Abstract
Sex identification provides important information for ecological and evolutionary studies, as well as benefiting snake conservation management. Traditional methods such as cloacal probing or cloacal popping are counterproductive for sex identification concerning very small species, resulting in difficulties in the management of their breeding programs. In this study, the nucleotide sequences of gametologous genes (CTNNB1 and WAC genes) were used for the development of molecular sexing markers in caenophidian snakes. Two candidate markers were developed with the two primer sets, and successfully amplified by a single band on the agarose gel in male (ZZ) and two bands, differing in fragment sizes, in female (ZW) of 16 caenophidian snakes for CTNNB1 and 12 caenophidian snakes for WAC. Another candidate marker was developed with the primer set to amplify the specific sequence for CTNNB1W homolog, and the PCR products were successfully obtained in a female‐specific 250‐bp DNA bands. The three candidate PCR sexing markers provide a simple sex identification method based on the amplification of gametologous genes, and they can be used to facilitate effective caenophidian snake conservation and management programs.
Keywords: CTNNB1, gametolog, sex chromosome, snake, WAC
1. INTRODUCTION
Sex identification is important for mating systems and sexual behavior in ecological and conservation research in vertebrates. Intriguingly, vertebrates display considerable diversity in their sex determination systems, especially in squamate reptiles which have both temperature‐dependent sex determination (TSD) and genotypic sex determination (GSD) with ZZ/ZW‐type, XX/XY‐type, or multiple (X1X2Y and Z1Z2W) sex chromosomes (Ezaz, Srikulnath, & Graves, 2017). Moreover, sex identification cannot be reliably determined in many squamate reptiles, because males and females have similar morphology, and morphological sexual dimorphism appears in the short period of time before mating (Frýdlová et al., 2011; Garland, 1985; Wangkulangkul, Thirakhupt, & Voris, 2005). Snakes (Serpentes) are a species‐rich lineage of extant reptiles that exhibit phenotypically diverse radiation (Castoe, Jiang, Gu, Wang, & Pollock, 2008; Castoe et al., 2009; Secor & Diamond, 1998), with distribution broadly found in arboreal, terrestrial, and aquatic habitats. The snakes are considered as excellent model organisms for biomedical research, and snake venom is extracted for developing antivenoms to treat snakebites or for chemotherapeutical development (Blackburn, 2006; Kerkkamp et al., 2016; Ratanabanangkoon et al., 2016). Two distinct groups of snakes are classified as follows: (i) Scolecophidia including “blind” snakes, and (ii) Alethinophidia comprising Henophidia (pythons, boas, and other “primitive” snakes) and Caenophidia (advanced snakes) (Pyron, Burbrink, & Wiens, 2013; Wiens et al., 2012). Snake ecology, population, and diversity have been widely studied (Laopichienpong et al., 2016, 2017; Liu et al., 2015; Supikamolseni et al., 2015). Accurate sex identification is important for snake management and breeding. Improvements in conservation programs are necessary to identify the sex of juveniles before the development of primary and secondary sexual characteristics to reduce the risk of extinction, or when samples are obtained without handling individuals (e.g., noninvasive sampling). This is a very important feature when working with endangered species (Waits & Paetkau, 2005). Sex identification of snakes is commonly conducted by observation of sexually dimorphic characters such as size between sexes, with males usually having larger body, length of tail, and body color as basal practical procedure (Laszlo, 1975). However, males and females have similar morphologies in several snake species such as Acrochordus spp., thereby making it difficult to identify sexes in snakes (Wangkulangkul et al., 2005). This has led to the development of alternative sexing methods based on observation of the sex organ (hemipenes). Several methods have been developed to observe the sex organs, such as cloacal probing or cloacal popping (Laszlo, 1975). However, these methods induce stress with potential injury and require specific skills. There is a crucial need for the development of a rapid, safe, and accurate method for sex identification in snakes.
Molecular and chromosomal processes underlying the sex determination of vertebrates have been extensively investigated, especially in mammals and birds (Ezaz et al., 2017). The homologous gene located in the nonrecombining region of differentiated sex chromosomes is known as a “gametologous gene.” In mammals, these are the ZFX and ZFY genes and in birds they are the CHDZ and CHDW, and ATP5A1Z and ATP5A1W genes which can be applied to determine the sex of individuals (Carmichael, Fridolfsson, Halverson, & Ellegren, 2000; Ellegren & Fridolfsson, 1997; García‐Moreno & Mindell, 2000; Lawson & Hewitt, 2002). Almost all caenophidian snakes exhibit GSD with ZZ/ZW‐type sex chromosomes. Z sex chromosomes which have homology with chicken chromosomes 2 and 27 is the fourth or fifth largest metacentric chromosome in the karyotypes of most snake species (Matsubara et al., 2006, 2012; Rovatsos, Vukić, Lymberakis, & Kratochvíl, 2015; Vicoso, Emerson, Zektser, Mahajan, & Bachtrog, 2013). Recently, sequence analysis of two gametologous genes as the catenin (cadherin‐associated protein) beta 1 (CTNNB1) and the WW domain containing adaptor with coiled‐coil (WAC) has provided information on the evolutionary process of sex chromosome differentiation in snakes (Laopichienpong et al., 2017; Matsubara, Nishida, Matsuda, & Kumazawa, 2016; Matsubara et al., 2006). Size and sequence differences between the Z and W homologs of these genes were found in many caenophidian snakes, leading us to develop molecular sexing markers to identify male and female individuals. In this study, we developed novel molecular sexing markers using three primer pairs to amplify fragments from the Z and W homologs of the genes with a clear size difference of PCR products. This assay provides a rapid and reliable method to identify genetic sex across different caenophidian snake species.
2. MATERIALS AND METHODS
2.1. Specimen and DNA extraction
Twenty‐two snake species containing both male and female individuals were examined, and detailed information was presented in Table 1. The sex of each species was identified morphologically and confirmed by mating observations and sexing probes that searched for the male hemipenes. Blood samples were collected from the ventral tail vein using a 25‐gauge needle attached to a 1‐ml disposable syringe containing 10 mm ethylenediaminetetraacetic acid (EDTA). Whole genomic DNA was extracted following the standard salting‐out protocol as described previously (Supikamolseni et al., 2015) and used as templates for polymerase chain reaction (PCR). DNA quality and concentration were determined using 1% agarose gel electrophoresis and spectrophotometric analysis. Animal care and all experimental procedures were approved by the Animal Experiment Committee, Kasetsart University, Thailand (approval no. ACKU00359) and conducted according to the Regulations on Animal Experiments at Kasetsart University.
Table 1.
Molecular sexing markers of 22 snake species with both males and females
| Species | Families | Abbreviation | CTNNB1 | Accession number | WAC | Accession number | CTNNB1W specific | No. of used snakes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA band patterns | Ta | Size (bp) | DNA band patterns | Ta | Size (bp) | DNA band patterns | Ta | Size (bp) | |||||||||
| Male | Female | Male | Female | Male | Female | ||||||||||||
| Cylindrophis ruffus | Cylindrophiidae | CRU | ++ | 55 | 750 | 750 | LC076763, LC076764 | ++ | 63 | 1,200 | 1,200 | LC213910, LC213921 | − | 65 | 1 male, 1 female | ||
| Epicrates maurus | Boidae | EMA | ++ | 55 | 750 | 750 | LC213019, LC213020 | ++ | 63 | 600 | 600 | LC213911, LC213912 | − | 65 | 1 male, 1 female | ||
| Xenopeltis unicolor | Xenopeltidae | XUN | ++ | 55 | 750 | 750 | LC076761, LC076762 | ++ | 63 | 2000 | 2000 | LC213922, LC213923 | − | 65 | 2 males, 2 females | ||
| Python bivittatus | Pythonidae | PBI | ++ | 55 | 750 | 750 | LC076765, LC076766 | ++ | 63 | 2000 | 2000 | LC213924, LC213925 | − | 65 | 1 male, 1 female | ||
| Python regius | Pythonidae | PRE | ++ | 55 | 750 | 750 | LC213021, LC213022 | ++ | 63 | 2000 | 2000 | LC213926, LC213927 | − | 65 | 4 males, 2 females | ||
| Acrochordus javanicus | Acrochordidae | AJA | ++ | 55 | 1,000 | 1,000 | LC213023, LC213024 | +++ | 63 | 2000 | 3,500, 1,000 | LC213928, LC213929, LC213930 | − | 65 | 1 male, 1 female | ||
| Daboia siamensis | Viperidae | DSI | +++ | 55 | 1,100 | 1,100, 700 | LC076767, LC076768, LC076769 | +++ | 63 | 1,200 | 2,200, 1,200 | LC213931, LC213933, LC213932 | + | 63 | 250 | 1 male, 1 female | |
| Enhydris enhydris | Homalopsidae | EEN | +++ | 55 | 3,000 | 3,000, 700 | LC213904, LC213905, LC213918 | +++ | 63 | 1,000 | 1,200, 1,000 | LC213934, LC213913, LC213935 | + | 65 | 250 | 2 males, 2 females | |
| Naja kaouthia | Elapidae | NKA | +++ | 55 | 1,100 | 1,100, 700 | LC076789, LC076790, LC076791 | ++ | 63 | 1,000 | 1,000 | LC213936, LC213937 | + | 63 | 250 | 1 male, 1 female | |
| Naja siamensis | Elapidae | NSI | +++ | 55 | 1,100 | 1,100, 700 | LC076792, LC076793, LC076794 | ++ | 63 | 1,000 | 1,000 | LC213938, LC213939 | + | 63 | 250 | 2 males, 1 female | |
| Ophiophagus hannah | Elapidae | OHA | +++ | 55 | 1,200 | 1,200, 700 | LC076795, LC076796, LC076797 | ++ | 63 | 1,000 | 1,000 | LC213940, LC213941 | + | 63 | 250 | 2 males, 1 female | |
| Bungarus candidus | Elapidae | BCA | +++ | 55 | 1,100 | 1,100, 700 | LC076798, LC086064, LC086065 | ++ | 63 | 1,000 | 1,000 | LC213942, LC213943 | + | 63 | 250 | 1 male, 1 female | |
| Bungarus flaviceps | Elapidae | BFL | +++ | 55 | 1,100 | 1,100, 700 | LC213025, LC213026, LC213027 | ++ | 63 | 1,000 | 1,000 | LC213944, LC213945 | + | 63 | 250 | 1 male, 1 female | |
| Leioheterodon madagascariensis | Lamprophiidae | LMA | +++ | 55 | 1,100 | 1,100, 700 | LC213028, LC213029, LC213030 | +++ | 63 | 1,000 | 1,000, 800 | LC213946, LC213947, LC213948 | + | 65 | 250 | 1 male, 1 female | |
| Oligodon fasciolatus | Colubridae | OFA | +++ | 55 | 1,500 | 1,500, 700 | LC076772, LC076773, LC076774 | +++ | 63 | 1,000 | 1,200, 1,000 | LC213949, LC213914, LC213950 | + | 65 | 250 | 2 males, 1 female | |
| Ahaetulla prasina | Colubridae | APR | +++ | 55 | 1,500 | 1,500, 700 | LC076775, LC076776, LC076777 | +++ | 63 | 2,500 | 2,500, 800 | LC213951, LC213952, LC213915 | + | 63 | 250 | 2 males, 2 females | |
| Boiga dendrophila | Colubridae | BDE | +++ | 55 | 1,700 | 1,700, 700 | LC076778, LC076779, LC076780 | +++ | 63 | 1,000 | 1,000, 800 | LC213953, LC213954, LC213955 | + | 65 | 250 | 2 males, 2 females | |
| Gonyosoma oxycephalum | Colubridae | GOX | +++ | 55 | 1,300 | 1,500, 700 | LC076781, LC076782, LC076783 | +++ | 63 | 2,500 | 2,500, 1,300 | LC213916, LC213917, LC213956 | + | 63 | 250 | 1 male, 1 female | |
| Coelognathus flavolineatus | Colubridae | CFL | +++ | 55 | 1,500 | 2000, 700 | LC076784, LC076785, LC076786 | +++ | 63 | 1,000 | 1,000, 800 | LC213957, LC213958, LC213959 | + | 63 | 250 | 1 male, 1 female | |
| Coelognathus radiatus | Colubridae | CRA | +++ | 55 | 2,200 | 2,200, 700 | LC213906, LC213907, LC213919 | +++ | 63 | 1,000 | 1,000, 800 | LC213960, LC213961, LC213962 | + | 63 | 250 | 3 males, 5 females | |
| Ptyas mucosa | Colubridae | PMU | +++ | 55 | 1,500 | 1,500, 700 | LC213908, LC213909, LC213920 | +++ | 63 | 1,000 | 1,500, 1,000 | LC213963, LC213965, LC213964 | + | 63 | 250 | 2 males, 1 female | |
| Pantherophis guttatus | Colubridae | PGU | +++ | 55 | 1,500 | 1,500, 700 | LC213031, LC213032, LC213033 | +++ | 63 | 1,000 | 1,000, 800 | LC213966, LC213967, LC213968 | + | 63 | 250 | 6 males, 3 females | |
“−” No DNA band in both male and female.
“+” One DNA band in female only.
“++” One DNA band in both male and female.
“+++” One DNA band in male and two DNA bands in female.
2.2. Molecular sexing marker development
For WAC genes, partial DNA fragments of exons 9–10 were amplified using target‐specific primers Eq‐WAC‐int9‐F: 5′‐CTCAGCCATCTAATCAGTCCCCAA‐3′ and Eq‐WAC‐int9‐R: 5′‐GAACGCTGAAGACTTCGAGGAG‐3′ (Matsubara et al., 2016). For the CTNNB1 gene, partial DNA fragments were amplified using PCR primers (Eq‐CTNNB1‐11‐F1: 5′‐AGAGACGTCCACAATCGGATTG‐3′ and Eq‐CTNNB1‐13‐R: 5′‐CAGACGTTTCTTATAATCTTGTGG‐3′) (Laopichienpong et al., 2017; Matsubara et al., 2016). The female‐specific PCR products with different size from the DNA fragments commonly found for both males and females (WACZ and CTNNB1Z) were expected to derive from the genes WACW and CTNNB1W (File S1). In addition, a new female‐specific primer, CTNNB1W‐F: 5′‐GACAAAGAAGCAGCTGAGTCAG‐3′, was designed, based on the nucleotide difference between the CTNNB1Z and CTNNB1W sequences at exon 11 identified in our previous study (Laopichienpong et al., 2017), to amplify a female‐specific PCR product using CTNNB1W‐F and Eq‐CTNNB1‐13‐R (File S1). The BDNF (brain derived neurotrophic factor) gene was used as a positive PCR control marker using primers BDNF‐F (5′‐GACCATCCTTTTCCTGACTATGGTTATTTCATACTT‐3′) and BDNF‐R (5′‐CTATCTTCCCCTTTTAATGGTCAGTGTACAAAC‐3′) (Leaché & McGuire, 2006). PCR amplification was performed using 20 μl of 1× ExTaq buffer containing 1.5 mm MgCl2, 0.2 mm dNTPs, 5.0 μm the primers, and 0.25 U of TaKaRa Ex Taq (TaKaRa Bio, Otsu, Japan), and 25 ng of genomic DNA. PCR conditions was as follows: an initial denaturation at 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 7 min. The nucleotide sequences of the DNA fragments derived from the PCR reaction of the primer sets Eq‐CTNNB1‐11‐F1 and Eq‐CTNNB1‐13‐R or Eq‐WAC‐int9‐F and Eq‐WAC‐int9‐R were only determined directly using the DNA sequencing services of First Base Laboratories Sdn Bhd (Seri Kembangan, Selangor, Malaysia). Nucleotide sequences of the Z and W homologs from each species were searched for homologies with the nucleotide sequences in the National Center for Biotechnology Information (NCBI) database to identify DNA fragments of the target gene, using the BLASTx and BLASTn programs. All the sequences were then deposited in the DNA Data Bank of Japan (DDBJ) (Table 1).
3. RESULTS
By agarose gel electrophoresis with the primer Eq‐CTNNB1‐11‐F1 and Eq‐CTNNB1‐13‐R, one same‐sized DNA fragment was observed in both males and females, and one additional DNA fragment was detected from females in each snake species: Daboia siamensis, Enhydris enhydris, Naja kaouthia, N. siamensis, Ophiophagus hannah, Bungarus candidus, B. flaviceps, Leioheterodon madagascariensis, Oligodon fasciolatus, Ahaetulla prasina, Boiga dendrophila, Coelognathus radiatus, Ptyas mucosa, and Pantherophis guttatus. This indicates that the same‐sized DNA bands and the female‐specific DNA bands were derived from the Z and W homologs, respectively. No female‐specific DNA fragments were observed in Cylindrophis ruffus, Epicrates maurus, Xenopeltis unicolor, Python bivittatus, P. regius, Acrochordus javanicus, Gonyosoma oxycephalum, and C. flavolineatus (Table 1; Figures 1, 2; Files S2–S4). However, there was a wide variation in the DNA fragment sizes between species, ranging from approximately 750 bp in C. ruffus, E. maurus, X. unicolor, P. bivittatus, and P. regius to 3,000 bp in E. enhydris for CTNNB1Z (Table 1). Individual size variation within the same species was also found in the Z‐derived fragments in two species: G. oxycephalum and C. flavolineatus (Table 1). In addition to the results from PCR with Eq‐CTNNB1‐11‐F1 and Eq‐CTNNB1‐13‐R primers, the primer pairs Eq‐WAC‐int9‐F and Eq‐WAC‐int9‐R yielded one similar‐sized DNA fragment for both males and females, and one additional DNA fragment for females in each snake species: D. siamensis, E. enhydris, L. madagascariensis, O. fasciolatus, A. prasina, B. dendrophila, G. oxycephalum, C. flavolineatus, C. radiatus, P. mucosa, and P. guttatus, but no female‐specific DNA fragments were observed in C. ruffus, E. maurus, X. unicolor, P. bivittatus, P. regius, A. javanicus, N. kaouthia, N. siamensis, O. hannah, B. candidus, and B. flaviceps. A wide size variation between species was found for both the Z‐ and W‐derived fragments. The Z‐derived fragments ranged from approximately 600 bp in E. maurus to 3,500 bp in A. javanicus, and the W‐derived fragments range from 800 bp in L. madagascariensis, A. prasina, B. dendrophila, C. flavolineatus, C. radiatus, and P. guttatus to 2,200 bp in D. siamensis (Table 1). Individual size variation within the same species was also found in Z‐derived fragments in one species: A. javanicus.
Figure 1.
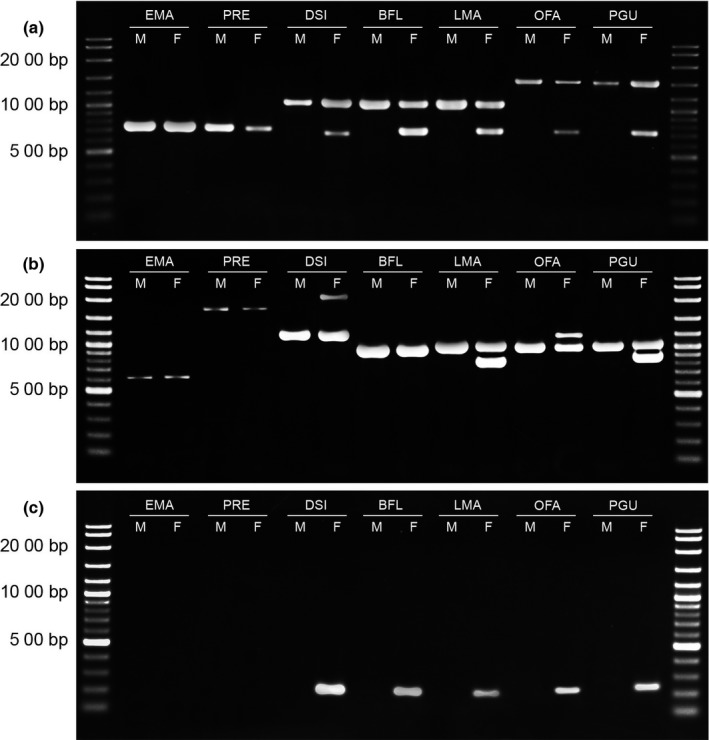
Agarose gel electrophoresis of PCR products in males and females of seven snake species using Eq‐CTNNB1‐11‐F1 and Eq‐CTNNB1‐13‐R (a), Eq‐WAC‐int9‐F and Eq‐WAC‐int9‐R (b), and CTNNB1W‐F and Eq‐CTNNB1‐13‐R (c). Molecular size of DNA is indicated in the left lane using VC 100‐bp Plus DNA ladder (Vivantis Technologies Sdn Bhd, Selangor Darul Ehsan, Malaysia). EMA, Epicrates maurus; PRE, Python regius; DSI, Daboia siamensis; BFL, Bungarus flaviceps; LMA, Leioheterodon madagascariensis; OFA, Oligodon fasciolatus; PGU, Pantherophis guttatus. M, male F, female
Figure 2.
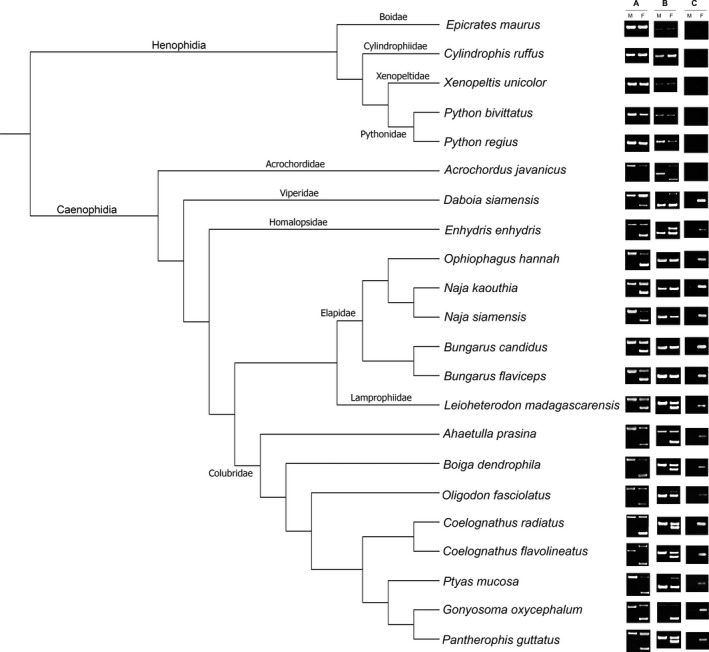
Phylogenetic relationships among sampled snake species illustrating the sex‐specific amplification of CTNNB1 and WAC genes using the primers: Eq‐CTNNB1‐11‐F1 and Eq‐CTNNB1‐13‐R; Eq‐WAC‐int9‐F and Eq‐WAC‐int9‐R; and CTNNB1W‐F and Eq‐CTNNB1‐13‐R. Phylogeny was partially derived from Vidal, Rage, Couloux, and Hedges (2009). Agarose gel electrophoresis of PCR products in males and females of twenty‐two snake species using three sexing markers: Eq‐CTNNB1‐11‐F1 and Eq‐CTNNB1‐13‐R (column A), Eq‐WAC‐int9‐F and Eq‐WAC‐int9‐R (column B), and CTNNB1W‐F and Eq‐CTNNB1‐13‐R (column C) are indicated at the right edge of the tree. M, male; F, female
Additionally, in PCR with the CTNNB1W‐specific primer set, amplification of DNA fragments was found only in females as a single 250‐bp DNA band (Figures 1 and 2). To distinguish the absences of PCR products in males from failures of PCR reactions, the BDNF primers were used separately as an internal control under the same PCR condition. The BDNF primers produced larger fragments than the CTNNB1W‐specific products. Samples were identified as female if two products of 250‐bp (CTNNB1W) and 750‐bp sized bands (BDNF) were observed and male if only the control band was amplified (Figure 2; File S5). If the control locus failed to amplify, then sex was not assigned.
4. DISCUSSION
Sex identification is very important, not only for the basic understanding of the ecology and behavior of endangered or protected animals but also for establishing management and conservation plans (Dubey et al., 2011; Webb, Brook, & Shine, 2002). To contribute to breeding programs, the sexes of the snakes must be precisely identified while avoiding snake injury and stress. Traditional methods of cloacal probing or cloacal popping are counterproductive in very small species, whereas PCR has the advantage of identifying sexually heteromorphic PCR products as genomic DNA from small quantities of tissue such as blood in this study, or applied for skin remnants, slough, or eggshell membranes left behind after hatching as shown in birds (Martín‐Gálvez et al., 2011). Based on the hypothesis of sex chromosome differentiation, the cessation of recombination between sex chromosomes leads to the accumulation of gene mutations, resulting in the occurrence of sexually antagonistic alleles or the functional inactivation of genes, followed by the partial deletion of the sex chromosomes (Ezaz et al., 2017). In snake lineages, Boidae and Pythonidae have morphologically homomorphic Z and W chromosomes (Matsubara et al., 2006; Olmo & Signorino, 2005). By contrast, the W chromosomes are highly degenerated in caenophidian snakes that predominantly showed heteromorphic ZW sex chromosomes (Beçak & Beçak, 1969; Beçak, Beçak, & Nazareth, 1964; Matsubara et al., 2006, 2016; O'Meally et al., 2010; Oguiura, Collares, Furtado, Ferrarezzi, & Suzuki, 2009; Ray‐Chaudhuri, Singh, & Sharma, 1971; Singh, 1972; Vicoso et al., 2013). Considering the practically routine technique, cytogenetic approaches to examine the sex chromosomes require large sample volume with long cell culture and chromosome preparation time. Therefore, they are not practical in terms of wildlife ecological studies and conservation programs. A molecular sexing method utilizing sex‐specific sequences is, thus, more advantageous than cytogenetic analyses to identify sex chromosome systems. We developed novel PCR‐based molecular sexing methods with three primer sets to identify individual caenophidian snake sex, based on the nucleotide sequence differences of two gametologous genes. In molecular sexing with two of the three sets, Eq‐CTNNB1‐11‐F1 and Eq‐CTNNB1‐13‐R, and Eq‐WAC‐int9‐F and Eq‐WAC‐int9‐R, the males with the homogametic sex chromosome (ZZ) were characterized by a single DNA fragment band from the two Z homologs, and the females with the heterogametic sex chromosome (ZW) were identified by two bands differing in fragment sizes from the one Z and one W homologs. The two primer sets Eq‐CTNNB1‐11‐F1—Eq‐CTNNB1‐13‐R and Eq‐WAC‐int9‐F—Eq‐WAC‐int9‐R were available for molecular sexing in 16 and 12 caenophidian snakes, respectively. These two markers exhibited co‐dominant DNA pattern type. This suggests that the Z and W forms of the CTNNB1 or WAC genes were differentiated by the cessation of recombination in these caenophidian lineages, which led to insertions and deletions of nucleotide sequences on the Z and W chromosomes.
No female‐specific PCR products of W homolog were found in A. javanicus for the CTNNB1 gene with the primer set Eq‐CTNNB1‐11‐F1 and Eq‐CTNNB1‐13‐R and N. kaouthia, N. siamensis, O. hannah, B. candidus, and B. flaviceps for the WAC gene with the primer set Eq‐WAC‐int9‐F and Eq‐WAC‐int9‐R, even though these species belong to Caenophidia. This suggests that the absence of the size differences with female‐specific PCR products was caused by the accumulation of mutations at existing primer sites, leading to failure of PCR reaction at the W homologs. Moreover, large insertions in the amplified target fragments could result in failure of PCR amplification for the extended W sequences. Alternatively, CTNNB1 and WAC may have been lost independently from the W chromosomes in the species. This result agreed with the chromosome and genomic map of snakes which showed the absence of these two genes in some snake lineages (Matsubara et al., 2006; Vicoso et al., 2013). The third explanation is that there is none or little difference in sizes of PCR products between the Z and W homologs, as a consequence of a very small region of the nonrecombining portion of the W chromosome which did not have time to diverge significantly from the Z chromosome in these genes. Snake W sex chromosomes are known to degenerate at varying rates and undergo substantial reorganization over short periods of evolutionary time (Oguiura et al., 2009). The observation of DNA bands of the two primer sets revealed high variability in length between species or individuals, which probably determined individual sex difficulty (Table 1; Figure 1). Such individual variation in the Z form of the gametologous gene within the same species was also reported by nucleotide analysis for the CHD1Z gene of birds (Casey, Jones, Sandercock, & Wisely, 2009; Friesen, Congdon, Walsh, & Birt, 1997; Trimbos et al., 2013) and for the CTNNB1 gene of snakes (Laopichienpong et al., 2017).
To avoid the misidentification of sexes through size variation, we designed the additional primer at the exon of the CTNNB1W gene which yielded a female‐specific 250‐bp PCR products in 16 caenophidian snakes and indicated dominant DNA pattern type. Internal PCR control with the BDNF gene was also used to avoid amplification failure which might result in a misidentification of a female as a male. The present results of the three sexing markers could also be applied to identify the sex of individuals in new snake species as a simple procedure for males and females. We highlight that all the three sexing markers, with an additional BDNF control marker, could be simultaneously used to identify sex with higher diagnostic accuracy.
In the two henophidian snakes P. bivittatus and B. constrictor, CTNNB1 and WAC genes were located on both Z and W chromosomes, and no sex‐specific sequence was detected for the two gametologous genes (Matsubara et al., 2006, 2016; Vicoso et al., 2013). In this study, five henophidian snakes (C. ruffus, E. maurus, X. unicolor, P. bivittatus, and P. regius) showed the absence of female‐specific PCR products in molecular sexing with our three primer sets. This implies that the CTNNB1 and WAC genes on the Z and W chromosomes have not been differentiated in henophidian snakes (Laopichienpong et al., 2017; Matsubara et al., 2006, 2016; Vicoso et al., 2013). However, more intensive sampling over a wider range of individuals or species within the same family is required to confirm the absence of CTNNB1 and WAC genes on the W chromosome. Simple PCR assay could not identify the sex of species with homomorphic sex chromosomes or highly similar Z and W sex chromosomes. Advanced molecular techniques are required to investigate cryptic sex‐specific sequences in these species.
Currently, only two PCR methods have been described to determine the sex in caenophidian snakes. Firstly, a simple PCR assay is used to amplify at the gametologous genes CTNNB1 and WAC on the Z and W chromosomes that showed size difference of DNA bands, or the simple presence/absence sex‐linked marker or a specific allele based on nucleotide substitution. The alternative approach uses a single set of primers with a quantitative real‐time PCR (qPCR) technique to amplify Z‐specific genes without homologs in the W chromosome (Rovatsos & Kratochvíl, 2017; Rovatsos et al., 2015). Males (ZZ) have twice as many copies of genes linked to the Z‐specific part of sex chromosomes than females (ZW), while genes in autosomal or pseudoautosomal regions should have equal copy numbers in both sexes. Although qPCR techniques offer a high‐throughput environment for routine genotyping, the cost of the qPCR reaction is high compared to the simple PCR assay and needs extra equipment. By contrast, simple PCR assay provides a rapid, reliable, sensitive, cost‐effective, and highly accurate result for sex identification. However, both the two PCR methods cannot be used for species with poorly differentiated sex chromosomes.
This study reveals the potential and usefulness of gametologous sequences which develop new candidate sexing markers in caenophidian snakes. Our work opens a new tool kit for further application with the genomic DNA of small and degraded tissues such as slough as noninvasive sampling. To facilitate application of this tool, we provide a “know‐how” guide to apply Loop‐mediated lsothermal Amplification (LAMP) or aptamer to conservation program where species identification or sex determination is needed in real time and in situ. Future work in this direction will enormously facilitate the work of wildlife managers, researchers, and exotic snake breeders with important conservationist, economic, and commercial benefits.
CONFLICT OF INTEREST
None declared.
Supporting information
ACKNOWLEDGMENTS
This study was financially supported by grants from Professor Motivation (PM) (No. PM4/2558), the Bilateral Research Cooperation (BRC) (No. BRC7/2557), Special Track Staff (STS) (No. STS1/2558), and Science Research Fund (ScRF) (No. ScRF‐S19‐2558) from the Faculty of Science, Kasetsart University, Center for Advanced Studies in Tropical Natural Resources, National Research University‐Kasetsart University (CASTNAR, NRU‐KU, Thailand) (No. 6/2558 and 9/2559), the Fellowship of Capacity Building for Kasetsart University on Internationalization, Thailand Research Fund (TRF) (Nos. MSD60I0035 and PHD60I0014), and Science Achievement Scholarship of Thailand (SAST) (No. 5917400296) from the Office of the Higher Education Commission, Thailand. We would like to thank Taksa Vasaruchapong (Snake Farm, Queen Saovabha Memorial Institute, Thai Red Cross Society, Thailand), Kanchit Deesomsak (Thailand Exotic Pet keepers Association), Khampee Pattanatanang (Kasetsart University and Real Zoo, Thailand), and the Conservation Research and Education Division, Zoological Park Organization, Dusit, Bangkok, Thailand for advising on sample preparation. We are also grateful to Siwapech Sillapaprayoon (Kasetsart University, Thailand) for helpful discussions.
Laopichienpong N, Tawichasri P, Chanhome L, et al. A novel method of caenophidian snake sex identification using molecular markers based on two gametologous genes. Ecol Evol. 2017;7:4661–4669. https://doi.org/10.1002/ece3.3057
Funding information
Professor Motivation (PM), Faculty of Science, Kasetsart University, Grant/Award Number: PM4/2558; the Bilateral Research Cooperation (BRC), Faculty of Science, Kasetsart University, Grant/Award Number: No. BRC7/2557; Special Track Staff (STS), Faculty of Science, Kasetsart University, Grant/Award Number: STS1/2558; Science Research Fund (ScRF), Faculty of Science, Kasetsart University, Grant/Award Number: ScRF‐S19‐2558; National Research University‐Kasetsart University (CASTNAR, NRU‐KU, Thailand), Grant/Award Number: 6/2558 and 9/2559; the Fellowship of Capacity Building for Kasetsart University on Internationalization; Thailand Research Fund (TRF), Grant/Award Number: MSD60I0035 and PHD60I0014; Science Achievement Scholarship of Thailand (SAST), Grant/Award Number: 5917400296.
REFERENCES
- Beçak, W. , & Beçak, M. L. (1969). Cytotaxonomy and chromosomal evolution in Serpentes. Cytogenetics and Genome Research, 8, 247–262. [DOI] [PubMed] [Google Scholar]
- Beçak, W. , Beçak, M. L. , & Nazareth, H. R. (1964). Close karyological kinship between the reptilian suborder serpentes and the class aves. Chromosoma, 15, 606–617. [DOI] [PubMed] [Google Scholar]
- Blackburn, D. G. (2006). Squamate reptiles as model organisms for the evolution of viviparity. Herpetological Monographs, 20, 131–146. [Google Scholar]
- Carmichael, A. N. , Fridolfsson, A. K. , Halverson, J. , & Ellegren, H. (2000). Male‐biased mutation rates revealed from Z and W chromosome‐linked ATP synthase alpha‐subunit (ATP5A1) sequences in birds. Journal of Molecular Evolution, 50, 443–447. [DOI] [PubMed] [Google Scholar]
- Casey, A. E. , Jones, K. L. , Sandercock, B. K. , & Wisely, S. M. (2009). Heteroduplex molecules cause sexing errors in a standard molecular protocol for avian sexing. Molecular Ecology Resources, 9, 61–65. [DOI] [PubMed] [Google Scholar]
- Castoe, T. A. , Daza, J. M. , Smith, E. N. , Sasa, M. M. , Kuch, U. , Campbell, J. A. , … Parkinson, C. L. (2009). Comparative phylogeography of pitvipers suggests a consensus of ancient Middle American highland biogeography. Journal of Biogeography, 36, 88–103. [Google Scholar]
- Castoe, T. A. , Jiang, Z. J. , Gu, W. , Wang, Z. O. , & Pollock, D. D. (2008). Adaptive evolution and functional redesign of core metabolic proteins in snakes. PLoS ONE, 3, e2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey, S. , Sumner, J. , Pike, D. A. , Keogh, J. S. , Webb, J. K. , & Shine, R. (2011). Genetic connectivity among populations of an endangered snake species from Southeastern Australia (Hoplocephalus bungaroides, Elapidae). Ecology and Evolution, 1, 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren, H. , & Fridolfsson, A. K. (1997). Male‐driven evolution of DNA sequences in birds. Nature Genetics, 17, 182–184. [DOI] [PubMed] [Google Scholar]
- Ezaz, T. , Srikulnath, K. , & Graves, J. A. M. (2017). Origin of amniote sex chromosomes: An ancestral super sex chromosome, or common requirements. Journal of Heredity, 108, 94–105. [DOI] [PubMed] [Google Scholar]
- Friesen, V. L. , Congdon, B. C. , Walsh, H. E. , & Birt, T. P. (1997). Intron variation in marbled murrelets detected using analyses of single‐stranded conformational polymorphisms. Molecular Ecology, 6, 1047–1058. [DOI] [PubMed] [Google Scholar]
- Frýdlová, P. , Velenský, P. , Šimková, O. , Cikánová, V. , Hnízdo, J. , Rehák, I. , & Frynta, D. (2011). Is body shape of mangrove‐dwelling monitor lizards (Varanus indicus; Varanidae) sexually dimorphic? Amphibia‐Reptilia, 32, 27–37. [Google Scholar]
- García‐Moreno, J. , & Mindell, D. P. (2000). Rooting a phylogeny with homologous genes on opposite sex chromosomes (gametologs): A case study using avian CHD . Molecular Biology and Evolution, 17, 1826–1832. [DOI] [PubMed] [Google Scholar]
- Garland, T. (1985). Ontogenetic and individual variation in size, shape and speed in the Australian agamid lizard Amphibolurus nuchalis . Journal of Zoology, 207, 425–439. [Google Scholar]
- Kerkkamp, H. M. , Kini, R. M. , Pospelov, A. S. , Vonk, F. J. , Henkel, C. V. , & Richardson, M. K. (2016). Snake genome sequencing: Results and future prospects. Toxins (Basel), 8, E360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laopichienpong, N. , Muangmai, N. , Chanhome, L. , Suntrarachun, S. , Twilprawat, P. , Peyachoknagul, S. , & Srikulnath, K. (2017). Evolutionary dynamics of the gametologous CTNNB1 gene on the Z and W chromosomes of snakes. Journal of Heredity, 108, 142–151. [DOI] [PubMed] [Google Scholar]
- Laopichienpong, N. , Muangmai, N. , Supikamolseni, A. , Twilprawat, P. , Chanhome, L. , Suntrarachun, S. , … Srikulnath, K. (2016). Assessment of snake DNA barcodes based on mitochondrial COI and Cytb genes revealed multiple putative cryptic species in Thailand. Gene, 594, 238–247. [DOI] [PubMed] [Google Scholar]
- Laszlo, J. (1975). Probing as a practical method of sex recognition in snakes. International Zoo Yearbook, 15, 178–179. [Google Scholar]
- Lawson, L. J. , & Hewitt, G. M. (2002). Comparison of substitution rates in ZFX and ZFY introns of sheep and goat related species supports the hypothesis of male‐biased mutation rates. Journal of Molecular Evolution, 54, 54–61. [DOI] [PubMed] [Google Scholar]
- Leaché, A. D. , & McGuire, J. A. (2006). Phylogenetic relationships of horned lizards (Phrynosoma) based on nuclear and mitochondrial data: Evidence for a misleading mitochondrial gene tree. Molecular Phylogenetics and Evolution, 39, 628–644. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Zhu, F. , Zhong, G. , Wang, Y. , Fang, M. , Xiao, R. , … Guo, P. (2015). COI‐based barcoding of Chinese vipers (Reptilia: Squamata: Viperidae). Amphibia‐Reptilia, 36, 361–372. [Google Scholar]
- Martín‐Gálvez, D. , Peralta‐Sánchez, J. M. , Dawson, D. A. , Martín‐Platero, A. M. , Martínez‐Bueno, M. , Burke, T. , & Soler, J. J. (2011). DNA sampling from eggshell swabbing is widely applicable in wild bird populations as demonstrated in 23 species. Molecular Ecology Resources, 11, 481–493. [DOI] [PubMed] [Google Scholar]
- Matsubara, K. , Kuraku, S. , Tarui, H. , Nishimura, O. , Nishida, C. , Agata, K. , … Matsuda, Y. (2012). Intra‐genomic GC heterogeneity in sauropsids: Evolutionary insights from cDNA mapping and GC3 profiling in snake. BMC Genomics, 13, 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara, K. , Nishida, C. , Matsuda, Y. , & Kumazawa, Y. (2016). Sex chromosome evolution in snakes inferred from divergence patterns of two gametologous genes and chromosome distribution of sex chromosome‐linked repetitive sequences. Zoological Letters, 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara, K. , Tarui, H. , Toriba, M. , Yamada, K. , Nishida‐Umehara, C. , Agata, K. , & Matsuda, Y. (2006). Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step‐wise differentiation of snake sex chromosomes. Proceedings of the National Academy of Sciences, 103, 18190–18195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguiura, N. , Collares, M. A. , Furtado, M. F. , Ferrarezzi, H. , & Suzuki, H. (2009). Intraspecific variation of the crotamine and crotasin genes in Crotalus durissus rattlesnakes. Gene, 446, 35–40. [DOI] [PubMed] [Google Scholar]
- Olmo, E. , & Signorino, G. (2005). Chromorep: A reptile chromosomes database; [cited 2016 Feb 16]. Available from: http://chromorep.univpm.it
- O'Meally, D. , Patel, H. R. , Stiglec, R. , Sarre, S. D. , Georges, A. , Graves, J. A. M. , & Ezaz, T. (2010). Non‐homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosome Research, 18, 787–800. [DOI] [PubMed] [Google Scholar]
- Pyron, R. A. , Burbrink, F. T. , & Wiens, J. J. (2013). A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evolutionary Biology, 13, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratanabanangkoon, K. , Tan, K. Y. , Eursakun, S. , Tan, C. H. , Simsiriwong, P. , Pamornsakda, T. , … Tan, N. H. (2016). A simple and novel strategy for the production of a pan‐specific antiserum against elapid snakes of Asia. PLOS Neglected Tropical Diseases, 10, e0004565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray‐Chaudhuri, S. P. , Singh, L. , & Sharma, T. (1971). Evolution of sex‐chromosomes and formation of W‐chromatin in snakes. Chromosoma, 33, 239–251. [DOI] [PubMed] [Google Scholar]
- Rovatsos, M. , & Kratochvíl, L. (2017). Molecular sexing applicable in 4000 species of lizards and snakes? From dream to real possibility. Methods in Ecology and Evolution. https://doi.org/10.1111/2041-210X.12714 [Google Scholar]
- Rovatsos, M. , Vukić, J. , Lymberakis, P. , & Kratochvíl, L. (2015). Evolutionary stability of sex chromosomes in snakes. Proceedings Biological sciences, 282, 20151992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secor, S. M. , & Diamond, J. (1998). A vertebrate model of extreme physiological regulation. Nature, 395, 659–662. [DOI] [PubMed] [Google Scholar]
- Singh, L. (1972). Evolution of karyotypes in snakes. Chromosoma, 38, 185–236. [DOI] [PubMed] [Google Scholar]
- Supikamolseni, A. , Ngaoburanawit, N. , Sumontha, M. , Chanhome, L. , Suntrarachun, S. , Peyachoknagul, S. , & Srikulnath, K. (2015). Molecular barcoding of venomous snakes and species‐specific multiplex PCR assay to identify snake groups for which antivenom is available in Thailand. Genetics and Molecular Research, 14, 13981–13997. [DOI] [PubMed] [Google Scholar]
- Trimbos, K. B. , Kentie, R. , van der Velde, M. , Hooijmeijer, J. C. E. W. , Poley, C. , Musters, C. J. M. , … Piersma, T. (2013). Intronic variation at the CHD1‐Z gene in Black‐tailed Godwits Limosa limosa limosa: Correlations with fitness components revisited. IBIS International Journal Avian Science, 155, 508–517. [Google Scholar]
- Vicoso, B. , Emerson, J. J. , Zektser, Y. , Mahajan, S. , & Bachtrog, D. (2013). Comparative sex chromosome genomics in snakes: Differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biology, 11, e1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal, N. , Rage, J. C. , Couloux, A. , & Hedges, S. B. (2009). Snake (Serpentes) In Hedges S. B., & Kumar S. (Eds.), The time tree of life (pp. 390–397). New York: Oxford University Press. [Google Scholar]
- Waits, L. P. , & Paetkau, D. (2005). Noninvasive genetic sampling tools for wildlife biologists: A review of applications and recommendations for accurate data collection. The Journal of Wildlife Management, 69, 1419–1433. [Google Scholar]
- Wangkulangkul, S. , Thirakhupt, K. , & Voris, H. K. (2005). Sexual size dimorphism and reproductive cycle of the little file snake Acrochordus granulatus in Phangnga Bay, Thailand. Science Asia, 31, 257–263. [Google Scholar]
- Webb, J. K. , Brook, B. W. , & Shine, R. (2002). Collectors endanger Australia's most threatened snake, the broad‐headed snake Hoplocephalus bungaroides, vol. 36 (pp. 170–181). Cambridge: Cambridge University Press. [Google Scholar]
- Wiens, J. J. , Hutter, C. R. , Mulcahy, D. G. , Noonan, B. P. , Townsend, T. M. , Sites, J. W. Jr. , & Reeder, T. W. (2012). Resolving the phylogeny of lizards and snakes (Squamata) with extensive sampling of genes and species. Biology Letters, 8, 1043–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


