Abstract
RNAs can begin to fold immediately after emerging from RNA polymerase during transcription. Interactions between nascent RNAs and ligands during cotranscriptional folding can direct the formation of alternative RNA structures, a feature exploited by non-coding RNAs called riboswitches to make gene regulatory decisions. Despite their importance, cotranscriptional folding pathways have yet to be uncovered with sufficient resolution to reveal how cotranscriptional folding governs RNA structure and function. To access cotranscriptional folding at nucleotide resolution, we extend selective 2’-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) to measure structural information of nascent RNAs during transcription. With cotranscriptional SHAPE-Seq, we determine how the B. cereus crcB fluoride riboswitch cotranscriptional folding pathway undergoes a ligand-dependent bifurcation that delays or promotes terminator formation via a series of coordinated structural transitions. Our results directly link cotranscriptional RNA folding to a genetic decision and establish a framework for cotranscriptional analysis of RNA structure at nucleotide resolution.
Introduction
As nascent RNA molecules exit RNA polymerase (RNAP), they transition through intermediate structural states that ultimately determine the final structure and function of an RNA1–4. Because RNA folding generally occurs faster than transcription, the 5’ to 3’ polarity of RNA synthesis directs an order of folding, or cotranscriptional ‘folding pathway’, that sets the structural stage for many types of interactions that govern cellular processes such as transcription, translation, and macromolecular assembly5–7.
Cotranscriptional folding is predicted to be particularly important for bacterial riboswitches8, a class of regulatory RNAs that control gene expression as a function of specific ligand concentration. Riboswitches contain two functional domains that can overlap structurally: a ligand-binding aptamer and an expression platform that makes regulatory decisions based on the structural state of the aptamer8,9. For riboswitches that regulate transcription, ligand binding must influence folding pathways within a short time window in order to commit the riboswitch to one of two mutually exclusive pathways: promote intrinsic terminator hairpin formation or prevent it10–14. A number of structural studies have revealed the details of RNA-ligand interactions for many aptamers8, and biochemical15,16 and biophysical 13,14 studies using actively transcribing RNAP have observed distinct RNA structural transitions during transcription. However, we still lack a complete, nucleotide-resolution understanding of how ligand binding influences the folding pathway of an entire riboswitch and enables it to regulate gene expression.
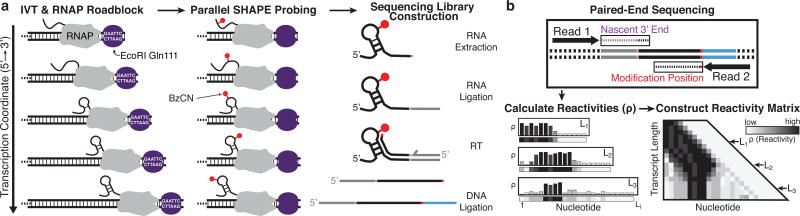
Here we introduce cotranscriptional SHAPE-Seq (selective 2’-hydroxyl acylation analyzed by primer extension sequencing), a method that couples in vitro RNAP arrest with high-throughput structure probing to characterize the structures of nascent RNA transcripts at single-nucleotide resolution (Fig. 1). Cotranscriptional SHAPE-Seq begins with in vitro transcription of a DNA template library that directs the synthesis of each intermediate length of a target RNA (Fig. 1a). Each template contains an EcoRI site at the 3’ end that when bound by the catalytically inactive EcoRI E111Q mutant (Gln111)17 establishes a transcription roadblock that halts RNAP 14 nt upstream of the EcoRI binding site18. Initiation of single-round transcription from this template library generates halted elongation complexes at all intermediate lengths of the target RNA that, after 30s of transcription, are rapidly modified with the fast-acting SHAPE reagent benzoyl cyanide (BzCN) (reaction t1/2 of 250 ms)19.
Fig. 1.
Cotranscriptional SHAPE-Seq overview. (a) A set of templates are generated that each contain an E. coli promoter, a variable length RNA template, and an EcoRI Gln111 roadblock site. Single-round in vitro transcription (IVT) with E. coli RNA polymerase (RNAP) is performed using a template library containing a roadblock site at each intermediate transcript length, followed by simultaneous SHAPE probing of the arrested complexes and preparation for sequencing. (b) Paired-end sequencing reveals the SHAPE modification position and the 3’ end of each nascent RNA transcript. Reads are binned by transcript length and used to calculate SHAPE reactivity profiles that are stacked to generate the reactivity matrix. Increases or decreases in reactivity between transcript lengths (rows) at particular nts (columns) of this matrix reveal cotranscriptional folding events.
Following chemical modification, RNAs are extracted and processed for paired-end sequencing according to our previously developed SHAPE-Seq v2.1 protocol20. Each paired-end read encodes the location of the halted RNAP (nascent RNA 3’ end) and the SHAPE modification position (Fig. 1b). Reads are bioinformatically binned by RNAP position and used to calculate a SHAPE-Seq reactivity spectrum for each intermediate length of the RNA21. These reactivities represent flexibilities for every nucleotide within each nascent RNA transcript length. High reactivities are indicative of unpaired bases and low reactivities indicate bases that are potentially involved in base pairing, stacking, or ligand interactions20,22. Comparison of reactivities at different points during transcription allows structural rearrangment events to be identified as transcription proceeds.
Results
Cotranscriptional folding of the E. coli SRP RNA
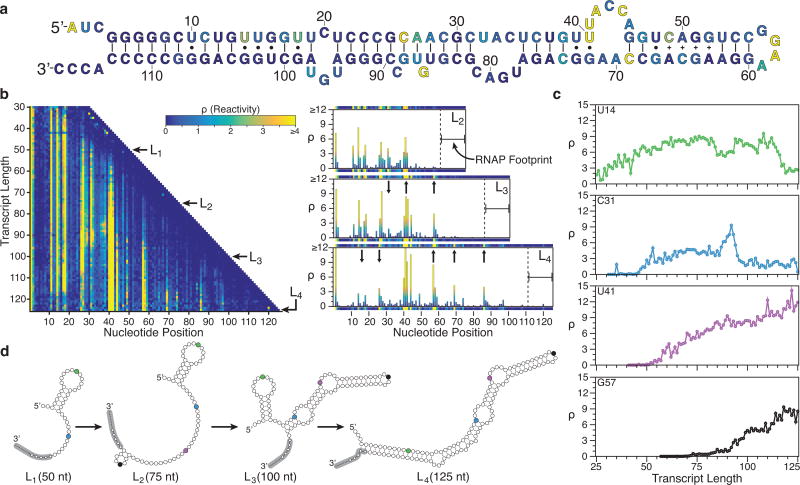
To validate cotranscriptional SHAPE-Seq, we first examined the signal recognition particle (SRP) RNA from E. coli. The final folded form of the SRP RNA is an extended helical structure containing interspersed inner loops (Fig. 2a)23. Biochemical studies have suggested that the 5’ end forms a labile hairpin structure early during transcription that rearranges into the extended helix only after the 3’ end is synthesized1. To test if we could observe this rearrangement, we used cotranscriptional SHAPE-Seq to obtain a matrix of reactivity spectra for the intermediate lengths of the nascent SRP RNA transcripts (Fig. 2b).
Fig. 2.
SRP RNA cotranscriptional folding. (a) Secondary structure of the final SRP RNA fold colored by reactivity intensity at length L4 (125 nts) drawn according to a crystal structure determined in Batey et al.23. (b) Cotranscriptional SHAPE-Seq reactivity matrix for the SRP RNA folding pathway (left). Lengths L1, L2, L3, and L4 correspond to 50, 75, 100, and 125 nts, respectively. Selected bar charts and corresponding matrix rows above and below (right) display reactivities for L2 to L4. Reactivity changes for L2 → L3 and for L3 → L4 are marked with arrows. (c) Reactivity values for positions U14, C31, U41, and G57 over the course of transcription. U14 undergoes a loop → helix transition at length 117. Similarly, C31 becomes paired at length 96. Plot colors correspond to the marked base positions in (d) outlining a proposed folding pathway of the SRP RNA that is consistent with these transitions. The 14 nt RNAP footprint24 for each length is indicated with gray, with the ~5 nts in the RNA exit channel marked as small circles. Results shown in b and c are n=1 and are representative of three biological replicates (Supplementary Fig. 9a).
Over the course of transcription, changes in nucleotide reactivity patterns (Fig. 2c) suggest a series of structural transitions that correspond to the early formation of a stem loop that ultimately rearranges into the elongated SRP RNA helical structure (Fig. 2d). Formation of the early stem-loop structure is observed as a cluster of highly reactive nucleotides (nts) across positions 11–18 in the loop. The pattern of high reactivity persists until the SRP RNA reaches a length of 117 nts when the sharp drop in reactivity at these positions indicates rearrangement into the elongated helical structure. We observed similar transitions when intermediate SRP RNA fragments were refolded at equilibrium before probing (Supplementary Fig. 1a), although the cotranscriptional transitions occur later due to the 14 nt RNAP footprint protecting the RNA 3’ ends24 (Supplementary Fig. 1b).
To further validate cotranscriptional SHAPE-Seq, we repeated our analysis of the SRP RNA using dimethyl sulfate (DMS), an RNA structure probe that preferentially modifies unstructured A and C bases25 (Supplementary Fig. 2). Overall the resulting reactivity matrix agrees with the BzCN data and similar structural transitions were observed in both datasets. However, the DMS dataset shows weaker reactivities at G and U bases, as expected, which limits the overall resolution of important structural transitions due to the lack of A and C residues in key regions of the SRP RNA sequence.
Ligand-mediated folding of the B. cereus fluoride riboswitch
Based on our SRP RNA results, we expected cotranscriptional SHAPE-Seq to possess the resolution necessary to reveal how alternative folding pathways are controlled during ligand-mediated transcription regulation by a riboswitch. To test this, we examined the B. cereus crcB fluoride riboswitch, which controls transcription by preventing the formation of an intrinsic terminator hairpin in the presence of fluoride26. Covariation and equilibrium structural analyses have suggested how the fluoride bound and unbound forms of the aptamer domain may interact with the downstream expression platform sequence (Fig. 3a)26,27. However, the specific mechanism by which fluoride binding directs or prevents the folding of the intrinsic terminator during transcription has yet to be elucidated.
Fig. 3.
B. cereus fluoride riboswitch cotranscriptional SHAPE-Seq data. (a) The antiterminated and terminated folds26 of the fluoride riboswitch, colored by the reactivity values at lengths 90 nt and 82 nt, respectively (bottom). (b) Cotranscriptional SHAPE-Seq reactivity matrices for the fluoride riboswitch transcribed with 10 mM (top) or 0 mM NaF (bottom). (c) Reactivity differences (Δρ) between the matrices in (b) annotated according to folding events during transcription. The reactivity changes that occur over the course of transcription suggest that the fluoride riboswitch traverses two cotranscriptional folding pathways depending on the presence of ligand. Results shown in b and c are n=1 and are representative of three biological replicates (Supplementary Fig. 9c,d).
To probe the OFF (terminated) and ON (antiterminated) structural states of the B. cereus fluoride riboswitch, we generated cotranscriptional SHAPE-Seq reactivity matrices in the presence of either 0 mM or 10 mM NaF, respectively (Fig. 3 and Supplementary Fig. 3a). Comparison of the matrices reveals both ligand-independent similarities in the initial aptamer folding followed by a bifurcation in the folding pathway that accompanies fluoride binding and fluoride-directed antitermination (Figs. 3c and 4a). Early in transcription, the B. cereus fluoride riboswitch folds into two hairpins that precede formation of the aptamer, regardless of fluoride concentration. The first hairpin forms within the first ~40 nts of transcription and is comprised of the P1 stem and a highly reactive loop between nts 11–16 (Supplementary Fig. 4a–c). The second hairpin forms shortly thereafter and is comprised of the P3 helix and a loop that exhibits a highly reactive position at U34 and low to moderate reactivities elsewhere (Supplementary Fig. 4d–f).
Fig. 4.
Changes in reactivities over transcript lengths for single nucleotides highlight structural transitions in the cotranscriptional folding pathway of the B. cereus fluoride riboswitch. (a) Single nucleotide reactivity trajectories for nucleotides involved in several key structural transitions when transcribed with either 0 mM (gray) or 10 mM NaF (black). The trajectories diverge at lengths were structural transitions occur. Results shown are extracted from the matrices presented in Fig. 3b. (b) As in (a), except that the RNAs were transcribed, extracted, denatured, and equilibrium refolded in transcription buffer with either 0 mM (gray) or 10 mM NaF (black) prior to SHAPE modification. The lack of divergence between the equilibrium refolding trajectories indicates that cotranscriptional folding is required to obtain alternate ligand-dependent structures. Results shown are extracted from the matrices presented in Supplementary Fig. 8a.
Folding of the P1 and P3 hairpins sets the stage for folding of the aptamer. At transcript length 58 we observe a fluoride-independent drop in the reactivity values at nts 12–16 in the P1 loop that signals the formation of pseudoknot PK1 between nts 12–17 and 42–47 as the latter emerge from the RNA exit channel (Fig. 4a and Supplementary Fig. 5a). Once PK1 is formed the aptamer is complete26,27, demonstrating that it first enters a pre-organized state independent of ligand, as has been observed for other aptamers28,29. From here, the fate of the aptamer structure is directed by the presence or absence of fluoride.
The first steps in the fluoride-dependent bifurcation of the folding pathway involve fluoride-mediated aptamer stabilization. In the presence of fluoride, the P1 loop reactivities continue to decrease until length 69, suggesting that fluoride binding stabilizes the pseudoknot (Fig. 4a and Supplementary Fig. 5a). Stabilization of the fluoride-binding pocket also requires a long-range non-canonical base pair between U38 and A10 (Fig. 3a)26,27, the latter of which is paired in the P1 stem prior to PK1 formation. In the presence of fluoride we first observe an increase in A10 reactivity, indicating that PK1 formation disrupts its base pair within the P1 stem. A10 then transitions to low reactivity at length 58, indicating the formation of a long range interaction with U38 following PK1 formation (Fig. 4a). In the absence of fluoride the sustained high reactivity at A10 suggests its interaction with U38 is not favored without ligand. Our observation of fluoride-induced aptamer stabilization is further supported by distinct reactivity changes in nts 22–27, which join the P1 and P3 helices but do not participate in any pairing interactions in the analogous T. petrophila aptamer domain27. Specifically, nts 24, 25, and 27 display lower reactivities in the presence of fluoride (Supplementary Fig. 5b) after length 58. In contrast, A22 undergoes a dramatic reactivity spike upon PK1 formation when fluoride is present, but only a modest increase in the absence of fluoride (Fig. 4a) revealing that A22 hyper-reactivity is a strong indicator of aptamer state. Taken together, these results support a model in which the pseudoknot forms the basis of the aptamer that can undergo further coordinated restructuring upon fluoride binding to form a more stable structure with additional interactions.
Following aptamer formation, the riboswitch follows one of two ligand-dependent folding trajectories that direct transcription termination or antitermination. When RNAP reaches length 69, the loop of the terminator hairpin begins to emerge from the RNA exit channel (Fig. 4a). Without fluoride, the terminator hairpin nucleates at length 77, observed as a decrease in reactivity in the upper terminator stem (nts 52–55) (Fig. 4a and Supplementary Fig. 5c). Increased reactivity in the P1 loop (nts 12–16) (Supplementary Fig. 5a) and decreased reactivity at A22 occur concurrently, indicating that PK1 opens, thereby dissolving the meta-stable aptamer (Fig. 4a).
With fluoride, the stabilized aptamer promotes antitermination in two ways: 1) disfavoring complete terminator formation by sequestering part of the terminator hairpin (Fig. 3a) and 2) delaying terminator hairpin nucleation until length 88, after RNAP has transcribed past the poly-U sequence (Fig. 4a and Supplementary Fig. 5c). When the terminator hairpin does begin to form, high reactivities at U48 and nts 69–74 indicate that only the top half of the terminator winds (Figs. 3 and 4a), leaving the ribosome binding site (RBS, nts 67–72) accessible for translation of the downstream fluoride transporter (Supplementary Fig. 5d)26. Thus, fluoride binding fundamentally alters the cotranscriptional folding pathway of the B. cereus fluoride riboswitch by stabilizing an RNA structure that promotes transcription via antitermination and translation by RBS exposure.
Riboswitch mutants alter folding pathways in defined ways
Previous work on the B. cereus fluoride riboswitch examined mutations in the aptamer and terminator stem26. Each mutation confers distinct changes in ligand binding and termination capability that correspond to its location in the riboswitch (Supplementary Fig. 3b–d). Therefore, we used these mutants to both corroborate our interpretations of the wt fluoride riboswitch cotranscriptional SHAPE-Seq data (Figs. 3 and 4) and uncover details of how individual mutations impact the cotranscriptional folding and function of the riboswitch (Fig. 5 and Supplementary Figs. 6 and 7).
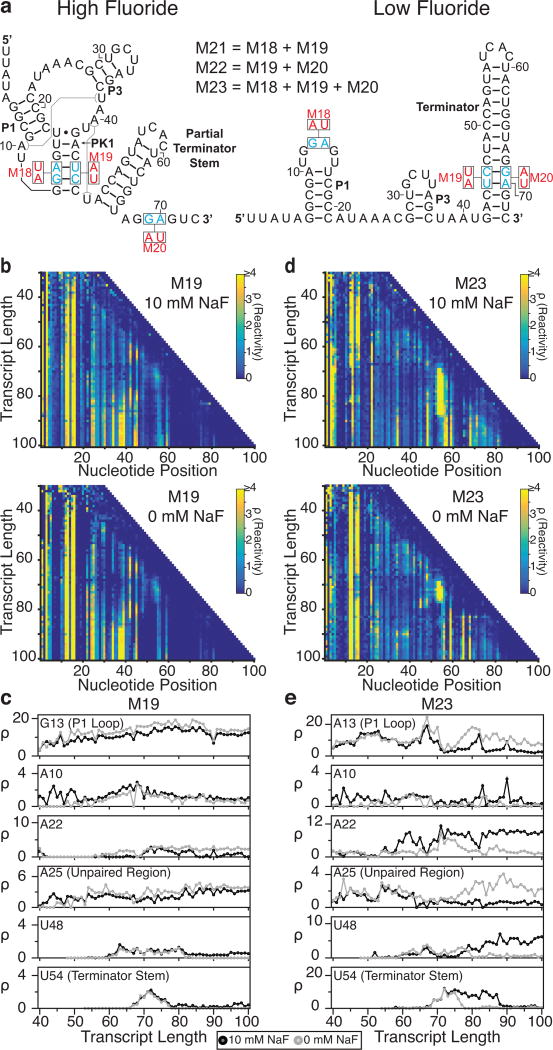
Fig. 5.
Cotranscriptional SHAPE-Seq analysis of B. cereus fluoride riboswitch mutants. (a) The locations of mutations M18-M23 from Baker et al.26 within the antiterminated (high fluoride) and terminated (low fluoride) secondary structures for the wild-type system (see Fig. 3a). (b) Reactivity matrices of the M19 mutant (pseudoknot and terminator base pairing disrupted) transcribed with either 10 mM (top) or 0 mM NaF (bottom). (c) Single nucleotide cotranscriptional reactivity trajectories for the same key nucleotides highlighted in Fig. 4 for the M19 mutant when transcribed with either 0 mM (gray) or 10 mM NaF (black). (d) As in (b), but for mutant M23 (pseudoknot and terminator base pairing restored through compensatory mutations). (e) As in (c), but for mutant M23. Results shown are n=1.
We first sought to corroborate our interpretation of structural transitions that lead to aptamer formation by analyzing mutants that disrupt PK1 formation. Mutant M19 (U45A/C46U) (Fig. 5b,c) interrupts base pairing in both PK1 and the terminator stem. Mutants M18 (G13A/A14U) (Supplementary Fig. 6a–c) and M22 (M19+M20) (Supplementary Fig.6d–f) disrupt PK1 folding but maintain base pairing in the terminator stem. Consequently, M18, M19, and M22 cannot properly form the fluoride aptamer and are therefore fluoride-insensitive as observed in functional in vitro termination assays (Supplementary Figure 3d). From a structural perspective, we observe this defect in aptamer formation as the lack of major differences in cotranscriptional SHAPE-Seq datasets for these mutants in the presence and absence of fluoride, and an absence of transitions that reflect aptamer formation and stabilization. The most notable signature of fluoride insensitivity is consistently high P1 loop reactivities in both the presence and absence of fluoride, indicating that PK1 does not form in these mutants. Furthermore, A22 remains weakly reactive compared to the wt riboswitch across all transcript lengths (Fig. 5b,c and Supplementary Fig. 6), further supporting the interpretation that reactivity measurements at A22 are a strong indicator of aptamer state. Together these data support the conclusion that aptamer formation is necessary for the ligand-dependent bifurcation of the fluoride riboswitch folding pathway that leads to the regulation of terminator formation.
We next sought to examine how interactions between the aptamer and intrinsic terminator coordinate structural changes that lead to bifurcation of the folding pathway. Mutant M20 (G69A/A70U) contains base substitutions in the 3’ terminator stem that render the intrinsic terminator nonfunctional but do not disrupt the wt aptamer domain (Supplementary Fig. 3b–d). Mutant M21 (M18+M19) restores PK1 base-pairing but, like M20, does not form a complete terminator stem. As anticipated, the M20 and M21 mutants are non-functional due to inactivation of the terminator but undergo all transitions associated with aptamer formation and fluoride binding (Supplementary Fig. 7a–c). Interestingly, M21 displays consistently high P1 loop reactivity in the absence of fluoride and shows delayed formation in the presence of fluoride, suggesting that that PK1 does not stabilize as readily for this mutant, likely due to the replacement of a G:C base pair in PK1 with A:U. However, the characteristic increase in A22 still occurs, suggesting that PK1 still forms, but for a smaller fraction of the folded population. Taken together, these results indicate that M20 and M21 undergo nearly all transitions as the wt system and are non-functional only because the mutations disrupt the final event in termination formation.
To further corroborate our observations of the wt system, we analyzed the M23 mutant (M18+M19+M20), which restores base pairing in both PK1 and the terminator stem. (Fig. 5d,e) M23 can form the fluoride aptamer and is therefore capable of binding fluoride and, in the absence of fluoride, terminating transcription at near-wt levels (Supplementary Fig. 3d). As expected, we observe similar reactivity patterns within the matrices from M23 and the wt riboswitches during the formation of the aptamer domain, indicating that restoring base pairing is sufficient to reproduce a near-native folding pathway (Fig. 5d,e). However, similar to M21, M23 contains weakened base pairing in PK1 and does not form a stable aptamer as readily as the wt riboswitch in the absence of fluoride as can be seen in the absence of an increase in reactivity at A10 near length 58 nts. The analysis of M23 demonstrates that the overall folding pathway and decision-making events of the fluoride riboswitch can be enacted by different RNA sequences.
Cotranscriptional SHAPE-Seq accesses kinetically trapped RNA structures
We next sought to assess whether cotranscriptional SHAPE-Seq accesses non-equilibrium, kinetically trapped folded states of nascent RNAs by directly comparing our results to an equilibrium analysis whereby RNAs are equilibrium refolded prior to probing. To equilibrium refold all fluoride riboswitch intermediates, we first generated all of the intermediate transcript lengths as done for cotranscriptional SHAPE-Seq, but extracted, denatured, and equilibrium refolded the RNAs in transcription buffer prior to chemical probing.
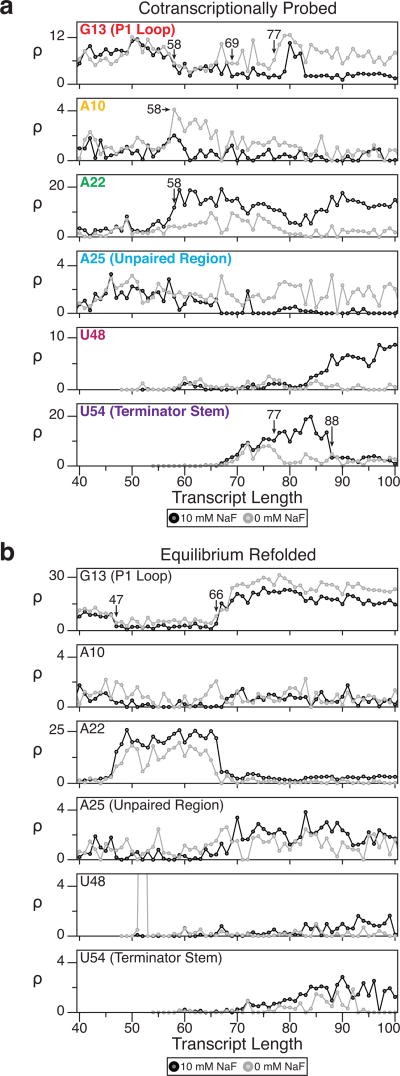
In our comparison we found two main differences between the cotranscriptional and equilibrium experiments that show that cotranscriptional experiments probe non-equilibrium folding states (Fig. 4b and Supplementary Fig. 8). Because cotranscriptional SHAPE-Seq experiments probe RNAs that exist as part of a transcription elongation complex, only folding events that involve nts that have left, or are in, the RNA exit channel of RNAP can be observed. During equilibrium refolding however, RNAP is not present to prevent the 3’ end of the transcript from folding with the rest of the RNA, causing the structural transitions to occur at shorter transcript lengths than we observe with cotranscriptional SHAPE-Seq. Specifically, we observe ligand-independent folding of PK1 (via decreases in P1 loop reactivities) at transcript length 47 with equilibrium refolding (Fig. 4b and Supplementary Fig. 8) as opposed to length 58 (Figs. 3 and 4a) when probing the arrested elongation complexes cotranscriptionally. In cotranscriptional SHAPE-Seq experiments, length 58 corresponds to a point when nt 47 is expected to be leaving the RNA exit channel. Shifts in structural transitions to earlier lengths during equilibrium refolding were also observed for the SRP RNA (Supplementary Fig. 1a,b).
The second piece of evidence supporting the capture of non-equilibrium RNA structures is the distinct fluoride-independent behavior of the P1 loop at longer transcript lengths in the equilibrium refolding experiments. Specifically, equilibrium refolding produces a sharp, fluoride-independent rise in reactivities of the P1 loop nucleotides over transcript lengths 66–69, indicating the complete opening of PK1 in both the absence and presence of ligand (Fig. 4b). However, in the cotranscriptional SHAPE-Seq experiment PK1 remains stabilized at longer transcript lengths in the presence of fluoride. The observed deviation from equilibrium structures indicates that the RNAs probed in cotranscriptional SHAPE-Seq are out of equilibrium.
Based on these results, we conclude that equilibrium refolding does not permit meaningful analysis of the cotranscriptional folding pathways of the fluoride riboswitch beyond length 66 because terminator hairpin formation is thermodynamically favored regardless of fluoride concentration. However, this observation is important, as it reveals the relative thermodynamic favorability of the terminator structure over the aptamer. Furthermore, in the terminated structure G68 pairs with C47, the latter of which participates in the last base pair of PK1 within the aptamer. Thus the loss of a single base pair in PK1 to pairing in the terminator stem is sufficient to preclude aptamer folding in favor of terminator folding. This thermodynamic tipping point corresponds well to the cotranscriptional opening of PK1 at length 77 in the absence of fluoride. During transcription without fluoride, the instability of PK1 could enable terminator base-pairing to extend into the RNA exit channel, thereby favoring the terminated structure.
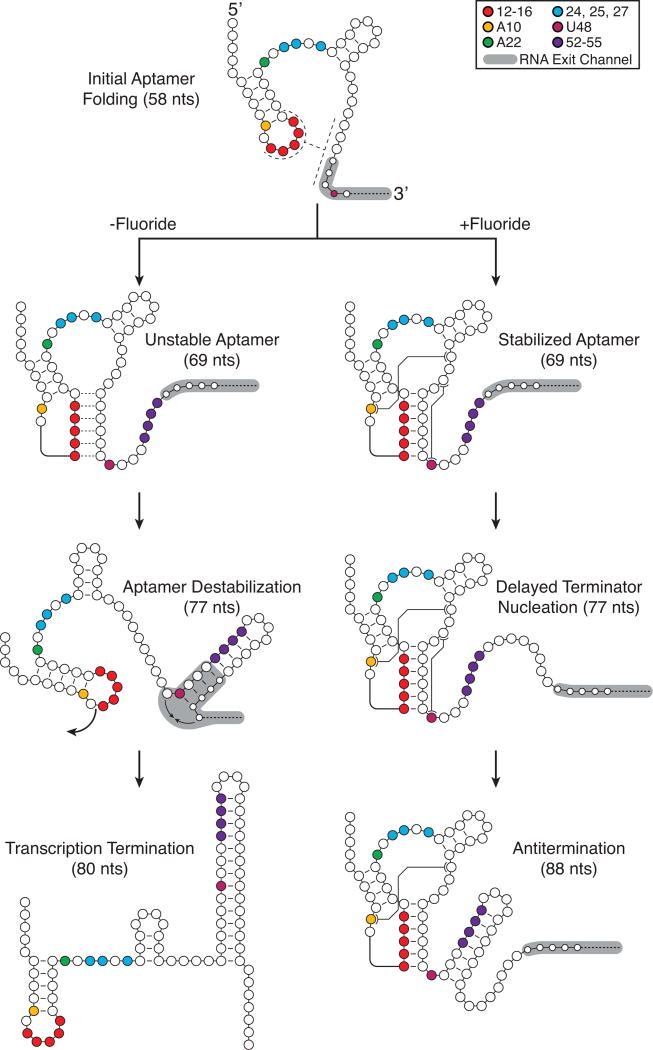
A model of the folding pathway and decision making process of the B. cereus fluoride riboswitch that combines these observations with the cotranscriptional analysis of the wt and mutant systems is shown in Fig. 6.
Fig. 6.
A model for ligand-dependent cotranscriptional folding of the B. cereus fluoride riboswitch. The folding pathway for the fluoride riboswitch begins with initial aptamer folding. If fluoride binds (top), the pre-folded aptamer then stabilizes through specific interactions, leading to delays in the early folding stages of the intrinsic terminator hairpin, which does not nucleate until RNAP has escaped the polyU tract. However, if there is no fluoride binding (bottom), the top of the terminator hairpin quickly folds disrupting the pseudoknot and reaching into the RNA exit channel to allow the full terminator hairpin to trigger transcription termination. Intermediate structural states are inferred from cotranscriptional SHAPE-Seq reactivities, covariation analysis26, and crystallographic data27.
Discussion
We have developed cotranscriptional SHAPE-Seq to facilitate the experimental characterization of cotranscriptional RNA folding at nucleotide resolution. Furthermore, we have demonstrated that cotranscriptional SHAPE-Seq interrogates the structure of non-equilibrium, kinetically trapped nascent RNAs to generate functionally meaningful structural information. Replicate datasets for the E. coli SRP RNA and the B. cereus crcB fluoride riboswitch show that the technique is highly reproducible (Supplementary Fig. 9).
When interpreting cotranscriptional SHAPE-Seq data it is important to consider the relative timescales of RNA folding and RNA synthesis as well as the advantages and limitations of chemical RNA structure probing measurements. Whereas the uninterrupted nucleotide addition cycle of E. coli RNAP occurs at 50–100 nt/s (10–20 ms/nt)30, simple RNA folding events such as base-pair melting occur on a microsecond to millisecond timescale31, with larger conformation changes occurring on the order of seconds to tens of seconds. While desirable, interrogation of RNA folding on the microsecond to millisecond timescale is inaccessible with even the most fast-acting SHAPE probe, BzCN, which reacts with a t1/2 of 250 ms19. This is further exacerbated by chemical probes like DMS that have reaction times on the orders of minutes32. Furthermore, in its current form, the temporal resolution of cotranscriptional SHAPE-Seq is restricted by the manual manipulation of samples and the need to halt transcription elongation complexes prior to RNA probing. Thus, the power of cotranscriptional SHAPE-Seq lies not in temporal sensitivity, but in the capability of capturing single-nucleotide resolution “snapshots” of kinetically trapped intermediate folds that have not equilibrated within the seconds timescale of the experiment. As such, it complements single-molecule force spectroscopy, which produces high temporal resolution but low spatial resolution of RNA structural changes during transcription14. Despite its limitations, as demonstrated above, the resulting SHAPE-Seq reactivity profiles facilitate the identification of distinct nucleotide signatures that indicate nascent RNA structural state and the observation of major conformational rearrangements that occur as the RNA sequence is synthesized.
While the cotranscriptional SHAPE-Seq experiment occurs on timescales much longer than the rate of transcription, this does not preclude its use in analyzing the role of transcription dynamics in RNA folding. Transcription pausing, which can occur on the seconds timescale, has been shown to play a critical role in RNA folding through studies of several RNAs, including the E. coli SRP RNA1 and the btuB coenzyme B12 riboswitch33. By itself, cotranscriptional SHAPE-Seq does not measure transcription pausing directly, as transcription roadblocking obscures the observation of pause distributions and the saturating NTP concentrations used are not conducive to precise measurement of transcription pausing34. However, the structural states probed at a specific length of RNA would reflect any folding afforded by a transcription pause upstream of that length. Therefore, the consequences of altering native transcription dynamics on RNA folding would be observable in cotranscriptional SHAPE-Seq datasets.
Finally, it is important to consider that the stochastic nature of cotranscriptional RNA folding produces an ensemble of structural conformations. Because SHAPE-Seq measurements are made in bulk, reactivity profiles reflect an average over this ensemble. Interestingly, this can be seen by comparing equilibrium refolded SHAPE-Seq matrices to cotranscriptional SHAPE-Seq matrices, the latter generally showing blurrier transitions that likely reflect the probing of a sub-population of molecules that have not yet made the specific transitions (compare Fig. 2b to Supplementary Fig. 1a, Fig. 3b to Supplementary Fig. 8a). Detection of modification sites by mutational profiling (MaP)35 may be able to access information about the folding pathways of sub-populations if the MaP strategy can robustly detect the modification of RNA by BzCN.
We have used cotranscriptional SHAPE-Seq to evaluate the RNA folding pathway of a fluoride riboswitch and in this context have identified molecular signatures of aptamer folding and ligand binding as well as the precise point at which the nascent RNA mediates a genetic decision (Fig. 6). In agreement with force spectroscopy analysis of pbuE adenine riboswitch cotranscriptional folding14 and biochemical analysis of the folding pathways of the btuB coenzyme B12 riboswitch33, we find that the crcB fluoride riboswitch is controlled by the kinetics of its cotranscriptional folding events. Despite this similarity, there are key differences in the cotranscriptional folding pathways of the adenine and fluoride riboswitches both at the level of ligand sensing and stability of the ligand bound aptamer that suggest they use different overall strategies to make genetic regulatory decisions. One clear difference is in aptamer folding. While the pbuE adenine aptamer was not observed to fold in the absence of adenine14, our data indicate that the crcB fluoride aptamer adopts an “unstable” folded state in the absence of fluoride on the timescales of our experiment (Fig. 4a and Supplementary Fig. 5a). The fate of the fluoride-bound crcB aptamer is also distinct from the fate of the adenine-bound pbuE aptamer, which rearranges into an aptamer-disrupted structure following antitermination. In contrast, the crcB fluoride riboswitch aptamer is kinetically trapped and remains stable for at least 30s in the presence of fluoride, even following terminator nucleation (Figs. 3 and 4a). The origin of this distinction could be functional in nature: whereas the pbuE adenine riboswitch functions at the transcriptional level, the crcB fluoride riboswitch could exert dual transcriptional and translational control over gene expression by simultaneously preventing complete terminator folding and exposing a ribosome binding site that would otherwise be sequestered within the terminator hairpin (Fig. 5d). These mechanistic distinctions between fluoride and adenine riboswitch folding suggest diversity in the mechanisms through which cotranscriptional RNA folding can direct targeted RNA functions, such as ligand sensing and genetic control. A fundamental understanding of the relationship between RNA folding and function could be achieved with a broad characterization of the cotranscriptional folding pathways of diverse riboswitches and comparative studies of the conservation of RNA folding pathways for specific riboswitch variants, both of which are now afforded by our cotranscriptional SHAPE-Seq technique.
The work presented here provides an experimental means with which to answer fundamental questions about how the cotranscriptional nature of RNA folding directs RNA structure and function. We anticipate that the integrated analysis of measurements made using cotranscriptional SHAPE-Seq and complementary biophysical, biochemical, and computational techniques will provide a powerful framework with which to understand the roles of cotranscriptional folding in regulating broader cellular processes.
Online Methods
Plasmids
Plasmids used for DNA template synthesis contained a chloramphenicol resistance gene, the p15A origin of replication, and a consensus E. coli σ70 promoter followed by a sequence encoding the RNA under study. The E. coli SRP RNA sequence (Genbank NC_000913.3, bases 476448 to 476561) with the sequence ATC appended at the 5’ end1 was cloned upstream of the antigenomic hepatitis δ ribozyme. The B. cereus crcB fluoride riboswitch (Genbank AE017194.1, bases 4763724 to 4763805) was cloned upstream of a consensus ribosome binding site and the superfolder green fluorescent protein (SFGFP) sequence. These non-native downstream sequences were used to allow transcription to proceed far enough such that the full length RNA of interest would emerge from RNA polymerase (RNAP). These sequences do not influence cotranscriptional SHAPE-Seq interpretations, as lengths where non-native RNA had emerged from RNAP were not used to draw conclusions. The fluoride riboswitch mutants were derived from the plasmid described above.
Proteins
EcoRI E111Q (Gln111) was a generous gift from Jeffrey Roberts and Joshua Filter (Cornell University, Ithaca NY).
Template preparation
DNA template libraries (Supplementary Table 3) for cotranscriptional SHAPE-Seq were prepared by combining individual PCR amplifications of each RNA template length. Each 25 µL PCR reaction included 20.4 µL H2O, 2.5 µL 10X Standard Taq Reaction Buffer (New England Biolabs), 0.5 µL 10 mM dNTPs, 0.25 µL 100 µM oligo J (forward primer, Supplementary Table 4), 0.15 µL plasmid DNA template, 0.25 µl Taq DNA polymerase (New England Biolabs), and 1 µL 25 µM reverse primer (Supplementary Tables 4 and 5). The reverse primer incorporated an EcoRI site. Reaction mixes were run using a standard thermal cycle program consisting of 30 cycles of amplification using an annealing temperature of 55 °C. After thermal cycling, PCR reactions were pooled, mixed, and split into 500 µL aliquots before addition of 50 µL 3 M NaOAc pH 5.5 and 1 mL of 100% EtOH for EtOH precipitation. Precipitated pellets were dried using a SpeedVac and pooled by dissolving all pellets in 30 µL H2O. The template was then run on a 1% agarose gel and extracted using the QIAquick Gel Extraction Kit (Qiagen). The concentration of the purified template was measured using the Qubit Fluorometer (Life Technologies) and the molarity of the template was calculated using the median template length.
Single-length DNA templates (Supplementary Table 3) were prepared by performing five 100 µL PCR reactions including 82.25 µL H2O, 10 µL 10X Standard Taq Reaction Buffer (New England Biolabs), 1.25 µL 10 mM dNTPs, 2.5 µL 10 µM oligo J (forward primer, Supplementary Table 4), 2.5 µl 10 µM oligo K (Supplementary Table 4), 0.5 µL plasmid DNA template, and 0.5 µL Taq DNA polymerase (New England Biolabs). Reactions were run with the thermal cycling program described above. After thermal cycling, reactions were pooled before addition of 50 µL 3M NaOAc pH 5.5 and 1 mL of 100% EtOH for EtOH precipitation. The precipitated pellet was dried using a SpeedVac and dissolved in 30 µL H2O. The template was then run on a 1% agarose gel and extracted using the QIAquick Gel Extraction Kit (Qiagen). The concentration of the purified template was measured using the Qubit 2.0 Fluorometer (Life Technologies).
in vitro transcription (single length, radiolabeled)
25 µL reaction mixtures containing 5 nM linear DNA template (see above) and 0.5 U of E. coli RNAP holoenzyme (New England Biolabs) were incubated in transcription buffer (20 mM Tris-HCl pH 8.0, 0.1 mM EDTA, 1 mM DTT and 50 mM KCl), 0.1 mg/mL bovine serum albumin, 200 µM ATP, GTP, CTP and 50 µM UTP containing 0.5 µCi/µL [α-32P]-UTP for 10 min at 37 °C to form open complexes. When present, NaF was included to a final concentration of 1 µM, 10 µM, 100 µM, 1 mM or 10 mM as indicated in Supplementary Fig. 3a. Single-round transcription reactions were initiated by addition of MgCl2 to 5 mM and rifampicin to 10 µg/mL. Transcription was stopped by adding 125 µL of stop solution (0.6 M Tris pH 8.0, 12 mM EDTA, 0.16 mg/mL tRNA).
RNA from stopped transcription reactions was purified by addition of 150 µL of phenol:chloroform:isoamyl alcohol (25:24:1, v/v), vortexing, centrifugation, and collection of the aqueous phase that was then ethanol precipitated by adding 450 µL of 100% ethanol to each reaction and storage at −20 °C overnight. Precipitated RNA was resuspended in transcription loading dye (1X transcription buffer, 80% formamide, 0.05% bromophenol blue and xylene cyanol). Reactions were fractionated by electrophoresis using 12% denaturing polyacrylamide gels containing 7.5 M urea (National Diagnostics, UreaGel). Reactive bases were detected using an Amersham Biosciences Typhoon 9400 Variable Mode Imager. Quantification of bands was performed using ImageQuant. For all experiments, individual bands were normalized for incorporation of [α-32P]-UTP by dividing band intensity by the number of Us in the transcript. %Readthrough was calculated by dividing the sum of run-off RNAs by the sum of all terminated and run-off products.
in vitro transcription (cotranscriptional SHAPE-Seq experiment)
50 µL total reaction mixtures containing 100 nM linear DNA template library (see above) and 4 U of E. coli RNAP holoenzyme (New England Biolabs) were incubated in transcription buffer (20 mM Tris-HCl pH 8.0, 0.1 mM EDTA, 1 mM DTT and 50 mM KCl), 0.2 mg/mL bovine serum albumin, and 500 µM NTPs for 7.5 min at 37 °C to form open complexes. When present, NaF was included to a final concentration of 10 mM. Following the first incubation, EcoRI Gln111 dimer was added to a final concentration of 500 nM and incubated at 37 °C for another 7.5 min. Immediately following the second incubation, single-round transcription reactions were initiated by addition of MgCl2 to 5 mM and rifampicin to 10 µg/ml. All transcription reactions were allowed to proceed for 30 seconds. Cotranscriptional experiments were then directly SHAPE modified (see RNA modification and purification below). Equilibrium refolding experiments were stopped by addition of 150 µL TRIzol solution (Life Technologies), purified, and equilibrium refolded in transcription buffer before SHAPE modification as described below (see RNA modification and purification).
in vitro transcription (single length, unlabeled)
in vitro transcription of single length, unlabeled RNA was performed as described above for cotranscriptional SHAPE-Seq, except at 25 µL total volume with 2 U of E. coli RNAP holoenzyme (New England Biolabs) and without Gln111 addition or SHAPE modification. The resulting RNAs were purified as described for cotranscriptional SHAPE-Seq and fractionated using a 10% denaturing polyacrylamide gel containing 7.0 M urea. The resulting gel was stained using SYBR Gold (Life Technologies) and imaged using a Bio-Rad ChemiDoc MP system and quantified using Image Lab (Bio-Rad). %Readthrough was calculated as described above.
RNA modification and purification
For cotranscriptional experiments the 30 second transcription products were immediately SHAPE modified by splitting the reaction and mixing half with 2.78 µL of 400 mM benzoyl cyanide (BzCN; Pfaltz & Bower) dissolved in anhydrous dimethyl sulfoxide (DMSO; (+) sample) or anhydrous DMSO only (Sigma Aldrich; (−) sample) for ~2 seconds before addition of 75 µL of TRIzol solution. DMS modification was performed by splitting the reaction and mixing half with 2.78 µL of 3.5% DMS in ethanol ((+) sample) or 100% ethanol only ((−) sample) and incubating at 37 °C for 3 min before quenching the reaction with 6.67 µL of β-mercaptoethanol, incubation at 37 °C for 1 min and addition of 75 µL of TRIzol solution. Transcription products for equilibrium refolding had 150 µL TRIzol added after in vitro transcription. Both were extracted according to the manufacturer’s protocol and dissolved in 20 µL total of 1X DNase I buffer (New England Biolabs) containing 1 U of DNase I enzyme. Digestion proceeded at 37 °C for 30 min, after which 30 µL of RNase-free H2O was added, followed by 150 µL TRIzol. The RNA samples were then extracted again according to the manufacturer’s protocol and dissolved in either: 10 µL 10% DMSO in H2O (cotranscriptional experiments) or 25 µL RNase-free H2O (equilibrium refolding experiments). Equilibrium refolding experiment samples were then heated to 95 °C for 2 min, snap cooled on ice for 1 min, and refolded in 1X folding buffer for 20 min at 37 °C (20 mM Tris-HCl pH 8.0, 0.1 mM EDTA, 1 mM DTT, 50 mM KCl, 0.2 mg/mL bovine serum albumin, and 500 µM NTPs), optionally containing 10 mM fluoride. RNA modification of the equilibrium refolding samples was performed as described above, followed by the addition of 30 µL RNase-free H2O and 150 µL TRIzol and extracted a third time according to the manufacturer’s instructions. The resulting pellet was dissolved in 10 µL 10% DMSO in H2O.
Linker preparation
The phosphorylated linker, oligonucleotide A (Supplementary Table 4), was purchased from Integrated DNA Technologies and adenylated with the 5’ DNA Adenylation Kit (New England Biolabs) according to the manufacturer’s protocol at a 20X scale, dividing the reactions into 50 µL aliquots. After completion of the reaction, 150 µL TRIzol was added and the reactions were extracted according to the manufacturer’s instructions, dissolving the products in 20 µL RNase-free H2O. The concentration of purified linker was measured using the Qubit Fluorometer (Life Technologies) and the molarity of the RNA was calculated using 6782.1g/mol as the molecular weight. The adenylation reaction was assumed to be 100% efficient. The linker was diluted to a 2 µM stock to be used later.
Linker ligation
To the modified and unmodified RNAs in 10% DMSO (see RNA modification and purification above), 0.5 µL of SuperaseIN (Life Technologies), 6 µL 50% PEG 8000, 2 µL 10X T4 RNA Ligase Buffer (New England Biolabs), 1 µL of 2 µM 5’-adenylated RNA linker, and 0.5 µL T4 RNA Ligase, truncated KQ (200 U/µL; New England Biolabs) were added to bring the total reaction volume to 20 µL. The reactions were mixed well and incubated overnight (>10 hours) at room temperature.
Reverse transcription
The completed linker ligations were brought to 150 µL with RNase-free H2O before addition of 15 µL 3 M NaOAc, 1 µL 20 mg/mL glycogen, and 450 µL EtOH for EtOH precipitation. Precipitated pellets were dissolved in 10 µL RNase-free H2O. Then, 3 µL of 0.5 µM reverse transcription primer, oligonucleotide B (Supplementary Table 4), were added. The resulting mix was then denatured completely by heating to 95 °C for 2 min, followed by an incubation at 65 °C for 5 min before placing on ice for ~30 seconds. Then, 7 µL of SSIII master mix was added, containing: 0.5 µL of Superscript III (Life Technologies), 4 µL 5X First Strand Buffer (Life Technologies), 1 µL 100 mM (DTT), 1 µL 10 mM dNTPs, and 0.5 µL RNase-free H2O. The reaction mix was further incubated at 42 °C for 1 min, then 52 °C for 25 min and deactivated by heating at 65 °C for 5 min. The RNA was then hydrolyzed by the addition of 1 µL of 4 M NaOH solution and heating at 95 °C for 5 min. The basic solution containing the cDNA was partially neutralized with 2 µL of 1 M HCl, then precipitated with 69 µL cold EtOH, using 15 min at −80 °C and 15 min of spinning at 4 °C at max speed to pellet the RNA before washing the pellet with 70% EtOH. The washed pellet, free of base, was dissolved in 22.5 µL of nuclease-free H2O.
Adapter ligation
To the cDNA, 3 µL 10X CircLigase Buffer (Epicentre), 1.5 µL 50 mM MnCl2, 1.5 µL 1 mM ATP, 0.5 µL 100 µM DNA adapter, oligonucleotide C (Supplementary Table 4), and 1 µL CircLigase I (Epicentre) were added. The reaction was incubated at 60 °C for 2 hr, then 80 °C for 10 min to inactivate the ligase. The ligated DNA was EtOH precipitated with 1 µL 20 mg/mL glycogen as a carrier and dissolved in 20 µL of nuclease-free H2O. Then the cDNA was purified using 36 µL of Agencourt XP beads (Beckman Coulter), according to manufacturer’s instructions and eluted with 20 µL TE buffer.
Quality analysis
For quality analysis (QA), a separate PCR reaction for each (+) and (−) sample was mixed by combining: 13.75 µL nuclease-free H2O, 5 µL 5X Phusion Buffer (New England Biolabs), 0.5 µL 10 mM dNTPs, 1.5 µL of 1 µM labeling primer (oligonucleotides D/E (Supplementary Table 4)), 1.5 µL of 1 µM primer PE_F (oligonucleotide F (Supplementary Table 4)), 1 µL of 0.1 µM selection primer (oligonucleotides G/H (Supplementary Table 4)), 1.5 µL ssDNA library (+ or -), and 0.25 µL Phusion DNA polymerase (New England Biolabs). Both fluorescent primers were purchased from Applied Biosystems and the selection primers were purchased from Integrated DNA Technologies. Asterisks represent phosphorothioate modifications to prevent the 3’→5’ exonuclease activity of Phusion polymerase. Amplification was performed for 15 cycles first, using an annealing temperature of 65 °C and an extension time of 15 seconds, excluding the PE_F primer. Then, the PE_F primer was added for an additional 10 cycles of amplification. To the complete reactions 50 µL nuclease-free H2O was added, and the diluted reaction was ethanol precipitated. The resulting pellet was dissolved in formamide and analyzed with an ABI 3730xl capillary electrophoresis device.
Library preparation and next generation sequencing
To construct sequencing libraries, a separate PCR for each (+) and (−) sample was mixed by combining: 33.5 µL nuclease-free H2O, 10 µL 5X Phusion Buffer (New England Biolabs), 0.5 µL 10 mM dNTPs, 0.25 µL of 100 µM TruSeq indexing primer (oligonucleotide I (Supplementary Table 4)), 0.25 µL of 100 µM primer PE_F, 2 µL of 0.1 µM selection primer (+ or −, as noted above), 3 µL ssDNA library (+ or −), and 0.5 µL Phusion DNA polymerase (New England Biolabs). Amplification was performed as indicated in ‘Quality analysis’ above. Completed reactions were chilled at 4 °C for 2 min before addition of 5 U exonuclease I (New England Biolabs) to remove unextended primer. The reactions were then incubated at 37 °C for 30 min. Following incubation, 90 µL of Agencourt XP beads (Beckman Coulter) were added for purification according to manufacturer’s instructions. The complete libraries were eluted with 20 µL TE buffer and quantified with the Qubit 2.0 Fluorometer (Life Technologies).
To prepare the libraries for sequencing, the average length of each sample was determined using the results from the quality analysis in order to calculate the molarity of each (+) or (−) separately. Sequencing pools were mixed to be equimolar, such that all of the sequencing libraries were present in the solution at the same level. Sequencing was performed on the Illumina HiSeq 2500 in either ‘rapid run’ or ‘high output’ mode, using 2×36 bp paired end reads. To help overcome the low-complexity of the linker region during sequencing 10–20% PhiX DNA was included.
Data Analysis
For a detailed description of the cotranscriptional SHAPE-Seq data analysis pipeline, see Supplementary Note 1.
Code Availability
Spats v1.0.1 can be accessed at https://github.com/LucksLab/spats/releases/. Scripts used in data processing are located at https://github.com/LucksLab/Cotrans_SHAPE-Seq_Tools/releases/.
Data Availability
Raw sequencing data that support the findings of this study have been deposited in the Small Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) with the BioProject accession code PRJNA342175. Individual BioSample accession codes are available in Supplementary Table 1. SHAPE-Seq Reactivity Spectra generated in this work have been deposited in the RNA Mapping Database (RMDB) (http://rmdb.stanford.edu/repository/) with the accession codes FLUORSW_BZCN_0001, FLUORSW_BZCN_0002, FLUORSW_BZCN_0003, FLUORSW_BZCN_0004, FLUORSW_BZCN_0005, FLUORSW_BZCN_0006, FLUORSW_BZCN_0007, FLUORSW_BZCN_0008, FLUORSW_BZCN_0009, FLUORSW_BZCN_0010, FLUORSW_BZCN_0011, FLUORSW_BZCN_0012, FLUORSW_BZCN_0013, FLUORSW_BZCN_0014, FLUORSW_BZCN_0015, FLUORSW_BZCN_0016, FLUORSW_BZCN_0017, FLUORSW_BZCN_0018, FLUORSW_BZCN_0019, FLUORSW_BZCN_0020, SRPECLI_BZCN_0001, SRPECLI_BZCN_0002, SRPECLI_BZCN_0003, SRPECLI_BZCN_0004, SRPECLI_DMS_0001, SRPECLI_DMS_0002, and SRPECLI_DMS_0003. Sample details are available in Supplementary Table 2. Source data for Figure 2–Figure 5 and Supplementary Figures 1, 2, and 4–9 are available with the paper online. All other data that support the findings of this paper are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank J. Roberts (Cornell University), J. Filter (Cornell University), and I. Artsimovitch (The Ohio State University) for EcoRI Gln111 protein, its expression plasmid, and thoughtful discussions. We also thank J. Liao for plasmid construction. This work was supported by the Tri-Institutional Training Program in Computational Biology and Medicine [NIH training grant T32GM083937 to AMY], a New Innovator Award through the National Institute of General Medical Sciences of the National Institutes of Health [grant number 1DP2GM110838 to JBL], the National Institute of General Medical Sciences of the National Institutes of Health [grant GM25232 to JTL], a National Science Foundation Graduate Research Fellowship Program [grant number DGE-1144153 to KEW], and Searle Funds at The Chicago Community Trust [to JBL]. KEW is a Fleming Scholar in the Robert F. Smith School of Chemical and Biomolecular Engineering at Cornell University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Accession Codes
Raw sequencing data that support the findings of this study have been deposited in the Small Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) with the BioProject accession code PRJNA342175. Individual BioSample accession codes are available in Supplementary Table 1. SHAPE-Seq Reactivity Spectra generated in this work have been deposited in the RNA Mapping Database (RMDB) (http://rmdb.stanford.edu/repository/) with the accession codes FLUORSW_BZCN_0001, FLUORSW_BZCN_0002, FLUORSW_BZCN_0003, FLUORSW_BZCN_0004, FLUORSW_BZCN_0005, FLUORSW_BZCN_0006, FLUORSW_BZCN_0007, FLUORSW_BZCN_0008, FLUORSW_BZCN_0009, FLUORSW_BZCN_0010, FLUORSW_BZCN_0011, FLUORSW_BZCN_0012, FLUORSW_BZCN_0013, FLUORSW_BZCN_0014, FLUORSW_BZCN_0015, FLUORSW_BZCN_0016, FLUORSW_BZCN_0017, FLUORSW_BZCN_0018, FLUORSW_BZCN_0019, FLUORSW_BZCN_0020, SRPECLI_BZCN_0001, SRPECLI_BZCN_0002, SRPECLI_BZCN_0003, SRPECLI_BZCN_0004, SRPECLI_DMS_0001, SRPECLI_DMS_0002, and SRPECLI_DMS_0003. Sample details are available in Supplementary Table 2.
Author Contributions
K.E.W., E.J.S., A.M.Y., J.T.L., and J.B.L. designed experiments. K.E.W. and E.J.S. performed experiments. K.E.W., E.J.S., A.M.Y., J.T.L., and J.B.L. analyzed data. K.E.W., E.J.S., A.M.Y., J.T.L., and J.B.L. wrote the manuscript
The authors declare no competing financial interests.
References
- 1.Wong TN, Sosnick TR, Pan T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc Natl Acad Sci U S A. 2007;104:17995–8000. doi: 10.1073/pnas.0705038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heilman-Miller SL, Woodson SA. Effect of transcription on folding of the Tetrahymena ribozyme. RNA. 2003;9:722–33. doi: 10.1261/rna.5200903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kramer FR, Mills DR. Secondary structure formation during RNA synthesis. Nucleic Acids Res. 1981;9:5109–24. doi: 10.1093/nar/9.19.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc Natl Acad Sci U S A. 1999;96:9545–50. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–75. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 6.Russell R, et al. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci U S A. 2002;99:155–60. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Hashimi HM, Walter NG. RNA dynamics: it is about time. Curr Opin Struct Biol. 2008;18:321–9. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garst AD, Edwards AL, Batey RT. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 11.Rieder U, Kreutz C, Micura R. Folding of a transcriptionally acting preQ1 riboswitch. Proc Natl Acad Sci U S A. 2010;107:10804–9. doi: 10.1073/pnas.0914925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 13.Haller A, Altman RB, Souliere MF, Blanchard SC, Micura R. Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proc Natl Acad Sci U S A. 2013;110:4188–93. doi: 10.1073/pnas.1218062110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frieda KL, Block SM. Direct observation of cotranscriptional folding in an adenine riboswitch. Science. 2012;338:397–400. doi: 10.1126/science.1225722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickiser JK. Kinetics of riboswitch regulation studied by in vitro transcription. Methods Mol Biol. 2009;540:53–63. doi: 10.1007/978-1-59745-558-9_5. [DOI] [PubMed] [Google Scholar]
- 16.Wong TN, Pan T. RNA folding during transcription: protocols and studies. Methods Enzymol. 2009;468:167–93. doi: 10.1016/S0076-6879(09)68009-5. [DOI] [PubMed] [Google Scholar]
- 17.Wright DJ, King K, Modrich P. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J Biol Chem. 1989;264:11816–21. [PubMed] [Google Scholar]
- 18.Pavco PA, Steege DA. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J Biol Chem. 1990;265:9960–9. [PubMed] [Google Scholar]
- 19.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat Protoc. 2009;4:1413–21. doi: 10.1038/nprot.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watters KE, Yu AM, Strobel EJ, Settle AH, Lucks JB. Characterizing RNA structures in vitro and in vivo with selective 2’-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Methods. 2016;103:34–48. doi: 10.1016/j.ymeth.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aviran S, Lucks JB, Pachter L. RNA structure characterization from chemical mapping experiments. Proc. 49th Allerton Conf. on Communication, Control, and Computing 1743–1750; Monticello, IL. 2011. [Google Scholar]
- 22.Bindewald E, et al. Correlating SHAPE signatures with three-dimensional RNA structures. RNA. 2011;17:1688–96. doi: 10.1261/rna.2640111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–9. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- 24.Komissarova N, Kashlev M. Functional topography of nascent RNA in elongation intermediates of RNA polymerase. Proc Natl Acad Sci U S A. 1998;95:14699–704. doi: 10.1073/pnas.95.25.14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehresmann C, et al. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–28. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker JL, et al. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335:233–5. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren A, Rajashankar KR, Patel DJ. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature. 2012;486:85–9. doi: 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali M, Lipfert J, Seifert S, Herschlag D, Doniach S. The ligand-free state of the TPP riboswitch: a partially folded RNA structure. J Mol Biol. 2010;396:153–65. doi: 10.1016/j.jmb.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ottink OM, et al. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism. RNA. 2007;13:2202–12. doi: 10.1261/rna.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts JW, Shankar S, Filter JJ. RNA polymerase elongation factors. Annu Rev Microbiol. 2008;62:211–33. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mustoe AM, Brooks CL, Al-Hashimi HM. Hierarchy of RNA functional dynamics. Annu Rev Biochem. 2014;83:441–66. doi: 10.1146/annurev-biochem-060713-035524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat Protoc. 2007;2:2608–23. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perdrizet GA, 2nd, Artsimovitch I, Furman R, Sosnick TR, Pan T. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc Natl Acad Sci U S A. 2012;109:3323–28. doi: 10.1073/pnas.1113086109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landick R, Wang D, Chan CL. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: his leader pause site as paradigm. Methods Enzymol. 1996;274:334–53. doi: 10.1016/s0076-6879(96)74029-6. [DOI] [PubMed] [Google Scholar]
- 35.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat Methods. 2014;11:959–65. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data that support the findings of this study have been deposited in the Small Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) with the BioProject accession code PRJNA342175. Individual BioSample accession codes are available in Supplementary Table 1. SHAPE-Seq Reactivity Spectra generated in this work have been deposited in the RNA Mapping Database (RMDB) (http://rmdb.stanford.edu/repository/) with the accession codes FLUORSW_BZCN_0001, FLUORSW_BZCN_0002, FLUORSW_BZCN_0003, FLUORSW_BZCN_0004, FLUORSW_BZCN_0005, FLUORSW_BZCN_0006, FLUORSW_BZCN_0007, FLUORSW_BZCN_0008, FLUORSW_BZCN_0009, FLUORSW_BZCN_0010, FLUORSW_BZCN_0011, FLUORSW_BZCN_0012, FLUORSW_BZCN_0013, FLUORSW_BZCN_0014, FLUORSW_BZCN_0015, FLUORSW_BZCN_0016, FLUORSW_BZCN_0017, FLUORSW_BZCN_0018, FLUORSW_BZCN_0019, FLUORSW_BZCN_0020, SRPECLI_BZCN_0001, SRPECLI_BZCN_0002, SRPECLI_BZCN_0003, SRPECLI_BZCN_0004, SRPECLI_DMS_0001, SRPECLI_DMS_0002, and SRPECLI_DMS_0003. Sample details are available in Supplementary Table 2. Source data for Figure 2–Figure 5 and Supplementary Figures 1, 2, and 4–9 are available with the paper online. All other data that support the findings of this paper are available from the corresponding author upon reasonable request.