Abstract
Non-coding RNAs (ncRNAs), that is, RNAs not translated into proteins, are crucial regulators of a variety of biological processes in plants. While protein-encoding genes have been relatively well-annotated in sequenced genomes, accounting for a small portion of the genome space in plants, the universe of plant ncRNAs is rapidly expanding. Recent advances in experimental and computational technologies have generated a great momentum for discovery and functional characterization of ncRNAs. Here we summarize the classification and known biological functions of plant ncRNAs, review the application of next-generation sequencing (NGS) technology and ribosome profiling technology to ncRNA discovery in horticultural plants and discuss the application of new technologies, especially the new genome-editing tool clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) systems, to functional characterization of plant ncRNAs.
Introduction
Horticultural plants (for example, such as fruits, vegetables, ornamental trees and flowers, herbs, and tea trees) have been domesticated to satisfy human’s food and aesthetical needs via various forms of hybridization breeding, mutation breeding, and transgenic breeding.1 Protein-coding genes related to specific target agricultural trait were chosen as major targets in the early time of transgenic breeding.2 Recently, non-coding RNAs (ncRNAs) have been shown to play key roles in the regulation of plant growth, development and response to environmental stresses at either transcriptional or post-transcriptional levels.3,4 Thus, ncRNAs are emerging as a spotlighted target materials to accelerate the domestication of horticultural crops.
Though discovery and functional characterization of ncRNAs have been carried out for more than half a century,5 their widespread occurrence and myriad functions in various organisms have not been truly appreciated until the post-genomics era. An unexpected finding from the annotation of sequenced genomes is that DNA sequences encoding proteins occupy only a small portion (2–25%) of the genomic space.6 The advent of next-generation sequencing (NGS) revolutionized the exploration of ncRNAs, and as a result, many novel ncRNAs have been recently discovered,7,8 which were highlighted by the new discovery of circular RNAs (circRNAs).7, 9,10,11,12 One of the big challenges in ncRNAs discovery is the determination of the coding potential of RNA sequences. Recent advances in ribosome profiling have shown a great potential for distinguishing between coding and non-coding transcripts and consequently improve the accuracy of ncRNA annotations.13,14
Molecular genetics approaches have been applied to functional characterization of ncRNAs via gain-of-function analysis or loss-of-function analysis.7,15,16 Precision genome engineering is a powerful tool for functional characterization of ncRNAs. Recently, a platform using RNA-guided engineered nucleases was developed for genome editing. The type II clustered, regularly interspaced, short palindromic repeat, (CRISPR)/CRISPR-associated protein 9 (Cas9) system found naturally occurring in Streptococcus pyogenes has been used to obtain rapid and efficient editing of genomes in plant species, and could facilitate the analysis of loss-of-function, gain-of-function and gene expression.17
In this review, we describe the classification and known functions of plant ncRNAs. Then, we review the application of NGS and ribosome profiling technology to ncRNAs discovery in horticultural plants, followed by a discussion of the new technologies for functional characterization of ncRNAs.
Classification and functions of plant ncrnas
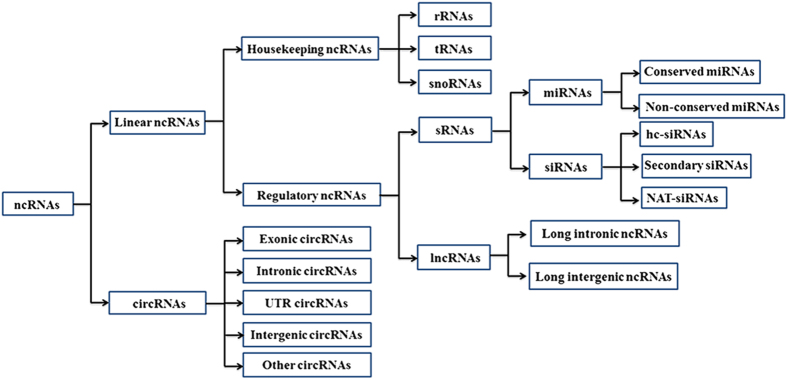
Based on the molecular structure, plant ncRNAs can be classified as linear ncRNAs and circular ncRNAs (circRNAs; Figure 1). The catalog of ncRNAs is currently dominated by linear ncRNAs compared with circRNAs that were just recently discovered as an emerging new class of ncRNAs.7, 9,10,11,12,18 On the basis of molecular function, linear ncRNAs can be divided into two categories: (1) housekeeping ncRNAs, including ribosomal RNAs (rRNAs), transfer RNAs (tRNAs) and small nucleolar RNAs (snoRNAs); and (2) regulatory ncRNAs, which can be further divided into two sub-categories: (a) small RNAs (sRNAs), including microRNAs (miRNAs) and small interfering RNAs (siRNAs) and (b) long ncRNAs (lncRNAs), including long intronic ncRNAs and long intergenic ncRNAs.3,19,20 On the basis of the genome region from which circRNAs arise, circRNAs can be divided into (1) exonic circRNAs, (2) intronic circRNAs, (3) UTR circRNAs, (4) intergenic circRNAs and (5) other circRNAs deriving from two or more genes (Figure 1).12 So far, functional characterization of ncRNAs has focused on sRNAs, lncRNAs and circRNAs. The known biological functions of these three types of ncRNAs are summarized as follows.
Figure 1.
Classification of plant non-coding RNAs (ncRNAs). circRNAs, circular ncRNAs; UTR, untranslated region; rRNAs, ribosomal RNAs; tRNAs, transfer RNAs; snoRNAs, small nucleolar RNAs; sRNAs, small RNAs; lncRNAs, long ncRNAs; miRNAs, microRNAs; siRNAs, small interfering RNAs; hc-siRNAs, heterochromatic siRNAs; NAT-siRNAs, natural antisense transcript siRNAs.
The function of sRNAs
sRNAs are involved in the regulation of plant growth, development and stress response via silencing endogenous gene expression at either transcriptional or post-transcriptional levels.21,22
miRNAs, derived from single-stranded hairpin RNAs,23 can be classified as conserved miRNAs and non-conserved miRNAs.24 Many miRNAs have been characterized from plants, which play important roles in different signaling pathways (Table 1). Usually conserved miRNAs are abundantly expressed, targeting transcription factors that directly regulate gene expression (Table 1). The relationships between conserved miRNAs and their targets have been considered to be stable during the evolution process,25 but it was recently reported that the targets of several conserved plant miRNAs (for example, miR396 and miR159) are somewhat flexible.26,27,28 In general, non-conserved miRNAs are weakly expressed and have been shown to occur in temporal patterns. Moreover, they are imprecisely processed without tractable targets and thus considered to be randomly evolved with a limited number of biological function.23 In addition, primary miRNAs of miR171b of Medicago truncatula and miR165a of Arabidopsis thaliana have been recently reported to produce peptides, which enhance the accumulation of their corresponding mature miRNAs.29
Table 1. Function of miRNAs validated by experiments in plants.
| miRNA | Conservation | Targets | Functions | References |
|---|---|---|---|---|
| miR156 | Conserved | SPL | Development | 102 |
| miR159 | Conserved | MYB, SGN-U567133 | Signaling pathway and development | 103,28 |
| miR160 | Conserved | ARF | Seed germination | 104 |
| miR163 | Non-conserved | PXMT1, FAMT | Metabolite biosynthesis | 105 |
| miR165/166 | Conserved | HD-ZIPIII | Leaf and vascular development | 106 |
| miR167 | Conserved | ARF | Signaling pathway, flower development | 107 |
| miR172 | Conserved | AP2 | Signaling pathway, flower development, stress response | 108 |
| miR173 | Non-conserved | TAS1, TAS2 | Uncharacterized | 109 |
| miR319 | Conserved | TCP | Flower development | 110 |
| miR390 | Conserved | TAS | Development | 111 |
| miR395 | Conserved | Sulfate transporter | Sulfate transport | 112 |
| miR396 | Conserved | GRF, bHLH74, HaWRKY6 | Leaf development, heat tolerance | 27,113,26 |
| miR400 | Non-conserved | PPR | Heat tolerance | 114 |
| miR408 | Conserved | Gene coding Copper ion binding protein | Copper homeostasis | 115 |
| miR444 | Non-conserved | MADS57 | Tillering development, nutrition accumulation | 115 |
| miR472 | Non-conserved | CNLs | Pathogen resistance | 116 |
| miR482 | Non-conserved | NBS-LRR | Pathogen resistance | 117 |
| miR820 | Non-conserved | DRM2 | Epigenetic silencing | 118 |
| miR824 | Non-conserved | AGL16 | Stomata development, plant flowering | 119 |
| miR828 | Non-conserved | MYB2 | Fiber development | 120 |
| miR842/846 | Non-conserved | Jacalin lectin | Vegetative storage | 121 |
| miR858 | Non-conserved | MYB2 | Fiber development | 120 |
| miR1512 | Non-conserved | Gene coding copine-like calmodulin-binding protein | Nodule development | 122 |
| miR1863 | Non-conserved | Os06g38480 | DNA methylation | 122 |
| miR4376 | Non-conserved | Ca2+-ATPase | Flower and fruit development | 123 |
| miR5200 | Non-conserved | FTL1/2 | Flowering initiation | 124 |
| miR6019 | Non-conserved | NB-LRR/LRR | Pathogen resistance | 125 |
| miR6020 | Non-conserved | NB-LRR/LRR | Pathogen resistance | 125 |
| miR7695 | Non-conserved | Nramp6 | Pathogen resistance | 126 |
Abbreviations: AGL16, agamous-like 16; AP2, apetala2-like transcription factor; ARF, auxin response factor; bHLH74, basic Helix-Loop-Helix 74; CNLs, coiled-coil nucleotide-binding leucine-rich repeat; DRM2, domains rearranged methyltransferase 2; FTL1/2, flowering locus T-like1/2; FAMT, farnesoic acid carboxyl methyltransferase; GRF, growth regulating factor; HD-ZIPIII, class III homeodomain-leucine zipper transcriptional factor; Nramp6, natural resistance-associated macrophage protein 6; NBS-LRR, nucleotide-binding site-leucine-rich repeat; PPR, pentatricopeptide repeat; SGN-U567133, encoding a protein with unknown function; SPL, squamosa-promoter binding protein-like; TAS, tasiRNA-generating; TCP, teosinte branched1/cycloidea/proliferating cell factor.
siRNAs, including heterochromatic siRNAs (hc-siRNAs), secondary siRNAs and natural antisense transcript siRNAs (NAT-siRNAs), are derived from Dicer-like (DCL)-catalyzed processing of double-stranded RNA (dsRNA) precursors.23 So far, siRNAs have been suggested to play roles in: (1) DNA methylation and chromatin modification mediated by hc-siRNAs,30 (2) repression of distinct mRNA targets by trans-acting siRNAs23,31,32,33 and (3) specific phenotypes, for example, proline accumulation,34 fertilization35 and bacterial infection,36 associated with NAT-siRNAs.
The function of lncRNAs
lncRNAs are linear ncRNAs of greater than 200 nt in length,37 which have been demonstrated to involve in multiple biological processes such as phosphate homeostasis, flowering, photomorphogenesis and fertility (Table 2). The molecular mechanisms underlying the biological function of plant lncRNAs include: (1) processing into shorter ncRNAs for functioning,38 (2) acting as the target mimics of miRNAs,39,40 (3) repressing histone-modifying activities and direct epigenetic silencing via interaction with specific chromatin domains,41,42,43,44 (4) acting as molecular cargo for protein re-localization45,46 and (5) post-translational regulation through protein modification and protein–protein interactions.6
Table 2. Function of the lncRNAs reported in plants.
| lncRNAs | Species | Biological function | Regulation mechanism | Refs |
|---|---|---|---|---|
| APOLO | Arabidopsis thaliana | Auxin-controlled development | Chromatin topology | 127 |
| ASCO-lncRNA | Arabidopsis thaliana | Lateral root development | Alternative splicing regulators | 127 |
| asHSFB2a | Arabidopsis thaliana | Gametophytic development | Antisense transcription | 128 |
| cis-NATPHO1;2 | Oryza sativa | Phosphate homeostasis | Translational enhancer | 129 |
| COLDAIR | Arabidopsis thaliana | Flowering | Histone modification | 42 |
| COOLAIR | Arabidopsis thaliana | Flowering | Histone modification | 43 |
| ENOD40 | Medicago truncatula, Glycine max | Nodule development | Protein re-localization | 130,131 |
| HID1 | Arabidopsis thaliana | Photomorphogenesis | Association with chromatin | 132 |
| HvCesA6 lnc-NAT | Hordeum vulgare | Cell wall biosynthesis | siRNA precursor | 133 |
| LDMAR | Oryza sativa | Photoperiod-sensitive male sterility | Promoter methylation | 134 |
| XLOC_057324 | Oryza sativa | Flowering and sterility | Unknown | 135 |
Abbreviations: APOLO, auxin-regulated promoter loop; ASCO, alternative splicing competitor; asHSFB2a, natural long non-coding antisense RNA of heat stress transcription factor B; PHO1;2, PHOSPHATE1;2; COLDAIR, cold-assisted intronic non-coding RNA; COOLAIR, cold induced long antisense intragenic RNAs; ENOD40, early nodulin 40; HID1, hidden treasure 1; CesA6 lncNAT, natural antisense of CesA6 cellulose synthase gene; IPS1, induced by phosphate starvation 1; LDMAR, long day-specific male-fertility-associated RNA.
The function of circRNAs
Discovery of thousands of circRNAs across a range of plant species have been summarized in other review paper,47 and recently demonstrated in horticultural plants, for example, Solanum lycopersicum48 and Actinidia chinensis.18 However, little is known about the function of circRNAs in plants. In Arabidopsis, Conn et al.49 reported that the circRNAs derived from exon 6 of the SEPALLATA3 (SEP3) gene can bind strongly to its cognate DNA locus, forming an RNA:DNA hybrid, or R-loop, whereas the linear RNA equivalent bound significantly more weakly to DNA. R-loop formation results in transcriptional pausing, in turn driving floral homeotic phenotypes. The function of circRNAs reported in mammalian may serve as an initial guidance for future studies on the function of plant circRNAs. For example, Hansen et al.50 reported that circular transcript ciRS-7 from human and Sry9 from mouse acts as a ‘molecular sponge’ of miR7 and miR138, respectively. The human circRNA ITCH was reported to act as a sponge for miR7, miR17 and miR214, respectively.51 Another circRNA ZNF91 containing 24 miR23 sites, as well as 39 additional sites for miR296, was discovered in mammals.52 Zhang et al.9 showed that an intronic circRNA, ci-ankrd52, positively involves in the regulation of RNA polymerase II transcription. Also, exon-intron circRNAs have been shown to enhance the expression of their parental genes in a cis configuration.7
Application of new technologies to discovery of ncrnas
A variety of experimental approaches have been used for discovering ncRNAs in plants, such as molecular cloning, microarray, next-generation sequencing (NGS), third-generation sequencing,53 epitope tagging, mass-spectrometry and ribosome profiling.54 These approaches heavily rely on bioinformatics tools, such as TopHat,55 Cufflinks,56 CIRCexplorer,57 CIRI,58 CPC59 and HMMER,60 for the discovery of ncRNAs. Recently, some new computational tools, for example, miRDeep-P,61 miRDeepFinder62 and miR-PREFeR63 were developed for the identification of plant miRNAs, which are often belong to large families with high-sequence similarity among the paralogous members. Moreover, these tools do not necessarily rely on a reference genome and are useful for species-specific ncRNA detection. A pipeline for discovery of ncRNAs in plants is illustrated in Figure 2. Most of above approaches for ncRNA discovery have been discussed in some recent review articles.7,15 Currently, more and more horticultural plant genomes and transcriptomes were decoded by third-generation sequencing such as Pacific Biosciences (PacBio, Menlo Park, CA, USA), Illumina Tru-seq Synthetic Long-Read technology (San Diego, CA, USA) and the Oxford Nanopore Technologies sequencing platform (Oxford, UK).53,64,65,66 These platforms offer longer read sequencing to facilitate the accurate de novo assembly of full-length RNAs without needs for mapping of the transcriptome sequencing reads to the reference genomes. Thus, while still under active development, the third-generation sequencing platforms will definitely accelerate the discovery of ncRNAs and their targets. In combination with appropriate bioinformatics tools such as PLEK,67 the ongoing and future efforts for transcriptome sequencing using third-generation sequencing technologies are expected to shed new light on the ncRNA landscape of horticultural plants without reference genomes. Here we focus on two frequently used technologies that offer potential for the discovery and characterization of ncRNAs in horticultural plants: that is, NGS and ribosome profiling.
Figure 2.
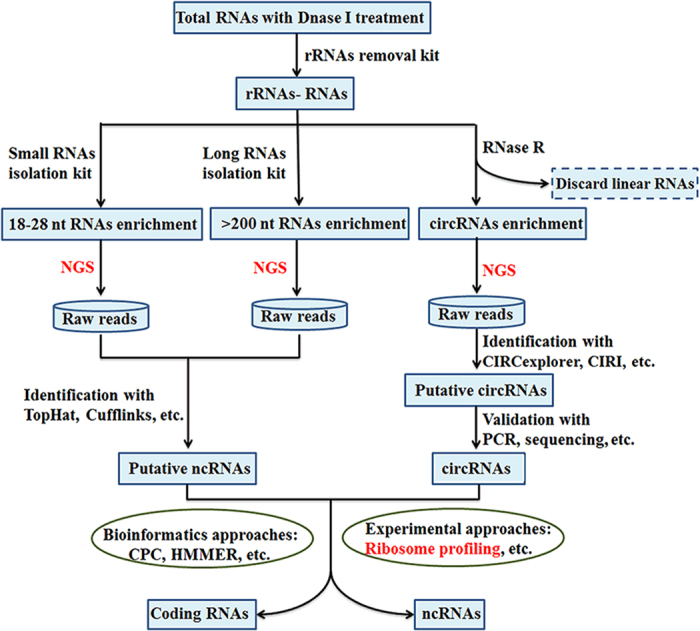
A pipeline for discovery of non-coding RNAs (ncRNAs) in plants. rRNAs, ribosomal RNAs; NGS, next-generation sequencing; CIRI, circular RNA identifier; circRNAs, circular ncRNAs; CPC, coding potential calculator; HMM, hidden markov models.
NGS as a new powerful tool for the prediction of ncRNAs
The ncRNAs can be identified through the direct detection of the transcribed RNAs.68 Initially, direct cloning approach has been used to discover ncRNAs in plants.69,70 Subsequently, the hybridization-based microarray technology has been used to discover a large number of ncRNAs in the intergenic regions of A. thaliana71,72 and rice.73 However, the ability of these hybridization-based technologies suffer several limitations such as reduced dynamic range, high false positives6 and difficultly defining splice junctions and connecting transcribed regions into transcript models.74,75
NGS overcomes the challenges related to microarray technology,76 providing a powerful tool for defining the ncRNA domain. For example, miRNAs were previously thought to be dominant members in the sRNAs landscape; however, recent global analysis of plant transcriptomes revealed millions of siRNAs, making them the most abundant class of sRNAs in plants.77 More recently, circRNAs were recognized as a large new category of RNAs with thousands of members in animals and plants through high-throughput transcriptome sequencing (RNA-Seq) followed by ncRNA prediction based on RNA-Seq data using new computational algorithms customized for ncRNAs (Figure 2).7,11,12,57,58 With advancement of NGS technology, many ncRNAs are being discovered in an expanding list of horticultural plant species (Table 3).
Table 3. Examples of the application of next-generation sequencing (NGS) technology to ncRNAs discovery in horticultural plants.
| Species | Type of ncRNAs | No. of ncRNAs | References |
|---|---|---|---|
| Actinidia chinensis | circRNAs | 3582 | 18 |
| Arachis hypogaea | miRNAs | 59 | 136 |
| Brassica campestris | miRNAs | 131 | 137 |
| Brassica napus | lncRNAs | 3181 | 138 |
| Cucumis sativus | lncRNAs | 3274 | 139 |
| Cucumis sativus | miRNAs, rRNAs, tRNAs, snoRNAs | 1400 | 140 |
| Fragaria×ananassa | miRNAs | 190 | 141 |
| Phalaenopsis aphrodite | miRNAs | 204 | 78 |
| Prunus persica | lncRNAs | 1417 | 142 |
| Rosa sp. | miRNAs | 267 | 143 |
| Vitis amurensis Rupr. | miRNAs | 232 | 144 |
| Solanum lycopersicum | circRNAs | 854 | 48 |
| Solanum lycopersicum | lncRNAs | 10774 | 145 |
| Solanum tuberosum | miRNAs | 259 | 146 |
Ribosome profiling as a new tool for the validation of ncRNA predictions
A key aspect of ncRNA validation is to determine the coding potential of predicted ncRNAs. The length of 18 to 30 nucleotides is the threshold commonly used for the prediction of miRNA78,79 whereas the length of greater than 200 nucleotides is often used as the threshold for lncRNAs prediction.80 Presence of an open-reading frame (ORF) of at least 100 amino acids (aa) is the threshold commonly used for defining a protein-coding transcript and as such, many important small proteins (<100 aa) were not annotated in plants.7, 81,82,83 More recently, a large number of protein sequences have been predicted by translation of the longest ORFs without any further experimental evidence.74 It is possible that some of the predicted protein-coding genes, based on an arbitrary ORF length, might be mis-annotated. For example, some well characterized human lncRNAs, such as H19, Hotair, Kcnq1ot1, Meg3 and Xist, contain ORFs of 100 aa or longer.84 Most of predicted lncRNAs contain putative ORFs, which may be translated into non-functional proteins or may be unable to be translated at all.74
Recently, ribosome profiling, which uses deep sequencing to monitor in vivo translation, has shown high potential for the genome-wide examination of protein-coding potential (Figure 2). Ribosome profiling has been used to segregate several hundred small proteins (<100 aa) from predicted lncRNAs in zebrafish and humans.13,14 Also,Pamudurti et al.85 demonstrated that a group of circRNAs was associated with translating ribosomes by performing ribosome profiling from fly heads and found a circRNA generated from the muscleblind locus encodes a protein. In Arabidopsis, 237 protein-encoding transcripts from the existing compendia of ncRNAs were found based on the ribosome profiling technology.86,87 Thus, the ribosome profiling technology can be used as a high-throughput tool for removing false positives in the ncRNAs predictions of horticultural plants.
Application of new technologies to functional characterization of ncrnas
Thanks to the advance in the aforementioned new technologies, the universe of ncRNAs is currently expanding at an increasing rate. However, the biological function of these ncRNAs remains largely unknown.16 Various approaches have been developed for functional studies of ncRNAs (Figure 3). The primary goal of functional studies on ncRNAs is to understand the biological processes in which the ncRNAs are involved. To achieve this goal, many researchers have used gain-of-function and loss-of-function mutants for functional characterization of ncRNA genes.7 CRISPR/Cas9, a new genome-editing technology, holds great potential for generating knockout and knock-in mutants in plants, as demonstrated in a range of plant species,17 and recently demonstrated in horticultural plant species, for example, Citrus sinensis,88 Malus pumila,89 Solanum lycopersicum90 and Solanum tuberosum.91 Compared with RNA inference (RNAi) that has several limitations such as incomplete gene knock-down and extensive off-target activities, CRISPR/Cas9 technology has the advantage of complete gene knockout with relatively low off-target activities.92 In addition, the action of RNAi is restricted in cytoplasm where RNA-induced silencing complexes are located.93 However, many ncRNAs have been shown to be localized in the nucleus, which cannot be manipulated in similar manner using RNAi.68,94 Thus, CRISPR/Cas9 provides an efficient and effective alternative to RNAi for characterizing the function of ncRNAs. In fact, this new genome-editing technology has been used to knockout several ncRNAs in animals such as humans, mouse, zebrafish,94–97 as well as in plants such as soybean.98 Once the CRISPR/Cas9-mediated knockout and knock-in mutation is created, the NGS technology, mentioned above, can be used to profile the expression of target transcripts and other downstream genes in the biological pathways (Figure 3).
Figure 3.
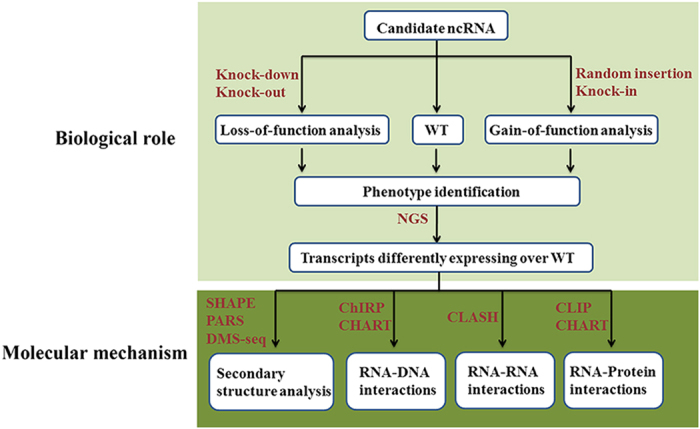
A pipeline for functional characterization of ncRNAs in plants. SHAPE, RNA-selective 2′-hydroxyl acylation and primer extension; PARS, parallel analysis of RNA structure; DMS-seq, dimethyl sulfate-modified RNA for sequencing; CHIRP, chromatin isolation by RNA purification; CHART, capture hybridization analysis of RNA targets, CLASH, crosslinking, ligation, sequencing of hybrids; CLIP, crosslinking immunoprecipitation.
After identification of the biological roles of ncRNAs, it is important to understand the molecular mechanism underlying these biological roles (Figure 3). Examination of the secondary structure of ncRNAs is informative in studying the function of ncRNAs at the molecular level. Several experimental approaches, such as selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE), parallel analysis of RNA structure (PARS) or dimethyl sulfate-modified RNA for sequencing (DMS-seq), can be used for deciphering of the secondary structure of ncRNAs.7,15 To understand where and how the ncRNAs function, chromatin isolation by RNA purification (CHIRP), capture hybridization analysis of RNA targets (CHART), crosslinking, ligation, sequencing of hybrids (CLASH) and crosslinking IP (CLIP) have been developed to detect the interactions between ncRNAs and DNA, RNA or protein.15,16 Recently, Shechner et al.99 used CRISPR/ dCas9, based on a catalytically dead variant of Cas9, to deploy lncRNAs cargos to DNA loci by incorporating the cargo into the sgRNA, thus providing initial insights into the utility of CRISPR/dCas9 for studying the function of ncRNAs. Besides its potential for validating ncRNA prediction, ribosome profiling can also be used to unravel the function of ncRNAs. For example, using ribosome profiling, Guo et al.100 studied the effects of miRNAs on protein production from their target mRNAs and found that the destabilization of target mRNAs by the miRNAs is the predominant reason for reduced protein output. Similarly, Bazzini et al.101 studied the impact of miR430 on endogenous mRNAs in zebrafish using ribosome profiling and found that this sRNA reduced translation. These technologies provide new approaches for functional characterization of ncRNAs in horticultural plants.
Concluding remarks
The discovery and functional characterization of ncRNAs could facilitate the domestication of horticultural plants, resulting in more nutritious, colorful, tasteful, and esthetic fruits, vegetables, and ornamental flowers and trees. While the number of protein-encoding genes is relatively less variable among plants, the ncRNA domain in plants is very dynamic, with increasingly more ncRNA members being discovered and characterized annually. In particular, recent advances in NGS and ribosome profiling technology have offered great potential for expediting the discovery of ncRNAs in horticultural plants. Also, the simplicity, robustness and versatility of the CRISPR/Cas9 systems make such systems attractive for functional characterization of ncRNAs in general and specifically to the process of accelerated domestication in horticultural crops. It is expected that these new technologies will be widely applied in ncRNA research while they become more cost-efficient and more technically mature in the near future.
Acknowledgments
We thank Dr Zong-Ming Cheng (University of Tennessee) and the reviewers for their critical review and instructive comments on our manuscript. This research is supported by the Department of Energy (DOE), Office of Science, Genomic Science Program under Award Number DE-SC0008834. Oak Ridge National Laboratory is managed by UT-Battelle, LLC for the US DOE under Contract Number DE-AC05–00OR22725. This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Footnotes
The authors declare no conflict of interest.
References
- Xiong J-S, Ding J, Li Y. Genome-editing technologies and their potential application in horticultural crop breeding. Hort Res 2015; 2: 15019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S-Y, Shin C. Regulatory non-coding RNAs in plants: potential gene resources for the improvement of agricultural traits. Plant Biotechnol Rep 2016; 10: 35–47. [Google Scholar]
- Matsui A, Nguyen AH, Nakaminami K, Seki M. Arabidopsis non-coding RNA regulation in abiotic stress responses. Int J Mol Sci 2013; 14: 22642–22654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Li Y-F, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci 2012; 17: 196–203. [DOI] [PubMed] [Google Scholar]
- Holley RW, Apgar J, Everett GA et al. Structure of a ribonucleic acid. Science 1965; 147: 1462–1465. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang H, Chua NH. Long noncoding RNA transcriptome of plants. Plant Biotechnol J 2015; 13: 319–328. [DOI] [PubMed] [Google Scholar]
- Pauli A, Valen E, Schier AF. Identifying (non‐) coding RNAs and small peptides: Challenges and opportunities. Bioessays 2015; 37: 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Reichel M, Li Y, Millar AA. The functional scope of plant microRNA-mediated silencing. Trends Plant Sci 2014; 19: 750–756. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang X-O, Chen T et al. Circular intronic long noncoding RNAs. Mol Cell 2013; 51: 792–806. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495: 333–338. [DOI] [PubMed] [Google Scholar]
- Wang PL, Bao Y, Yee M-C et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 2014; 9: e90859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye CY, Chen L, Liu C, Zhu QH, Fan L. Widespread noncoding circular RNAs in plants. New Phytol 2015; 208: 88–95. [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Johnstone TG, Christiano R et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 2014; 33: 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Norris ML, Valen E et al. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science 2014; 343: 1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Non-coding RNA. A new frontier in regulatory biology. Natl Sci Rev 2014; 1: 190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Spitale RC, Chang HY. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat Struct Mol Biol 2015; 22: 29–35. [DOI] [PubMed] [Google Scholar]
- Liu D, Hu R, Palla KJ, Tuskan GA, Yang X. Advances and perspectives on the use of CRISPR/Cas9 systems in plant genomics research. Curr Opin Plant Biol 2016; 30: 70–77. [DOI] [PubMed] [Google Scholar]
- Wang Z, Liu Y, Li D et al. Identification of circular RNAqs in kiwifruit and their species-specific response to bacterial canker pathogen invasion. Front Plant Sci 2017; 8: 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J et al. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012; 24: 4333–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santosh B, Varshney A, Yadava PK. Non‐coding RNAs: biological functions and applications. Cell Biochem Funct 2014; 33: 14–22. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 2009; 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikit S, Zhai J, Meyers BC. Biogenesis and function of rice small RNAs from non-coding RNA precursors. Curr Opin Plant Biol 2013; 16: 170–179. [DOI] [PubMed] [Google Scholar]
- Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 2013; 64: 137–159. [DOI] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. Evolution and functional diversification of MIRNA genes. Plant Cell 2011; 23: 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW. Conservation and divergence in plant microRNAs. Plant Mol Biol 2012; 80: 3–16. [DOI] [PubMed] [Google Scholar]
- Giacomelli JI, Weigel D, Chan RL, Manavella PA. Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol 2012; 195: 766–773. [DOI] [PubMed] [Google Scholar]
- Debernardi JM, Rodriguez RE, Mecchia MA, Palatnik JF. Functional specialization of the plant miR396 regulatory network through distinct microRNA–target interactions. PLoS Genet 2012; 8: e1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxdorf K, Hendelman A, Stav R, Lapidot M, Ori N, Arazi T. Identification and characterization of a novel miR159 target not related to MYB in tomato. Planta 2010; 232: 1009–1022. [DOI] [PubMed] [Google Scholar]
- Lauressergues D, Couzigou J-M, San Clemente H et al. Primary transcripts of microRNAs encode regulatory peptides. Nature 2015; 520: 90–93. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 2010; 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Guo M, Nogueira FT, Timmermans MC. Establishing leaf polarity: the role of small RNAs and positional signals in the shoot apex. Development 2007; 134: 813–823. [DOI] [PubMed] [Google Scholar]
- Luo Q-J, Mittal A, Jia F, Rock CD. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol Biol 2012; 80: 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005; 121: 207–221. [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu J-K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 2005; 123: 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Saez MA, Williams LE, Fletcher JC, McCormick S. Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev 2010; 24: 1010–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D et al. A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA 2006; 103: 18002–18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S-Y, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet 2013; 29: 461–468. [DOI] [PubMed] [Google Scholar]
- Amor BB, Wirth S, Merchan F et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res 2009; 19: 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR. MicroRNA399 is a long‐distance signal for the regulation of plant phosphate homeostasis. Plant J 2008; 53: 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K, Lin S-I, Wu C-C, Huang Y-T, Su C-l, Chiou T-J. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 2006; 141: 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci USA 2000; 97: 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011; 331: 76–79. [DOI] [PubMed] [Google Scholar]
- Marquardt S, Raitskin O, Wu Z, Liu F, Sun Q, Dean C. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol Cell 2014; 54: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q-H, Wang M-B. Molecular functions of long non-coding RNAs in plants. Genes 2012; 3: 176–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campalans A, Kondorosi A, Crespi M. Enod40, a short open reading frame–containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell 2004; 16: 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey M, Complainville A, Charon C et al. Phytohormonal responses in enod40‐overexpressing plants of Medicago truncatula and rice. Physiol Plant 2004; 120: 132–139. [DOI] [PubMed] [Google Scholar]
- Lee S-M, Kong HG, Ryu C-M. Are circular RNAs new kids on the block? Trends Plant Sci 2017; 22: 357–360. [DOI] [PubMed] [Google Scholar]
- Zuo J, Wang Q, Zhu B, Luo Y, Gao L. Deciphering the roles of circRNAs on chilling injury in tomato. Biochem Biophys Res Commun 2016; 479: 132–138. [DOI] [PubMed] [Google Scholar]
- Conn VM, Hugouvieux V, Nayak A et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants 2017; 3: 17053. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495: 384–388. [DOI] [PubMed] [Google Scholar]
- Li F, Zhang L, Li W et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 2015; 6: 6001–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014; 15: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao W-B, Schneeberger K. The impact of third generation genomic technologies on plant genome assembly. Curr Opin Plant Biol 2017; 36: 64–70. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009; 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protocol 2012; 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X-O, Wang H-B, Zhang Y, Lu X, Chen L-L, Yang L. Complementary sequence-mediated exon circularization. Cell 2014; 159: 134–147. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 2015; 16: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye Z-Q et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 2007; 35: W345–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform 2009; 23: 205–211. [PubMed] [Google Scholar]
- Yang X, Li L. miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics 2011; 27: 2614–2615. [DOI] [PubMed] [Google Scholar]
- Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol 2012; 80: 75–84. [DOI] [PubMed] [Google Scholar]
- Lei J, Sun Y. miR-PREFeR: an accurate, fast and easy-to-use plant miRNA prediction tool using small RNA-Seq data. Bioinformatics 2014: ; 30: 2837–2839. [DOI] [PubMed] [Google Scholar]
- Hoshino A, Jayakumar V, Nitasaka E et al. Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat Commun 2016; 7: 13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne SL, Nagy I, Pfeifer M et al. A synteny‐based draft genome sequence of the forage grass Lolium perenne. Plant J 2015; 84: 816–826. [DOI] [PubMed] [Google Scholar]
- Nowak MD, Russo G, Schlapbach R, Huu CN, Lenhard M, Conti E. The draft genome of Primula veris yields insights into the molecular basis of heterostyly. Genome Biol 2015; 16: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Zhang J, Zhou Z. PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics 2014; 15: 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014; 15: 7–21. [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 2002; 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev 2002; 16: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T et al. Arabidopsis tiling array analysis to identify the stress-responsive genes. In Sunkar R, ed, Plant Stress Tolerance: Methods and Protocols. Humana Press, Totowa, NJ, pp 141–155. [DOI] [PubMed]
- Rehrauer H, Aquino C, Gruissem W et al. AGRONOMICS1: a new resource for Arabidopsis transcriptome profiling. Plant Physiol 2010; 152: 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang X, Stolc V et al. Genome-wide transcription analyses in rice using tiling microarrays. Nat Genet 2006; 38: 124–129. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013; 154: 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A, Koppstein D, Rozowsky J et al. Comparison and calibration of transcriptome data from RNA-Seq and tiling arrays. BMC Genomics 2010; 11: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 2011; 12: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Cubas C, Palomar M, Arteaga-Vázquez M, Reyes JL, Covarrubias AA. Non-coding RNAs in the plant response to abiotic stress. Planta 2012; 236: 943–958. [DOI] [PubMed] [Google Scholar]
- Chao Y-T, Su C-L, Jean W-H, Chen W-C, Chang Y-CA, Shih M-C. Identification and characterization of the microRNA transcriptome of a moth orchid Phalaenopsis aphrodite. Plant Mol Biol 2014; 84: 529–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Lei B, Niu M, Huang Y, Kong Q, Bie Z. High throughput sequencing of small RNAs in the two Cucurbita germplasm with different sodium accumulation patterns identifies novel MicroRNAs involved in salt stress response. PLoS ONE 2015; 10: e0127412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkel JM. Visiting ‘noncodarnia’. Biotechniques 2013; 54: 301–304. [DOI] [PubMed] [Google Scholar]
- Costa LM, Marshall E, Tesfaye M et al. Central cell–derived peptides regulate early embryo patterning in flowering plants. Science 2014; 344: 168–172. [DOI] [PubMed] [Google Scholar]
- Hanada K, Higuchi-Takeuchi M, Okamoto M et al. Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc Natl Acad Sci USA 2013; 110: 2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XH, Tschaplinski TJ, Hurst GB et al. Discovery and annotation of small proteins using genomics, proteomics, and computational approaches. Genome Res 2011; 21: 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’conundrum. Genes Dev 2007; 21: 11–42. [DOI] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M et al. Translation of circRNAs. Mol Cell 2017; 66: 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Girke T, Bazin J, Bailey-Serres J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA 2014; 111: E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Calviello L, Wu H-YL et al. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc Natl Acad Sci USA 2016; 113: E7126–E7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Wang N. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 2014; 9: e93806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani C, Hirai N, Komori S et al. Efficient genome editing in apple using a CRISPR/Cas9 system. Sci Rep 2016; 6: 31481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C, Nekrasov V, Lippman ZB, Van Eck J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol 2014; 166: 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler NM, Atkins PA, Voytas DF, Douches DS. Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS ONE 2015; 10: e0144591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 2015; 16: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 2000; 101: 25–33. [DOI] [PubMed] [Google Scholar]
- Ho T-T, Zhou N, Huang J et al. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res 2014; 43: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 2015; 161: 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Miller S, Filipiak W, Mell B, Saunders T, Joe B. A CRISPR/Cas9 system based targeted disruption of a novel rat long non-coding RNA in a rat genetic model of hypertension. FASEB J 2015; 29: 814.811. [Google Scholar]
- Xiao A, Wang Z, Hu Y et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res 2013: ; 41: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA. Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol 2015; 15: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods 2015; 12: 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010; 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 2012; 336: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-W, Schwab R, Czech B, Mica E, Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 2008; 20: 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Peral MM, Li J, Li Y et al. The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol 2010; 154: 757–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post‐germination stages. Plant J 2007; 52: 133–146. [DOI] [PubMed] [Google Scholar]
- Ng DW, Zhang C, Miller M et al. cis-and trans-Regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. Plant Cell 2011; 23: 1729–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi J, Watanabe Y. miR165/166 and the development of land plants. Dev Growth Differ 2012; 54: 93–99. [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I, Weigel D. Coordination of flower maturation by a regulatory circuit of three microRNAs. PLoS Genet 2013; 9: e1003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 2003; 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Yoo SJ, Fahlgren N et al. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci USA 2008; 105: 20055–20062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, King S, Jack T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 2009; 106: 22534–22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon EK, Yang JH, Lim J, Kim SH, Kim S-K, Lee WS. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res 2009: ;38: 1382–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima CG, Yoshimoto N, Maruyama‐Nakashita A. Sulphur starvation induces the expression of microRNA‐395 and one of its target genes but in different cell types. Plant J 2009; 57: 313–321. [DOI] [PubMed] [Google Scholar]
- Mecchia MA, Debernardi JM, Rodriguez RE, Schommer C, Palatnik JF. MicroRNA miR396 and RDR6 synergistically regulate leaf development. Mech Dev 2013; 130: 2–13. [DOI] [PubMed] [Google Scholar]
- Yan K, Liu P, Wu C-A et al. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol Cell 2012; 48: 521–531. [DOI] [PubMed] [Google Scholar]
- Guo S, Xu Y, Liu H et al. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun 2013; 4: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccara M, Sarazin A, Thiebeauld O et al. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP-and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathogens 2014; 10: e1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen H-M, Patel K, Bond DM, Santos BA, Baulcombe DC. A microRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell 2012; 24: 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosaka M, Itoh J-I, Nagato Y, Ono A, Ishiwata A, Sato Y. Role of transposon-derived small RNAs in the interplay between genomes and parasitic DNA in rice. PLoS Genet 2012; 8: e1002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter C, Schöb H, Stadler M, Meins F, Si-Ammour A. MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 2007; 19: 2417–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Pang M, Nah G et al. miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Commun 2014; 5: 3050. [DOI] [PubMed] [Google Scholar]
- Jia F, Rock CD. MIR846 and MIR842 comprise a cistronic MIRNA pair that is regulated by abscisic acid by alternative splicing in roots of Arabidopsis. Plant Mol Biol 2013; 81: 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q et al. DNA methylation mediated by a microRNA pathway. Mol Cell 2010; 38: 465–475. [DOI] [PubMed] [Google Scholar]
- Wang Y, Itaya A, Zhong X et al. Function and evolution of a microRNA that regulates a Ca2+-ATPase and triggers the formation of phased small interfering RNAs in tomato reproductive growth. Plant Cell 2011; 23: 3185–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. Plant microRNAs and development. J Genet Genomics 2013; 40: 217–230. [DOI] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C et al. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 2012; 109: 1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo S, Peris‐Peris C, Siré C et al. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance‐associated macrophage protein 6) gene involved in pathogen resistance. New Phytol 2013; 199: 212–227. [DOI] [PubMed] [Google Scholar]
- Bardou F, Ariel F, Simpson CG et al. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev Cell 2014; 30: 166–176. [DOI] [PubMed] [Google Scholar]
- Wunderlich M, Groß-Hardt R, Schöffl F. Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol Biol 2014; 85: 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 2013; 25: 4166–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Katinakis P, Hendriks P et al. Characterization of GmENOD40, a gene showing novel patterns of cell‐specific expression during soybean nodule development. Plant J 1993; 3: 573–585. [DOI] [PubMed] [Google Scholar]
- Sousa C, Johansson C, Charon C et al. Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame-containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol Cell Biol 2001; 21: 354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fan X, Lin F et al. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci USA 2014; 111: 10359–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held MA, Penning B, Brandt AS et al. Small-interfering RNAs from natural antisense transcripts derived from a cellulose synthase gene modulate cell wall biosynthesis in barley. Proc Natl Acad Sci USA 2008; 105: 20534–20539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Lu Q, Ouyang Y et al. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci USA 2012; 109: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-C, Liao J-Y, Li Z-Y et al. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol 2014; 15: 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C-Z, Xia H, Frazier TP et al. Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.). BMC Plant Biol 2010; 10: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jiang D, Zhang C et al. Genome-wide identification of turnip mosaic virus-responsive microRNAs in non-heading Chinese cabbage by high-throughput sequencing. Gene 2015; 571: 178–187. [DOI] [PubMed] [Google Scholar]
- Joshi RK, Megha S, Basu U, Rahman MH, Kav NN. Genome wide identification and functional prediction of long non-coding RNAs responsive to Sclerotinia sclerotiorum infection in Brassica napus. PLoS ONE 2016; 11: e0158784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z, Fan C, Cheng T, Su Y, Wei Q, Li G. Genome-wide identification, characterization and evolutionary analysis of long intergenic noncoding RNAs in cucumber. PLoS ONE 2015; 10: e0121800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li R, Zhang Z et al. The genome of the cucumber, Cucumis sativus L. Nat Genet 2009; 41: 1275–1281. [DOI] [PubMed] [Google Scholar]
- Ge A, Shangguan L, Zhang X et al. Deep sequencing discovery of novel and conserved microRNAs in strawberry (Fragaria×ananassa). Physiol Plant 2013; 148: 387–396. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhao S, Gu C et al. Deep RNA-Seq uncovers the peach transcriptome landscape. Plant Mol Biol 2013; 83: 365–377. [DOI] [PubMed] [Google Scholar]
- Kim J, Park JH, Lim CJ et al. Small RNA and transcriptome deep sequencing proffers insight into floral gene regulation in Rosa cultivars. BMC Genomics 2012; 13: 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Han J, Liu C et al. Identification of microRNAs from Amur grape (Vitis amurensis Rupr.) by deep sequencing and analysis of microRNA variations with bioinformatics. BMC Genomics 2012; 13: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarano D, Rao R, Corrado G. In Silico identification and annotation of non-coding RNAs by RNA-seq and de novo assembly of the transcriptome of tomato fruits. PLoS ONE 2017; 12: e0171504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Marshall D, Bryan GJ, Hornyik C. Identification and characterization of miRNA transcriptome in potato by high-throughput sequencing. PLoS ONE 2013; 8: e57233. [DOI] [PMC free article] [PubMed] [Google Scholar]