Upon DNA damage, histone modifications are reshaped to accommodate DNA damage signaling and repair. Gong et al. report that the histone demethylase KDM5A promotes loading of the chromatin remodeling complex ZMYND8–NuRD to double-strand DNA breaks through H3K4me3 demethylation, thereby allowing repair of the lesion.
Abstract
Upon DNA damage, histone modifications are dynamically reshaped to accommodate DNA damage signaling and repair within chromatin. In this study, we report the identification of the histone demethylase KDM5A as a key regulator of the bromodomain protein ZMYND8 and NuRD (nucleosome remodeling and histone deacetylation) complex in the DNA damage response. We observe KDM5A-dependent H3K4me3 demethylation within chromatin near DNA double-strand break (DSB) sites. Mechanistically, demethylation of H3K4me3 is required for ZMYND8–NuRD binding to chromatin and recruitment to DNA damage. Functionally, KDM5A deficiency results in impaired transcriptional silencing and repair of DSBs by homologous recombination. Thus, this study identifies a crucial function for KDM5A in demethylating H3K4 to allow ZMYND8–NuRD to operate within damaged chromatin to repair DSBs.
Introduction
Maintaining genome and epigenome integrity is vital for preserving DNA-templated processes, including transcription, replication, and DNA repair (Branzei and Foiani, 2010; Kim and Jinks-Robertson, 2012; Helmrich et al., 2013). DNA damage can disrupt these processes, resulting in genome instability, which can contribute to diseases, including cancer (Jackson and Bartek, 2009; Negrini et al., 2010). DNA damage occurs within chromatin, thereby necessitating the engagement of chromatin-based processes to detect, signal, and repair the damage. The multifaceted pathways that handle DNA damage are collectively referred to as the DNA damage response (DDR; Jackson and Bartek, 2009; Ciccia and Elledge, 2010). The DDR relies on posttranslational modifications of histone and nonhistone proteins, which act to dynamically regulate DNA repair activities within chromatin (Lukas et al., 2011; Miller and Jackson, 2012; Gong and Miller, 2013; Jackson and Durocher, 2013; Gong et al., 2016).
Histone posttranslational modifications, including phosphorylation, acetylation, and methylation, modulate chromatin structure and also provide recognition signals that are bound by DDR factors to promote their localization and function at DNA damage sites (Polo and Jackson, 2011). Histone acetyltransferases, histone deacetylases (HDACs), and the acetylation reader proteins that bind acetylated marks have been identified as key participants of the DDR (Ogiwara et al., 2011; Gong and Miller, 2013; Kakarougkas et al., 2014; Gong et al., 2015, 2016). For example, acetylated H4K16 by TIP60 promotes homologous recombination (HR) repair, whereas deacetylated H4K16 by HDAC1/HDAC2 facilitates nonhomologous end joining (Miller et al., 2010; Tang et al., 2013). The bromodomain protein ZMYND8 is also an important DDR factor that recruits the NuRD (nucleosome remodeling and histone deacetylation) complex to damage chromatin, where it represses transcription and promotes DNA repair (Gong et al., 2015; Savitsky et al., 2016; Spruijt et al., 2016). Methylated histones also participate in the DDR (Klose and Zhang, 2007; van Attikum and Gasser, 2009; Lukas et al., 2011; Greer and Shi, 2012; Miller and Jackson, 2012). For example, H4K20 methylation, along with H2A ubiquitylation, creates dual docking sites for the DDR factor 53BP1 (Botuyan et al., 2006; Fradet-Turcotte et al., 2013), and SETD2-mediated H3K36me3 promotes DNA double-strand break (DSB) repair within actively transcribed chromatin (Aymard et al., 2014; Pfister et al., 2014). Similarly to acetylation signaling, numerous “writers” and “erasers” of histone methylation are recruited to DNA damage (Mosammaparast et al., 2013; Young et al., 2013; Khoury-Haddad et al., 2014; Li et al., 2014; Dimitrova et al., 2015), suggesting the coordinated efforts of chromatin-modifying enzymes that remodel the chromatin landscape to allow DNA damage signaling and repair. Thus, histone modifications represent crucial epigenetic components of chromatin-based DNA damage signaling and repair pathways.
Here, we identify the histone demethylase KDM5A as a new DDR factor that regulates ZMYND8–NuRD DDR activities. We demonstrate that KDM5A is recruited to DNA damage, where it demethylates H3K4me3. We find that demethylation of H3K4me3 by KDM5A promotes interactions between damaged chromatin and ZMYND8–NuRD, which facilitates repression of active transcription and the repair of DSBs by HR. Thus, our work highlights the temporal reshaping of histone modifications by chromatin modifiers that occurs at DNA damage sites, which functions to coordinate DDR processes within chromatin to ensure the maintenance of genome and epigenome integrity.
Results
Identification of ZMYND8–NuRD DDR regulators
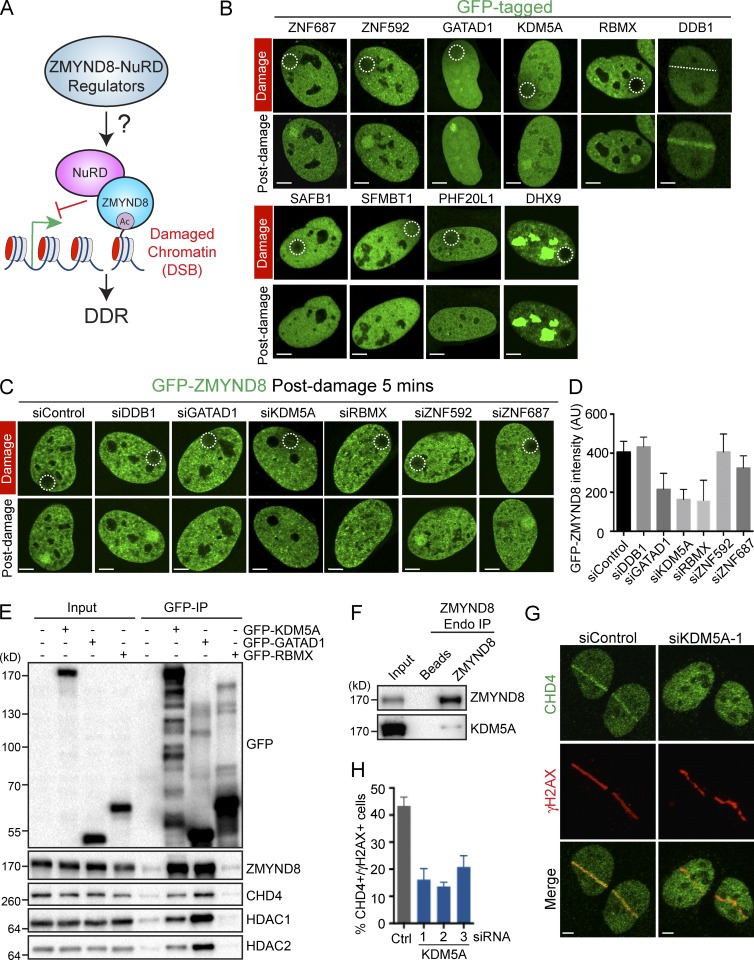
We recently determined that ZMYND8–NuRD is recruited to DNA damage within actively transcribing chromatin to repress transcription and promote the DDR (Fig. 1 A; Gong et al., 2015). To further delineate regulatory steps of this pathway, we surveyed DNA damage localization of putative ZMYND8-interacting factors identified previously by mass spectrometry (Gong et al., 2015). DNA damage recruitment analysis of ten GFP-tagged candidate proteins identified six that exhibited robust recruitment to DNA damage under these experimental conditions, which confirmed previous results of DNA damage recruitment for several of these factors (Fig. 1 B; Wang et al., 2006; Seiler et al., 2011; Adamson et al., 2012; Spruijt et al., 2016). We also observed exclusion of DHX9 from DNA damage sites, a phenotype previously ascribed to some DDR factors (Fig. 1 B; Beli et al., 2012; Gong et al., 2015). This screen identified six candidate ZMYND8-interacting proteins (i.e., ZNF687, ZNF592, RBMX, DDB1, GATAD1, and KDM5A) that were responsive to DNA damage.
Figure 1.
Identification of ZMYND8–NuRD DDR regulators, including the demethylase KDM5A. (A) Scheme for interrogating the ZMYND8–NuRD DDR pathway. (B) Potential ZMYND8-interacting factors were GFP tagged and screened for damage-dependent relocalization by laser damage with live-cell confocal microscopy. (C) Proteins identified in B were analyzed for their involvement in promoting GFP-ZMYND8 damage localization. U2OS cells stably expressing GFP-ZMYND8 were treated with the indicated siRNAs and analyzed as in B. (D) Quantification of C. Fluorescence intensity (in arbitrary units [AU]) of GFP-ZMYND8 in damaged versus undamaged regions at 5 min after damage is plotted. One representative experiment out of two is shown (error bars indicate SEM, n > 10 cells per condition). (E) GFP-tagged proteins were expressed in HEK293T cells and purified with GFP-TRAP beads. Samples were analyzed by WB with the indicated antibodies. (F) Analysis of endogenous ZMYND8 and KDM5A interactions. ZMYND8 was immunoprecipitated from 293T cells, and samples were analyzed as in E. (G) Recruitment of CHD4 to laser damage in siControl and siKDM5A U2OS cells. (H) Quantification of G. Data represent analysis of >50 cells; n = 2. Error bars represent SEM. Regions damaged are indicated by dotted white lines/circles. Bars, 5 µm.
Recruitment to DNA damage is often observed for DDR factors. Given their potential to interact with ZMYND8, we queried the involvement of these factors in regulating ZMYND8 localization to DNA damage. For this approach, we individually depleted each damage-recruited factor identified in Fig. 1 B by siRNA (depletion validated in Fig. S1, A and B) and analyzed the recruitment of ZMYND8 to DNA damage in such settings. These experiments revealed that ZMYND8 recruitment to damage sites was dependent on KDM5A, GATAD1, and RBMX (Fig. 1 C; quantified in Fig. 1 D). This analysis identified several new factors required for ZMYND8 recruitment to DNA damage sites.
KDM5A promotes ZMYND8–NuRD localization to damaged chromatin
Our analysis revealed that ZMYND8 recruitment to DNA damage was dependent on KDM5A, GATAD1, and RBMX. KDM5A and GATAD1 reside in the same complex (Vermeulen et al., 2010), suggesting these two factors may function in concert to regulate ZMYND8. To detect whether these genetic interactions between these three factors and ZMYND8 were likely to be direct, we tested if these factors interacted with ZMYND8. In coimmunoprecipitation assays, GFP-KDM5A or GFP-GATAD1, but not GFP-RBMX, readily pulled down endogenous ZMYND8 and the NuRD components HDAC1, HDAC2, and CHD4 (Fig. 1 E). Reciprocal immunoprecipitations in several cancer cell lines confirmed these results (Fig. S1, C–E). As KDM5A belongs to the KDM5 demethylase family (KDM5A, KDM5B, KDM5C, and KDM5D; Christensen et al., 2007; Klose et al., 2007; Blair et al., 2011), we analyzed interactions between ZMYND8 and other KDM5 demethylases. We observed by immunoprecipitation and Western blot analysis that KDM5A was the principal KDM5 demethylase that interacted with ZMYND8 in 293T and U2OS cells (Fig. S1, C–E). We also observed an interaction between endogenously expressed ZMYND8 and KDM5A, which was moderately enhanced upon DNA damage (Fig. 1 F; and Fig. S1, F and G). The requirement of KDM5A for ZMYND8 damage recruitment was confirmed using several independent siRNAs to rule out off-target effects (Fig. S1 H; and Fig. S2, A and B). Previously, we and others showed that ZMYND8 promoted NuRD recruitment to DNA damage (Gong et al., 2015; Spruijt et al., 2016). Consistent with KDM5A regulation of ZMYND8 and NuRD, KDM5A deficiency by several independent siRNAs impaired damage localization of the NuRD core component CHD4 (Fig. 1, G and H). Collectively, these data demonstrate that KDM5A–GATAD1 interacts with ZMYND8–NuRD and promotes their localization to DNA damage.
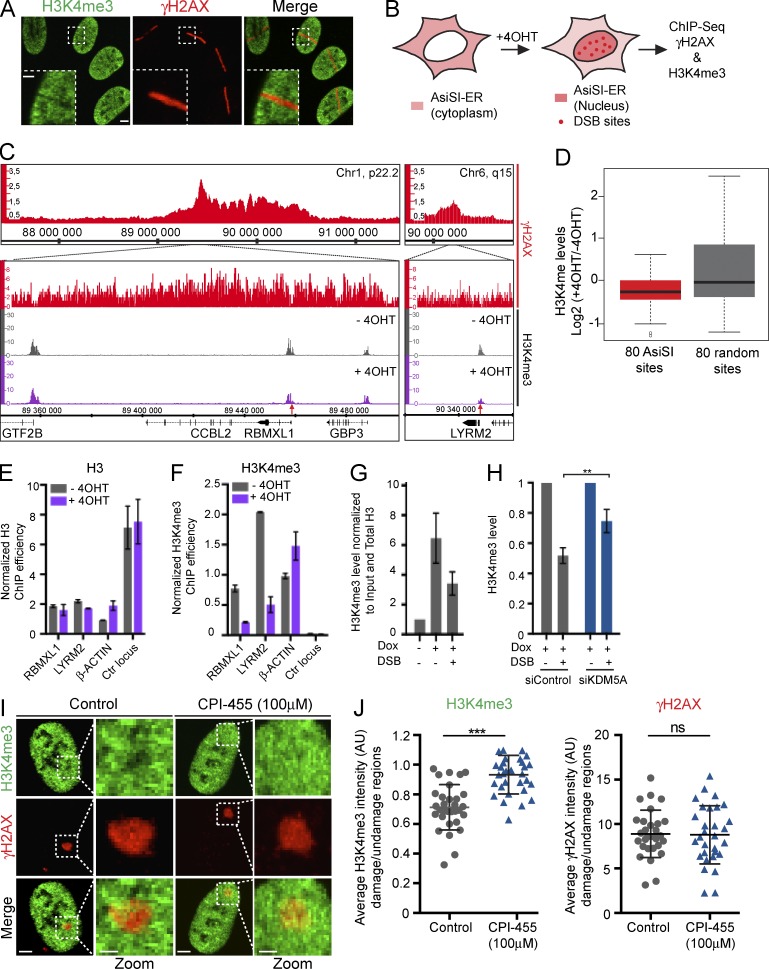
KDM5A demethylates H3K4me3 at DSBs
KDM5A is a lysine demethylase that primarily demethylates H3K4me2/me3 and is involved in transcriptional regulation (Christensen et al., 2007; Klose et al., 2007). Having established that KDM5A is recruited to DNA damage, we next determined if H3K4me3 demethylation occurred at DSBs. Laser damage exhibited reductions in H3K4me3 signal in regions positive for the DNA damage marker γ-H2AX, which is consistent with the reduction in H3K4me3 that has been observed at ionizing radiation (IR)–induced DNA damage foci by immunofluorescence (Seiler et al., 2011; Fig. 2 A). To analyze H3K4me3 levels at higher resolution directly at DSBs, we used the DIvA (DSB inducible via AsiSI) system (Iacovoni et al., 2010; Aymard et al., 2014; Caron et al., 2015). This system consists of a U2OS cell line that expresses a regulatable AsiSI restriction enzyme fused with the ligand-binding domain of ER (Fig. 2 B). Upon 4-hydroxytamoxifen (4-OHT) treatment, AsiSI-ER becomes nuclear, where it induces multiple DSBs throughout the genome that can be analyzed by genome-wide methodologies, including chromatin immunoprecipitation (ChIP) followed by massively parallel DNA sequencing (ChIP-seq; Iacovoni et al., 2010; Aymard et al., 2014; Caron et al., 2015). Upon 4-OHT treatment, we observed reduced H3K4me3 signals adjacent to AsiSI-induced DSB sites by ChIP-seq (Fig. 2 C). Analysis of the averaged H3K4me3 signal ±500 bp around 80 DSBs revealed a highly significant reduction of this mark after DSB induction compared with nondamage conditions or at 80 non–AsiSI-associated locations, randomly chosen for comparison (Fig. 2 D). We further validated our ChIP-seq data by ChIP-qPCR, which confirmed that H3K4me3 levels were decreased at gene promoters (RBMXL1 and LYRM2) adjacent to AsiSI-DSB sites, but not at the β-actin gene promoter or a random genomic region (Ctr locus) that is not adjacent to an AsiSI site (Fig. 2, E and F). Analysis of H3K4me3 on genes previously identified as up-regulated by IR (Rashi-Elkeles et al., 2014) detected increased levels of H3K4me3 on these genes, which further validated our ChIP-seq analysis of H3K4me3 and was consistent with H3K4me3 known association with active transcription (Fig. S2 C; Barski et al., 2007). Collectively, our results from two independent DNA damage methods demonstrated that H3K4me3 levels are decreased upon DNA damage specifically at DSBs.
Figure 2.
H3K4me3 is reduced at DNA damage sites by the demethylase KDM5A. (A) Analysis of H3K4me3 at laser damage sites marked by γ-H2AX. U2OS cells were laser damaged and analyzed by immunofluorescence with the indicated antibodies. (B) Scheme for analysis with the DIvA (AsiSI-ER-U2OS) site-specific DNA damage system. (C) ChIP-seq analysis in DlvA cells ±4-OHT (4-h treatment) with γ-H2AX and H3K4me3 antibodies. Profiles for γ-H2AX (+4-OHT) and H3K4me3 with and without 4-OHT at two AsiSI-induced DSB sites are shown. Red arrow indicates an AsiSI site. (D) Box plot of H3K4me3 changes occurring after 4-OHT treatment at the best 80 AsiSI-induced DSB and 80 random genomic loci. H3K4me3 levels ±500 bp surrounding DSB or non-DSB random loci were averaged and plotted as log2 ratio (+4OHT/−4OHT). H3K4me3 reduction at AsiSI DSB after4-OHT treatment was highly significant (P = 7.8503e-06) compared with non–AsiSI-associated loci (P = 0.038; nonparametric Mann–Whitney test). Center line, median; box limits, first and third quartiles; whiskers, maximum and minimum without outliers; points, outliers (see Materials and methods). (E and F) Validation of ChIP-qPCR analysis of H3 and H3K4me3 ±4-OHT at two genes adjacent to AsiSI-induced DSBs. β-Actin and Ctr loci, distant from any AsiSI site, act as controls. ChIP efficiency is represented as percentage of input, normalized against TAF12. Mean and SEM (n = 4 technical replicates) from a representative experiment out of two are shown. (G) ChIP analysis of H3K4me3 in the transgene promoter region within the U2OS FokI-induced DSB reporter system (Tang et al., 2013). Transcription and DSBs were induced by doxycycline (Dox) and Shield-1 + 4-OHT, respectively. ChIP-qPCR analysis upon transcription and/or DSB induction. H3K4me3 levels represent ChIP signal normalized to input and H3 ChIP. (H) H3K4me3 levels were analyzed by ChIP as in G in siControl and siKDM5A cells. Error bars represent SEM; n = 4. (I) H3K4me3 levels by immunofluorescence at laser damage sites in untreated and 100 µM CPI-455–treated U2OS cells. (J) Quantification of I (n > 30 cells; error bars represent SD). P-value was determined by Student’s t test (**, P < 0.01; ***, P < 0.001; ns, not significant). AU, arbitrary units. Bars: 10 µm; (magnified images) 2 µm.
Given our observations that H3K4me3 levels are reduced upon DNA damage and KDM5A is recruited to such sites, we sought to test whether the reduction of H3K4me3 observed within damaged chromatin was dependent on KDM5A. To this end, we used the FokI-inducible DSB reporter system, which allows regulated transcription of a reporter gene and induction of DSBs upstream of this gene (Tang et al., 2013). As expected, induction of transcription increased H3K4me3 levels in the proximity of the reporter gene, as measured by ChIP-qPCR (i.e., −Dox vs. +Dox; Fig. 2 G). Consistent with our observation from laser- and AsiSI-induced DNA damage, we detected a reduction of H3K4me3 levels at FokI-induced DSBs compared with undamaged conditions (Fig. 2 G). Importantly, the reduction of H3K4me3 levels observed at DSBs was impaired in KDM5A-depleted cells (Fig. 2 H). Furthermore, treatment of cells with a specific KDM5A inhibitor, CPI-455 (Vinogradova et al., 2016), also reduced the loss of H3K4me3 within damage sites (Fig. 2, I and J). Collectively, these results establish that H3K4me3 levels are reduced at DSBs and that KDM5A participates in the removal of this mark at these DNA lesions.
H3K4me3 inhibits ZMYND8–NuRD binding
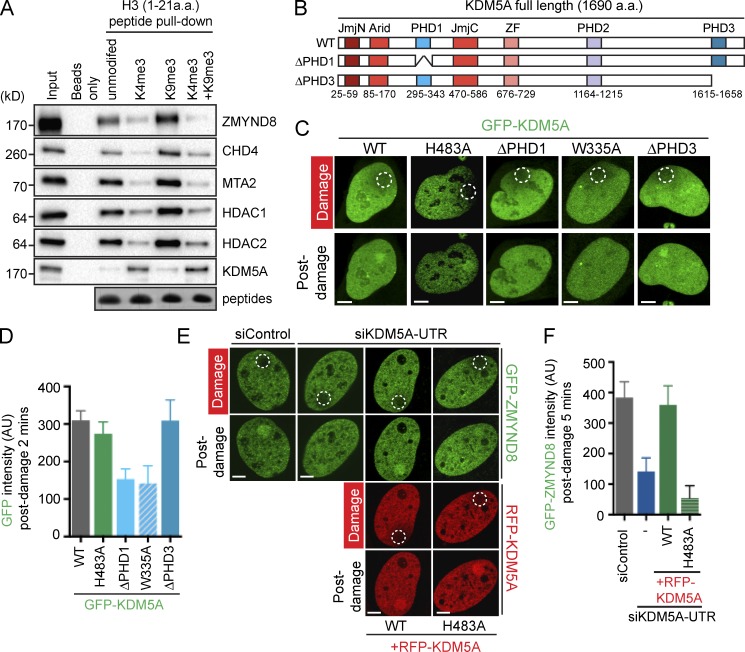
We previously determined that the ZMYND8–NuRD complex is recruited to DNA damage within actively transcribed chromatin (Gong et al., 2015). However, H3K4me3 has been shown to strongly inhibit the NuRD complex from binding histone H3 (Nishioka et al., 2002; Zegerman et al., 2002; Eberl et al., 2013). We reproduced these results using peptide pull-down assays of both unmodified and modified H3 peptides, finding in addition that H3K4me3 reduced the interaction between H3 and ZMYND8 (Figs. 3 A and S2 D). The inhibitory effects of H3 methylation appeared to be primarily through trimethylation, as di- and monomethylated H3K4 minimally reduced ZMYND8-NuRD interactions with H3 compared with H3K4me3 (Fig. S2 D). Conversely, we readily detected interactions between KDM5A and H3K4me3, consistent with previous studies showing that KDM5A binds its substrate (Fig. 3 A; Christensen et al., 2007; Klose et al., 2007). In addition to binding unmodified H3, the NuRD complex and ZMYND8, unlike KDM5A, can bind H3K9me3 (Fig. 3 A; Mansfield et al., 2011; Eberl et al., 2013; Savitsky et al., 2016), a repressive mark that occurs in heterochromatin as well as at DSBs (Ayrapetov et al., 2014). We observed similar binding of ZMYND8 and NuRD (i.e., CHD4) to unmodified and modified H3 peptides in WT extracts, as well as in ZMYND8 knockout (KO), CHD4 KO, and siRNA-depleted extracts (Fig. S2, E–G). These results are consistent with ZMYND8 and NuRD having binding capabilities toward unmodified and methylated H3. Interestingly, doubly modified trimethylated H3 peptides at K4 and K9 did not bind ZMYND8 but were still able to bind KDM5A (Fig. 3 A). These data also revealed that the inhibitory effects of H3K4me3 on ZMYND8 and NuRD binding to H3 occurred even in the presence of H3K9me3 (Zegerman et al., 2002). These biochemical studies provide a potential explanation for why demethylation of H3K4me3 is required for ZMYND8–NuRD binding, even in the event that H3K9me3 is present. Collectively, these data suggest that the demethylation of H3K4me3 by KDM5A must ensue rapidly at DSBs to allow ZMYND8–NuRD to bind and function within damaged chromatin.
Figure 3.
KDM5A regulates ZMYND8–NuRD damage localization. (A) Chromatin binding of factors in HEK293T cell lysates using H3 N-terminal peptides either unmodified or methylated at K4 and/or K9. Purified samples were analyzed by Western blotting with the indicated antibodies. Coomassie blue staining confirms equal peptide concentrations. (B) KDM5A domain organization. Full-length wild type (WT), ΔPHD1, and ΔPHD3 KDM5A mutants are indicated. (C) Recruitment of WT and mutant GFP-KDM5A derivatives to laser-induced DNA damage. Experiments performed as in Fig. 1 C. (D) Quantification of C from one representative experiment out of two as in Fig. 1 D (error bars represent SEM, n > 10 cells). (E) Complementation of defective GFP-ZMYND8 damage recruitment in KDM5A-depleted cells. U2OS cells stably expressing GFP-ZMYND8 were treated with siControl or siKDM5A-UTR after ectopic expression of RFP-KDM5A WT or enzyme-dead (H483A) mutant. Live imaging assays were performed as in Fig. 1 C. (F) GFP-ZMYND8 quantification of E from one representative experiment out of two as in Fig. 1 D. Data collection was performed in GFP or GFP+RFP–positive cells (error bars represent SEM, n > 10 cells). AU, arbitrary units. Bars, 5 µm.
Recruitment of KDM5A requires its PHD1 domain and PARP
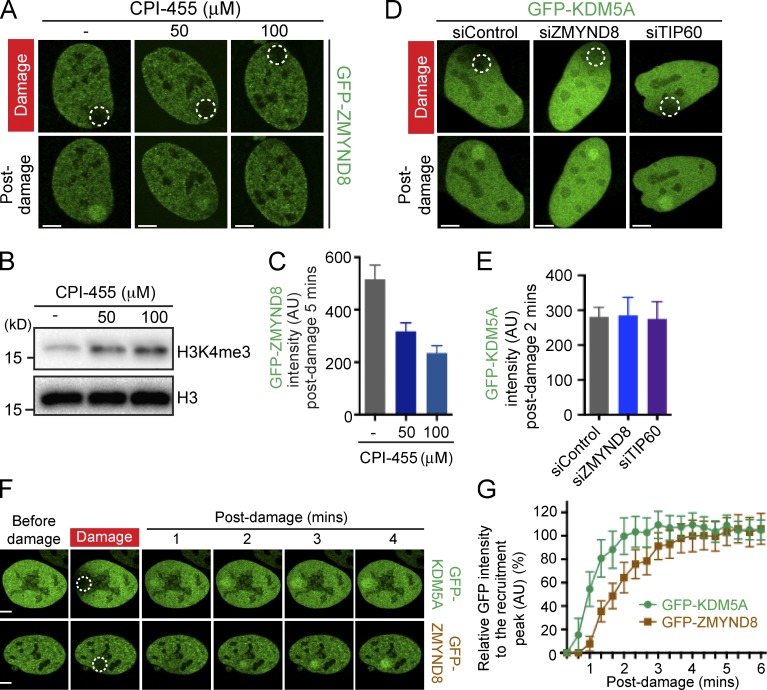
The plant homeodomain (PHD) fingers in KDM5 demethylases are binding modules that regulate its chromatin occupancy and enzymatic activity (Klein et al., 2014; Torres et al., 2015). KDM5A contains several domains, including three PHD domains, the first being reported as a binding module for unmodified H3 and the third functioning in recognition and demethylation of H3K4me3 (Fig. 3 B; Torres et al., 2015). Key residues, including H483 within the JmjC domain, are required for enzymatic activity of KDM5A (Christensen et al., 2007; Klose et al., 2007). To analyze the DDR functions of these chromatin-interaction domains and the enzymatic activity with KDM5A, we created deletion constructs of each PHD domain and the enzymatic inactivation mutation within the JmjC domain (i.e., H483A). Expression analysis revealed that deletion of PHD2 destabilized the protein (Fig. S3 A), which prohibited further analysis of this mutant. We observed that damage recruitment of KDM5A did not require PHD3 or its enzymatic activity, but required PHD1 as deletion or an inactivating point mutation in PHD1 (W335A) significantly reduced the association of KDM5A with damaged chromatin (Fig. 3 C; quantified in Fig. 3 D). We next sought to test the involvement of these domains and activity of KDM5A for promoting downstream signaling events, including supporting the recruitment of ZMYND8 to DNA damage sites. For this analysis, we characterized and validated a siRNA targeting the 3′ UTR of KDM5A, which resulted in reduced endogenous protein levels and defective ZMYND8 recruitment to DNA damage sites in siKDM5A-UTR–treated cells (Fig. 3, E and F; and Fig. S3 B). These findings allowed us to perform complementation assays using this approach. Ectopic expression of siRNA-resistant KDM5A rescued the ZMYND8 damage-recruitment defect in cells deficient for endogenous KDM5A, whereas expression of PHD1, PHD3, or enzymatic-dead H483A mutants of KDM5A did not (Fig. 3, E and F; and Fig. S2, C and D). Consistent with these results, treatment with the KDM5A inhibitor, CPI-455, although increasing global H3K4me3 levels, also reduced the association of ZMYND8 with DNA damage sites (Fig. 4, A–C). Our results reveal the first PHD domain of KDM5A as a critical domain involved in promoting interactions with damaged chromatin and show that once recruited to DNA damage, both PHD1 and PHD3, along with its enzymatic activity, are required for KDM5A to promote ZMYND8 localization to DNA damage.
Figure 4.
KDM5A acts upstream of ZMYND8 and requires its catalytic activity to promote ZMYND8 damage recruitment. (A) Damage recruitment of GFP-ZMYND8 in U2OS cells ±CPI-455 treatment. Experiments performed as in Fig. 1 C. (B) Western blot analysis of H3K4me3 levels in CPI-455–treated cells (24 h). (C) Quantification of A as in Fig. 1 D. (D) GFP-KDM5A damage recruitment in siRNA-treated cells as in A. (E) Quantification of D as in C. (F) DNA damage recruitment analysis of GFP-KDM5A and GFP-ZMYND8 in U2OS cells. Experiments performed as in Fig. 3 (C and E). (G) Quantification of F. Mean fluorescence intensity of GFP-KDM5A and GFP-ZMYND8 between damaged and undamaged areas is plotted as percentage of maximum fluorescence intensity. Quantification graphs (C, E, and G) in this figure were shown as one representative experiment out of two (error bars represent SEM; n > 10 cells per condition). Regions of laser damage are indicated by dotted white circles. AU, arbitrary units. Bars, 5 µm.
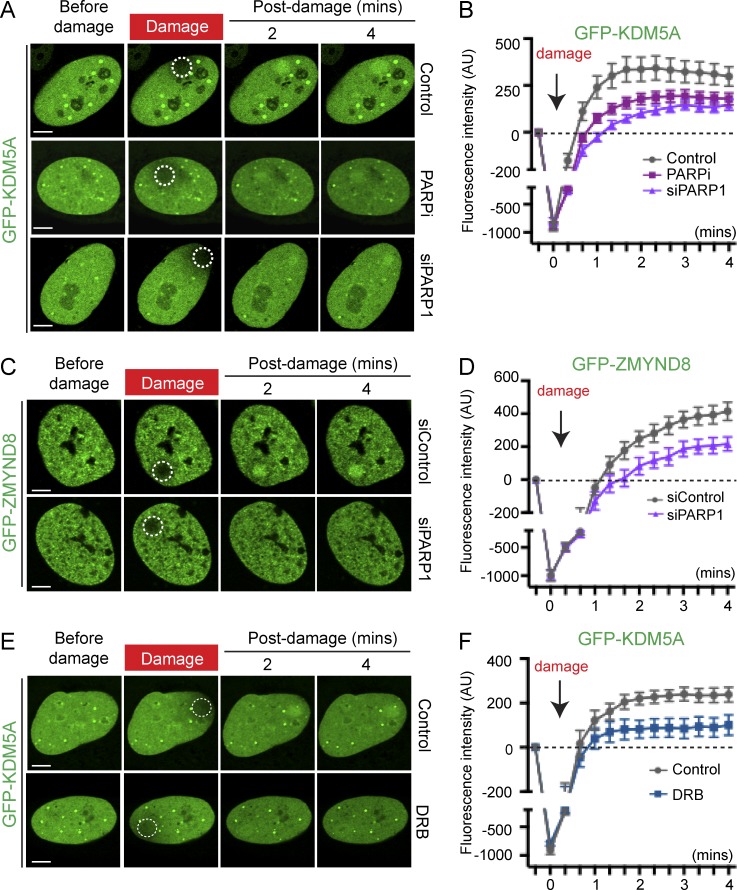
Our previous work reported that the NuRD complex required both ZMYND8 and histone acetyltransferase TIP60 for damage recruitment (Gong et al., 2015). In addition, poly-ADP ribosylation by PARP has also been shown to be an important signaling event for the damage localization of both NuRD and ZMYND8 (Chou et al., 2010; Polo et al., 2010; Spruijt et al., 2016). Our identification of KDM5A as a regulator of ZMYND8–NuRD dynamics at damaged chromatin prompted us to investigate the potential involvement of these pathways in controlling KDM5A localization. Using a combination of siRNAs and/or CRISPR/Cas9-mediated gene KOs, we tested the involvement of TIP60, ZMYND8, NuRD, and PARP for promoting damage recruitment of GFP-KDM5A. ZMYND8 siRNA depletion or KO did not affect KDM5A recruitment to DNA damage (Fig. 4, D and E; and Fig. S4, A–C). Similarly, depletion of TIP60 by siRNAs or KO of the NuRD component CHD4 did not alter KDM5A localization to DNA damage (Fig. 4, D and E; and Fig. S4, D–F). However, live-cell confocal microscopy of damage recruitment dynamics revealed that GFP-KDM5A was recruited more rapidly than ZMYND8 to laser damage (Fig. 4, F and G). Similar to ZMYND8–NuRD, PARP1 depletion by siRNAs or inhibition by a small molecule inhibitor (Olaparib) reduced significantly the recruitment of KDM5A (Fig. 5, A and B), as well as ZMYND8, to DNA damage (Fig. 5, C and D). Thus, in addition to PARP, our findings highlight a role for the demethylase KDM5A as a key regulator of the ZMYND8–NuRD DDR node and suggest that KDM5A acts upstream of ZMYND8–NuRD in this pathway.
Figure 5.
KDM5A is reliant on PARP and active transcription for damage recruitment. (A and B) GFP-KDM5A damage recruitment was analyzed in untreated, PARP inhibitor (Opaparib) and siPARP1 treated U2OS cells with quantification in B. Experiment were performed as in (Fig. S2, A and B). (C and D) Damage recruitment of GFP-ZMYND8 was analyzed in siControl and siPARP1-treated cells. Experiments were performed and quantified (D) as in A and B in stably expressing GFP-ZMYND8 U2OS cells. (E and F) U2OS cells expressing GFP-KDM5A with control or 5,6-dichlorobenzimidazole riboside (DRB) treatment were damaged, imaged and quantified as in A and B. Quantification graphs (B, D, and F) in this figure were shown as one representative experiment out of two (error bars represent SEM; n > 10 cells per condition). AU, arbitrary units. Bars, 5 µm.
KDM5A mediates damage-induced transcriptional repression and HR repair
Having delineated the molecular events that orchestrate KDM5A-dependent recruitment of ZMYND8–NuRD to DNA damage, we next sought to address the DDR functions of KDM5A. Upon DNA damage, ZMYND8 recruits the NuRD complex to damage within active chromatin, which represses transcription and promotes DNA DSB repair by homologous recombination (Gong et al., 2015). Interestingly, inhibition of transcription abolishes the recruitment of ZMYND8–NuRD (Gong et al., 2015). Similarly, inhibition of transcription impaired the accumulation of KDM5A at damage sites (Fig. 5, E and F). These data corroborate the important relationship between transcription and the DDR functions of KDM5A and ZMYND8–NuRD.
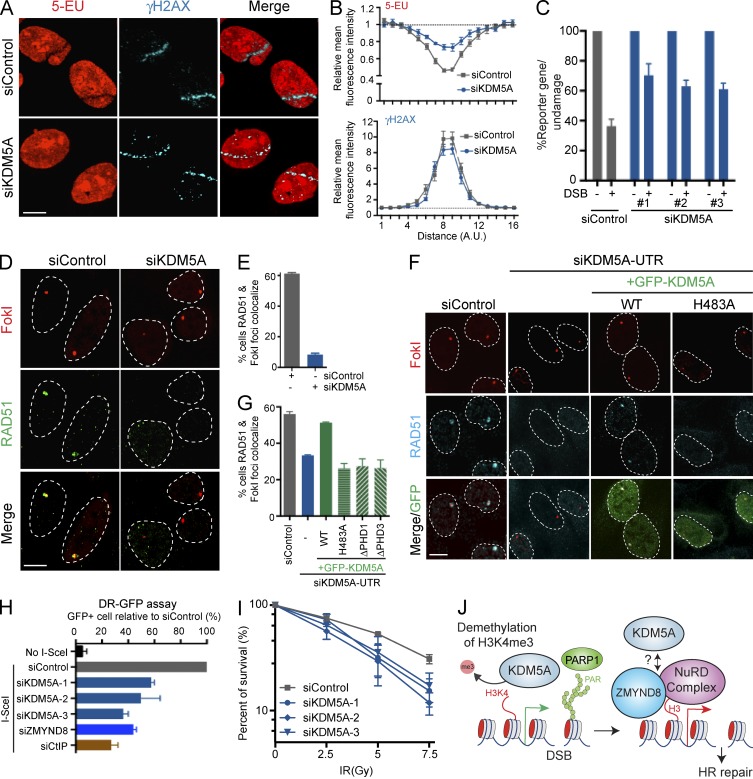
To further understand the connection between transcription and KDM5A in the DDR, we determined whether KDM5A is involved in repressing transcription after DNA damage. We previously developed an assay involving fluorescent labeling of 5-ethynyl uridine to monitor nascent transcription at laser-induced DNA damage to show that ZMYND8 and CHD4 promoted damage-induced transcriptional silencing (Gong et al., 2015). Consistent with the role of KDM5A as a ZMYND8–CHD4 mediator, depletion of KDM5A reduced the transcriptional silencing that occurs after DNA damage by laser- and FokI-induced DSBs (Fig. 6, A–C; Tang et al., 2013). We also observed reduced loading of the HR factor RAD51 onto FokI-mediated DSBs in KDM5A-deficient cells (Fig. 6, D and E) or in response to KDM5A inhibition using the small-molecule inhibitor CPI-455 (Fig. S5, A–C), which is in line with previous studies reporting a role for ZMYND8–NuRD in HR (Gong et al., 2015; Spruijt et al., 2016). These results were not caused by reduced protein levels of the HR factor RAD51 or ZMYND8–NuRD, as KDM5A deficiency did not reduce their protein levels (Fig. S5 D). We were also able to rescue RAD51 loading defects in KDM5A-deficient cells by ectopic expression of WT KDM5A, but not PHD mutant or an enzymatic-dead KDM5A (Fig. 6, F and G), which is in accord with these derivatives affecting ZMYND8 damage accumulation (Fig. 3 E and F; Fig. S3, C and D). To measure the frequency of HR repair more directly, we used the DR-GFP HR reporter system that monitors HR-dependent repair of I-SceI–induced DSBs (Pierce et al., 1999). KDM5A-depleted cells, as assayed with multiple individual siRNAs to control for off-target effects, displayed reduced HR levels (Fig. 6 H). For comparison, the decreased levels of HR between KDM5A- and ZMYND8-deficient cells were similar, albeit not as severe as in CtIP-depleted cells, a known facilitator of HR (Fig. 6 H; Sartori et al., 2007). KDM5A-depleted cells did not display appreciable differences in cell cycle distribution, ruling out potential cell cycle effects on HR (Fig. S5 E). Consistent with a role for KDM5A in DSB repair, we found that KDM5A depletion by three independent siRNAs reduced survival to IR compared with control cells (Fig. 6 I). We also observed an epistatic relationship between KDM5A and ZMYND8-NuRD as codepletion of KDM5A in ZMYND8 or CHD4 KO cells resulted in similar IR sensitivities as the cell lines deficient for only one factor (Fig. S5 F). Altogether, this study has identified the demethylase KDM5A as a DDR factor involved in the ZMYND8–NuRD DDR axis. These results suggest retooling of damaged chromatin by KDM5A creates a suitable environment for ZMYND8–NuRD binding and function, which acts to repress transcription and promote HR repair of DSBs (Fig. 6 H).
Figure 6.
KDM5A mediates transcriptional repression, HR repair, and cellular resistance to DNA damage. (A) Effects of DNA damage on nascent transcription in siControl and siKDM5A cells. Fluorescent 5-ethynyluridine (5-EU) monitors nascent transcription and γ-H2AX marks DNA damage as previously described (Gong et al., 2015). (B) Quantification of 5-EU and γ-H2AX fluorescence intensity from A. Values were normalized to undamaged regions (n > 10). (C) Transcription ±DSB formation was analyzed in FokI-inducible DSB reporter cells in siControl or siKDM5A (three unique siRNAs)–treated cells (Tang et al., 2013). (D) RAD51 loading at FokI-induced DSBs in siControl and siKDM5A treated cells (Tang et al., 2013). Rad51 and FokI restriction enzyme localization was analyzed by immunofluorescence 3 h after FokI induction. (E) Quantification of D (n = 3). (F) Experiments were performed as in D to analyze RAD51 loading at FokI-induced DSBs in siControl and siKDM5A-UTR conditions with or without ectopic expression of WT or GFP-KDM5A derivatives. (G) Quantification of F (n = 2). For conditions with siKDM5A-UTR and WT/mutant GFP-KDM5A expression, data were only obtained from GFP-positive cells. (H) HR efficiency by DR-GFP. HR efficiency was determined in control and siRNA-treated cells by FACS. GFP+ cells indicate repair event and plotted values were normalized to control (n = 4). siCtIP is a positive control. (I) Clonogenic survival assays of siControl and siKDM5A (three independent siRNAs) to IR. Graphs are means ± SEM; n = 3. (J) Model for KDM5A regulation of the ZMYND8-NuRD DDR pathway. Error bars indicate SEM for all graphs unless otherwise indicated. Bars, 10 µm.
Discussion
Bromodomain-containing proteins including ZMYND8 are key epigenetic readers of acetylation signaling involved in the DDR (Gong et al., 2015, 2016). Here, we report the molecular events governing the recruitment and function of ZMYND8–NuRD within damaged chromatin. Our study identifies the histone demethylase KDM5A and GATAD1 as DNA damage recruitment factors that regulate ZMYND8–NuRD DDR function. Molecular studies of modified chromatin revealed DNA damage–induced demethylation of H3K4me3 by KDM5A and inhibition of ZMYND8–NuRD chromatin binding by H3K4me3. We demonstrate that KDM5A acts upstream of ZMYND8–NuRD to promote its association with DNA damage and DDR activities within damaged chromatin, including repressing transcription and promoting HR repair (Fig. 6 H). Collectively, these results establish a KDM5A-dependent mechanism that regulates the ZMYND8–NuRD DDR pathway. Our findings suggest that demethylation of H3K4me3 by KDM5A, in addition to PARP signaling, represents a key initial step in this pathway, which allows ZMYND8–NuRD to bind and function within damaged chromatin.
The KDM5A–ZMYND8–NuRD DDR axis contains several chromatin interaction modules that constitute the molecular basis for its recognition of damaged chromatin. Recruitment of KDM5A to DNA damage was independent from ZMYND8–NuRD but dependent on PARP1 signaling. In addition, deletion of PHD3 domain of KDM5A, which binds its target H3K4me3, did not affect damage accumulation, suggesting additional interactions with chromatin and/or PARylated substrates for localization. Given that H3K4me3 prevents NuRD and ZMYND8 binding to chromatin (Fig. 3 A; Nishioka et al., 2002; Zegerman et al., 2002; Eberl et al., 2013), we speculate that an important function of KDM5A is to remove this inhibitory mark by demethylating H3K4me3 to allow chromatin binding of this complex. As we observed a partial dependency for KDM5A in demethylating H3K4me3, additional methyltransferases, including KDM5B, which has been shown to promote genome stability, may also participate in this pathway (Li et al., 2014). Although we favor demethylation of H3K4me3 by KDM5A as the mechanism whereby this mark is decreased at damage sites, we cannot rule out that histone exchange also participates in this process. For example, turnover of H3.3 occurs at DSBs and UVC damage to regulate nonhomologous end joining and UV responses, respectively (Adam et al., 2013; Luijsterburg et al., 2016). Interestingly, the PHD–BRD–PWWP domain of ZMYND8 can read both unmodified H3 and H3K14Ac (Li et al., 2016b; Savitsky et al., 2016). Thus demethylation of H3K4me3 could also provide a binding interface for ZMYND8. In addition, increases in H3K14 and H4 acetylations are observed at DSBs, which could act collectively as binding platforms for ZMYND8 (Lee et al., 2010; Miller et al., 2010; Tang et al., 2013). Indeed, the histone acetyltransferase TIP 60 is vital for promoting the recruitment of ZMYND8–NuRD, but not KDM5A, to damage sites (Fig. 4, D and E; Gong et al., 2015). Thus, acetylation is also important in regulating this pathway. ZMYND8 has been shown to interact with several different acetylation sites including N-terminal H4 acetylations and H3K14 (Gong et al., 2015; Adhikary et al., 2016; Li et al., 2016b; Savitsky et al., 2016). Binding to H4Acs is dependent on the BRD of ZMYND8 (Gong et al., 2015; Savitsky et al., 2016). Upon DNA damage, maximal recruitment to DNA damage sites requires the BRD of ZMYND8, although other chromatin and DNA-binding modules of ZMYND8 are also involved (Gong et al., 2015; Savitsky et al., 2016). Interestingly, the requirement of the BRD of ZMYND8 to localize to DNA damage is not shared among all variants of ZMYND8. For example, ZMYND8 contains numerous variants, and isoform 1, but not isoform 17, of ZMYND8 requires the BRD for damage recruitment (Gong et al., 2015; Spruijt et al., 2016). In addition, other ZNF proteins also promote NuRD recruitment to damage sites, suggesting the presence of other interactions with DNA and/or factors that stabilize this complex on damaged chromatin (Gong et al., 2015; Savitsky et al., 2016; Spruijt et al., 2016). Although direct interactions have been demonstrated between ZMYND8 and the NuRD complex (Spruijt et al., 2016), it is yet unclear the extent to which KDM5A interacts with this complex. Although we observed a weak interaction between KDM5A and ZMYND8 in cells, whether or not this is direct or occurs exclusively at DNA damage sites will require additional studies. Based on our findings and previous studies, we hypothesize that the numerous chromatin interactions involved in damage recognition by KDM5A and ZMYND8–NuRD act in concert to ensure that the chromatin-modifying and reader activities of these factors are tightly controlled at damage sites. Although multivalent chromatin interacting modules are common in transcription, their involvement in DNA damage recognition and DDR functions are less well understood. Additional mechanistic studies are therefore required to further elucidate how DNA damage alters chromatin to promote interactions with DDR factors, which collectively govern DNA damage signaling and repair to maintain genome integrity.
Our identification of KDM5A as a regulator of ZMYND8–NuRD in the DDR has potential implications in cancer and its treatments. Mutations or aberrant expression of KDM5A and other members of this demethylase family have been reported to promote cancer and drug tolerance (Sharma et al., 2010; Hou et al., 2012; Kooistra and Helin, 2012; Teng et al., 2013; Yamamoto et al., 2014; Shen et al., 2016). ZMYND8 and CHD4 are also found to be dysregulated or mutated in several cancers (Eichmuller et al., 2001; Lai and Wade, 2011; Le Gallo et al., 2012; Panagopoulos et al., 2013; Wada et al., 2014; Li et al., 2016b; Savitsky et al., 2016; Shen et al., 2016). Interactions between these pathways in cancer have started to emerge. ZMYND8–KDM5C modulates enhancer activity in breast cancer cells (Shen et al., 2016). Loss of either ZMYND8 or KDM5C resulted in increased tumorigenic capabilities, suggestive a tumor suppressive function for these factors. Similarly, ZMYND8 functions with the male specific KDM5D to repress metastasis-promoting genes in prostate cancer (Li et al., 2016b). Interestingly, KDM5D is frequently lost or down-regulated in metastatic prostate cancer (Li et al., 2016a). The DDR functions of KDM5A–ZMYND8–NuRD reported here, as well as in other studies (Gong et al., 2015; Spruijt et al., 2016), could contribute to the etiology of these cancers. Our study also highlights the potential use of DNA-damaging agents and PARP inhibitors in cancer settings deficient in the KDM5A-ZMYND8–NuRD pathway, given the therapeutic benefit associated with these treatments in DDR-deficient cancers (Helleday et al., 2008; Chou et al., 2010; Polo et al., 2010; Spruijt et al., 2016). These studies have emphasized the importance of understanding the interplay between the DDR and epigenetic pathways, including those regulated by KDM5 histone demethylases, which can impact the DDR, cancer, and its treatments (Li et al., 2014; Gong et al., 2016). Collectively, our work provides new mechanistic insights into the DDR functions of the epigenetic regulators, KDM5A–ZMYND8–NuRD, which coordinates transcriptional responses and DNA repair activities within damaged chromatin to promote genome integrity.
Materials and methods
Cell culture and treatments
U2OS and HEK293T cell lines were maintained in DMEM supplemented with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen). U2OS cells stably expressing GFP-ZMYND8 were maintained in medium with 10 µg/ml blasticidin S HCl (Invitrogen). U2OS Flp-In T-REx cell lines expressing indicated genes were selected and maintained in medium with 200 µg/ml hygromycin B (Invitrogen) following standard protocol (Invitrogen). 1 µg/ml doxycycline (Sigma-Aldrich) was added to the medium for at least 24 h to induce the expression of the indicated genes. The U2OS-DSB reporter cell lines were provided by R. Greenberg (University of Pennsylvania, Philadelphia, PA). DSBs in these cells were induced by adding 0.5 mM Shield-1 (Takara Bio Inc.) and 1 µM 4-OHT (H7904; Sigma-Aldrich) for at least 3 h. Transcription of the transgene was induced by adding doxycycline for at least 3 h. DIvA (AsiSI-ER-U2OS) was cultured in DMEM supplemented with antibiotics, 10% FCS (Invitrogen), and 1 µg/ml puromycin at 37°C under a humidified atmosphere with 5% CO2. For AsiSI-dependent DSB induction, DIvA cells were treated with 300 nM 4-OHT (H7904; Sigma-Aldrich) for 4 h. IR was generated by a Faxitron x-ray machine. 5 µM PARP inhibitor (Olaparib; Selleckchem) was used to treat cells for 2 h before DNA damage analysis as indicated. For inhibiting transcription, cells were treated with 100 µM 5,6-dichlorobenzimidazole riboside for 2 h before DNA damage, and analysis was performed as indicated. For detecting damage induced transcription repression, cells were laser microirradiated first and then incubated in medium containing 1 mM 5-ethynyluridine for 1 h. Nascent transcription after damage was analyzed by detection with the Click-iT RNA imaging kit (Invitrogen) according to the manufacturer’s instructions. CPI-455 (Axon Medchem; Vinogradova et al., 2016) was used at 50 µM and 100 µM to treat U2OS cells for 24 h before performing assays and analyses.
Plasmid and siRNA transfection
Mammalian expression vectors were transfected into indicated cell lines by Fugene HD (Promega) according to the manufacturer’s instructions. siRNA transfection was performed with Lipofectamine RNAiMax (Invitrogen) following the manufacturer’s instructions. The sequences of the siRNAs used in this study are as follows: siControl, SMARTpool; siZMYND8, 5′-GAACAUAGAUGAAUGAAA-3′; siCHD4, 5′-CCCAGAAGAGGAUUUGUCA-3′; siTIP60, SMARTpool; siKDM5A-1, 5′-GGAACUGGGUCUCUUUUGA-3′; siKDM5A-2, 5′-GCAAAUGAGACAACGGAAA-3′; siKDM5A-3, 5′-UGACAAUGGUGGACCGCAU-3′; siKDM5A-UTR (targeting the 3′-UTR region of KDM5A), 5′-GAUAGUAGUUAGAGGCUUA-3′; siPARP1, 5′-GGGCAAGCACAGUGUCAAA-3′ (Polo et al., 2010); siGATAD1, 5′-GGAAAAGGGAGAAGACAUA-3′; siRBMX, 5′-UAUGGUAACUCACGUAGUG-3′ (Adamson et al., 2012); siDDB1, 5′-GCAAGGACCUGCUGUUUAU-3′; siZNF592, SMARTpool; siZNF687, SMARTpool. SMARTpool siRNAs were purchased from GE Healthcare. Other siRNAs were purchased from Sigma-Aldrich. siRNA knockdown efficiencies were validated by Western blotting (WB) or quantitative RT-PCR.
Cloning and plasmids
cDNAs of human genes used in this study were amplified by PCR from HEK293T or U2OS cells and cloned into pDONR201 vector by gateway cloning. W335A and H483A mutant forms of KDM5A in pDONR201 were generated by site-directed mutagenesis following standard protocols (Agilent Technologies). The ΔPHD3 form of KDM5A was generated by PCR-amplifying KDM5A cDNA without 4,846–5,970 bp (regions to encode the C-terminus 1615–1690 amino acids, including PHD3). ΔPHD1 and ΔPHD2 forms of KDM5A were generated by forward and reverse primers containing complementary sequence to the regions flanking the area to be deleted, following standard site-directed mutagenesis protocols to obtain vectors lacking deleted regions in pDONR201. Mammalian expression vectors with indicated human genes were generated by subcloning the pDONR vectors into Gateway destination vectors containing the epitope tags as indicated. N-RFP DEST Gateway empty vector was generated by replacing the EmGFP sequence of pcDNA6.2/N-EmGFP DEST vector (Invitrogen) with the RFP sequence through regular PCR and ligation cloning. FRT/TO/N-EmGFP DEST Gateway empty vector, which was used for the Flp-In T-REx system, was obtained by ligation of the N-EmGFP sequence and Gateway recombination cassette into the polylinker of pcDNA5/FRT/TO (Invitrogen).
Protein extraction and WB
Whole-cell extracts were obtained from cells washed with PBS, collected with Laemmli buffer (4% [vol/vol] SDS, 20% [vol/vol] glycerol, and 120 mM Tris-HCl, pH 6.8), and then sonicated with a Diagenode Bioruptor for 10 min followed by 5 min of boiling at 95°C before loading. Samples were resolved by SDS-PAGE and analyzed by standard WB protocols. Signals of the Western blots were detected by standard chemiluminescence (GE Healthcare) and analyzed using a ChemiDoc XRS+ system (Bio-Rad Laboratories). Primary antibodies used were rabbit anti-ZMYND8 (A302-089; Bethyl Laboratories, Inc.), rabbit anti-CHD4 (39289; Active Motif), rabbit anti-HDAC1 (ab19845; Abcam), rabbit anti-HDAC2 (ab7029; Abcam), rabbit anti-MTA2 (A300-395; Bethyl Laboratories, Inc.), mouse anti-KDM5A (ab78322; Abcam), rabbit anti-KDM5B (A301-813; Bethyl Laboratories, Inc.), rabbit anti-KDM5C (A301-034; Bethyl Laboratories, Inc.), rabbit anti-H3 (ab1791; Abcam), rabbit anti-H3K4me3 (9751; Cell Signaling Technology), rabbit anti–β-tubulin (ab6046; Abcam), rabbit anti-GFP (A11122; Invitrogen), mouse anti-Flag (F1804; Sigma-Aldrich), rabbit anti-ZNF592 (A301-530; Bethyl Laboratories, Inc.), rabbit anti-ZNF687 (A303-278; Bethyl Laboratories, Inc.), rabbit anti-RBMX (14794; Cell Signaling Technology), rabbit anti-DDB1 (A300-462; Bethyl Laboratories, Inc.), and mouse anti-RAD51 (ab88572; Abcam).
Immunoprecipitation analysis
Cells were collected and lysed with NETN buffer (150 mM NaCl, 20 mM Tris-Cl, pH 8.0, 1 mM EDTA, and 0.5% NP-40 [vol/vol]) containing 1 mM MgCl2 and TurboNuclease (Accelagen) for 1 h at 4°C. Cell lysates were cleared by 15,000 rpm centrifugation for 30 min at 4°C. Overexpressed SFB (S-protein, 2XFlag, Streptavidin-binding peptide)-tagged proteins were pulled down with 30 µl Streptavidin beads (GE Healthcare) after mixing by rotation for >1 h at 4°C. Overexpressed GFP-tagged proteins were pulled down with 30 µl GFP-Trap beads (ChromoTek) after rotating at 4°C for >2 h. Endogenous proteins were immunoprecipitated by 1 µg of the indicated antibodies and 30 µl protein A Agarose (EMD Millipore) after overnight rotating at 4°C. After indicated incubation and rotation, beads were washed at least four times with NETN buffer. Protein mixtures were eluted by boiling with Laemmli buffer and then subjected to standard WB analysis with the indicated antibodies.
Laser microirradiation and microscopy analysis
Laser microirradiation and quantification of live-cell imaging of laser damage recruitment were performed with FV-10 ASW3.1 software on a Fluoview 1000 confocal microscope (Olympus) following standard protocols designed in previous studies (Gong et al., 2015). In brief, cells were seeded on glass-bottomed dishes (Willco wells). All cells used for laser microirradiation and after the experiments (live-cell imaging or immunofluorescence) were presensitized by adding 10 µM BrdU in regular DMEM medium for 20 h before damage treatment. Live-cell imaging experiments were performed at 37°C and 5% CO2 conditions maintained by a heated incubation system on the microscope (TOKAI HIT). All images were captured using an Olympus 60× oil objective lens (NA 1.42). A 405-nm laser beam (60%) was used to generate laser microirradiation. For quantification of live-cell imaging after laser microirradiation, the fluorescence intensity at the damage site and an undamaged control region of the same size from the same cell were directly measured and recorded using FV-10 ASW3.1 software. Variation of the fluorescence intensity was quantified as the difference between the mean fluorescence intensity in the damaged region versus the mean fluorescence intensity in an undamaged region from the same cell. For all live-cell imaging experiments in this study, two to four independent experiments were performed. For each experiment, quantification data were collected from >10 cells under each condition. Plots shown in figures are from one representative experiment.
Immunofluorescence analysis
Immunofluorescence was performed essentially as previously described (Leung et al., 2014). In brief, after indicated treatments, cells were pre-extracted with CSK buffer (10 mM PIPES, pH 6.8, 100 NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.5% [vol/vol] Triton X-100) for 5 min on ice after a 15-min fixation by 2% paraformaldehyde at room temperature. Fixed cells were washed three times with PBS and blocked with 3% BSA in PBS for 15 min at room temperature. Cells were then incubated with indicated primary antibodies for 1 h at room temperature or overnight at 4°C. After three PBS washes, cells were incubated with secondary antibodies and Hoechst for 1 h at room temperature in the dark. Cells were then washed three times with PBS and mounted on coverslips (or glass-bottom dishes) with Vectashield. Cells were imaged and analyzed with the Z-Stacked setting under the FV-10 ASW3.1 software on the Fluoview 1000 confocal microscope (Olympus). The color, brightness, and contrast of presented images were adjusted using FV-10 ASW3.1 software before being exported as TIFF files. Exported images were further adjusted to the proper size using Image J software and organized in the final figures by Adobe Illustrator CS5.1 (Adobe). For quantification of RAD51 loading at FokI-induced DSBs, data from >100 cells were collected under each condition from two or three independent experiments. Primary antibodies used for immunofluorescence were mouse anti–γ-H2AX (05–636; EMD Millipore), rabbit anti-H3K4me3 (9751; Cell Signaling Technology), rabbit anti-CHD4 (39289; Active Motif), and mouse anti-RAD51 (ab88572; Abcam). Secondary antibodies used were Alexa Fluor 488 goat anti–rabbit IgG (A11034; Invitrogen), Alexa Fluor 488 goat anti–mouse IgG (A11029; Invitrogen), Alexa Fluor 594 goat anti–mouse IgG (A11032; Invitrogen), and Alexa Fluor 647 goat anti–mouse IgG (A21236; Invitrogen).
ChIP and ChIP-seq methods for DIvA cells
ChIP assays were performed according to a protocol described previously (Iacovoni et al., 2010). 10 µg chromatin was immunoprecipitated using 1 µg anti-H3K4me3 (8580, validated in ChIP by the provider; Abcam), anti-H3 (1791; Abcam) or without antibody (mock). Immunoprecipitated DNA and input DNA were analyzed in duplicate by quantitative RT-PCR and normalized against TAF12, using the following primers: RBMXL1_FW 5′-GATTGGCTATGGGTGTGGAC-3′; RBMXL1_REV 5′-CATCCTTGCAAACCAGTCCT-3′; LYRM2_FW 5′-TGCCGGTCTCCTAGAAGTTG-3′; LYRM2_REV 5′-GCGCTTGATTTCCCTGAGT-3′; β-ACTIN_FW 5′-AGCCGGGCTCTTGCCAAT-3′; β-ACTIN_REV 5′-AGTTAGCGCCCAAAGGACCA-3′; 884 (CTRL_FW) 5′-CCCATCTCAACCTCCACACT-3′; 885 (CTRL_REV) 5′-CTTGTCCAGATTCGCTGTGA-3′; TAF12_FW 5′-GCTGAGACGAACGCTTCACT-3′; TAF12_REV 5′-CCTTCGAACACTGACCCACT-3′.
For ChIP-seq, sequencing libraries were prepared by using 10 ng of purified DNA (mean size, 250–300 bp), and subjected to high-throughput sequencing (single-end read) by the Beijing Genomics Institute using a HiSeq 2000 sequencing system. ChIP-seq data were aligned against the hg19 human genome using BWA (http://bio-bwa.sourceforge.net/) software. Only uniquely mapped reads were then kept and the potential reads belonging to PCR duplicate were removed using SAMtools. Each sample was normalized by the final number of reads. Data were visualized with the Integrated Genome Browser (www.bioviz.org). All further analysis (figures and statistical tests) were done using R and Bioconductor packages. The list of the 80 best cleaved AsiSI site was determined in 4-OHT–treated DIvA cells using direct in situ BLESS (break labeling, enrichment on Streptavidin and next-generation sequencing), a technique developed to capture and identify DSBs at a genome-wide level (Crosetto et al., 2013). A list of 80 random positions on the genome was also generated. The mean of the normalized number of reads aligned for each sample was computed for each AsiSI or non-AsiSI site. Log2 (+4OHT/-4OHT) was computed for each sites and displayed as a box plot. To test whether the mean of the distribution is significantly different from 0, we applied a nonparametric Mann–Whitney test. For comparisons of H3K4me3 levels in DDR-regulated genes, we used a list of 296 genes significantly up-regulated 4 h after irradiation in CAL51 cells (Rashi-Elkeles et al., 2014). For these genes, we selected the normalized number of reads aligned for each sample in windows of ±3 kb around each gene to generate metagene profiles (homemade software, available as supplemental material).
ChIP analysis in FokI-induced DSB reporter cells
After the indicated treatments, cells were cross-linked with 1% (vol/vol) formaldehyde for 10 min after 125 mM glycine treatment for 5 min to stop the cross-linking. Cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, pH 8.0, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with protease inhibitor cocktail (Roche). Cell lysates were sonicated by Bioruptor (Diagenode) to obtain the chromatin solution for each sample. After normalization, 1 ml chromatin solution (with a DNA concentration of 5 ng/µl) for each sample was incubated with primary antibodies overnight at 4°C. After this overnight incubation, 25 µl IgG + IgA Dynabeads (Invitrogen) was added to each sample to incubate 3 h with rotation at 4°C. Beads were washed with standard ChIP washing buffers in the following order: TSE-150 (1% Triton X-100, 0.1% SDS, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, and 150 mM NaCl), TSE-500 (1% Triton X-100, 0.1% SDS, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, and 500 mM NaCl), LiCl detergent (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.0), and TE (10 mM Tris-HCl, pH 8.0, and 0.1 mM EDTA). After washing, beads were resuspended with 100 µl elution buffer (1% SDS, 0.1 M NaHCO3) following 65°C overnight de-cross-linking treatment. DNA samples were purified with QIAquick PCR purification kit (QIAGEN). Quantitative PCR was performed on the StepOnePlus as described previously (Gong et al., 2015). ChIP-qPCR primers used here were as described previously (forward, 5′-GGAAGATGTCCCTTGTATCACCAT-3′; reverse, 5′-TGGTTGTCAACAGAGTAGAAAGTGAA-3′; Tang et al., 2013). Antibodies used for this experiment were rabbit anti-H3 (ab1791; Abcam) and rabbit anti-H3K4me3 (9751; Cell Signaling Technology).
Quantitative RT-PCR
Quantitative RT-PCR was performed as previously described (Gong et al., 2015). Primers used in this study were: GATAD1 forward, 5′-TCCCATCAAAGCTCCTGAGT-3′; GATAD1 reverse, 5′-TCAATCACAGAAACAACATCACC-3′; GAPDH forward, 5′-CAATGACCCCTTCATTGACC-3′; and GAPDH reverse, 5′-GATCTCGCTCCTGGAAGATG-3′ (Gong et al., 2015).
Peptide pull-down assays
The protocol for the peptide pull-down assay was modified from a previous description (Gong et al., 2015). Peptides used for this study were purchased from AnaSpec. Different histone peptides were immobilized on Dynabeads MyOne Streptavidin T1 (Invitrogen) in NETN buffer by rotating at 4°C for 1 h after three washes with NETN buffer. For preparing cell extracts, 293T and U2OS cells were lysed with NETN buffer containing 1 mM MgCl2 and TurboNuclease (Accelagen) for 1 h at 4°C, followed by a 30 min 15,000 rpm centrifugation to obtain the cell extract. Cell lysates were diluted into equal volumes to mix with each peptide and rotated at 4°C for 1 h. After the indicated incubation, beads were washed at least four times with NETN buffer. Proteins pulled down by the indicated peptides were eluted by boiling with Laemmli buffer and then subjected to standard WB analysis with the indicated antibodies.
Clonogenic cell survival assays
72 h after siRNA treatments, U2OS cells were treated with indicated IR doses. Cells were left to form colonies for 10 to 14 d. Colonies were washed with PBS and stained with 0.5% (wt/vol) crystal violet and 20% (vol/vol) ethanol for 30 min at room temperature. After staining, plates were gently rinsed with water, and colonies were counted. Results were normalized to plating efficiencies of untreated cells for each siRNA.
CRISPR/Cas9 targeting for gene KO
ZMYND8 and CHD4 gene KOs were generated in U2OS cells using CRISPR/Cas9 following standard protocols (Ran et al., 2013). The single-guide RNA (sgRNA) sequences used were: ZMYND8, 5′-GGATATCTCTACTCGCTCCA-3′; CHD4, 5′-AGCCTGCCCCCACCCCACCC-3′. Indicated sgRNA sequences were cloned into pSpCas9 (BB)-2A-Puro (62988; Addgene) following the standard protocol (Ran et al., 2013). Cloned vectors were validated by sequencing. To generate U2OS KO cell lines, plasmids containing sgRNA targeting the indicated gene were transfected into cells with Fugene HD. 48 h after transfection, cells were selected with medium containing 2 µg/ml puromycin (Invitrogen) for 24 h. After puromycin selection, cells were recovered in DMEM medium for 2–3 d. After recovering, cells were seeded into 96-well plate with a mean of one cell per well. Cells were allowed to grow until an individual colony in one well was formed. Individual colonies were then expanded and screened by WB. Successful KO of the indicated genes was validated by WB analysis.
DR-GFP assay
An integrated HR reporter DR-GFP–containing U2OS cell line was used as described previously (Pierce et al., 1999). 1 d after indicated siRNA treatments, U2OS DR-GFP cells were transfected with I-SceI–expressing vector (pCAG-I-SceI) or control vector (pCAG). 48 h after I-SceI transfection, cells were trypsinized, washed once with PBS, and then resuspended in sodium citrate solution without fixation. Resuspended cells were placed into Falcon 5-ml polystyrene round-bottom tubes through the cell-strainer caps. The percentage of GFP-positive cells was determined by an Accuri Flow Cytometer (BD). All samples were normalized with the siControl sample transfected with pCAG-I-SceI vector.
FACS analysis
U2OS cells were harvested 72 h after treatment (indicated siRNA or drug) and fixed with 80% ethanol overnight at 4°C. The fixed samples were washed three times with PBS and stained with propidium iodide and analyzed by Accuri C6 flow cytometer to obtain cell cycle profiles.
Statistics
Statistics were performed using GraphPad Prism (version 6.0). Unpaired two-tailed Student’s t test was applied to compare individual conditions of the indicated experiment. P < 0.05 indicates significant difference. Asterisks indicate p-values (*, P < 0.05; **, P < 0.01; ***, P < 0.001). N represents the number of events or experimental/biological replicates as described for each experiment.
Online supplemental material
Fig. S1 is related to Fig. 1 and shows knockdown efficiency of indicated siRNA and additional data for KDM5A and ZMYND8 interactions. Fig. S2 shows DNA damage recruitment of ZMYND8 upon multiple KDM5A siRNA treatments (related to Fig. 1). It also shows H3K4me3 levels (±4-OHT) on promoter regions of IR-induced genes (related to Fig. 2). In addition, it contains peptide pull-down assays as in Fig. 3 A with additional peptides and different conditions. Fig. S3 is related to Fig. 3 and shows expression levels of WT or mutant KDM5A vectors by Western blots and rescue experiments for damage recruitment of ZMYND8 with indicated WT and mutant KDM5A expression upon siKDM5A-UTR. Fig. S4 is related to Fig. 4 and provides data for independence of damage recruitment of KDM5A from ZMYND8-NuRD. Fig. S5 is related to Fig. 6. It analyzes RAD51 loading at FokI-DSB sites upon CPI-455 treatment and provides protein expression levels and cell cycle distribution of U2OS cells in siControl and siKDM5A conditions. A supplemental .txt file provides source code for computational methods used to draw Fig. 2 D and Fig. S2 C (see Materials and methods).
Supplementary Material
Acknowledgments
We thank Jenny Brodbelt and her laboratory (The University of Texas at Austin [UT Austin]) for Mass Spectrometry and Roger Greenberg for providing the FokI DSB cell lines. We are grateful to Nadima Uprety for assistance in the initial cloning and analysis of some ZMYND8 interactors. We are thankful for comments on the manuscript from Blerta Xhemalce (UT Austin).
Funding in G. Legube’s laboratory was provided by grants from the European Research Council (ERC-2014-CoG 647344), Agence Nationale pour la Recherche (ANR-14-CE10-0002-01 and ANR-13-BSV8-0013), and the Ligue Nationale contre le Cancer. This research in the K.M. Miller laboratory was supported by start-up funds from UT Austin, Cancer Prevention Research Institute of Texas (CPRIT, R1116), and the National Institutes of Health, National Cancer Institute (RO1 CA201268 and CA198279).
The authors declare no competing financial interests.
Author contributions: F. Gong performed experiments, and F. Gong and K.M. Miller designed and analyzed the experiments, unless indicated otherwise. T. Clouaire, M. Aguirrebengoa, and G. Legube performed and analyzed all experiments with DIvA cells. K.M. Miller conceived the study and supervised the project. F. Gong and K.M. Miller wrote the manuscript with input and discussions from all authors.
Footnotes
Abbreviations used:
- 4-OHT
- 4-hydroxytamoxifen
- ChIP
- chromatin immunoprecipitation
- DDR
- DNA damage response
- DSB
- DNA double-strand break
- HDAC
- histone deacetylase
- IR
- ionizing radiation
- KO
- knockout
- NuRD
- nucleosome remodeling and histone deacetylation
- PHD
- plant homeodomain
- sgRNA
- single-guide RNA
- WB
- Western blotting
References
- Adam S., Polo S.E., and Almouzni G.. 2013. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 155:94–106. 10.1016/j.cell.2013.08.029 [DOI] [PubMed] [Google Scholar]
- Adamson B., Smogorzewska A., Sigoillot F.D., King R.W., and Elledge S.J.. 2012. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 14:318–328. 10.1038/ncb2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikary S., Sanyal S., Basu M., Sengupta I., Sen S., Srivastava D.K., Roy S., and Das C.. 2016. Selective recognition of H3.1K36 dimethylation/H4K16 acetylation facilitates the regulation of all-trans-retinoic acid (ATRA)-responsive genes by putative chromatin reader ZMYND8. J. Biol. Chem. 291:2664–2681. 10.1074/jbc.M115.679985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F., Bugler B., Schmidt C.K., Guillou E., Caron P., Briois S., Iacovoni J.S., Daburon V., Miller K.M., Jackson S.P., and Legube G.. 2014. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 21:366–374. 10.1038/nsmb.2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrapetov M.K., Gursoy-Yuzugullu O., Xu C., Xu Y., and Price B.D.. 2014. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc. Natl. Acad. Sci. USA. 111:9169–9174. 10.1073/pnas.1403565111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., and Zhao K.. 2007. High-resolution profiling of histone methylations in the human genome. Cell. 129:823–837. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Beli P., Lukashchuk N., Wagner S.A., Weinert B.T., Olsen J.V., Baskcomb L., Mann M., Jackson S.P., and Choudhary C.. 2012. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell. 46:212–225. 10.1016/j.molcel.2012.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair L.P., Cao J., Zou M.R., Sayegh J., and Yan Q.. 2011. Epigenetic regulation by lysine demethylase 5 (KDM5) enzymes in cancer. Cancers (Basel). 3:1383–1404. 10.3390/cancers3011383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan M.V., Lee J., Ward I.M., Kim J.E., Thompson J.R., Chen J., and Mer G.. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 127:1361–1373. 10.1016/j.cell.2006.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., and Foiani M.. 2010. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11:208–219. 10.1038/nrm2852 [DOI] [PubMed] [Google Scholar]
- Caron P., Choudjaye J., Clouaire T., Bugler B., Daburon V., Aguirrebengoa M., Mangeat T., Iacovoni J.S., Álvarez-Quilón A., Cortés-Ledesma F., and Legube G.. 2015. Non-redundant functions of ATM and DNA-PKcs in response to DNA double-strand breaks. Cell Reports. 13:1598–1609. 10.1016/j.celrep.2015.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiácovo M.P., and Elledge S.J.. 2010. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA. 107:18475–18480. 10.1073/pnas.1012946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J., Agger K., Cloos P.A., Pasini D., Rose S., Sennels L., Rappsilber J., Hansen K.H., Salcini A.E., and Helin K.. 2007. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 128:1063–1076. 10.1016/j.cell.2007.02.003 [DOI] [PubMed] [Google Scholar]
- Ciccia A., and Elledge S.J.. 2010. The DNA damage response: Making it safe to play with knives. Mol. Cell. 40:179–204. 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N., Mitra A., Silva M.J., Bienko M., Dojer N., Wang Q., Karaca E., Chiarle R., Skrzypczak M., Ginalski K., et al. 2013. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods. 10:361–365. 10.1038/nmeth.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova E., Turberfield A.H., and Klose R.J.. 2015. Histone demethylases in chromatin biology and beyond. EMBO Rep. 16:1620–1639. 10.15252/embr.201541113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl H.C., Spruijt C.G., Kelstrup C.D., Vermeulen M., and Mann M.. 2013. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell. 49:368–378. 10.1016/j.molcel.2012.10.026 [DOI] [PubMed] [Google Scholar]
- Eichmuller S., Usener D., Dummer R., Stein A., Thiel D., and Schadendorf D.. 2001. Serological detection of cutaneous T-cell lymphoma-associated antigens. Proc. Natl. Acad. Sci. USA. 98:629–634. 10.1073/pnas.98.2.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradet-Turcotte A., Canny M.D., Escribano-Díaz C., Orthwein A., Leung C.C., Huang H., Landry M.C., Kitevski-LeBlanc J., Noordermeer S.M., Sicheri F., and Durocher D.. 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 499:50–54. 10.1038/nature12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F., and Miller K.M.. 2013. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutat. Res. 750:23–30. 10.1016/j.mrfmmm.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Gong F., Chiu L.Y., Cox B., Aymard F., Clouaire T., Leung J.W., Cammarata M., Perez M., Agarwal P., Brodbelt J.S., et al. 2015. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 29:197–211. 10.1101/gad.252189.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F., Chiu L.Y., and Miller K.M.. 2016. Acetylation reader proteins: Linking acetylation signaling to genome maintenance and cancer. PLoS Genet. 12:e1006272 10.1371/journal.pgen.1006272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E.L., and Shi Y.. 2012. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13:343–357. 10.1038/nrg3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T., Petermann E., Lundin C., Hodgson B., and Sharma R.A.. 2008. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 8:193–204. 10.1038/nrc2342 [DOI] [PubMed] [Google Scholar]
- Helmrich A., Ballarino M., Nudler E., and Tora L.. 2013. Transcription-replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol. 20:412–418. 10.1038/nsmb.2543 [DOI] [PubMed] [Google Scholar]
- Hou J., Wu J., Dombkowski A., Zhang K., Holowatyj A., Boerner J.L., and Yang Z.Q.. 2012. Genomic amplification and a role in drug-resistance for the KDM5A histone demethylase in breast cancer. Am. J. Transl. Res. 4:247–256. [PMC free article] [PubMed] [Google Scholar]
- Iacovoni J.S., Caron P., Lassadi I., Nicolas E., Massip L., Trouche D., and Legube G.. 2010. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 29:1446–1457. 10.1038/emboj.2010.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., and Bartek J.. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078. 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., and Durocher D.. 2013. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell. 49:795–807. 10.1016/j.molcel.2013.01.017 [DOI] [PubMed] [Google Scholar]
- Kakarougkas A., Ismail A., Chambers A.L., Riballo E., Herbert A.D., Künzel J., Löbrich M., Jeggo P.A., and Downs J.A.. 2014. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol. Cell. 55:723–732. 10.1016/j.molcel.2014.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury-Haddad H., Guttmann-Raviv N., Ipenberg I., Huggins D., Jeyasekharan A.D., and Ayoub N.. 2014. PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proc. Natl. Acad. Sci. USA. 111:E728–E737. 10.1073/pnas.1317585111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., and Jinks-Robertson S.. 2012. Transcription as a source of genome instability. Nat. Rev. Genet. 13:204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein B.J., Piao L., Xi Y., Rincon-Arano H., Rothbart S.B., Peng D., Wen H., Larson C., Zhang X., Zheng X., et al. 2014. The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Reports. 6:325–335. 10.1016/j.celrep.2013.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R.J., and Zhang Y.. 2007. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8:307–318. 10.1038/nrm2143 [DOI] [PubMed] [Google Scholar]
- Klose R.J., Yan Q., Tothova Z., Yamane K., Erdjument-Bromage H., Tempst P., Gilliland D.G., Zhang Y., and Kaelin W.G. Jr. 2007. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 128:889–900. 10.1016/j.cell.2007.02.013 [DOI] [PubMed] [Google Scholar]
- Kooistra S.M., and Helin K.. 2012. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 13:297–311. [DOI] [PubMed] [Google Scholar]
- Lai A.Y., and Wade P.A.. 2011. Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat. Rev. Cancer. 11:588–596. 10.1038/nrc3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.S., Park J.H., Kim S.J., Kwon S.J., and Kwon J.. 2010. A cooperative activation loop among SWI/SNF, γ-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 29:1434–1445. 10.1038/emboj.2010.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gallo M., O’Hara A.J., Rudd M.L., Urick M.E., Hansen N.F., O’Neil N.J., Price J.C., Zhang S., England B.M., Godwin A.K., et al. NIH Intramural Sequencing Center (NISC) Comparative Sequencing Program . 2012. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat. Genet. 44:1310–1315. 10.1038/ng.2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J.W., Agarwal P., Canny M.D., Gong F., Robison A.D., Finkelstein I.J., Durocher D., and Miller K.M.. 2014. Nucleosome acidic patch promotes RNF168- and RING1B/BMI1-dependent H2AX and H2A ubiquitination and DNA damage signaling. PLoS Genet. 10:e1004178 10.1371/journal.pgen.1004178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Dhar S.S., Chen T.Y., Kan P.Y., Wei Y., Kim J.H., Chan C.H., Lin H.K., Hung M.C., and Lee M.G.. 2016a JARID1D is a suppressor and prognostic marker of prostate cancer invasion and metastasis. Cancer Res. 76:831–843. 10.1158/0008-5472.CAN-15-0906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Li Y., Lv J., Zheng X., Wen H., Shen H., Zhu G., Chen T.Y., Dhar S.S., Kan P.Y., et al. 2016b ZMYND8 reads the dual histone mark H3K4me1-H3K14ac to antagonize the expression of metastasis-linked genes. Mol. Cell. 63:470–484. 10.1016/j.molcel.2016.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu L., Yang S., Song N., Zhou X., Gao J., Yu N., Shan L., Wang Q., Liang J., et al. 2014. Histone demethylase KDM5B is a key regulator of genome stability. Proc. Natl. Acad. Sci. USA. 111:7096–7101. 10.1073/pnas.1324036111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg M.S., de Krijger I., Wiegant W.W., Shah R.G., Smeenk G., de Groot A.J., Pines A., Vertegaal A.C., Jacobs J.J., Shah G.M., and van Attikum H.. 2016. PARP1 Links CHD2-Mediated Chromatin Expansion and H3.3 Deposition to DNA Repair by Non-homologous End-Joining. Mol. Cell. 61:547–562. 10.1016/j.molcel.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J., Lukas C., and Bartek J.. 2011. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13:1161–1169. 10.1038/ncb2344 [DOI] [PubMed] [Google Scholar]
- Mansfield R.E., Musselman C.A., Kwan A.H., Oliver S.S., Garske A.L., Davrazou F., Denu J.M., Kutateladze T.G., and Mackay J.P.. 2011. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J. Biol. Chem. 286:11779–11791. 10.1074/jbc.M110.208207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.M., and Jackson S.P.. 2012. Histone marks: Repairing DNA breaks within the context of chromatin. Biochem. Soc. Trans. 40:370–376. 10.1042/BST20110747 [DOI] [PubMed] [Google Scholar]
- Miller K.M., Tjeertes J.V., Coates J., Legube G., Polo S.E., Britton S., and Jackson S.P.. 2010. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 17:1144–1151. 10.1038/nsmb.1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N., Kim H., Laurent B., Zhao Y., Lim H.J., Majid M.C., Dango S., Luo Y., Hempel K., Sowa M.E., et al. 2013. The histone demethylase LSD1/KDM1A promotes the DNA damage response. J. Cell Biol. 203:457–470. 10.1083/jcb.201302092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S., Gorgoulis V.G., and Halazonetis T.D.. 2010. Genomic instability: An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11:220–228. 10.1038/nrm2858 [DOI] [PubMed] [Google Scholar]
- Nishioka K., Chuikov S., Sarma K., Erdjument-Bromage H., Allis C.D., Tempst P., and Reinberg D.. 2002. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16:479–489. 10.1101/gad.967202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara H., Ui A., Otsuka A., Satoh H., Yokomi I., Nakajima S., Yasui A., Yokota J., and Kohno T.. 2011. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. 30:2135–2146. 10.1038/onc.2010.592 [DOI] [PubMed] [Google Scholar]
- Panagopoulos I., Micci F., Thorsen J., Haugom L., Buechner J., Kerndrup G., Tierens A., Zeller B., and Heim S.. 2013. Fusion of ZMYND8 and RELA genes in acute erythroid leukemia. PLoS One. 8:e63663 10.1371/journal.pone.0063663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister S.X., Ahrabi S., Zalmas L.P., Sarkar S., Aymard F., Bachrati C.Z., Helleday T., Legube G., La Thangue N.B., Porter A.C., and Humphrey T.C.. 2014. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Reports. 7:2006–2018. 10.1016/j.celrep.2014.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A.J., Johnson R.D., Thompson L.H., and Jasin M.. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13:2633–2638. 10.1101/gad.13.20.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S.E., and Jackson S.P.. 2011. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25:409–433. 10.1101/gad.2021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S.E., Kaidi A., Baskcomb L., Galanty Y., and Jackson S.P.. 2010. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29:3130–3139. 10.1038/emboj.2010.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashi-Elkeles S., Warnatz H.J., Elkon R., Kupershtein A., Chobod Y., Paz A., Amstislavskiy V., Sultan M., Safer H., Nietfeld W., et al. 2014. Parallel profiling of the transcriptome, cistrome, and epigenome in the cellular response to ionizing radiation. Sci. Signal. 7:rs3 10.1126/scisignal.2005032 [DOI] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., and Jackson S.P.. 2007. Human CtIP promotes DNA end resection. Nature. 450:509–514. 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky P., Krojer T., Fujisawa T., Lambert J.P., Picaud S., Wang C.Y., Shanle E.K., Krajewski K., Friedrichsen H., Kanapin A., et al. 2016. Multivalent histone and DNA engagement by a PHD/BRD/PWWP triple reader cassette recruits ZMYND8 to K14ac-rich chromatin. Cell Reports. 17:2724–2737. 10.1016/j.celrep.2016.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler D.M., Rouquette J., Schmid V.J., Strickfaden H., Ottmann C., Drexler G.A., Mazurek B., Greubel C., Hable V., Dollinger G., et al. 2011. Double-strand break-induced transcriptional silencing is associated with loss of tri-methylation at H3K4. Chromosome Res. 19:883–899. 10.1007/s10577-011-9244-1 [DOI] [PubMed] [Google Scholar]
- Sharma S.V., Lee D.Y., Li B., Quinlan M.P., Takahashi F., Maheswaran S., McDermott U., Azizian N., Zou L., Fischbach M.A., et al. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 141:69–80. 10.1016/j.cell.2010.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Xu W., Guo R., Rong B., Gu L., Wang Z., He C., Zheng L., Hu X., Hu Z., et al. 2016. Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell. 165:331–342. 10.1016/j.cell.2016.02.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt C.G., Luijsterburg M.S., Menafra R., Lindeboom R.G., Jansen P.W., Edupuganti R.R., Baltissen M.P., Wiegant W.W., Voelker-Albert M.C., Matarese F., et al. 2016. ZMYND8 co-localizes with NuRD on target genes and regulates poly(ADP-ribose)-dependent recruitment of GATAD2A/NuRD to sites of DNA damage. Cell Reports. 17:783–798. 10.1016/j.celrep.2016.09.037 [DOI] [PubMed] [Google Scholar]
- Tang J., Cho N.W., Cui G., Manion E.M., Shanbhag N.M., Botuyan M.V., Mer G., and Greenberg R.A.. 2013. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 20:317–325. 10.1038/nsmb.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y.C., Lee C.F., Li Y.S., Chen Y.R., Hsiao P.W., Chan M.Y., Lin F.M., Huang H.D., Chen Y.T., Jeng Y.M., et al. 2013. Histone demethylase RBP2 promotes lung tumorigenesis and cancer metastasis. Cancer Res. 73:4711–4721. 10.1158/0008-5472.CAN-12-3165 [DOI] [PubMed] [Google Scholar]
- Torres I.O., Kuchenbecker K.M., Nnadi C.I., Fletterick R.J., Kelly M.J., and Fujimori D.G.. 2015. Histone demethylase KDM5A is regulated by its reader domain through a positive-feedback mechanism. Nat. Commun. 6:6204 10.1038/ncomms7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., and Gasser S.M.. 2009. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 19:207–217. 10.1016/j.tcb.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Vermeulen M., Eberl H.C., Matarese F., Marks H., Denissov S., Butter F., Lee K.K., Olsen J.V., Hyman A.A., Stunnenberg H.G., and Mann M.. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 142:967–980. 10.1016/j.cell.2010.08.020 [DOI] [PubMed] [Google Scholar]
- Vinogradova M., Gehling V.S., Gustafson A., Arora S., Tindell C.A., Wilson C., Williamson K.E., Guler G.D., Gangurde P., Manieri W., et al. 2016. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat. Chem. Biol. 12:531–538. 10.1038/nchembio.2085 [DOI] [PubMed] [Google Scholar]
- Wada Y., Matsuura M., Sugawara M., Ushijima M., Miyata S., Nagasaki K., Noda T., and Miki Y.. 2014. Development of detection method for novel fusion gene using GeneChip exon array. J. Clin. Bioinforma. 4:3 10.1186/2043-9113-4-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhai L., Xu J., Joo H.Y., Jackson S., Erdjument-Bromage H., Tempst P., Xiong Y., and Zhang Y.. 2006. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell. 22:383–394. 10.1016/j.molcel.2006.03.035 [DOI] [PubMed] [Google Scholar]
- Yamamoto S., Wu Z., Russnes H.G., Takagi S., Peluffo G., Vaske C., Zhao X., Moen Vollan H.K., Maruyama R., Ekram M.B., et al. 2014. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell. 25:762–777. 10.1016/j.ccr.2014.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L.C., McDonald D.W., and Hendzel M.J.. 2013. Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following γ-irradiation. J. Biol. Chem. 288:21376–21388. 10.1074/jbc.M113.491514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P., Canas B., Pappin D., and Kouzarides T.. 2002. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J. Biol. Chem. 277:11621–11624. 10.1074/jbc.C200045200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.