Abstract
Antibiotic treatment has two conflicting effects: the desired, immediate effect of inhibiting bacterial growth and the undesired, long-term effect of promoting the evolution of resistance. Although these contrasting outcomes seem inextricably linked, recent work has revealed several ways by which antibiotics can be combined to inhibit bacterial growth while, counterintuitively, selecting against resistant mutants. Decoupling treatment efficacy from the risk of resistance can be achieved by exploiting specific interactions between drugs, and the ways in which resistance mutations to a given drug can modulate these interactions or increase the sensitivity of the bacteria to other compounds. Although their practical application requires much further development and validation, and relies on advances in genomic diagnostics, these discoveries suggest novel paradigms that may restrict or even reverse the evolution of resistance.
Antibiotics are among the most important tools in medicine, but their efficacy is threatened by the evolution of resistance. Since the earliest days of antibiotics, resistance has been observed and recognized as a challenge (1). Today, many first-generation antibiotics are all but ineffective (2). We have thus far avoided a crisis through the continued modification of existing compounds and the discovery of new antibiotic classes. However, while resistance rates continue to rise, the rate of antibiotic discovery has dropped substantially (3, 4). Today, resistance claims over 25,000 lives in the European Union and 23,000 lives in the United States every year (2,5). In addition to discovering new antibiotics, we must therefore prioritize the development of methods addressing the evolution of resistance (6, 7). In particular, we need to devise new strategies for antimicrobial treatments that could limit, redirect, and perhaps even reverse the course of resistance evolution.
Bacteria evolve resistance to antibiotics by one of two routes: spontaneous mutation and horizontal gene transfer. Spontaneous mutations can confer resistance to an antibiotic by modifying the antibiotic’s target or its expression level, or by up-regulating resistance genes, such as those encoding efflux pumps (8–10). Alternatively, bacteria can acquire dedicated resistance genes through horizontal gene transfer. These genes may encode specialized antibiotic degradation enzymes, efflux pumps, target protection proteins, or bypass pathways (e.g., supply mechanisms for alternative cell wall synthesis pathways) (8, 9). Once they have acquired the resistance gene or mutation, bacteria can continue to grow in the presence of antibiotics, while the growth of sensitive bacteria is halted. Resistant mutants quickly outnumber sensitive bacteria and thus rapidly spread throughout a population, eventually rendering the drug ineffective.
It has been hoped that in the absence of antibiotic pressure, the physiological cost of maintaining resistance would be strong enough to select for loss of the resistance allele, eventually leading to resensitization. In practice, such loss of resistance has not been widely observed for four reasons. First, with few exceptions (11), the fitness cost of resistance is often not large enough to be appreciably selected against, and thus resistance genes can remain in the population for years after removal of the drug (12–15). Second, even when the cost of resistance is large, it can be neutralized by compensatory mutations, or through genetic regulatory mechanisms that activate resistance only in the presence of the drug (16, 17). Third, sustained selection for the presence of a resistance gene with an antibiotic can lead to the accumulation of mutations that not only compensate for the cost of the resistance gene, but make its presence essential for growth even in the absence of the antibiotic (18). Finally, antibiotic resistance mutations can, in certain cases, confer increased virulence, giving the resistant mutant a fitness advantage in the absence of antibiotic selection (19, 20).
Thus, as a Sisyphean consequence of their desired short-term inhibition of growth, antibiotics ultimately lead to long-term selection for resistance. Recent theoretical and experimental studies indicate that with particular combinations of compounds, we could decouple the conflicting effects of antibiotic therapy. Thus, it should be possible to develop strategies that use combinations of antibiotics and other compounds to inhibit bacterial growth while minimizing or reversing selection for resistance to the individual components.
Several mechanisms have been studied for minimizing or inverting the selective advantage of antibiotic resistance. The most established approach so far is to administer antibiotics with molecules that inhibit a particular resistance mechanism, thus neutralizing the evolutionary advantage of resistant strains. More recent work has developed strategies that go beyond neutralizing resistance to actively selecting against it using evolutionary and physiological interactions between drugs (see Box 1 and Fig. 1). First, combinations of drugs that physiologically interact to have different effects when coadministered can be used to slow and even invert the evolution of resistance. Second, drug interactions that change as resistance evolves can be exploited to select against resistant mutants. Finally, there has been a resurgence of interest in evolutionary trade-offs between resistances to individual compounds, in which one compound may channel evolution toward increased sensitivity to another compound. Below, we review these strategies and their potential for inhibiting the evolution of resistance. Several complementary strategies for evolutionarily robust bacterial inhibition have been suggested, including suppression of virulence (21–23), persistence (24), and quorum sensing (25, 26), as well as novel targeting strategies to allow higher effective doses (27); however, they are outside the scope of this review.
Box 1. Physiological and evolutionary drug interactions.
Physiological interactions: synergy, antagonism, and suppression
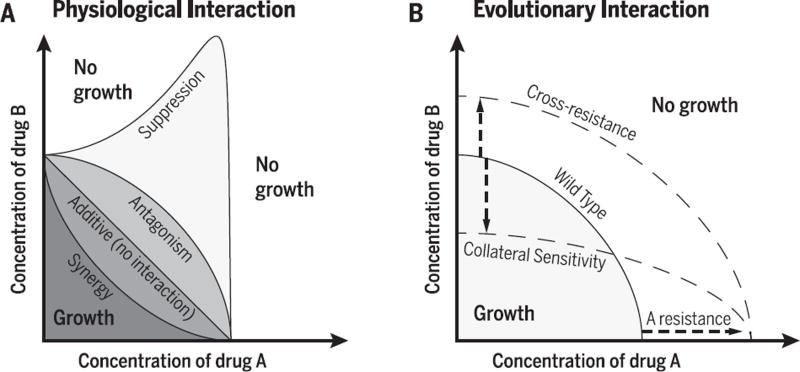
Antibiotics used in combination can interact to synergize, antagonize, or even to suppress each other’s effects (Fig. 1A). Antibiotic drug interactions appear when the combined inhibitory effect of two drugs is larger or smaller than expected based on an additive model (76,104,105). A common way to understand drug interactions is to consider the shape of the line in the two-drug concentration space, beyond which the bacteria are fully inhibited (Fig. 1A) (46). The null expectation for no interaction is that this isobole appears straight, i.e., the inhibitory power of the combinations depends only on the sum of the two antibiotics’ concentrations. Synergistic drug combinations are more inhibitory than this null expectation, whereas antagonistic combinations require higher concentrations to achieve the same degree of inhibition (concave and convex lines, respectively; Fig. 1A). An extreme type of antagonism appears when the combined effect of two drugs is weaker not only compared to the null additive expectation, but also weaker than the effect of one of the drugs alone. These are termed “suppressive” drug interactions and appear as a nonmonotonic inhibitory line, where the addition of a drug can in fact relieve growth inhibition (Fig. 1A) (106).
Recent technical developments have enabled broad systematic efforts to identify drug interactions and their underlying mechanisms. These studies have revealed synergistic, antagonistic, and suppressive interactions among pairs of antibiotics (106–108), as well as interactions between antibiotics and compounds with little or no antimicrobial activity (31,109–114). Analysis of pairwise interaction networks between multiple drugs have shown that drugs with the same mode of action have broadly the same interaction profile (106), suggesting that drug interactions operate through the core physiology of the cell and not through direct chemical interaction of the compounds. Although for many drug combinations the specific mechanism of interaction is not fully understood, recent work has elucidated the mechanisms behind several interactions: Simultaneous inhibition of different steps in a pathway causes a synergistic interaction between trimethoprim and sulfa drugs (115), nonoptimal gene regulation in the face of antibiotic inhibition results in a suppressive interaction between tetracyclines and fluoroquinolones (116), and mutations in polysaccharide and adenosine 5´-triphosphate (ATP) synthesis reshape a variety of interactions between antibiotic pairs (44).
Evolutionary interactions: cross-resistance and collateral sensitivity
Spontaneous mutations or acquired genes conferring resistance to one antibiotic can increase or decrease the resistance to another antibiotic (Fig. 1B) (67, 68,117). These positive and negative evolutionary interactions between antibiotics are termed “cross-resistance” and “collateral sensitivity” (or negative cross-resistance), respectively. For spontaneous resistance mutations, these evolutionary interactions have been mapped systematically, and both positive and negative cross-resistance interactions have been found between many pairs of antibiotics (56, 57). This phenomenon is not unique to bacteria and antibiotics; it has been seen in malaria (118, 119), HIV therapies (120), cancer treatments (121), and pesticide resistance in plants (122,123). Importantly, unlike physiological interactions, cross-resistance does not require drugs to be applied in combination, but is a function of the evolutionary response to a single antibiotic.
Recent surveys of cross-resistance interactions found that, as expected, the cross-resistance interactions between drugs in the same class tend to be positive (56, 57), although there are important exceptions (70, 71). Negative cross-resistance is seen frequently with resistance to aminoglycoside antibiotics, resulting from a change in the proton motive force associated with resistance (56, 57). More broad principles of when and in what environments cross-resistance interactions should occur, are not yet known.
Fig. 1. Physiological interactions and cross-resistance.
(A) Isoboles of minimum inhibitory concentration (MIC) are shown in the two-drug concentration space for different drug interactions. The MIC of each drug alone occurs where the isobole intersects each drug axis. When the effect of the two drugs is equal to the effect expected when combining two identical drugs, the shape of the MIC line is linear and the drugs are said to be noninteracting (104). Synergistic drugs require less-than-expected concentrations, corresponding to a concave MIC line, whereas antagonistic interactions require higher drug concentrations, producing a convex line. Finally, drug interactions are suppressive when their effect in combination is less than that of one of the drugs alone, appearing as a nonmonotonic isobole. (B) Cross-resistance and collateral sensitivity: A mutation or acquired gene conferring resistance to drug A can also increase resistance (positive cross-resistance) or decrease resistance (negative cross-resistance or collateral sensitivity) to drug B without otherwise changing the shape of the interaction.
Resistance mechanism inhibitors
Perhaps the most direct way to bypass resistance is to block the resistance mechanism. Resistance is frequently conferred by dedicated efflux pumps or antibiotic-degrading enzymes, which in turn can be countered by compounds that inhibit the resistance machinery (28, 29). To use this strategy therapeutically, an antibiotic is delivered concurrently with resistance-inhibiting compounds; for example, a β-lactam antibiotic paired with an inhibitor of β-lactamase (a resistance enzyme that degrades β-lactams). This allows the antibiotic to kill both resistant and susceptible strains, thereby potentiating the efficacy of the drug and diminishing the selective advantage of the resistance gene (28).
Compounds have been discovered that inhibit a large variety of resistance mechanisms (29). The most clinically successful examples are the pairings of amoxicillin-clavulanic acid, ampicillin-sulbactam, and pipericillin-tazobactam to block serine β-lactamases (28, 30). Recently, this principle has been expanded to metallo-β-lactamases with the discovery that aspergillomarasmine A inhibits NDM-1 and VIM-2, two clinically important enzymes that degrade β-lactams, including carbapenem antibiotics (31). Inhibitors of aminoglycoside-modifying enzymes have also been synthesized (32, 33). Further, high-throughput screening (34) and medicinal chemistry efforts (35) have yielded leads for inhibitors of ErmC methyltransferase, which confers macrolide resistance. Lastly, several studies have identified inhibitors of different efflux pumps, a major mode of resistance across antibiotic classes (36–41).
Some of these inhibitors are produced by the same bacterial species that synthesize the antibiotic whose resistance mechanism they inhibit, suggesting an evolutionary advantage to their use in combination. Clavulanic acid, a β-lactamase inhibitor, is produced by Streptomyces clavuligerus, which also produces several β-lactam antibiotics (42). Similarly, Berberis fremontii makes both the antibiotic berberine and the efflux pump inhibitor, 5′-methoxyhydnocarpin, that blocks berberine export (40). It is intriguing to hypothesize that these species evolved not only antibiotic production, but also ways to preserve the activity of those antibiotics by suppressing resistance mechanisms. Hence, known antibiotic producers may be fruitful sources of resistance mechanism inhibitors.
Although inhibitors have been found for several resistance mechanisms, in clinical practice the success of resistance mechanism inhibitors has been limited to serine β-lactamases (28, 30). Several barriers limit the broader application of resistance inhibitors, including drug toxicity, pharmacokinetic differences between the inhibitor and the antibiotic, and an inhibitor’s specificity for a particular resistance mechanism. Even when successfully implemented, these approaches are not resilient to the evolutionary process.
While resistance mechanism inhibitors can suppress or bypass specialized bacterial resistance machinery, they are themselves subject to resistance. Most resistance mechanism inhibitors are specific to one class of degradation enzymes or pumps, and therefore their widespread use can select for inhibitor-resistant variants within the class, or for alternate resistance mechanisms (28). For example, the β-lactamase inhibitors clavulanic acid, sulbactam, and tazobactam are ineffective against AmpC β-lactamases and metallo-β-lactamases, and over the years have selected for inhibitor-resistant variants of TEM β-lactamases (28, 43).
Critically, resistance mechanism inhibitors, while neutralizing the advantage of resistant bacteria, do not necessarily put them at a competitive disadvantage. Thus, while these inhibitors restore the efficacy of the antibiotic, they do not reduce the relative prevalence of resistance within a patient or in the population. Without negative selective pressure, the resistant strain will remain in the population, even in the absence of the antibiotic (12–15).
Selection inversion
There are several strategies to combine multiple physiologically or evolutionarily interacting antibiotics, not only to neutralize the selective advantage of resistance but also to impose a direct cost on resistance. The evolution of multidrug resistance often requires the sequential accumulation of resistance to each of the individual drugs. It is therefore important to find out whether single-drug-resistance steps would be selected for or against in a multidrug environment. The result critically depends on the physiological and evolutionary interactions between the drugs (Box 1).
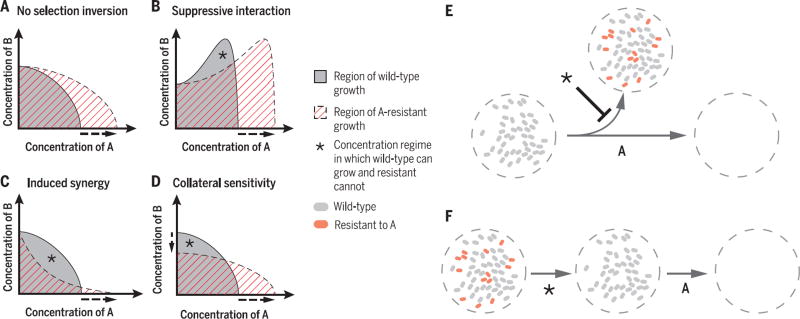
With some notable exceptions, single-drug-resistant mutants maintain the same drug interactions and the same resistances to other drugs as their drug-sensitive parents (Fig. 2A) (44). This is because, to a first approximation, bacteria acquiring resistance to one drug (e.g., via a spontaneous target mutation or a horizontally transferred drug efflux pump) behave as if they were exposed to less of that drug. The region of growth for resistant bacteria in a two-drug concentration space would therefore be similar in shape to the region of growth for their drug-sensitive parent, except stretched toward higher concentrations along the drug axis to which the bacteria are resistant (Fig. 2, A and B, horizontal axis). Thus, normally the regime where the resistant bacteria can grow fully encompasses the regime where the sensitive bacteria can grow (Fig. 2A); there is no combination of the two drugs in which the sensitive bacteria outcompete the resistant mutants.
Fig. 2. Selection inversion approaches and potential strategies.
(A) In typical drug interactions, the region of growth of a single-drug–resistant mutant (e.g., A-resistant, dashed area) completely covers the region of growth of the drug-sensitive wild type (gray area), and the mutant therefore always outcompetes the wild type. (B to D) There are three principal ways for establishing a concentration regime (*) that selects against resistance: (B) When drug A suppresses drug B, the MIC isobole is nonmonotonic, and so scaling it along the A axis because of resistance leaves a selection-inverting regime. (C) An antagonistic interaction can become synergistic with the acquisition of resistance, making the mutant more sensitive to the combination. (D) Collateral sensitivity, when the MIC of drug B decreases as a result of resistance to drug A, allowing selection against resistance even in the absence of A. (E) Using selection inversion approaches on a nonresistant population can decrease the probability of resistance evolution and make long-term therapy more likely to succeed. (F) Selection against resistance can also be used as part of a two-phase strategy against a population with resistant mutants. The drug-resistant mutants are selected out of the population in the first phase, allowing a previously ineffective antibiotic to be used in the second.
There are, however, three primary ways by which antibiotic combinations can impose a direct cost on resistance and thus select against drug-resistant strains. First, when one drug suppresses another, bacteria becoming resistant to the first drug lose its protective effect and can thus be inhibited more strongly by the second drug than their sensitive ancestors (45). In such a suppressive drug pair, the region of concentration allowing growth has a nonmonotonic shape, and therefore stretching this region toward higher drug concentration as a result of resistance to that drug leaves behind a range of concentrations in which the sensitive bacteria can grow and the resistant bacteria cannot (Fig. 2B). Second, if the mutation conferring resistance to one drug also increases the synergy between the two drugs, the mutant can again be inhibited more than its sensitive parent (46). This appears as a shape change in the resistant mutant’s region of growth, generating a combination of the drugs in which the wild type can grow while the mutant cannot (Fig. 2C). Finally, there may be an evolutionary trade-off, such that resistance to one drug generates sensitivity to the other (Fig. 2D, vertical axis). In this case, sensitivity to the second drug decreases with resistance to the first, allowing selection against resistance with the second drug alone.
Selection inversion using suppressive drug interactions
Suppressive drug interactions can select against single-drug-resistant mutants by creating a concentration regime that inhibits resistant, but not sensitive, bacterial growth. If drug A suppresses drug B, the line of inhibition in the two-drug space becomes nonmonotonic, such that the bacteria grow better at high concentrations of drug B when drug A is present. When the bacteria become resistant to drug A, this inhibitory line is stretched into higher concentrations of drug A (Fig. 2B). This leaves behind a concentration regime where the bacteria sensitive to A can grow, benefiting from the suppression of the effect of drug B by drug A, while the A-resistant ones cannot benefit. Therefore, within certain concentration regimes, suppressive combinations can cause drug-resistant mutants to lose out in competition with their drug-sensitive parental strains (45) (Fig. 2B).
Although suppressive interactions can fully invert the selective advantage of resistance, less extreme antagonistic interactions can also reduce, although not invert, the selective advantage of resistant mutants. Such antagonistically interacting drugs can therefore slow the rate of resistance evolution (47, 48). Conversely, synergistic interactions increase the selective advantage of resistant mutants, as becoming resistant to one drug relieves not only its own inhibitory effect but also its synergistic effect on the other drug (47, 48).
The evolutionary benefits of antagonistic or suppressive antibiotic combinations may not always warrant their additional costs. While reducing or inverting selection for resistance, antagonistic and suppressive combinations require higher doses of drugs and longer treatment time. This poses toxicity issues and can increase the potential for accumulating additional resistance mutations (49–53). Conversely, synergistic combinations increase the selective advantage of resistance mutations, but can also clear infections faster using less drug, reducing toxicity and the time in which resistance can arise (52). Thus, there is an optimal level of drug interaction, depending on the context of the infection, that balances clearance and prevention of resistance (52).
Deploying combination therapies clinically is likely to be complicated by the need to fine-tune concentrations of the drugs and by potential changes in their interactions. Differential absorption and penetration of the two drugs limit our ability to control their ratio in vivo and can create single-drug compartments that select for resistance (54). Further, interactions between the drugs can change within the body or as the target bacteria evolve, and there is no guarantee that interactions will remain suppressive (55).
Selection inversion using synergy-inducing drug pairs
Drug combinations can invert the selective pressure to favor sensitivity if their interaction becomes more synergistic in resistant mutants than in the sensitive parental strain. Although, as discussed above, the type of interaction between two drugs typically does not change with the acquisition of resistance (Fig. 2, A and B), in certain cases a resistant allele may not only confer resistance to a given drug but also change its interaction with other drugs (46, 55). If the drug interaction becomes more synergistic, there may exist a concentration regime where the susceptible strain can grow while the resistant strain cannot (Fig. 2C). A comprehensive study examining the pair-wise interactions of six antibiotics on a library of nonessential Escherichia coli gene deletion mutants showed that the shape of interactions is sometimes modified by inhibition of cellular functions, suggesting that antibiotic interactions can indeed change with the acquisition of particular mutations (44). This principle has been established in other contexts—for example, in non-small-cell lung cancer lines, the drugs gefitinib and 17-AAG interact antagonistically in susceptible cells, but interact synergistically when the cells gain gefitinib resistance, and therefore this combination may reduce the emergence of gefitinib-resistant mutants (46).
Applying this approach to antimicrobial therapy is currently only hypothetical. Unlike the suppressive drug pairs discussed above, or collaterally sensitive drug pairs discussed below, we currently do not know of a specific drug pair for which resistance to one drug consistently induces synergy to the combination. Further, should we find such pairs of antibiotics, the application of the approach will be challenging because of the need to fine tune the concentrations of two drugs simultaneously.
Selection inversion using collateral sensitivity
Collateral sensitivity, whereby resistance to one drug confers sensitivity to another (Box 1), provides a third mechanism for selection against resistance. Unlike suppression-based selection and synergy-inducing resistance mutations, which require the coadministration of two drugs for inverting the selective advantage of resistance, selection against resistance by collateral sensitivity occurs without coapplication of drugs (Fig. 2D, vertical axis), opening avenues for alternating drugs within the treatment of a single patient, or cycling drugs in a broader population context (56). Recent attention has been focused on the use of collateral sensitivity to select against spontaneous resistance mutations (56–59); its value in countering horizontally transferred resistance has been less explored (60–66).
Selection against spontaneous resistance
The pioneering work of Szybalski and Bryson in the early 1950s, testing whether spontaneous mutants resistant to different drugs grow in a range of other drugs, showed that cross-resistance and collateral sensitivity between antibiotics are common (67, 68). More recently, there have been several systematic screens for cross-resistance and collateral sensitivity (56–59). Some drug pairs show unidirectional collateral sensitivity, where resistance to one drug causes sensitivity to another, but not the other way around (56–59). In others, reciprocal collateral sensitivity appears, in which selection for resistance to either of the two drugs causes sensitivity to the other (56).
Collateral sensitivity can occur directly through mutations in the antibiotic target or indirectly through mutations in other cellular mechanisms (69). Drug pairs that target the same protein, such as quinolones and novobiocin (both DNA gyrase inhibitors), often show collateral sensitivity or cross-resistance (70, 71). Here, the interaction occurs directly through the target: The amino acid changes that provide resistance to one drug increase or decrease sensitivity to the other. Collateral sensitivity can also occur through less direct means. Several recent studies have highlighted the prevalent collateral sensitivity between aminoglycosides and other antibiotic classes (56, 57–59). Both the import of aminoglycosides and the export of multiple antibiotics through intrinsic efflux pumps require the proton motive force (72, 73). Therefore, when a strain evolves resistance to aminoglycosides by diminishing the proton motive force, it becomes more susceptible to other antibiotics, such as β-lactams, quinolones, and tetracyclines, that are normally exported by the proton-force–dependent pumps (57, 59, 74). By adapting to the presence of one antibiotic, bacteria effectively specialize and can become less resilient to other antibiotics.
Cross-resistance and collateral sensitivity may affect the potential for and change the rate of evolution of multidrug resistance (47, 75, 76). Collaterally sensitive drugs can be applied concurrently to reduce the selective advantage of single-drug resistant mutants, or alternatingly to either select for the wild-type over resistance or for de novo mutations that lose resistance (56). Concurrent or alternating application of drugs that have unidirectional collateral sensitivity can reduce the evolution of spontaneous resistance, compared with either of the drugs alone (55, 77). Furthermore, coadministration of drugs that have both synergy and collateral sensitivity can restore the activity of defunct antibiotics against resistant strains while preventing the evolution of further resistance (71).
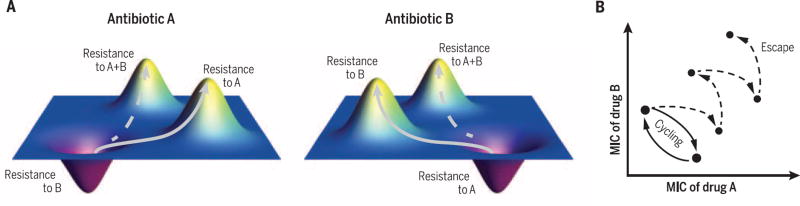
Considerable practical challenges stand in the way of exploiting collateral sensitivities between compounds to suppress resistance. All potential methods are likely to fail in the presence of mutations that confer resistance to both drugs (Fig. 3A) (78). Further, alternating application of a reciprocally collaterally sensitive pair of drugs suffers from two additional failure modes. First, if collaterally sensitive mutations confer more resistance to one drug than sensitivity to the other drug, such mutations would gradually accumulate resistance to both drugs (Fig. 3B). Second, mutations may not combine additively, and it is possible that the likelihood of collaterally sensitive mutations is reduced following an initial resistance mutation. Translating these methods into effective clinical strategies will require a more comprehensive and precise accounting of the spectrum of resistance mutations and their interactions.
Fig. 3. The efficacy and potential failure of cycling collaterally sensitive antibiotics.
(A) Fitness landscapes in collaterally sensitive antibiotics. Genotypes that are resistant to drug A or drug B appear as fitness peaks when the environment contains the drug to which they are resistant but as fitness valleys in the other drug treatment. In principle, alternating the drugs can lead to a cycle of evolution switching between these genotypes (solid arrows). However, doubly resistant mutants can evade this trap (dashed arrows). (B) Two possible evolutionary trajectories in the MICs of component drugs in antibiotic cycling. Ideally, resistance will alternate between two states (solid arrows). However, repeated accumulation of resistance mutations can also create double-resistance, even in the case where each individual mutation induces collateral sensitivity (dashed arrow).
Preliminary attempts to clinically deploy sequential antibiotic therapies have seen mixed results (78–81). Both theoretical and clinical studies have found that strict regimens of hospital-wide antibiotic cycling do not improve clinical outcomes (78, 80, 81). Results have been more promising within the context of treating a single patient. An “adjustable cycling” protocol, in which antibiotics are changed based on the patient’s condition, can be effective in reducing the evolution of resistance (79). Beyond pairs of drugs, more complex regimens, with multiple steps and decisions based on the specific mutations that emerge, will likely be needed to fully invert the selection for antibiotic resistance.
Selection against acquired resistance
Whereas collateral sensitivity mediated through spontaneous mutations has been mapped extensively, there are fewer examples of collateral sensitivity caused by dedicated resistance genes and cassettes spread by horizontal gene transfer (60–66). A resistance mechanism confers an advantage in the presence of the antibiotic it targets, but may lead to sensitivity to other compounds. Almost all existing examples of such selection-inverting compounds center on the tet efflux pumps, which confer tetracycline resistance, but also make bacteria more susceptible to aminoglycosides, salt stress, and fusaric acid (61–65). A recent screen for selection-inverting compounds, measuring the relative growth of competing resistant and sensitive strains, found that some soil microbes produce compounds that select against tetA (60). The generality of this screening technique will potentially allow systematic identification of compounds that can select against other horizontally transferable antibiotic resistance mechanisms. Furthermore, the same mobility that allows resistance genes to spread rapidly would also accelerate their loss, relative to spontaneous mutations (73, 82, 83). New selection-inverting compounds would therefore open up possibilities for novel treatment regimens that can convert a population carrying a resistance cassette back to drug sensitivity, potentially increasing sensitivity to other antibiotics whose resistance genes were on the same genetic cassette.
Translation and the need for anticipatory diagnostics
One can envision several different ways in which selection-inverting approaches can be used. For example, since becoming resistant to the combined treatment often requires the stepwise accumulation of mutations providing resistance to each of the individual components, using methods that select against these single-drug resistant mutants can reduce the chance that a doubly resistant mutant appears during treatment (Fig. 2E) (52). These approaches can also be used in a “one-two punch” treatment strategy: First select against resistance to eliminate single-drug resistant mutants from a population, and then follow up with the now-effective classical antibiotic (Fig. 2F). It is conceivable that even when a resistant allele is fully fixed in the population, applying a selective pressure against it would select for mutations that delete it or disrupt its function and lead to its long-term loss. However, for selection inversion strategies to be practical, improved diagnostics of resistance mechanisms are needed.
To deploy the correct strategy against a specific mechanism of resistance, we must be able to differentiate at the point of diagnosis what antibiotics an infection is already resistant to and the potential it has to develop resistance. With a few notable exceptions (84–88), the diagnosis of microbial infections has not changed conceptually over the past several decades: The pathogen is cultured and its growth in the presence of a panel of antibiotics is tested. This culturing-phenotyping approach is not only slow, but only assesses the current abilities of the microbe and not its evolutionary potential. State-of-the-art diagnostic technologies are currently too cumbersome for clinical practice, and substantial technical challenges must be overcome to enable their widespread use. However, with anticipated improvements, technologies for the rapid genomic sequencing of pathogens have the potential to enable faster diagnosis and prediction of antibiotic resistance.
Genomic analysis can potentially detect both the resistance profile of the bug and the specific resistance genes involved (89), allowing more targeted use of approaches that inhibit or select against the specific resistance mechanism. As more clinical samples are phenotyped and sequenced, we expect that machine learning techniques will rapidly improve in their ability to predict antibiotic resistance from genomic data. It is possible that by correlating genotypes and phenotypes on a large scale, these approaches will be able to identify resistance conferred not only by known resistance genes, but also to identify novel resistance genotypes. Furthermore, it is possible that such approaches may be able to predict not only what drugs a pathogen is currently capable of resisting, but also its past exposures and future capacity to evolve resistance. As pathogens evolve at the population level, and even within a single patient, the resulting diversity of accumulated mutations allows reconstruction of their phylogeny and can reveal their past history of adaptation to antibiotics and other selective pressures (90, 91). Diagnostics may even be able to predict future evolution and the potential to evolve resistance. For example, if a pathogen’s genome is a few mutations away from resistance, we might predict that, while it is not currently resistant, it could become resistant if certain drugs are used. The predictability of the evolutionary process likely varies between drugs, but for some, spontaneous resistance appears to evolve through a limited range of pathways (91–93). However, higher-order interactions among mutations could make long-term evolution substantially harder to predict (93).
Beyond predictive diagnostics at the single-patient level, sequencing-based diagnostics may allow us to predict evolution at a population or epidemic level, ideally before a resistance mechanism even appears in a clinic (4). To do this, we must monitor for the emergence and spread of resistance mechanisms (4, 94, 95). Widespread antibiotic use in agriculture, cosmetics, and medicine can bias the genetic content of the ambient microbiome toward the prevalence of resistance genes, increasing the chance of horizontal gene transfer of resistance to clinical pathogens (96–98). This can foster low-level resistance, which decreases the additional resistance required to reach clinically significant levels and makes high levels of resistance possible via more evolutionary paths (99, 100). Through sequence-based predictive diagnostic techniques, we should be able to detect the emergence of resistance, its source, and its rate of spread and possibly begin to fight emerging resistance before it enters the clinic.
Challenges and outlook
The practical application of selection-inverting strategies faces major challenges. The majority of multidrug interaction and collateral sensitivity studies have been performed in vitro with Escherichia coli and need to be validated in animal models and clinical isolates. The specific uses of these strategies split broadly into two types of clinical case: that of a single patient with a long-term infection (e.g., tuberculosis or methicillin-resistant Staphylococcus aureus) or a resistant pathogen circulating in the population (e.g., vancomycin-resistant enterococci or cephalosporin-resistant gonorrhea), each with its own practical and ethical challenges. In a long-term single-patient infection, we must balance the risk of prolonging treatment against the risk of treatment failure from the evolution of resistance. In the case of a pathogen circulating in the population, the balance is between the efficacy of clearance and the population-level need for long-term suppression of resistance. Thus, even if a treatment strategy can suppress the evolution of resistance, it is unlikely to be widely adopted clinically unless it also provides increased survival on a per-patient basis. It could therefore be advantageous to begin trials of these approaches for suppressing circulating resistance in a veterinary setting, where the health of the herd, rather than of an individual animal, is of primary concern and the ethical concerns are less acute than for human clinical applications. Another difficulty lies in the simultaneous delivery of multicompound treatments; unequal absorption and penetration may lead to pockets of single-drug exposure and thereby promote resistance (54). To circumvent this difficulty, hybrid antibiotics linking existing compounds have been proposed, but their in vivo efficacy and evolutionary effects have undergone only limited testing (101, 102). Further, selection against resistance is dependent on the consistency of drug interactions. As microbes face vastly different environments in a host than in vitro, with different nutrient supplies, a range of immune responses, competition with other microbes, phenotypic variability [e.g., persister cells (24, 103)], and their own evolution, there is no immediate guarantee that the interactions observed in vitro are sufficiently stable to reliably direct evolution in vivo.
Ultimately, treating resistance will require a portfolio of strategies including drug discovery, resistance monitoring, and combinations of novel methods to invert the selection for resistance. We are in dire need of techniques to channel pathogens toward less evolvable genotypes. It is no longer sustainable nor sufficient to treat antibiotic-resistant infections simply in response to their current resistance phenotype. Rather, antimicrobial strategies are required that anticipate the evolutionary potential of the infection, and both treat and channel it away from multi-drug resistance. We can be certain bacteria will adapt to our treatments, and so our strategies of combatting resistance must also evolve to remain one step ahead.
Acknowledgments
We thank T. Bollenbach, W. P. Hanage, M. Elowitz, J. Jiao, and D. Van Valen for their thoughtful comments and input. This work was supported in part by U.S. National Institutes of Health grant R01-GM081617, European Research Council FP7 ERC Grant 281891, and F. Hoffmann-La Roche Ltd.
References
- 1.Abraham EP, et al. Further observations on penicillin. Lancet. 1941;238:177–189. [Google Scholar]
- 2.Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013. 2013 www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf.
- 3.Laxminarayan R. Antibiotic effectiveness: Balancing conservation against innovation. Science. 2014;345:1299–1301. doi: 10.1126/science.1254163. pmid: 25214620. [DOI] [PubMed] [Google Scholar]
- 4.President’s Council of Advisors on Science, Technology, “Report to the President on Combating Antibiotic Resistance”. Executive Office of the President; 2014. pp. 1–78. [Google Scholar]
- 5.World Health Organization. Antimicrobial resistance: Global report on surveillance. World Health Organization; Geneva, Switzerland: 2014. [Google Scholar]
- 6.McClure NS, Day T. A theoretical examination of the relative importance of evolution management and drug development for managing resistance. Proc. Biol. Sci. 2014;281:20141861–20141861. doi: 10.1098/rspb.2014.1861. pmid: 25377456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush K, et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011;9:894–896. doi: 10.1038/nrmicro2693. pmid: 22048738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. pmid: 25435309. [DOI] [PubMed] [Google Scholar]
- 9.Wright GD. Molecular mechanisms of antibiotic resistance. Chem. Commun. (Camb.) 2011;47:4055–4061. doi: 10.1039/c0cc05111j. pmid: 21286630. [DOI] [PubMed] [Google Scholar]
- 10.Palmer AC, Kishony R. Opposing effects of target overexpression reveal drug mechanisms. Nat. Commun. 2014;5:4296. doi: 10.1038/ncomms5296. pmid: 24980690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vincent BM, Lancaster AK, Scherz-Shouval R, Whitesell L, Lindquist S. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLOS Biol. 2013;11:e1001692–e17. doi: 10.1371/journal.pbio.1001692. pmid: 24204207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sjölund M, Wreiber K, Andersson DI, Blaser MJ, Engstrand L. Long-term persistence of resistant Enterococcus species after antibiotics to eradicate Helicobacter pylori. Ann. Intern. Med. 2003;139:483–487. doi: 10.7326/0003-4819-139-6-200309160-00011. pmid: 13679325. [DOI] [PubMed] [Google Scholar]
- 13.Sjölund M, Tano E, Blaser MJ, Andersson DI, Engstrand L. Persistence of resistant Staphylococcus epidermidis after single course of clarithromycin. Emerg. Infect. Dis. 2005;11:1389–1393. doi: 10.3201/eid1109.050124. pmid: 16229767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Gelder L, et al. Combining mathematical models and statistical methods to understand and predict the dynamics of antibiotic-sensitive mutants in a population of resistant bacteria during experimental evolution. Genetics. 2004;168:1131–1144. doi: 10.1534/genetics.104.033431. pmid: 15579675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersson DI, Hughes D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. pmid: 20208551. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen TN, Phan QG, Duong LP, Bertrand KP, Lenski RE. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol. Biol. Evol. 1989;6:213–225. doi: 10.1093/oxfordjournals.molbev.a040545. pmid: 2560115. [DOI] [PubMed] [Google Scholar]
- 17.Foucault M-L, Courvalin P, Grillot-Courvalin C. Fitness cost of VanA-type vancomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2009;53:2354–2359. doi: 10.1128/AAC.01702-08. pmid: 19332680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenski RE, Simpson SC, Nguyen TT. Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J. Bacteriol. 1994;176:3140–3147. doi: 10.1128/jb.176.11.3140-3147.1994. pmid: 8195066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skurnik D, et al. Enhanced in vivo fitness of carbapenem-resistant oprD mutants of Pseudomonas aeruginosa revealed through high-throughput sequencing. Proc. Natl. Acad. Sci. U.S.A. 2013;110:20747–20752. doi: 10.1073/pnas.1221552110. pmid: 24248354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roux D, et al. Fitness cost of antibiotic susceptibility during bacterial infection. Sci. Transl. Med. 2015;7:297ra114. doi: 10.1126/scitranslmed.aab1621. pmid: 26203082. [DOI] [PubMed] [Google Scholar]
- 21.Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: Can we make evolution-proof drugs? Nat. Rev. Microbiol. 2014;12:300–308. doi: 10.1038/nrmicro3232. pmid: 24625893. [DOI] [PubMed] [Google Scholar]
- 22.Ross-Gillespie A, Weigert M, Brown SP, Kümmerli R. Gallium-mediated siderophore quenching as an evolutionary robust antibacterial treatment. Evol Med Public Health. 2014;2014:18–29. doi: 10.1093/emph/eou003. pmid: 24480613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swoboda JG, et al. Discovery of a small molecule that blocks wall teichoic acid biosynthesis in Staphylococcus aureus. ACS Chem. Biol. 2009;4:875–883. doi: 10.1021/cb900151k. pmid: 19689117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith PA, Romesberg FE. Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat. Chem. Biol. 2007;3:549–556. doi: 10.1038/nchembio.2007.27. pmid: 17710101. [DOI] [PubMed] [Google Scholar]
- 25.Rutherford ST, Bassler BL. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harbor Perspect. Med. 2012;2:a012427–a012427. doi: 10.1101/cshperspect.a012427. pmid: 23125205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen G, et al. A strategy for antagonizing quorum sensing. Mol. Cell. 2011;42:199–209. doi: 10.1016/j.molcel.2011.04.003. pmid: 21504831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehar SM, et al. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature. 2015;527:323–328. doi: 10.1038/nature16057. pmid: 26536114. [DOI] [PubMed] [Google Scholar]
- 28.Drawz SM, Bonomo RA. Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. pmid: 20065329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright GD. Resisting resistance: New chemical strategies for battling superbugs. Chem. Biol. 2000;7:R127–R132. doi: 10.1016/s1074-5521(00)00126-5. pmid: 10873842. [DOI] [PubMed] [Google Scholar]
- 30.Ball P. Conclusions: The future of antimicrobial therapy-Augmentin® and beyond. Int. J. Antimicrob. Agents. 2007;30:139–141. doi: 10.1016/j.ijantimicag.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 31.King AM, et al. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature. 2014;510:503–506. doi: 10.1038/nature13445. pmid: 24965651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen NE, Alborn WE, Jr, Hobbs JN, Jr, Kirst HA. 7-Hydroxytropolone: An inhibitor of aminoglycoside-2″-O-adenylyltransferase. Antimicrob. Agents Chemother. 1982;22:824–831. doi: 10.1128/aac.22.5.824. pmid: 6185088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roestamadji J, Grapsas I, Mobashery S. Mechanism-based inactivation of bacterial aminoglycoside 3′-phosphotransferases. J. Am. Chem. Soc. 1995;117:80–84. [Google Scholar]
- 34.Clancy J, et al. Assays to detect and characterize synthetic agents that inhibit the ErmC methyltransferase. J Antibiot. 1995;48:1273–1279. doi: 10.7164/antibiotics.48.1273. pmid: 8557568. [DOI] [PubMed] [Google Scholar]
- 35.Hajduk PJ, et al. Novel inhibitors of Erm methyltransferases from NMR and parallel synthesis. J. Med. Chem. 1999;42:3852–3859. doi: 10.1021/jm990293a. pmid: 10508434. [DOI] [PubMed] [Google Scholar]
- 36.Nelson ML, et al. Inhibition of the tetracycline efflux antiport protein by 13-thio-substituted 5-hydroxy-6-deoxytetracyclines. J. Med. Chem. 1993;36:370–377. doi: 10.1021/jm00055a008. pmid: 8426364. [DOI] [PubMed] [Google Scholar]
- 37.Renau TE, et al. Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J. Med. Chem. 1999;42:4928–4931. doi: 10.1021/jm9904598. pmid: 10585202. [DOI] [PubMed] [Google Scholar]
- 38.Neyfakh AA, Borsch CM, Kaatz GW. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob. Agents Chemother. 1993;37:128–129. doi: 10.1128/aac.37.1.128. pmid: 8431010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markham PN, Neyfakh AA. Inhibition of the multidrug transporter NorA prevents emergence of norfloxacin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1996;40:2673–2674. doi: 10.1128/aac.40.11.2673. pmid: 8913490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stermitz FR, Lorenz P, Tawara JN, Zenewicz LA, Lewis K. Synergy in a medicinal plant: Antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1433–1437. doi: 10.1073/pnas.030540597. pmid: 10677479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pagès J-M, Amaral L. Mechanisms of drug efflux and strategies to combat them: Challenging the efflux pump of Gram-negative bacteria. Biochim. Biophys. Acta. 2009;1794:826–833. doi: 10.1016/j.bbapap.2008.12.011. pmid: 19150515. [DOI] [PubMed] [Google Scholar]
- 42.Reading C, Cole M. Clavulanic acid: A beta-lactamase-inhiting beta-lactam from Streptomyces clavuligerus. Antimicrob. Agents Chemother. 1977;11:852–857. doi: 10.1128/aac.11.5.852. pmid: 879738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaïbi EB, Sirot D, Paul G, Labia R. Inhibitor-resistant TEM beta-lactamases: Phenotypic, genetic and biochemical characteristics. J. Antimicrob. Chemother. 1999;43:447–458. doi: 10.1093/jac/43.4.447. pmid: 10350372. [DOI] [PubMed] [Google Scholar]
- 44.Chevereau G, Bollenbach T. Systematic discovery of drug interaction mechanisms. Mol. Syst. Biol. 2015;11:807. doi: 10.15252/msb.20156098. pmid: 25924924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chait R, Craney A, Kishony R. Antibiotic interactions that select against resistance. Nature. 2007;446:668–671. doi: 10.1038/nature05685. pmid: 17410176. [DOI] [PubMed] [Google Scholar]
- 46.Wood KB, Wood KC, Nishida S, Cluzel P. Uncovering scaling laws to infer multidrug response of resistant microbes and cancer cells. Cell Rep. 2014;6:1073–1084. doi: 10.1016/j.celrep.2014.02.007. pmid: 24613352. [DOI] [PubMed] [Google Scholar]
- 47.Hegreness M, Shoresh N, Damian D, Hartl D, Kishony R. Accelerated evolution of resistance in multidrug environments. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13977–13981. doi: 10.1073/pnas.0805965105. pmid: 18779569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michel JB, Yeh PJ, Chait R, Moellering RC, Jr, Kishony R. Drug interactions modulate the potential for evolution of resistance. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14918–14923. doi: 10.1073/pnas.0800944105. pmid: 18815368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chow JW, Yu VL. Combination antibiotic therapy versus monotherapy for gram-negative bacteraemia: A commentary. Int. J. Antimicrob. Agents. 1999;11:7–12. doi: 10.1016/s0924-8579(98)00060-0. pmid: 10075272. [DOI] [PubMed] [Google Scholar]
- 50.Safdar N, Handelsman J, Maki DG. Does combination antimicrobial therapy reduce mortality in Gram-negative bacteraemia? A meta-analysis. Lancet. 2004;4:519–527. doi: 10.1016/S1473-3099(04)01108-9. pmid: 15288826. [DOI] [PubMed] [Google Scholar]
- 51.Tamma PD, Cosgrove SE, Maragakis LL. Combination therapy for treatment of infections with gram-negative bacteria. Clin. Microbiol. Rev. 2012;25:450–470. doi: 10.1128/CMR.05041-11. pmid: 22763634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torella JP, Chait R, Kishony R. Optimal drug synergy in antimicrobial treatments. PLOS Comput. Biol. 2010;6:e1000796. doi: 10.1371/journal.pcbi.1000796. pmid: 20532210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peña-Miller R, Lähnemann D, Schulenburg H, Ackermann M, Beardmore R. The optimal deployment of synergistic antibiotics: A control-theoretic approach. J. R. Soc. Interface. 2012;9:2488–2502. doi: 10.1098/rsif.2012.0279. pmid: 22628215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moreno-Gamez S, et al. Imperfect drug penetration leads to spatial monotherapy and rapid evolution of multidrug resistance. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E2874–E2883. doi: 10.1073/pnas.1424184112. pmid: 26038564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Munck C, Gumpert HK, Wallin AIN, Wang HH, Sommer MOA. Prediction of resistance development against drug combinations by collateral responses to component drugs. Sci. Transl. Med. 2014;6:262ra156. doi: 10.1126/scitranslmed.3009940. pmid: 25391482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imamovic L, Sommer MOA. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 2013;5:204ra132. doi: 10.1126/scitranslmed.3006609. pmid: 24068739. [DOI] [PubMed] [Google Scholar]
- 57.Lázár V, et al. Bacterial evolution of antibiotic hypersensitivity. Mol. Syst. Biol. 2013;9:700. doi: 10.1038/msb.2013.57. pmid: 24169403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oz T, et al. Strength of selection pressure is an important parameter contributing to the complexity of antibiotic resistance evolution. Mol. Biol. Evol. 2014;31:2387–2401. doi: 10.1093/molbev/msu191. pmid: 24962091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki S, Horinouchi T, Furusawa C. Prediction of antibiotic resistance by gene expression profiles. Nat. Commun. 2014;5:5792. doi: 10.1038/ncomms6792. pmid: 25517437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chait R, Shrestha S, Shah AK, Michel J-B, Kishony R. A differential drug screen for compounds that select against antibiotic resistance. PLOS ONE. 2010;5:e15179. doi: 10.1371/journal.pone.0015179. pmid: 21209699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bochner BR, Huang HC, Schieven GL, Ames BN. Positive selection for loss of tetracycline resistance. J. Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. pmid: 6259126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Podolsky T, Fong ST, Lee BT. Direct selection of tetracycline-sensitive Escherichia coli cells using nickel salts. Plasmid. 1996;36:112–115. doi: 10.1006/plas.1996.0038. pmid: 8954882. [DOI] [PubMed] [Google Scholar]
- 63.Merlin TL, Davis GE, Anderson WL, Moyzis RK, Griffith JK. Aminoglycoside uptake increased by tet gene expression. Antimicrob. Agents Chemother. 1989;33:1549–1552. doi: 10.1128/aac.33.9.1549. pmid: 2684011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merlin TL, Corvo DL, Gill JH, Griffith JK. Enhanced gentamicin killing of Escherichia coli by tet gene expression. Antimicrob. Agents Chemother. 1989;33:230–232. doi: 10.1128/aac.33.2.230. pmid: 2655531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Griffith JK, Kogoma T, Corvo DL, Anderson WL, Kazim AL. An N-terminal domain of the tetracycline resistance protein increases susceptibility to aminoglycosides and complements potassium uptake defects in Escherichia coli. J. Bacteriol. 1988;170:598–604. doi: 10.1128/jb.170.2.598-604.1988. pmid: 3276661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Q, et al. NB2001, a novel antibacterial agent with broad-spectrum activity and enhanced potency against beta-lactamase-producing strains. Antimicrob. Agents Chemother. 2002;46:1262–1268. doi: 10.1128/AAC.46.5.1262-1268.2002. pmid: 11959554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szybalski W, Bryson V. Genetic studies on microbial cross resistance to toxic agents. I. Cross resistance of Escherichia coli to fifteen antibiotics. J. Bacteriol. 1952;64:489–499. doi: 10.1128/jb.64.4.489-499.1952. pmid: 12999676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szybalski W. Genetic studies on microbial cross resistance to toxic agents. IV. Cross resistance of Bacillus megaterium to forty-four antimicrobial drugs. Appl. Microbiol. 1954;2:57–63. doi: 10.1128/am.2.2.57-63.1954. pmid: 13149144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lázár V, et al. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat. Commun. 2014;5:4352. doi: 10.1038/ncomms5352. pmid: 25000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chao L. An unusual interaction between the target of nalidixic acid and novobiocin. Nature. 1978;271:385–386. doi: 10.1038/271385a0. pmid: 340962. [DOI] [PubMed] [Google Scholar]
- 71.Gonzales PR, et al. Synergistic, collaterally sensitive β-lactam combinations suppress resistance in MRSA. Nat. Chem. Biol. 2015;11:855–861. doi: 10.1038/nchembio.1911. pmid: 26368589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taber HW, Mueller JP, Miller PF, Arrow AS. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 1987;51:439–457. doi: 10.1128/mr.51.4.439-457.1987. pmid: 3325794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. pmid: 17382878. [DOI] [PubMed] [Google Scholar]
- 74.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. pmid: 8550435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dragosits M, Mozhayskiy V, Quinones-Soto S, Park J, Tagkopoulos I. Evolutionary potential, cross-stress behavior and the genetic basis of acquired stress resistance in Escherichia coli. Mol. Syst. Biol. 2013;9:643. doi: 10.1038/msb.2012.76. pmid: 23385483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yeh PJ, Hegreness MJ, Aiden AP, Kishony R. Drug interactions and the evolution of antibiotic resistance. Nat. Rev. Microbiol. 2009;7:460–466. doi: 10.1038/nrmicro2133. pmid: 19444248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim S, Lieberman TD, Kishony R. Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance. Proc. Natl. Acad. Sci. U.S.A. 2014;111:14494–14499. doi: 10.1073/pnas.1409800111. pmid: 25246554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bonhoeffer S, Lipsitch M, Levin BR. Evaluating treatment protocols to prevent antibiotic resistance. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12106–12111. doi: 10.1073/pnas.94.22.12106. pmid: 9342370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abel zur Wiesch P, Kouyos R, Abel S, Viechtbauer W, Bonhoeffer S. Cycling empirical antibiotic therapy in hospitals: Meta-analysis and models. PLOS Pathog. 2014;10:e1004225–e13. doi: 10.1371/journal.ppat.1004225. pmid: 24968123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kollef MH. Is antibiotic cycling the answer to preventing the emergence of bacterial resistance in the intensive care unit? Clin. Infect. Dis. 2006;43(Suppl 2):S82–S88. doi: 10.1086/504484. pmid: 16894520. [DOI] [PubMed] [Google Scholar]
- 81.Pakyz AL, Farr BM. Rates of Stenotrophomonas maltophilia colonization and infection in relation to antibiotic cycling protocols. Epidemiol. Infect. 2009;137:1679–1683. doi: 10.1017/S0950268809002830. pmid: 19874637. [DOI] [PubMed] [Google Scholar]
- 82.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. pmid: 10817707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chopra I, Roberts M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. pmid: 11381101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Azimi L, et al. Tracing of false negative results in phenotypic methods for identification of carbapenemase by Real-time PCR. Gene. 2016;576:166–170. doi: 10.1016/j.gene.2015.10.008. pmid: 26456106. [DOI] [PubMed] [Google Scholar]
- 85.van der Zee A, et al. Multi-centre evaluation of real-time multiplex PCR for detection of carbapenemase genes OXA-48, VIM, IMP, NDM and KPC. BMC Infect. Dis. 2014;14:27. doi: 10.1186/1471-2334-14-27. pmid: 24422880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.De Beenhouwer H, et al. Rapid detection of rifampicin resistance in sputum and biopsy specimens from tuberculosis patients by PCR and line probe assay. Tuber. Lung Dis. 1995;76:425–430. doi: 10.1016/0962-8479(95)90009-8. pmid: 7496004. [DOI] [PubMed] [Google Scholar]
- 87.Telenti A, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–651. doi: 10.1016/0140-6736(93)90417-f. pmid: 8095569. [DOI] [PubMed] [Google Scholar]
- 88.Jonas D, Speck M, Daschner FD, Grundmann H. Rapid PCR-based identification of methicillin-resistant Staphylococcus aureus from screening swabs. J. Clin. Microbiol. 2002;40:1821–1823. doi: 10.1128/JCM.40.5.1821-1823.2002. pmid: 11980967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bradley P, et al. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat. Commun. 2015;6:10063. doi: 10.1038/ncomms10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lieberman TD, et al. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat. Genet. 2014;46:82–87. doi: 10.1038/ng.2848. pmid: 24316980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lieberman TD, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat. Genet. 2011;43:1275–1280. doi: 10.1038/ng.997. pmid: 22081229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Toprak E, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 2012;44:101–105. doi: 10.1038/ng.1034. pmid: 22179135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Palmer AC, et al. Delayed commitment to evolutionary fate in antibiotic resistance fitness landscapes. Nat. Commun. 2015;6:7385. doi: 10.1038/ncomms8385. pmid: 26060115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nordahl Petersen T, et al. Meta-genomic analysis of toilet waste from long distance flights; a step towards global surveillance of infectious diseases and antimicrobial resistance. Sci. Rep. 2015;5:11444. doi: 10.1038/srep11444. pmid: 26161690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Martínez JL, Coque TM, Baquero F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 2015;13:116–123. doi: 10.1038/nrmicro3399. pmid: 25534811. [DOI] [PubMed] [Google Scholar]
- 96.Wegener HC. Antibiotics in animal feed and their role in resistance development. Curr. Opin. Microbiol. 2003;6:439–445. doi: 10.1016/j.mib.2003.09.009. pmid: 14572534. [DOI] [PubMed] [Google Scholar]
- 97.Hammerum AM, Heuer OE. Human health hazards from antimicrobial-resistant Escherichia coli of animal origin. Clin. Infect. Dis. 2009;48:916–921. doi: 10.1086/597292. pmid: 19231979. [DOI] [PubMed] [Google Scholar]
- 98.Vieira AR, et al. Association between antimicrobial resistance in Escherichia coli isolates from food animals and blood stream isolates from humans in Europe: An ecological study. Foodborne Pathog. Dis. 2011;8:1295–1301. doi: 10.1089/fpd.2011.0950. pmid: 21883007. [DOI] [PubMed] [Google Scholar]
- 99.Chang Q, Wang W, Regev-Yochay G, Lipsitch M, Hanage WP. Antibiotics in agriculture and the risk to human health: How worried should we be? Evol Appl. 2015;8:240–247. doi: 10.1111/eva.12185. pmid: 25861382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kouyos RD, et al. The path of least resistance: Aggressive or moderate treatment? Proc. Biol. Sci. 2014;281:20140566–20140566. doi: 10.1098/rspb.2014.0566. pmid: 25253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pokrovskaya V, Baasov T. Dual-acting hybrid antibiotics: A promising strategy to combat bacterial resistance. Expert Opin. Drug Discov. 2010;5:883–902. doi: 10.1517/17460441.2010.508069. pmid: 22823262. [DOI] [PubMed] [Google Scholar]
- 102.Wang KK, et al. A hybrid drug limits resistance by evading the action of the multiple antibiotic resistance pathway. Mol. Biol. Evol. 2015 doi: 10.1093/molbev/msv243. doi: 10.1093/molbev/msv243; pmid: 26538141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature. 2014;513:418–421. doi: 10.1038/nature13469. pmid: 25043002. [DOI] [PubMed] [Google Scholar]
- 104.Loewe S. Die quantitation probleme der pharmakologie. Ergeb. Physiol. 1928;27:47–187. [Google Scholar]
- 105.Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat. Rev. Drug Discov. 2005;4:71–78. doi: 10.1038/nrd1609. pmid: 15688074. [DOI] [PubMed] [Google Scholar]
- 106.Yeh P, Tschumi AI, Kishony R. Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 2006;38:489–494. doi: 10.1038/ng1755. pmid: 16550172. [DOI] [PubMed] [Google Scholar]
- 107.Cokol M, et al. Large-scale identification and analysis of suppressive drug interactions. Chem. Biol. 2014;21:541–551. doi: 10.1016/j.chembiol.2014.02.012. pmid: 24704506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cokol M, et al. Systematic exploration of synergistic drug pairs. Mol. Syst. Biol. 2011;7:544. doi: 10.1038/msb.2011.71. pmid: 22068327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. pmid: 21562562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. pmid: 20811456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vega NM, Allison KR, Samuels AN, Klempner MS, Collins JJ. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. U.S.A. 2013;110:14420–14425. doi: 10.1073/pnas.1308085110. pmid: 23946425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Malik M, et al. Lethal synergy involving bicyclomycin: An approach for reviving old antibiotics. J. Antimicrob. Chemother. 2014;69:3227–3235. doi: 10.1093/jac/dku285. pmid: 25085655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ejim L, et al. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 2011;7:348–350. doi: 10.1038/nchembio.559. pmid: 21516114. [DOI] [PubMed] [Google Scholar]
- 114.Taylor PL, Rossi L, De Pascale G, Wright GD. A forward chemical screen identifies antibiotic adjuvants in Escherichia coli. ACS Chem. Biol. 2012;7:1547–1555. doi: 10.1021/cb300269g. pmid: 22698393. [DOI] [PubMed] [Google Scholar]
- 115.Nichols RJ, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. pmid: 21185072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bollenbach T, Quan S, Chait R, Kishony R. Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell. 2009;139:707–718. doi: 10.1016/j.cell.2009.10.025. pmid: 19914165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Szybalski W, Bryson V. Genetic studies on microbial cross-resistance to toxic agents. III. Cross-resistance of Mycobacterium ranae to twenty-eight antimycobacterial agents. Am. Rev. Tuberc. 1954;69:267–279. doi: 10.1164/art.1954.69.2.267. pmid: 13114644. [DOI] [PubMed] [Google Scholar]
- 118.Hastings MD, Sibley CH. Pyrimethamine and WR99210 exert opposing selection on dihydrofolate reductase from Plasmodium vivax. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13137–13141. doi: 10.1073/pnas.182295999. pmid: 12198181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lukens AK, et al. Harnessing evolutionary fitness in Plasmodium falciparum for drug discovery and suppressing resistance. Proc. Natl. Acad. Sci. U.S.A. 2014;111:799–804. doi: 10.1073/pnas.1320886110. pmid: 24381157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Deeks SG. Treatment of antiretroviral-drug-resistant HIV-1 infection. Lancet. 2003;362:2002–2011. doi: 10.1016/S0140-6736(03)15022-2. pmid: 14683662. [DOI] [PubMed] [Google Scholar]
- 121.Pluchino KM, Hall MD, Goldsborough AS, Callaghan R, Gottesman MM. Collateral sensitivity as a strategy against cancer multidrug resistance. Drug Resist. Updat. 2012;15:98–105. doi: 10.1016/j.drup.2012.03.002. pmid: 22483810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gressel J, Segel LA. Negative cross-resistance; a possible key to atrazine resistance management: A call for whole plant data. Z. Naturforsch. 1990;45c:470–473. [Google Scholar]
- 123.Gadamski G, Ciarka D, Gressel J, Gawronski SW. Negative cross-resistance in triazine-resistant biotypes of Echinochloa crusgalli and Conyza canadensis. Weed Sci. 2000;48:176–180. [Google Scholar]