Abstract
Drug-induced phospholipidosis (DIPL) is characterized by an increase in the phospholipid content of the cell and the accumulation of drugs and lipids inside the lysosomes of affected tissues, including in the liver. Although of uncertain pathological significance for patients, the condition remains a major impediment for the clinical development of new drugs. Human Sandhoff disease (SD) is caused by inherited defects of the β subunit of lysosomal β-hexosaminidases (Hex) A and B, leading to a large array of symptoms, including neurodegeneration and ultimately death by the age of 4 in its most common form. The substrates of Hex A and B, gangliosides GM2 and GA2, accumulate inside the lysosomes of the CNS and in peripheral organs. Given that both DIPL and SD are associated with lysosomes and lipid metabolism in general, we measured the hepatic lipid profiles in rodent models of these two conditions using untargeted LC/MS to examine potential commonalities. Both model systems shared a number of perturbed lipid pathways, notably those involving metabolism of cholesteryl esters, lysophosphatidylcholines, bis(monoacylglycero)phosphates, and ceramides. We report here profound alterations in lipid metabolism in the SD liver. In addition, DIPL induced a wide range of lipid changes not previously observed in the liver, highlighting similarities with those detected in the model of SD and raising concerns that these lipid changes may be associated with underlying pathology associated with lysosomal storage disorders.
Keywords: ceramides, lipidomics, lysophospholipid, lysosome, mass spectrometry, storage diseases, toxicology
Drug-induced phospholipidosis (DIPL) is a condition caused by drug treatment, leading to an increase in the cellular phospholipid content. It was first detected in 1948 in rats after long-term treatment with chloroquine, an antimalarial agent (1). DIPL has been observed in vitro and in vivo in animals and humans. In this disorder, cells can appear vacuolated when observed by light microscopy, although this is not always evident, and there is an accumulation of foamy macrophages containing phagocytosed material (2, 3). The key characteristic feature of DIPL is the observation, by electron microscopy, of intracellular unicentric or multicentric lamellar bodies (4). Lamellar bodies, also called myelin bodies, contain mostly undegraded phospholipids and the administered drug that caused DIPL (5).
Several hundred cationic amphiphilic drugs (CADs) have the potential to cause DIPL (2, 6–8). These agents belong to different drug classes, including antidepressants such as Prozac, antibiotics, antipsychotics, and cholesterol-lowering drugs, such as simvastatin. However, they all share a common structure: one or several hydrophobic rings and a hydrophilic chain containing an amine group positively charged at neutral pH (6, 9). CADs have been intensely developed by pharmaceutical industries because they can easily cross biological membranes, including the blood-brain barrier, due to their amphiphilic character. Because they are weak bases, CADs are protonated in the acidic environment of the lysosome and can no longer cross membranes; they are therefore trapped within the organelle (6). Although they have been studied for more than 65 years, the potential toxicity and mechanisms of DIPL remain unclear.
Progress has been made to identify biomarkers of DIPL; bis(monoacylglycero)phosphate (BMP; also known as lyso-bisphosphatidic acid) (16:0/18:1) has been suggested as a biomarker in tissues from rats treated with amiodarone (10), and BMPs as a class were found to be increased in concentration after DIPL (10, 11). BMPs are negatively charged phospholipids principally found in the inner membranes of late endosomes and lysosomes (12) with an unusual sn-1:sn-1′ configuration in which each glycerol moiety is esterified by a fatty acid. They stimulate the degradation of glycosphingolipids in these organelles (13) and participate in the transport of cellular cholesterol and its degradation (14–16).
The two main hypotheses to explain the increase in cellular phospholipid content after induction of DIPL are impaired degradation of phospholipids by direct or indirect inhibition of phospholipase activity (7, 17–20) and increased phospholipid synthesis (21, 22). However, other metabolic pathways have also been found to be upregulated in DIPL, including stimulation of lipogenesis in a time- and dose-dependent manner (22), increased cholesterol biosynthesis (19, 22, 23), and concerted changes in phospholipid degradation and cholesterol and fatty acid syntheses (19). Moreover, in this latter study, N-acylsphingosine amidohydrolase 1, also called lysosomal acid ceramidase, was identified as a marker transcript of DIPL (19).
DIPL shares common features with some lysosomal storage disorders (LSDs), including Niemann-Pick type C (NPC) and Fabry diseases. In NPC disease, caused by a defect in NPC 1 or 2 proteins that are responsible for the cellular traffic of cholesterol, both cholesterol and sphingolipids accumulate (24). A similar observation has been made in the liver of rats treated with chloroquine (25) and treatment with amiodarone in vitro (26). Fabry disease is characterized by the accumulation of globotriasylceramide due to a deficiency in α-galactosidase. A misdiagnosis of Fabry disease has been reported in a rheumatoid arthritis patient treated with chloroquine, due to the similar morphological features observed in biopsies from this lysosomal disease to those in DIPL (27). Furthermore, the concentration of several BMP species has also been found to be increased in a variety of lysosomal diseases (28, 29), including BMP (22:6/22:6) in the brain of a mouse model of neuronal ceroid lipofuscinosis (29).
Sandhoff disease (SD), another LSD, is caused by a mutation in the HEXB gene, which encodes for the β-subunit of β-hexosaminidases (Hexs), leading to a deficiency in Hex A and B (30). Hexs are lysosomal acid hydrolases that cleave N-acetylglucosamine and N-acetylgalactosamine residues from a variety of substrates, mainly glycopeptides, glycolipids, glycosaminoglycans, and ganglioside GM2 (30, 31). In SD patients, accumulation of gangliosides GM2 and the sphingolipid GA2 is detected throughout the CNS, as well as peripherally to a lesser degree. Gangliosides are complex sphingolipids composed of a ceramide (Cer) backbone, polysaccharide units, and, for most species, a sialic acid moiety. They are principally abundant in the CNS (32, 33), with their concentration being one to two orders of magnitude lower in other tissues (34). In visceral tissues, increases in globoside concentrations have been detected previously (35). There are a large array of symptoms in SD patients, including decrease in motor, mental, and visual abilities; alteration of vision; macrocephaly; seizures; liver enlargement; slight bone deformation; and neurodegeneration, ultimately leading to death by 4 years of age in the most severe form of the disease (30, 36, 37). Hexb−/− mice are a widely used model to study the pathogenesis of and develop treatment for SD (38–41).
In the present study, we have explored the metabolic alterations in the liver of Hexb−/− mice and those induced by treatment with chloroquine, a hepatic phospholipidosis-inducer, in rats (300 mg/kg/day for 5 days) using an untargeted lipidomic approach. Comparison of the two lysosomal lipidoses by exploring their lipid profiles has facilitated the identification of perturbed biochemical pathways, which immediately suggests common pathological mechanisms.
MATERIALS AND METHODS
Animal studies
All animal studies were ethically reviewed and carried out in accordance with the UK Animals (Scientific Procedures) Act 1986 and the GSK Policy on the Care, Welfare and Treatment of Animals (for the DIPL study).
Rats for the study of DIPL.
A 7-day study was conducted on male Sprague-Dawley rats (n = 9 per group, 10 weeks old at the beginning of the study). The control group was orally dosed with vehicle (1% aqueous methylcellulose), and the chloroquine group received an oral dose of 300 mg/kg/day. The dose was chosen because it was known to produce lesions associated with DIPL in liver tissue (2, 6, 25, 42, 43). Animals were killed by exsanguination via the abdominal aorta under isoflurane anesthesia. Rats given 1% aqueous methylcellulose were killed at day 7, whereas rats treated with chloroquine were killed at day 5, due to intercurrent deaths and clinical signs observed up to that point.
Hexb−/− mice.
The congenic SD mouse model (strain: B6; 129S-Hexbtm1Rlp), developed by disruption of the Hexb gene (44), was obtained from the Jackson Laboratory and had been interbred for more than 20 generations. To maintain the colony, homozygous males were crossed with heterozygous females, or heterozygous animals were intercrossed. Mice (n = 5 for both the SD and control groups) were fed ad libitum and killed by asphyxiation at 4 months of age, the humane end point. Tissues were dissected and frozen immediately on dry ice.
Metabolite extraction
For the study of DIPL in rats, 100 mg of liver tissue was used for extraction, whereas 50 mg of liver tissue from mice was taken for the investigation of SD. Metabolites were extracted according to the following procedure described by Le Belle et al. (45). As an initial extraction solvent, 6.0 ml of a methanol/chloroform mixture in a 2:1 ratio (v/v) was used per gram of tissue. The mixture was homogenized in a TissueLyser (Qiagen, Hilden, Germany) and sonicated for 15 min at room temperature (22°C). To the mixture was added 2.0 ml of chloroform and 2.0 ml of water per gram of tissue to form an emulsion. The samples were then centrifuged for 20 min at 20,844 g at 22°C to generate distinct organic and aqueous fractions and a protein pellet. The organic and aqueous layers were individually removed and transferred to separate tubes. The entire extraction process was repeated on the protein pellet and any remaining aqueous or organic fraction in order to perform a double extraction to maximize recovery. Fractions prepared during the second extraction were combined with those from the first. The organic layer was placed in a fume hood under a stream of nitrogen gas to evaporate the solvent and, once dry, stored at −80°C until analysis.
LC/MS analysis
The open profiling of lipids was performed by using the dried lipid fraction obtained after metabolite extraction (10 mg of tissue equivalent). Samples were injected onto an LC column before MS to provide compound separation by chromatography.
For the DIPL sample analysis, chromatography was performed on an Acquity UPLC® (Waters, Inc., Milford, MA) by using an Acquity UPLC® High Strength Silica T3 column (1.8 µm × 2.1 mm × 100 mm) (Waters, Inc.) at 55°C. The lipid fraction was dissolved in methanol/chloroform mixture (1:1, v/v), and 5 µl of this was injected onto the column.
For the analysis of the SD samples, chromatography was performed on an Acquity UPLC® Charged Surface Hybrid C18 column (1.7 µm × 2.1 mm × 100 mm) (Waters, Inc.) at 55°C. The lipid fraction from each sample was diluted in a mixture of isopropanol/acetonitrile/water (2:1:1 v/v/v), and 2 µl of this was injected onto the column.
The LC parameters were the same for both studies. The flow rate was 0.4 ml/min. Mobile phase A was composed of 60% acetonitrile and 40% water, with the addition of 10 mM ammonium formate to improve ionization. Mobile phase B was 10% acetonitrile and 90% isopropanol with 10 mM ammonium formate. The chromatographic gradient progressed from 40% mobile phase B up to 99% B over 18 min, followed by 2 min of equilibration at 40% B.
MS was performed on a Xevo G2 Q-TOF mass spectrometer (Waters, Inc.) in positive- and negative-ion modes by using a scan time of 0.2 s, a collision energy of 6 V for each single scan, and a collision ramp from 25 to 40 V for the fragmentation function in positive-ion mode and from 30 to 50 V in negative-ion mode. Capillary voltage was 2 kV, sampling cone was 30 V, extraction cone was 3.5 V, and source temperature was 120°C. Leucine enkephalin was used as a lockmass to improve mass accuracy throughout the analysis, and 5 mM sodium formate was used to calibrate the instrument before analysis (the maximum threshold for mass difference between the measured mass and the exact mass was set at 5 parts per million).
Chromatograms were processed by using the MarkerLynx XS tool from the MassLynx software (version 4.1, Waters, Inc.) using peak detection analysis with the following parameters: retention time: 0–18 min; mass range: 50–1,800 kDa for the DIPL study and 50–2,300 kDa for the SD study; tolerance for the mass value: 0.05 kDa; and minimum intensity to consider for a spectral peak: 2,000 counts. Data were deisotoped and normalized to the internal standard [phosphatidylcholine (PC) 17:0/17:0] intensity, and metabolites were identified by using the following online databases: Lipid Metabolites and Pathways Strategies (www.lipidmaps.org), the Human Metabolome Database (www.hmdb.ca), and the Kyoto Encyclopedia of Genes and Genomes (www.genome.jp/kegg/).
Gene expression measurement using quantitative PCR
The extraction of RNA from 15 mg of rat liver was carried out by using the RNeasy lipid tissue mini kit (Qiagen). Briefly, liver tissue was homogenized with 1 ml of QIAzol Lysis Reagent by using a TissueLyser (Qiagen) operating at 20 Hz for 2 min. The lysate was then used for RNA extraction. After the addition of 200 µl of chloroform to each homogenate, each sample was centrifuged at 12,000 g for 15 min in a Centrifuge 5424 (Eppendorf, Hauppage, NY). After collection of the upper aqueous phase, one volume of 70% ethanol was added to each sample and vortex-mixed. Each sample was transferred to an RNeasy Mini Spin column and centrifuged for 15 s at 12,000 g. The column was then washed with 350 µl of Buffer RW1 and centrifuged for 15 s at 12,000 g. The DNA was eliminated with 80 µl of DNase 1 incubation mix directly placed onto the RNeasy spin column membrane and incubated at room temperature (22°C) for 15 min. The RNeasy Mini Spin column was then washed with 350 µl of Buffer RW1 and twice with 500 µl of Buffer RPE, followed by 15 s of centrifugation at 12,000 g and 2 min of centrifugation. RNA was then eluted twice with 50 µl of RNase-free water applied directly to the column, followed by centrifugation for 1 min at 16,000 g. The amount of RNA yielded was subsequently measured on a NanoDrop spectrophotometer (Thermo Fisher Scientific, Oakwood Village, OH).
Reverse transcription was carried out following the protocol supplied by the manufacturer. From the RNA isolated in the previous step, 1 µg was used as template. Genomic DNA was eliminated from the sample during 2 min of incubation in a TC-3000 PCR Thermal cycler (Techne, Staffordshire, UK) with 2 µl of gDNA wipeout buffer at 42°C. Reverse transcription was carried out during a further 15 min of incubation with 1 µl of Quantiscript reverse transcriptase, 4 µl of Quantiscript RT buffer, and 1 µl of RT primer mix at 42°C, and then stopped by applying a temperature of 95°C for 3 min. It was assumed that the totality of the mRNA was converted to cDNA through reverse transcription.
cDNA synthesized through the reverse transcription of mRNA was diluted in RNase-free water to 50 ng of cDNA as a template for amplification. The PCR experiment was carried out by using TaqMan technology on a StepOnePlus instrument (Applied Biosystems, Foster City, CA). The following method was used for DNA amplification: holding cycle at 50°C for 2 min, followed by 95°C for 10 min; amplification (40 cycles) at 95°C for 15 s, followed by 60°C for 1 min.
Data were analyzed by using the StepOne software (version 2.2, Applied Biosystems). Expression levels were normalized to the level of expression of 45S in all the control samples, according to the comparative CT real-time PCR method by calculating the ΔCT value. The ΔCT mean value was calculated as the average of the ΔCT for every replicate of each sample.
Multivariate statistics
Multivariate data analysis was performed by using SIMCA-P+ (versions 12.0 and 13.0, Umetrics AB, Malmö, Sweden). Datasets were unit variance scaled before analysis. The pattern recognition methods used were principal components analysis and partial least squares discriminate analysis.
Univariate statistics
Univariate statistics were conducted by using the GraphPad Prism package (version 4, GraphPad software). The unpaired two-tailed Student’s t test was used to compare Hexb null mice versus controls and chloroquine-treated rats versus controls. The significance value was set at P ≤ 0.05 to identify statistically significant changes.
RESULTS
To achieve the greatest coverage of the lipid species present in liver tissue, we applied a nontargeted lipidomic approach using LC/MS. Several lipid classes were commonly affected by DIPL and SD. Lysophosphatidylcholines (LPCs), cholesteryl esters (CEs), and Cers were analyzed in positive-ion mode, whereas BMPs were detected by using negative-ion mode. LPCs were eluted between 0.9 and 1.6 min, CEs between 15.2 and 16 min, and Cers between 7.4 and 14 min (Fig. 1). BMP species were detected between 2.9 and 4.8 min (supplemental Figs. S1, S2). The lipid species were annotated by using their exact mass, the fragmentation spectrum, and the elution time. Moreover, we analyzed standards for each of the lipid classes described, and the spectral data and retention times of the standards were used to confirm our annotations. Although we did not have standards for all of the species reported, we are at the level 2 of identification (putatively annotated compounds) according to the standards set by the Metabolomics Standards Initiative (46).
Fig. 1.
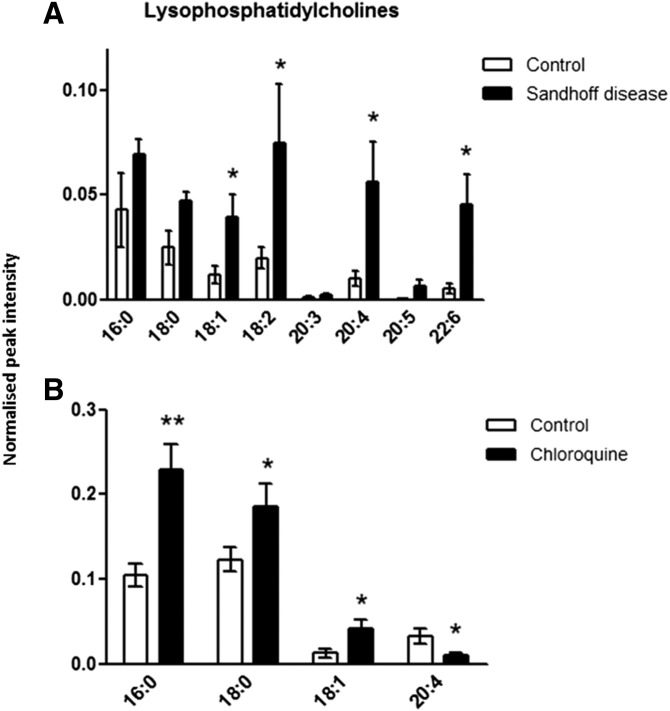
LPC concentrations in liver tissue. Metabolites were extracted from homogenized tissue, and LPCs were detected by using positive-ion mode LC/MS in control and Hexb−/− mice (A) and in control and chloroquine-treated rats (B). Results are mean ± SEM. Significance level is quoted for Student’s t test: * P ≤ 0.05; ** P ≤ 0.01.
LPCs
The concentrations of four unsaturated LPC species were significantly increased in the liver of Hexb−/− mice compared with controls (Fig. 1). In rat liver, after a 5-day treatment with chloroquine to induce phospholipidosis, we detected an increase in the concentrations of LPCs 16:0, 18:0, and 18:1, whereas the concentration of LPC 20:4 was decreased. Furthermore, the total LPC concentration was significantly increased by more than 3-fold in Hexb−/− mice and almost 2-fold in DIPL rats compared with their respective controls.
BMPs
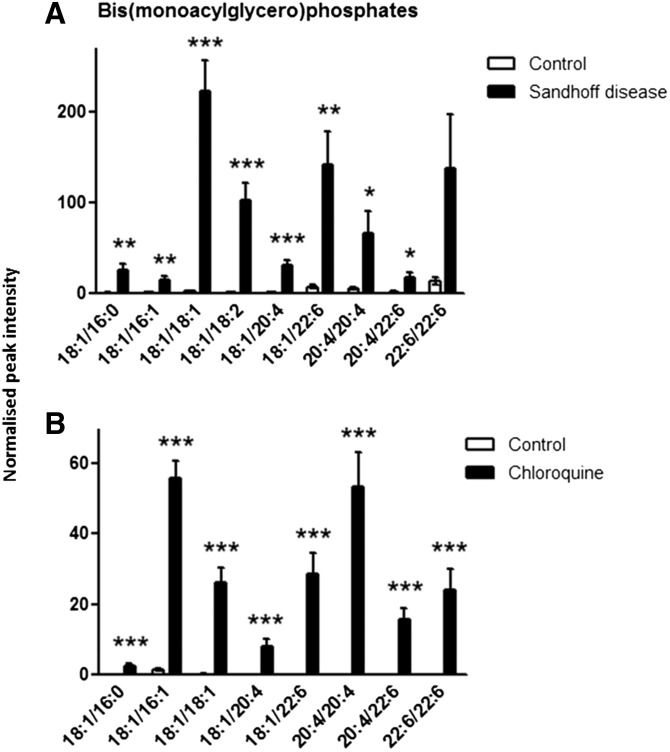
The same BMP species were detected in liver from mice and rats, except for BMP (18:1/18:2), which was exclusively found in mice (Fig. 2). The concentrations of all the BMPs were significantly increased in the liver of Hexb−/− mice and DIPL rats compared with the control groups, except for the concentration of BMP (22:6/22:6) in Hexb−/− mice, which showed particularly large variations in the SD group.
Fig. 2.
BMP concentrations in liver tissue. Metabolites were extracted from homogenized tissue, and BMPs were detected by using negative-ion mode LC/MS in control and Hexb−/− mice (A) and in control and chloroquine-treated rats (B). Results are mean ± SEM. Significance level is quoted for Student’s t test: * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001.
CEs
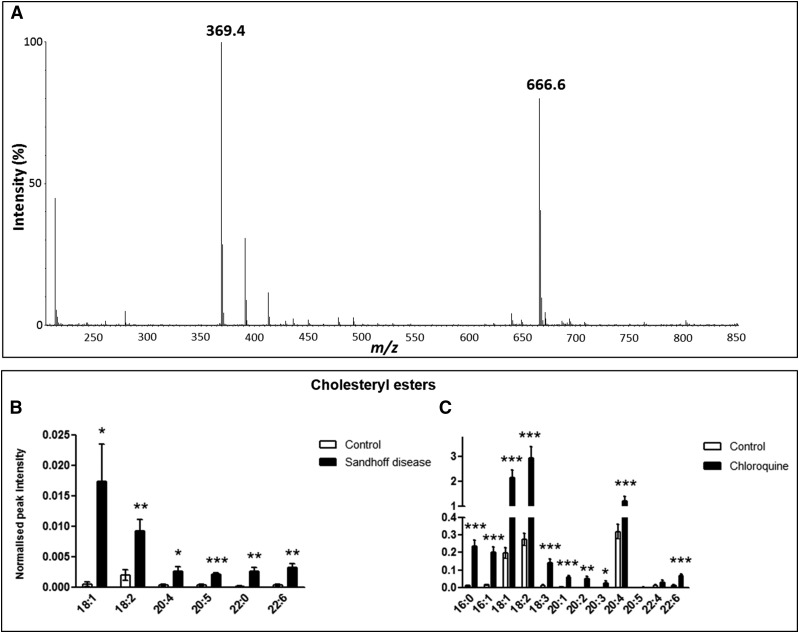
The identification of CEs was facilitated by the presence of a m/z 369 ion in the MS spectrum of the CEs, regardless of their acyl composition, associated with the cholesteryl head group (47, 48) (Fig. 3A). In the Hexb−/− mice, all the CE species detected were significantly increased compared with the control mice (Fig. 3B). In rat liver after DIPL, the concentrations of CEs were all significantly increased, except for those of CEs 20:5 and 22:4 (Fig. 3C). In addition, the total hepatic CE content was increased more than 10-fold in Hexb−/− mice and more than 8-fold in rats after DIPL.
Fig. 3.
CE concentrations in liver tissue. Mass spectrum of CE 18:2 represented by (M+NH4)+ (m/z 666.6) (A). The typical MS source fragment representative of CEs is also present (m/z 369.4). Metabolites were extracted from homogenized tissue, and CEs were detected by using positive-ion mode LC/MS in control and Hexb−/− mice (B) and in control and chloroquine-treated rats (C). Results are mean ± SEM. Significance level is quoted for Student’s t test: * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001.
Cers
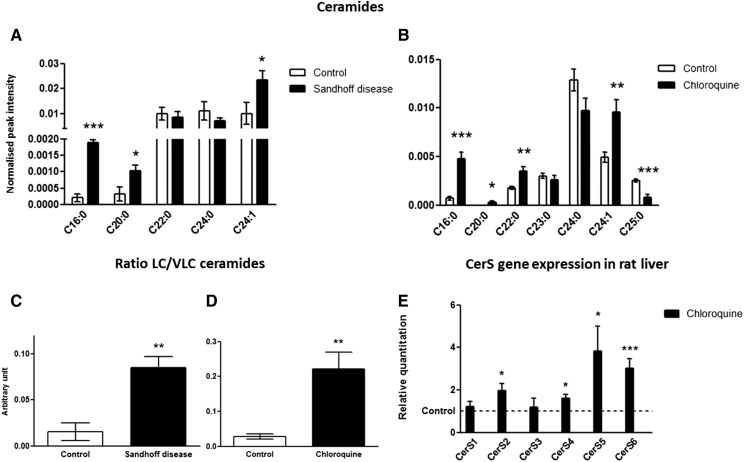
The assignment of Cers was confirmed by the presence of a fragment ion of m/z 264 in the MS/MS spectrum, which is typical of the fragmentation of Cers releasing their sphingoid base, sphingosine, as a doubly dehydrated form. The concentrations of three Cer species, C16:0-, C20:0-, and C24:1-Cer, were significantly increased in Hexb−/− mice compared with control animals. In rat liver after DIPL, the concentrations of C16:0-, C20:0-, C22:0-, and C24:1-Cer were significantly increased, whereas the C25:0-Cer concentration was decreased. In both disorders, C16:0-Cer was the most highly increased Cer species (Fig. 4A, B).
Fig. 4.
Cers in liver tissue. Metabolites were extracted from homogenized tissue, and Cers were detected by using positive-ion mode LC/MS in control and Hexb−/− mice (A) and in control and chloroquine-treated rats (B). Ratio of long-chain to VLC Cers in liver tissue from control and Hexb−/− mice (C) and from control and chloroquine-treated rats (D). Fold change in CerS mRNA expression in the liver of rats treated with chloroquine compared with controls (set to 1 for each transcript) as measured by real-time qPCR (E). Results are mean ± SEM. Significance level is quoted for Student’s t test: * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001.
Moreover, in the liver of both SD and DIPL animals, the ratio of long-chain to very-long-chain (VLC) Cers was significantly increased compared with the controls, indicating that the composition of the Cer pool was altered in a similar manner (Fig. 4C, D).
To investigate whether the variations in the concentrations of Cer species were associated with alterations to the expression of Cer synthases (CerSs), quantitative real time-PCR (qPCR) was performed on the six different CerSs in liver samples from control and chloroquine-treated rats. CerS2, CerS4, CerS5, and CerS6 gene expressions were significantly upregulated after treatment with amiodarone and chloroquine compared with controls (Fig. 4E). According to the enzyme specificities, the relative increases in the expression of the different CerSs in liver tissue after treatment with chloroquine are consistent with the alterations in Cer species after DIPL measured by LC/MS. In SD liver tissue, however, the gene expression of CerS1, CerS2, CerS4, CerS5, and CerS6 was not different from that of controls, whereas the expression of CerS3 was too low to be measured reliably (supplemental Fig. S3).
DISCUSSION
In the present study, we used a lipidomic approach to characterize the metabolic alterations occurring in the liver of two rodent models of lysosomal lipidoses, SD and DIPL. The use of LC/MS has allowed for the sensitive detection of several lipid species that were similarly affected in these two models. We measured increases in BMPs, LPCs, and CEs, and a remodeling of the Cer pool in favor of long-chain Cers, suggesting that both LSDs and DIPL have a number of common features.
DIPL has been associated with a loss of lysosomal phospholipase A2 (LPLA2) function (49, 50). In SD, the concentration of several species of LPCs was increased, although a different mechanism seemed to be responsible for these elevations; furthermore, the gene expression of the LPLA2 was found to be increased in an in vitro model of DIPL that we have previously investigated (data not shown).
Our results show that most BMP species detected were significantly increased in the liver, independently of their chain length, when Hexb was knocked out or after DIPL. This demonstrates that increases in BMPs are not specific to DIPL or one particular LSD, but, instead, are typical of impairments of the endo-lysosomal system, as other reports have suggested (14–16, 28, 29).
Mesens et al. (51) have shown that cholesterol esterification was one of the pathways principally affected in liver tissue from rats treated with amiodarone. These data perfectly concur with our findings, showing that there is a general increase in the concentrations of CEs in rats after exposure to chloroquine. Moreover, this increase was also detected in Hexb−/− mice, showing accumulation of CEs to be closely related to lysosomal storage.
The increases in BMPs and CEs and the changes in Cer concentrations suggest that there is disruption of the endomembrane system. This system is involved in the import of molecules in endosomes and lysosomes and their export through the endoplasmic reticulum and the Golgi apparatus. Cholesterol and sphingolipids localize together in lipid-ordered domains, called lipid rafts, at the plasma membrane (52–54). These domains can be endocytosed to be degraded or recycled. The increase in the concentrations of BMP species, markers of the late endosome and the lysosome, tends to indicate that there is an accumulation of late endosomes in the liver of rodents affected by DIPL and SD. This accumulation might be due to the impairment of fusion with the lysosome because of the storage of material in this organelle (drug, phospholipids, or gangliosides). Because the endomembrane system is highly dynamic and there is a continual exchange of lipid components between the organelles (55), dysfunctional fusion processes would cause a “traffic jam” in the cell. Moreover, if the flux through the endomembrane system is impaired, this could affect both cholesterol and the diverse sphingolipid species present in lipid-ordered domains, similarly to what occurs in NPC disease (24), and might lead to the changes in Cer pool composition that were observed.
It is well known that BMPs stimulate the degradation of sphingolipids; their presence in high proportion in the inner lysosomal membrane facilitates interactions between glycosphingolipids and their degrading enzymes (15, 56, 57). Therefore, the increase in BMP concentrations in these two lysosomal lipidoses could be partly due to an increase in BMP synthesis in order to stimulate the catabolism of the accumulated lipids, as reported by Wilkening et al. (56, 58).
In addition, in liver tissue from Hexb−/− mice and DIPL rats, a striking remodeling of the Cer content was detected. Although an alteration of sphingolipid metabolism has been hypothesized (59), it is the first time, to our knowledge, that Cer concentrations have been shown to be affected in DIPL. Changes in Cer concentrations have been linked to many diseases, including neurodegenerative diseases such as Alzheimer’s disease (60–63). Cers are known to regulate many cellular processes, including the balance between cell survival and apoptosis (64–67). However, our understanding of the specific functions of individual Cer species according to their chain length remains limited. Pinto et al. (68) have investigated the effects of Cer species with different chain lengths and degrees of saturation on PC membrane models. Notably, they have shown that C16:0-Cer stimulates the ability of C24:1-Cer to form gel phase domains, and both these Cer species, found increased in the two lysosomal lipidoses studied, segregate into specific “flower-shaped” domains. Moreover, Cers are also the precursors of sphingomyelins, known to colocalize with cholesterol in lipid rafts (69), and, therefore, one can presume that the alterations in the composition of the Cer pool would affect the formation of microdomains and membrane properties in SD and DIPL animals.
CerSs catalyze the production of Cers of specific chain length from sphingosine in the salvage pathway of Cer synthesis, or produce dihydroceramides from sphinganine through the de novo synthesis of Cers (70, 71). There are six different CerSs with overlapping functions and relative specificity toward acyl-CoAs, producing Cers with specific fatty acyl chains. CerS2 catalyzes the synthesis of VLC Cers (22–26 carbons) (70, 72, 73). CerS3 shows specificity for the synthesis of long-chain Cers from 18 to 36 carbons (70, 71, 73, 74). CerS5 catalyzes the synthesis of C16:0-Cer (70, 73) and has been suggested to participate in the production of C18:0-Cer (71). CerS4 and CerS1 show a common preference for C18:0- and C20:0-Cer (70, 71). Finally, CerS6 catalyzes the synthesis of C14:0-, C16:0-, and C18:0-Cer (64, 65, 73).
The alterations in CerS gene expression measured by qPCR are consistent with the changes observed in the concentration of Cer species in liver tissue after DIPL. The stimulation of the expression of CerS2, which is responsible for the synthesis of VLC Cers, after treatment with chloroquine in liver may explain the increase in the concentrations of C22:0- and C24:1-Cer measured by LC/MS. In addition, the increase in the expression of CerS5 and CerS6 may lead to the observed increase in C16:0-Cer. The upregulation in the expression of CerS4 may be responsible for the increase in the concentration of C20:0-Cer. However, one cannot speculate about the composition of the Cer pool solely by looking at the activity or expression of a specific CerS because the deficiency in one CerS can lead to an increase in nonrelated Cer species. This has been shown by using CerS2−/− mice and in knockdown experiments in vitro (65, 75–77). In SD, however, regulation of Cer species is not caused by changes in CerS gene expression, because no difference was detected between control and SD groups.
In addition, the results of the present study have shown that DIPL and SD share common lipid alterations, such as an accumulation of BMPs and an increase in CEs and in several LPC species, as well as alterations to Cer metabolism. There are also similar characteristics between DIPL and various LSDs, including Fabry and NPC diseases, that can lead to misdiagnosis (26, 27).
Shayman and Abe (49) compared DIPL to an acquired lysosomal disorder, describing a loss of LPLA2 activity due to the incorporation of CADs in the lysosomal membrane. LPLA2 binds to anionic phospholipids, such as BMPs, that are present in lysosomal membranes, along with its substrates, phospholipids. This interaction between the enzyme and its substrates results in phospholipid degradation. According to their theory, during drug treatment, CADs are integrated to the lysosomal membrane and reduce the negative charge of the membrane, perturbing the interactions between LPLA2 and its substrates. This leads to a reduction in phospholipid degradation, resulting in DIPL (49). The authors based their conclusion mainly on results obtained from the generation of Lpla2−/− mice (50). These animals are deficient in LPLA2 and exhibit an increase in phospholipid concentrations in alveolar and peritoneal macrophages, accompanied by the presence of foamy macrophages and lamellar bodies, which are characteristic of DIPL.
Lysosomal diseases are caused by a mutation in the gene coding for a lysosomal hydrolase, its activator, a lysosomal membrane protein, or a transporter. Although all the biochemical processes and cascades explaining the large array of symptoms associated with these diseases have not been identified so far, the primary insult is always the deficiency in a lysosomal enzyme. Various metabolic pathways affected in DIPL models have been identified recently by using microarray experiments (19, 23) and confirmed in the present work to be associated with both DIPL and SD. However, the mechanism(s) responsible for DIPL are not known for certain, although some authors have identified it as a loss of LPLA2 activity (49, 50), as stated above. However, until we can provide evidence for the impairment of LPLA2 activity by CADs in vivo as the primary insult with the alteration of other metabolic pathways after DIPL as a consequence of this event, we cannot assert that DIPL is an acquired LSD. Further investigation needs to demonstrate that the storage of phospholipids in DIPL and the ultrastructural changes observed are caused by a deficiency in a single enzyme, LPLA2, to affirm that it is an acquired LSD. Until then, two main hypotheses regarding the mechanism of DIPL remain valid; the alteration of LPLA2 activity by insertion of CAD in the lysosomal membrane, leading to phospholipid accumulation and dysregulation of various metabolic pathways, or simultaneous alteration of multiple pathways by the drug.
In conclusion, the data reported indicate that DIPL and SD share common lipid alterations in liver tissue. Further assessment is required to more clearly define the mechanism of DIPL. In line with previous data (19, 20, 51), we observed increases in the concentration of BMPs and CEs in rats after DIPL; similar changes were detected in Hexb−/− mice. Moreover, in both disorders, there was a general increase in LPC concentrations and changes in Cers in favor of LC Cers. The identification of metabolic pathways revealed to be altered after DIPL demonstrates that further mechanistic investigation is needed and that DIPL should be considered to be a much more complex disorder than is currently the case.
Supplementary Material
Acknowledgments
The authors thank Steven A. Murfitt for his technical help and reviewing of the manuscript and Helene Mobbs.
Footnotes
Abbreviations:
- BMP
- bis(monoacylglycero)phosphate
- CAD
- cationic amphiphilic drug
- CE
- cholesteryl ester
- Cer
- ceramide
- DIPL
- drug-induced phospholipidosis
- LPC
- lysophosphatidylcholine
- LPLA2
- lysosomal phospholipase A2
- LSD
- lysosomal storage disorder
- NPC
- Niemann-Pick type C
- qPCR
- quantitative PCR
- SD
- Sandhoff disease
- VLC
- very long chain
This work was funded by a Medical Research Council integrative toxicology training partnership grant with financial support from GlaxoSmithKline. The work on Sandhoff mice was supported by Sparks. J.L.G.’s laboratory is supported by Wellcome Trust equipment grant 093,148/Z/10/Z and Medical Research Council grants G0801841 and UD99999906.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Nelson A. A., and Fitzhugh O. G.. 1948. Chloroquine (SN-7618) pathologic changes observed in rats which for 2 years had been fed various proportions. Arch. Pathol. (Chic). 45: 454–462. [PubMed] [Google Scholar]
- 2.Kodavanti U. P., and Mehendale H.. 1990. Cationic amphiphilic drugs and phospholipid storage disorder. Pharmacol. Rev. 42: 327–354. [PubMed] [Google Scholar]
- 3.Reasor M. J., and Kacew S.. 2001. Drug-induced phospholipidosis: are there functional consequences? Exp. Biol. Med. (Maywood) 226: 825–830. [DOI] [PubMed] [Google Scholar]
- 4.Dake M. D., Madison J. M., Montgomery C. K., Shellito J. E., Hinchcliffe W., Winkler M. L., and Bainton D. F.. 1985. Electron microscopic demonstration of lysosomal inclusion bodies in lung, liver, lymph nodes, and blood leukocytes of patients with amiodarone pulmonary toxicity. Am. J. Med. 78: 506–512. [DOI] [PubMed] [Google Scholar]
- 5.Nonoyama T., and Fukuda R.. 2008. Drug-induced phospholipidosis -pathological aspects and its prediction. J. Toxicol. Pathol. 21: 9–24. [Google Scholar]
- 6.Halliwell W. H. 1997. Cationic amphiphilic drug-induced phospholipidosis. Toxicol Pathol 25: 53–60. [DOI] [PubMed] [Google Scholar]
- 7.Lüllmann H., Lüllmann-Rauch R., and Wassermann O.. 1978. Lipidosis induced by amphiphilic cationic drugs. Biochem. Pharmacol. 27: 1103–1108. [DOI] [PubMed] [Google Scholar]
- 8.Liu N., Tengstrand E. A., Chourb L., and Hsieh F. Y.. 2014. Di-22:6-bis(monoacylglycerol)phosphate: a clinical biomarker of drug-induced phospholipidosis for drug development and safety assessment. Toxicol. Appl. Pharmacol. 279: 467–476. [DOI] [PubMed] [Google Scholar]
- 9.Joshi U. M., Kodavanti P. R., Coudert B., Dwyer T. M., and Mehendale H. M.. 1988. Types of interaction of amphiphilic drugs with phospholipid vesicles. J. Pharmacol. Exp. Ther. 246: 150–157. [PubMed] [Google Scholar]
- 10.Mortuza G. B., Neville W. A., Delaney J., Waterfield C. J., and Camilleri P.. 2003. Characterisation of a potential biomarker of phospholipidosis from amiodarone-treated rats. Biochim. Biophys. Acta. 1631: 136–146. [DOI] [PubMed] [Google Scholar]
- 11.Kasama K., Yoshida K., Takeda S., Akeda S., and Kawai K.. 1974. Bis-(monoacylglyceryl)phosphate and acyl phosphatidylglycerol isolated from human livers of lipidosis induced by 4,4′-diethylaminoethoxyhexesterol. Lipids. 9: 235–243. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi T., Stang E., Fang K. S., de Moerloose P., Parton R. G., and Gruenberg J.. 1998. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 392: 193–197. [DOI] [PubMed] [Google Scholar]
- 13.Kolter T., and Sandhoff K.. 2005. Principles of lysosomal membrane digestion: stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu. Rev. Cell Dev. Biol. 21: 81–103. [DOI] [PubMed] [Google Scholar]
- 14.Chevallier J., Chamoun Z., Jiang G., Prestwich G., Sakai N., Matile S., Parton R. G., and Gruenberg J.. 2008. Lysobisphosphatidic acid controls endosomal cholesterol levels. J. Biol. Chem. 283: 27871–27880. [DOI] [PubMed] [Google Scholar]
- 15.Hullin-Matsuda F., Luquain-Costaz C., Bouvier J., and Delton-Vandenbroucke I.. 2009. Bis(monoacylglycero)phosphate, a peculiar phospholipid to control the fate of cholesterol: implications in pathology. Prostaglandins Leukot. Essent. Fatty Acids. 81: 313–324. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi T., Beuchat M. H., Lindsay M., Frias S., Palmiter R. D., Sakuraba H., Parton R. G., and Gruenberg J.. 1999. Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat. Cell Biol. 1: 113–118. [DOI] [PubMed] [Google Scholar]
- 17.Pappu A., and Hostetler K. Y.. 1984. Effect of cationic amphiphilic drugs on the hydrolysis of acidic and neutral phospholipids by liver lysosomal phospholipase A. Biochem. Pharmacol. 33: 1639–1644. [DOI] [PubMed] [Google Scholar]
- 18.Kubo M., Gardner M. F., and Hostetler K. Y.. 1986. Binding of propranolol and gentamicin to small unilamellar phospholipid vesicles. Contribution of ionic and hydrophobic forces. Biochem. Pharmacol. 35: 3761–3765. [DOI] [PubMed] [Google Scholar]
- 19.Sawada H., Takami K., and Asahi S.. 2005. A toxicogenomic approach to drug-induced phospholipidosis: analysis of its induction mechanism and establishment of a novel in vitro screening system. Toxicol. Sci. 83: 282–292. [DOI] [PubMed] [Google Scholar]
- 20.Lowe R., Mussa H. Y., Nigsch F., Glen R. C., and Mitchell J. B.. 2012. Predicting the mechanism of phospholipidosis. J. Cheminform. 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leli U., and Hauser G.. 1987. Modifications of phospholipid metabolism induced by chlorpromazine, desmethylimipramine and propranolol in C6 glioma cells. Biochem. Pharmacol. 36: 31–37. [DOI] [PubMed] [Google Scholar]
- 22.Chen G. L., Sutrina S. L., Frayer K. L., and Chen W. W.. 1986. Effects of lysosomotropic agents on lipogenesis. Arch. Biochem. Biophys. 245: 66–75. [DOI] [PubMed] [Google Scholar]
- 23.Nioi P., Perry B. K., Wang E-J., Gu Y-Z., and Snyder R. D.. 2007. In vitro detection of drug-induced phospholipidosis using gene expression and fluorescent phospholipid based methodologies. Toxicol. Sci. 99: 162–173. [DOI] [PubMed] [Google Scholar]
- 24.Liscum L. 2000. Niemann-Pick type C mutations cause lipid traffic jam. Traffic. 1: 218–225. [DOI] [PubMed] [Google Scholar]
- 25.Matsuzawa Y., and Hostetler K. Y.. 1980. Studies on drug-induced lipidosis: subcellular localization of phospholipid and cholesterol in the liver of rats treated with chloroquine or 4,4′-bis (diethylaminoethoxy)alpha, beta-diethyldiphenylethane. J. Lipid Res. 21: 202–214. [PubMed] [Google Scholar]
- 26.Piccoli E., Nadai M., Caretta C. M., Bergonzini V., Del Vecchio C., Ha H. R., Bigler L., Dal Zoppo D., Faggin E., Pettenazzo A., et al. . 2011. Amiodarone impairs trafficking through late endosomes inducing a Niemann-Pick C-like phenotype. Biochem. Pharmacol. 82: 1234–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albay D., Adler S. G., Philipose J., Calescibetta C. C., Romansky S. G., and Cohen A. H.. 2005. Chloroquine-induced lipidosis mimicking Fabry disease. Mod. Pathol. 18: 733–738. [DOI] [PubMed] [Google Scholar]
- 28.Meikle P. J., Duplock S., Blacklock D., Whitfield P. D., Macintosh G., Hopwood J. J., and Fuller M.. 2008. Effect of lysosomal storage on bis(monoacylglycero)phosphate. Biochem. J. 411: 71–78. [DOI] [PubMed] [Google Scholar]
- 29.Jabs S., Quitsch A., Käkelä R., Koch B., Tyynelä J., Brade H., Glatzel M., Walkley S., Saftig P., Vanier M. T., et al. . 2008. Accumulation of bis(monoacylglycero)phosphate and gangliosides in mouse models of neuronal ceroid lipofuscinosis. J. Neurochem. 106: 1415–1425. [DOI] [PubMed] [Google Scholar]
- 30.Mahuran D. J. 1999. Biochemical consequences of mutations causing the GM2 gangliosidoses. Biochim. Biophys. Acta. 1455: 105–138. [DOI] [PubMed] [Google Scholar]
- 31.Neufeld E. F. 1989. Natural history and inherited disorders of a lysosomal enzyme, beta-hexosaminidase. J. Biol. Chem. 264: 10927–10930. [PubMed] [Google Scholar]
- 32.Sonderfeld S., Conzelmann E., Schwarzmann G., Burg J., Hinrichs U., and Sandhoff K.. 1985. Incorporation and metabolism of ganglioside GM2 in skin fibroblasts gangliosidosis subjects from normal and GM2. Eur. J. Biochem. 149: 247–255. [DOI] [PubMed] [Google Scholar]
- 33.Zeller C. B., and Marchase R. B.. 1992. Gangliosides as modulators of cell function. Am. J. Physiol. 262: C1341–C1355. [DOI] [PubMed] [Google Scholar]
- 34.Kolter T. 2012. Ganglioside biochemistry. ISRN Biochem. 2012: 506160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandhoff K., Harzer K., Wassle W., and Jatzkewitz H.. 1971. Enzyme alterations and lipid storage in three variants of Tay-Sachs disease. J. Neurochem. 18: 2469–2489. [DOI] [PubMed] [Google Scholar]
- 36.Kolter T., and Sandhoff K.. 2006. Sphingolipid metabolism diseases. Biochim. Biophys. Acta. 1758: 2057–2079. [DOI] [PubMed] [Google Scholar]
- 37.Saouab R., Mahi M., Abilkacem R., Boumdin H., Chaouir S., Agader O., Amil T., and Hanine A.. 2011. A case report of Sandhoff disease. Clin. Neuroradiol. 21: 83–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cachón-González M. B., Wang S. Z., Lynch A., Ziegler R., Cheng S. H., and Cox T. M.. 2006. Effective gene therapy in an authentic model of Tay-Sachs-related diseases. Proc. Natl. Acad. Sci. USA. 103: 10373–10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J. Q., Trasler J. M., Igdoura S., Michaud J., Hanal N., and Gravel R. A.. 1997. Apoptotic cell death in mouse models of GM2 gangliosidosis and observations on human Tay-Sachs and Sandhoff diseases. Hum. Mol. Genet. 6: 1879–1885. [DOI] [PubMed] [Google Scholar]
- 40.Sargeant T. J., Wang S., Bradley J., Smith N. J. C., Raha A. A., McNair R., Ziegler R. J., Cheng S. H., Cox T. M., and Cachón-González M. B.. 2011. Adeno-associated virus-mediated expression of β-hexosaminidase prevents neuronal loss in the Sandhoff mouse brain. Hum. Mol. Genet. 20: 4371–4380. [DOI] [PubMed] [Google Scholar]
- 41.Wada R., Tifft C. J., and Proia R. L.. 2000. Microglial activation precedes acute neurodegeneration in Sandhoff disease and is suppressed by bone marrow transplantation. Proc. Natl. Acad. Sci. USA. 97: 10954–10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson N., and Borlak J.. 2006. Drug-induced phospholipidosis. FEBS Lett. 580: 5533–5540. [DOI] [PubMed] [Google Scholar]
- 43.Hostetler K. Y., Reasor M., and Yazaki P. J.. 1985. Chloroquine-induced phospholipid fatty liver. J. Biol. Chem. 260: 215–219. [PubMed] [Google Scholar]
- 44.Sango K., Yamanaka S., Hoffmann A., Okuda Y., Grinberg A., Wstphal H., McDonal M. P., Crawley J. N., Sandhoff K., Suzuki K., et al. . 1995. Mouse models of Tay–Sachs and Sandhoff diseases differ in neurologic phenotype and ganglioside metabolism. Nat. Genet. 11: 170–176. [DOI] [PubMed] [Google Scholar]
- 45.Le Belle J. E., Harris N. G., Williams S. R., and Bhakoo K. K.. 2002. A comparison of cell and tissue extraction techniques using high-resolution 1 H-NMR spectroscopy. NMR Biomed. 15: 37–44. [DOI] [PubMed] [Google Scholar]
- 46.Sumner L. W., Amberg A., Barrett D., Beale M. H., Beger R., Daykin C. A., Fan T. W-M., Fiehn O., Goodacre R., Griffin J. L., et al. . 2007. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics. 3: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hutchins P. M., Moore E. E., and Murphy R. C.. 2011. Electrospray MS/MS reveals extensive and nonspecific oxidation of cholesterol esters in human peripheral vascular lesions. J. Lipid Res. 52: 2070–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butovich I. A. 2009. Cholesteryl esters as a depot for very long chain fatty acids in human meibum. J. Lipid Res. 50: 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shayman J. A., and Abe A.. 2013. Drug induced phospholipidosis: an acquired lysosomal storage disorder. Biochim. Biophys. Acta. 1831: 602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiraoka M., Abe A., Lu Y., Yang K., Han X., Gross R. W., and Shayman J. A.. 2006. Lysosomal phospholipase A2 and phospholipidosis. Mol. Cell. Biol. 26: 6139–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mesens N., Desmidt M., Verheyen G. R., Starckx S., Damsch S., De Vries R., Verhemeldonck M., Van Gompel J., Lampo A., and Lammens L.. 2012. Phospholipidosis in rats treated with amiodarone: serum biochemistry and whole genome micro-array analysis supporting the lipid traffic jam hypothesis and the subsequent rise of the biomarker BMP. Toxicol. Pathol. 40: 491–503. [DOI] [PubMed] [Google Scholar]
- 52.van Meer G., and Sprong H.. 2004. Membrane lipids and vesicular traffic. Curr. Opin. Cell Biol. 16: 373–378. [DOI] [PubMed] [Google Scholar]
- 53.Pike L. J. 2003. Lipid rafts: bringing order to chaos. J. Lipid Res. 44: 655–667. [DOI] [PubMed] [Google Scholar]
- 54.Wang T. Y., and Silvius J. R.. 2000. Different sphingolipids show differential partitioning into sphingolipid/cholesterol-rich domains in lipid bilayers. Biophys. J. 79: 1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lippincott-Schwartz J., and Phair R.. 2010. Lipids and cholesterol as regulators of traffic in the endomembrane system. Annu. Rev. Biophys. 39: 559–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkening G., Linke T., and Sandhoff K.. 1998. Degradation on vesicular membrane surfaces Enhanced glucosylceramide degradation by lysosomal anionic lipids and activators. J. Biol. Chem. 273: 30271–30278. [DOI] [PubMed] [Google Scholar]
- 57.Simons K., and Gruenberg J.. 2000. Jamming the endosomal system: lipid rafts and lysosomal storage diseases. Trends Cell Biol. 10: 459–462. [DOI] [PubMed] [Google Scholar]
- 58.Wilkening G., Linke T., Uhlhorn-Dierks G., and Sandhoff K.. 2000. Degradation of membrane-bound ganglioside GM1. Stimulation by bis(monoacylglycero)phosphate and the activator proteins SAP-B and GM2-AP. J. Biol. Chem. 275: 35814–35819. [DOI] [PubMed] [Google Scholar]
- 59.Bocchini N., Giantin M., Crivellente F., Ferraresso S., Faustinelli I., Dacasto M., and Cristofori P.. 2015. Molecular biomarkers of phospholipidosis in rat blood and heart after amiodarone treatment. J. Appl. Toxicol. 35: 90–103. [DOI] [PubMed] [Google Scholar]
- 60.Filippov V., Song M., and Zhang K.. 2012. Increased ceramide in brains with Alzheimer’s and other neurodegenerative diseases. J. Alzheimers Dis. 29: 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cutler R. G., Kelly J., Storie K., Pedersen W. A., Tammara A., Hatanpaa K., Troncoso J. C., and Mattson M. P.. 2004. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 101: 2070–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ben-David O., and Futerman A. H.. 2010. The role of the ceramide acyl chain length in neurodegeneration: involvement of ceramide synthases. Neuromolecular Med. 12: 341–350. [DOI] [PubMed] [Google Scholar]
- 63.Jana A., Hogan E. L., and Pahan K.. 2009. Ceramide and neurodegeneration: susceptibility of neurons and oligodendrocytes to cell damage and death. J. Neurol. Sci. 278: 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Senkal C. E., Ponnusamy S., Manevich Y., Meyers-Needham M., Saddoughi S. A., Mukhopadyay A., Dent P., Bielawski J., and Ogretmen B.. 2011. Alteration of ceramide synthase 6/C16-ceramide induces activating transcription factor 6-mediated endoplasmic reticulum (ER) stress and apoptosis via perturbation of cellular Ca2+ and ER/Golgi membrane network. J. Biol. Chem. 286: 42446–42458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Senkal C. E., Ponnusamy S., Bielawski J., Hannun Y. A., and Ogretmen B.. 2010. Antiapoptotic roles of ceramide-synthase-6-generated C16-ceramide via selective regulation of the ATF6/CHOP arm of ER-stress-response pathways. FASEB J. 24: 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng W., Kollmeyer J., Symolon H., Momin A., Munter E., Wang E., Kelly S., Allegood J. C., Liu Y., Peng Q., et al. . 2006. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim. Biophys. Acta. 1758: 1864–1884. [DOI] [PubMed] [Google Scholar]
- 67.Cutler R. G., and Mattson M. P.. 2001. Sphingomyelin and ceramide as regulators of development and lifespan. Mech. Ageing Dev. 122: 895–908. [DOI] [PubMed] [Google Scholar]
- 68.Pinto S. N., Silva L. C., Futerman A. H., and Prieto M.. 2011. Effect of ceramide structure on membrane biophysical properties: the role of acyl chain length and unsaturation. Biochim. Biophys. Acta. 1808: 2753–2760. [DOI] [PubMed] [Google Scholar]
- 69.Slotte J. P., and Ramstedt B.. 2007. The functional role of sphingomyelin in cell membranes. Eur. J. Lipid Sci. Technol. 109: 977–981. [Google Scholar]
- 70.Grösch S., Schiffmann S., and Geisslinger G.. 2012. Chain length-specific properties of ceramides. Prog. Lipid Res. 51: 50–62. [DOI] [PubMed] [Google Scholar]
- 71.Mullen T. D., Hannun Y. A., and Obeid L. M.. 2012. Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem. J. 441: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laviad E. L., Albee L., Pankova-Kholmyansky I., Epstein S., Park H., Merrill A. H. Jr., and Futerman A. H.. 2008. Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 283: 5677–5684. [DOI] [PubMed] [Google Scholar]
- 73.Levy M., and Futerman A. H.. 2010. Mammalian ceramide synthases. IUBMB Life. 62: 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jennemann R., Rabionet M., Gorgas K., Epstein S., Dalpke A., Rothermel U., Bayerle A., van der Hoeven F., Imgrund S., Kirsch J., et al. . 2012. Loss of ceramide synthase 3 causes lethal skin barrier disruption. Hum. Mol. Genet. 21: 586–608. [DOI] [PubMed] [Google Scholar]
- 75.Park W-J., Park J-W., Erez-Roman R., Kogot-Levin A., Bame J. R., Tirosh B., Saada A., Merrill A. H. Jr., Pewzner-Jung Y., and Futerman A. H.. 2013. Protection of a ceramide synthase 2 null mouse from drug-induced liver injury: role of gap junction dysfunction and connexin 32 mislocalization. J. Biol. Chem. 288: 30904–30916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spassieva S. D., Mullen T. D., Townsend D. M., and Obeid L. M.. 2009. Disruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. Biochem. J. 424: 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mullen T. D., Spassieva S., Jenkins R. W., Kitatani K., Bielawski J., Hannun Y. A., and Obeid L. M.. 2011. Selective knockdown of ceramide synthases reveals complex interregulation of sphingolipid metabolism. J. Lipid Res. 52: 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.