress (Fig. 4A). Ntr regulation is further coupled with RpoS via the stringent response and the (p)ppGpp signaling alarmone (18, 43). It is likely that the ybaST operon is subject to a complex network of regulatory controls not unlike those identified for the gadA/BC genes for Glu-dependent AR. Further studies are required to elucidate how these hierarchies of regulation are coordinated.
Finally, we noted that Cu stress in our experimental conditions was sufficient to induce ybaS and ybaT but not the suf pathway for the repair of damaged iron–sulfur clusters (SI Appendix, Fig. S13). Hence, we propose that up-regulation of ybaS and ybaT represents subtle metabolic changes that may occur in the graded response to low physiological levels of Cu encountered in the host.
Methods
Strains and Culture Conditions.
E. coli EC958, UTI89, and MG1655 were used in this study as indicated. All strains were propagated from frozen glycerol stocks on antibiotic-free LB agar or, where specified, in liquid M9 medium. Strains carrying pSU2718 plasmids were propagated in the presence of chloramphenicol (30 µg/mL). Liquid cultures were prepared in modified M9 medium (3 g/L KH2PO4, 0.5 g/L NaCl, 1 mM MgSO4, 0.1 mM CaCl2, 16.5 µg/mL thiamine, 25 mg/mL nicotinamide) using glucose (1 g/L) as carbon source and ammonium chloride (1 g/L) as nitrogen source. The medium was buffered at pH 5 with Na-Mes (50 mM) or at pH 7 with Na-Mops (50 mM). These pH buffers do not form a stable complex with Cu2+ ions (44). The medium was used without any metal purification step. Cultures were inoculated to an initial OD600 of 0.01 and grown at 37 °C with shaking at 200 rpm.
Construction of Mutants.
Deletion mutants of copA, cueR, and the ybaST operon were constructed by λ-Red mediated homologous recombination using the cat or kan cassettes from plasmid pKD3 or pKD4, respectively, as the selection marker and primers listed in SI Appendix, Table S1. The antibiotic marker was excised using a pCP20-Gm or pCP20-Amp plasmid encoding the FLP recombinase. Complemented mutants were generated by cloning the gene of interest into plasmid pSU2718 between BamHI and XbaI cut sites and subsequent transformation.
Growth Assays.
Bacterial growth was monitored in U-bottomed 96-well microtiter plates using an automated microplate shaker and reader (FluoStar Optima, BMG Labtech). Each well contained 200 µL of culture supplemented with Cu and/or L-amino acids. Stocks of amino acids were prepared immediately before use. The microplate was sealed with a sterile gas-permeable polyurethane membrane (Sigma). OD600 values were recorded up to 16 h. Microplates were shaken at 200 rpm in the orbital mode between readings.
Biochemical Analyses.
Batch cultures (50 mL) were prepared in acid-washed glass flasks. Cu was added to the desired final concentration. Bacteria were cultured to the midexponential phase (OD600 ∼0.3, approximately four doublings) and harvested by centrifugation (4,000 × g, 4 °C). The final pellets were processed further for the measurements of intracellular Glu contents, enzyme activities, and gene expression levels. Details are available in SI Appendix, SI Methods.
Statistical Analysis.
All statistical analyses were performed using two-way ANOVA in GraphPad Prism7. Results were not corrected for multiple comparisons.
Supplementary Material
Acknowledgments
We thank A. Turner (University of Queensland) for critical reading of this paper and R. Borthwick and attendees of the 10th International Biometals Symposium for insightful discussions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620232114/-/DCSupplemental.
References
- 1.Djoko KY, Ong CL, Walker MJ, McEwan AG. The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J Biol Chem. 2015;290:18954–18961. doi: 10.1074/jbc.R115.647099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shafeeq S, et al. The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol Microbiol. 2011;81:1255–1270. doi: 10.1111/j.1365-2958.2011.07758.x. [DOI] [PubMed] [Google Scholar]
- 3.Ward SK, Abomoelak B, Hoye EA, Steinberg H, Talaat AM. CtpV: A putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol Microbiol. 2010;77:1096–1110. doi: 10.1111/j.1365-2958.2010.07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachman MA, et al. Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. MBio. 2015;6:e00775. doi: 10.1128/mBio.00775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White C, Lee J, Kambe T, Fritsche K, Petris MJ. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem. 2009;284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rensing C, Grass G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev. 2003;27:197–213. doi: 10.1016/S0168-6445(03)00049-4. [DOI] [PubMed] [Google Scholar]
- 7.Lu P, et al. L-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res. 2013;23:635–644. doi: 10.1038/cr.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown G, et al. Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis. Biochemistry. 2008;47:5724–5735. doi: 10.1021/bi800097h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol. 2004;186:6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moriel DG, et al. 2016. A novel protective vaccine antigen from the core Escherichia coli genome. mSphere 1: pii: e00326-16.
- 11.Williamson NR, Simonsen HT, Harris AK, Leeper FJ, Salmond GP. Disruption of the copper efflux pump (CopA) of Serratia marcescens ATCC 274 pleiotropically affects copper sensitivity and production of the tripyrrole secondary metabolite, prodigiosin. J Ind Microbiol Biotechnol. 2006;33:151–158. doi: 10.1007/s10295-005-0040-9. [DOI] [PubMed] [Google Scholar]
- 12.Djoko KY, et al. Copper(II)-Bis(Thiosemicarbazonato) complexes as antibacterial agents: Insights into their mode of action and potential as therapeutics. Antimicrob Agents Chemother. 2015;59:6444–6453. doi: 10.1128/AAC.01289-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci USA. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fung DKC, Lau WY, Chan WT, Yan A. Copper efflux is induced during anaerobic amino acid limitation in Escherichia coli to protect iron-sulfur cluster enzymes and biogenesis. J Bacteriol. 2013;195:4556–4568. doi: 10.1128/JB.00543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreau PL. The lysine decarboxylase CadA protects Escherichia coli starved of phosphate against fermentation acids. J Bacteriol. 2007;189:2249–2261. doi: 10.1128/JB.01306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roe AJ, McLaggan D, Davidson I, O’Byrne C, Booth IR. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J Bacteriol. 1998;180:767–772. doi: 10.1128/jb.180.4.767-772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Heeswijk WC, Westerhoff HV, Boogerd FC. Nitrogen assimilation in Escherichia coli: Putting molecular data into a systems perspective. Microbiol Mol Biol Rev. 2013;77:628–695. doi: 10.1128/MMBR.00025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan D. Protection of the glutamate pool concentration in enteric bacteria. Proc Natl Acad Sci USA. 2007;104:9475–9480. doi: 10.1073/pnas.0703360104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan G, et al. Copper binding in IscA inhibits iron-sulphur cluster assembly in Escherichia coli. Mol Microbiol. 2014;93:629–644. doi: 10.1111/mmi.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chillappagari S, et al. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J Bacteriol. 2010;192:2512–2524. doi: 10.1128/JB.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fonda ML. L-Glutamate decarboxylase from bacteria. Methods Enzymol. 1985;113:11–16. doi: 10.1016/s0076-6879(85)13005-3. [DOI] [PubMed] [Google Scholar]
- 23.Tramonti A, De Biase D, Giartosio A, Bossa F, John RA. The roles of His-167 and His-275 in the reaction catalyzed by glutamate decarboxylase from Escherichia coli. J Biol Chem. 1998;273:1939–1945. doi: 10.1074/jbc.273.4.1939. [DOI] [PubMed] [Google Scholar]
- 24.Thu Ho NA, Hou CY, Kim WH, Kang TJ. Expanding the active pH range of Escherichia coli glutamate decarboxylase by breaking the cooperativeness. J Biosci Bioeng. 2013;115:154–158. doi: 10.1016/j.jbiosc.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Djoko KY, et al. Phenotypic characterization of a copA mutant of Neisseria gonorrhoeae identifies a link between copper and nitrosative stress. Infect Immun. 2012;80:1065–1071. doi: 10.1128/IAI.06163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Djoko KY, McEwan AG. Antimicrobial action of copper is amplified via inhibition of heme biosynthesis. ACS Chem Biol. 2013;8:2217–2223. doi: 10.1021/cb4002443. [DOI] [PubMed] [Google Scholar]
- 27.Azzouzi A, et al. Coproporphyrin III excretion identifies the anaerobic coproporphyrinogen III oxidase HemN as a copper target in the Cu+-ATPase mutant copA− of Rubrivivax gelatinosus. Mol Microbiol. 2013;88:339–351. doi: 10.1111/mmi.12188. [DOI] [PubMed] [Google Scholar]
- 28.Tottey S, et al. Cyanobacterial metallochaperone inhibits deleterious side reactions of copper. Proc Natl Acad Sci USA. 2012;109:95–100. doi: 10.1073/pnas.1117515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson MD, Kehl-Fie TE, Rosch JW. Copper intoxication inhibits aerobic nucleotide synthesis in Streptococcus pneumoniae. Metallomics. 2015;7:786–794. doi: 10.1039/c5mt00011d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: From enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 31.Subashchandrabose S, et al. Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc Natl Acad Sci USA. 2014;111:18327–18332. doi: 10.1073/pnas.1415959112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stein WH, Moore S. The free amino acids of human blood plasma. J Biol Chem. 1954;211:915–926. [PubMed] [Google Scholar]
- 33.Windmueller HG, Spaeth AE. Uptake and metabolism of plasma glutamine by the small intestine. J Biol Chem. 1974;249:5070–5079. [PubMed] [Google Scholar]
- 34.Newsholme P. Why is L-glutamine metabolism important to cells of the immune system in health, postinjury, surgery or infection? J Nutr. 2001;131:2515S–2522S; discussion 2523S-2514S. doi: 10.1093/jn/131.9.2515S. [DOI] [PubMed] [Google Scholar]
- 35.McClelland M, et al. Comparison of the Escherichia coli K-12 genome with sampled genomes of a Klebsiella pneumoniae and three Salmonella enterica serovars, Typhimurium, Typhi and Paratyphi. Nucleic Acids Res. 2000;28:4974–4986. doi: 10.1093/nar/28.24.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lund PA, Ford SJ, Brown NL. Transcriptional regulation of the mercury-resistance genes of transposon Tn501. J Gen Microbiol. 1986;132:465–480. doi: 10.1099/00221287-132-2-465. [DOI] [PubMed] [Google Scholar]
- 37.De Wulf P, McGuire AM, Liu X, Lin EC. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J Biol Chem. 2002;277:26652–26661. doi: 10.1074/jbc.M203487200. [DOI] [PubMed] [Google Scholar]
- 38.Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: Transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol Microbiol. 2008;70:965–982. doi: 10.1111/j.1365-2958.2008.06458.x. [DOI] [PubMed] [Google Scholar]
- 39.Tucker DL, et al. Genes of the GadX-GadW regulon in Escherichia coli. J Bacteriol. 2003;185:3190–3201. doi: 10.1128/JB.185.10.3190-3201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo SW, Kim D, O’Brien EJ, Szubin R, Palsson BO. Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat Commun. 2015;6:7970. doi: 10.1038/ncomms8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macomber L, Rensing C, Imlay JA. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol. 2007;189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto K, Ishihama A. Transcriptional response of Escherichia coli to external copper. Mol Microbiol. 2005;56:215–227. doi: 10.1111/j.1365-2958.2005.04532.x. [DOI] [PubMed] [Google Scholar]
- 43.Brown DR, Barton G, Pan Z, Buck M, Wigneshweraraj S. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat Commun. 2014;5:4115. doi: 10.1038/ncomms5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mash HE, Chin YP, Sigg L, Hari R, Xue H. Complexation of copper by zwitterionic aminosulfonic (Good) buffers. Anal Chem. 2003;75:671–677. doi: 10.1021/ac0261101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.