Abstract
Human fertility is dependent upon the correct establishment and differentiation of the germline. This is because no other cell type in the body is capable of passing a genome and epigenome from parent to child. Terminally differentiated germline cells in the adult testis and ovary are called gametes. However, the initial specification of germline cells occurs in the embryo around the time of gastrulation. Most of our knowledge regarding the cell and molecular events that govern human germline specification involves extrapolating scientific principles from model organisms, most notably the mouse. However, recent work using next generation sequencing, gene editing and differentiation of germline cells from pluripotent stem cells has revealed that the core molecular mechanisms that regulate human germline development are different from rodents. Here, we will discuss the major molecular pathways required for human germline differentiation and how pluripotent stem cells have revolutionized our ability to study the earliest steps in human embryonic lineage specification to understand human fertility.
Human Reproduction
The United Nations Populations Division estimates that there are more than 7 billion people alive on earth today. By the middle of this century, it is estimated that the human population will reach 9 billion. At face value, these numbers suggest that the biology of human reproduction is sound. However, the United Nations Department of Economic and Social Affairs has signaled that the human population is in fertility decline, with a clear trend towards fewer children born per woman. Furthermore, the United States Centers for Disease Control and Prevention estimates that 12% of the reproductive age population (aged 15–44 years) has difficulty getting pregnant, or carrying a baby to term (CDC, 2012). Therefore, fertility decline from the point of view of population growth is most likely due to a combination of improved access to contraceptive methods, education and outreach, together with a stable but relatively high incidence of infertility. Therefore, we argue that studying the biology of human reproduction, and uncovering cell and molecular causes of human infertility is of paramount importance to human health, and the wellbeing of society.
Human Germ cells
Infertility is caused by a range of health problems, including underlying genetic mutations, cancer, obesity, hormonal imbalance, structural malformations of the urogenital tract or injury. However, a lack of germline cells guarantees infertility because only the germline is capable of transmitting genetic and epigenetic information from parent to child. Similarly a reduction in the quality or number of germ cells produced by an individual could also have a significant impact on a person’s fertility, as well as child health in the next generation.
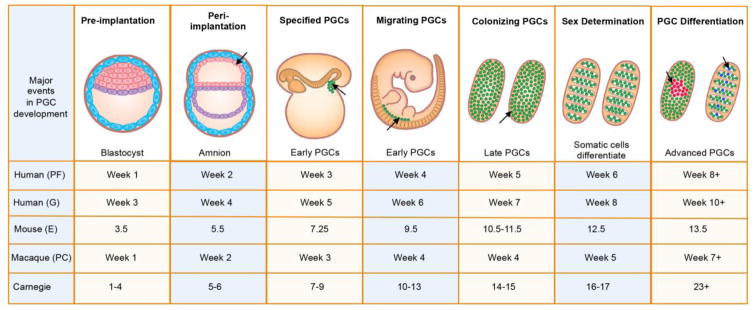
In humans, the pioneering germ cells in the embryo are called primordial germ cells (PGCs). These primitive embryonic cells are responsible for making the entire human germline, therefore the appropriate specification and allocation of PGCs is critical to promoting human reproductive health. PGCs develop very early in embryonic life, and are first observed at around 21 days post-fertilization, with the newly specified PGCs called “early PGCs” (Figure 1).
Figure 1. Time line of PGC development in humans.
Early PGCs (green) are identified in the yolk sac followed by the hindgut and then ultimately the genital ridge. Once PGCs exit the hindgut and begin expressing VASA they are called “late PGCs”. Late PGCs colonize the genital ridges beginning at week 5. Advanced PGCs develop at the conclusion of the Carnegie stages from 60–77 days with the emergence of male and female-specific transcriptional programs. In humans development is sometimes referred to as gestation (G), which refers to time since last menstrual cycle. PF = post fertilization, E = embryonic day, PC = post-coitus. The timing of mouse and macaque (rhesus) PGC development is shown for comparison.
Once specified, early PGCs are committed and have only one fate – that is to become either oogonia that differentiate into oocytes in girls, or spermatogonia, that differentiate into mature sperm in boys. Studies of monozygotic monoamniotic identical twins where the incidence of discordant primary ovarian insufficiency is high. lends support to the hypothesis that the window of PGC specification in humans is very narrow (Silber et al., 2008). Monozygotic monoamniotic twins are created by embryo splitting in the peri-implantation period after the formation of the amniotic sac. In these women, it is speculated that one twin receives the majority of PGC precursors and will have normal fertility, while the other twin will be deficient in PGC precursors, and will therefore become infertile. Put another way, once the window for germline potential has passed, the embryo cannot specify new germ cells and infertility is guaranteed.
Specification of the mammalian germ cell lineage is an inductive process
In model organisms such as Drosophila, C.elegans, Xenopus and Danio (zebrafish), PGCs are created each generation through a process known as pre-formation. This is a process driven by RNAs and proteins inherited from the oocyte, that selectively control translation of RNAs to endow a small number of transcriptionally quiescent cells in the embryo with PGC fate (Extavour and Akam, 2003). In contrast in mice, germ cell formation is induced by growth factor signaling from adjacent tissues leading to the expression of transcription factors that establish PGC fate.
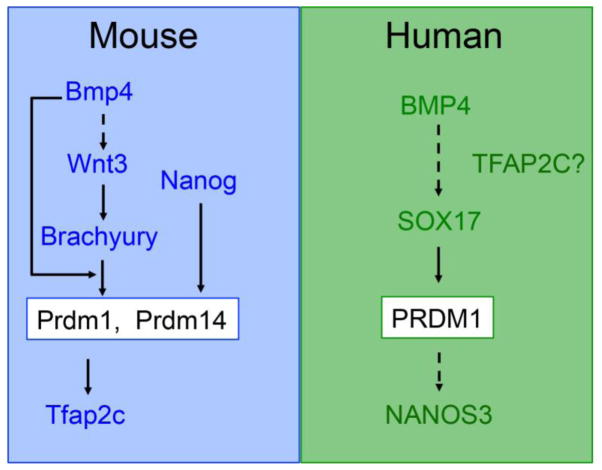
In the mouse, elegant lineage tracing and transplantation studies have shown that mouse PGCs are specified by bone morphogenetic protein 4 (BMP4) signaling from the extra-embryonic ectoderm to a Wnt3-primed proximal posterior epiblast at around embryonic (E) day E6.25 (Figure 2). Lineage restricted PGCs are formed from these precursors during the next 24 hours, around the late primitive streak/no-bud (LS/0B) to the early bud (EB) stage of mouse embryo development (E7.25) (Kurimoto et al., 2008; Lawson et al., 1999; Lawson and Hage, 1994; Vincent et al., 2005). The transcription factor network that controls mouse PGC specification downstream of BMP4 involves three transcription factors called transcriptional repressor PR domain 1 (Prdm1), Prdm14 and Transcription Factor AP-2 gamma (Tfap2c) (Kurimoto et al., 2008; Magnusdottir et al., 2013; Vincent et al., 2005; Weber et al., 2009; Yamaji et al., 2008). Using mouse pluripotent stem cells, PRDM14 can induce PGCs directly from epiblast-like cells in the absence of BMP4 signaling (Nakaki et al., 2013). More recently, the transcription factor NANOG was also found to induce PGC formation in the absence of BMP4 (Murakami et al., 2013). The model proposed from this work follows that NANOG functions upstream of both PRDM14 and PRDM1 by binding to their enhancers in the germline-competent in vitro epiblast-like cells to promote PGC fate (Murakami et al., 2013).
Figure 2. Major signaling pathways and transcription factors in mouse and human PGC specification.
The mouse has been invaluable for identifying the signaling pathways required for PGC specification. The finding that NANOG can induce PGCLC formation independent from BMP4 was discovered using in vitro differentiation into epiblast-like cells followed by induction of PGCLCs. Although the information on human PGC development is limited, initial experiments using pluripotent stem cell differentiation indicate that the mechanisms of human PGC development are different from the mouse.
Prior to BMP4 induction, the proximal epiblast must become responsive to PGC specification through the actions of Wnt3 (Figure 2). However, Wnt3 acts not only to prime PGC fate, but also to regulate primitive streak formation (Liu, 1999) For example, deletion of Wnt3 or β-catenin in the mouse abolishes primitive streak formation, and also disrupts induction of Prdm1 positive PGCs (Liu, 1999; Ohinata et al., 2009). A critical transcription factor that acts downstream of Wnt3 is Brachyury. Deleting Brachyury also leads to defects in primitive streak formation, and a failure to generate Prdm1 positive PGCs (Aramaki et al., 2013; Beddington et al., 1992). The relationship between Wnt3, Brachyury and BMP is complex, with the current model suggesting that Brachyury functions downstream of Wnt3 by directly inducing Prdm1, but only in the presence of BMP4 (Figure 2). Without BMP4, Brachyury is still induced downstream of Wnt3 in the epiblast, however it is unable to induce expression of Prdm1 or Prdm14 to promote PGC fate. The one way that BMP4 can be rendered unnecessary is with the in vitro stem cell model using Epiblast like cells, where Brachyury can be forced into the nucleus where it induces Prdm1 in the absence of BMP4.
Specification of germ cells in humans
The earliest identification of PGCs in the yolk sac was made by Fuss using transmission electron microscopy (TEM) during week 3 of life (Figure 1). This work revealed that human PGCs are large spherical cells located amongst the smaller cells of the yolk sac endoderm (Figure 1, arrow). For review see (Felici, 2013). The discovery of PGCs in the yolk sac lead to the original hypothesis that human PGCs are specified from the yolk sac endoderm (Witschi, 1948). If true, this could imply a potential PGC precursor in the hypoblast given that the yolk sac endoderm originates from hypoblast cells of the bilaminar embryo (Moore, 1977). Others have suggested that PGCs arise from the extra-embryonic mesoderm which lines the yolk sac, this would imply an origin from either the trophoblast or hypoblast, as both are speculated to give rise to extra embryonic mesodermal cells (Luckett, 1978). Others have hypothesized that human PGCs originate in the posterior epiblast similar to the mouse (Irie et al., 2014).
Recently using the cynomolgus macaque, PGCs were first identified in the amnion of the peri-implantation embryo at day 11 (Sasaki et al., 2016). The amnion develops coincident with the generation of the amniotic cavity and formation of the bilaminar embryo (Figure 1, arrow points to amnion cells in the peri-implantation embryo). In humans, any of the above developmental origins of PGCs could be possible, and the answer may be unveiled with the recently reported method extended human blastocyst culture, which results in the formation of bilaminar embryos with amniotic cavities and yolk sacks (Deglincerti et al., 2016; Shahbazi et al., 2016). However, the extended human blastocyst culture will only be useful if PGCs can be distinguished before 14 days or primitive streak formation in humans, as this is the ethical limit for culturing human embryos in vitro.
After PGC specification, and temporary residence in the yolk sac, the journey of PGCs through the embryo and into the gonad has been carefully documented using TEM by Witschi in 1948, and confirmed using Alkaline Phosphatase staining by Chiquoine in 1954 (Chiquoine, 1954; Witschi, 1948). These studies revealed that PGCs migrate through the yolk sac endoderm and mesoderm entering the hindgut endoderm and surrounding mesenchyme at around 25 days post-fertilization. By day 28, the first PGCs escape into the dorsal mesentery of the hindgut, and enter into the genital ridge. During this time PGCs will express the RNA helicase gene VASA (Castrillon et al., 2000), and are now called “late PGCs” to distinguish them from the VASA negative “early PGCs” in the peri-implantation embryo, yolk sac and hindgut endoderm. The early and late PGCs are promiscuous in their migration, and will migrate not only through the dorsal mesentery but also into a rudimentary structure known as the mesonephros (Park et al., 2009). Correctly localized late PGCs complete gonadal colonization over the next two weeks with sex determination occurring between the 6th and 7th week post fertilization.
To develop a more detailed map of human PGC development, we developed strategies for sorting late and advanced PGCs from single embryos followed by RNA-Seq (Gkountela et al., 2013). Our initial study involved sorting male and female cKIT positive advanced PGCs at 16 weeks post fertilization, which revealed a unique sex-specific germline signature that was distinct from undifferentiated hESCs (Gkountela et al., 2013), including confirmation of that PGCs lacked SOX2 expression (Perrett et al., 2008). A second major difference from hESCs and mouse PGCs discovered by us and others, was the notably low expression levels of PRDM14 relative to undifferentiated hESCs (Gkountela et al., 2015; Guo et al., 2015; Irie et al., 2015; Tang et al., 2015). We also showed that male and female cKIT-positive PGCs were transcriptionally similar to each other until week 8, and after this, female PGCs begin to adopt a strong female-specific molecular program including the up-regulation of meiotic genes around day 60 post-fertilization (Gkountela et al., 2015). This occurs at the end of the last Carnegie Stage (stage 23) of embryo development. In males, the discrimination of a male-specific identity from an indifferent PGC begins after the Carnegie stages have finished, between day 70–77 post-fertilization (Figure 1). The emerging male-specific program is associated with up-regulation of genes encoding proteins required for genome defense such as MORC1, MOV10L1, TDRD5, PIWIL3 (Gkountela et al., 2015). We refer to these sex-specific male and female PGCs as “advanced PGCs” because they still express cKIT and OCT4, but are no longer transcriptionally equivalent to the sexually indifferent late PGCs found in the gonad prior to 8 weeks. Taken together, unbiased gene expression mapping of human PGCs has identified three stages of primate PGC development, “early”, “late” and “advanced”. These studies also revealed that the molecular identity of human PGCs has diverged from the mouse with notably differences including the repression of PRDM14 and SOX2 and expression of SOX17 (Gkountela et al., 2015; Guo et al., 2015; Irie et al., 2015; Tang et al., 2015). Therefore, understanding the molecular basis for human germline development and human fertility requires new models, including the analysis of species where embryos develop from a disk rather than a cylinder, as well as evaluating species with a more closely related genome.
One of these closely related model organisms is the old world monkey Macaca (mulatta or fasicularis), which follows a Carnegie staging system that is highly correlated with the timing of human development, particularly in the beginning (Figure 1). Notably, formation of the primitive streak in macaques occurs at 14–15 days post fertilization (Luckett, 1978), making the macaques an excellent model for studying human embryo development, germ cell specification and fertility. Towards this goal, a recent study used single cell RNA-Seq of peri-implantation Macaca fasicularis (cynomolgus macaque) embryos up to embryonic day 17 was used to identify progenitor populations that emerge in the peri-implantation period, around the time of primitive streak formation in the epiblast (Nakamura et al., 2016). This work identified single cells with an early PGC identity (PRDM1+ SOX17+ TFAP2C+ and SOX2−) (Nakamura et al., 2016). However, subsequent studies have indicated that the epiblast may not be the origin of PGCs, and instead nascent early PGCs originate prior to primitive streak formation in the allantois (Sasaki et al., 2016).
Taken together, the body of descriptive knowledge on human and nonhuman primate germline development is continuing to expand. However, functional assessment of these pathways requires an in vitro model, for which the most appropriate is the differentiation of germline cells in vitro from pluripotent stem cells.
Differentiation of human germ cells in vitro from pluripotent stem cells
The first hint that human pluripotent stem cells had germline potential was revealed more than a decade ago using spontaneous differentiation of hESCs as three-dimensional aggregates in fetal bovine serum to create Embryoid Bodies (EBs) (Clark et al., 2004). Although the initial studies indicated that DAZL and VASA were expressed in EBs (Clark et al., 2004), single-cell, multi-gene analysis of sorted germ cells differentiated in vitro revealed that hESC-derived PGCs co-expressed genes associated with early PGCs such as NANOS3, BLIMP1 and OCT4, while being negative for DAZL and VASA (Gkountela et al., 2013). This paradoxical result was explained by single cell analysis revealing that DAZL and VASA are rare transcripts, found in only a small fraction of cells, with the majority of PGCs being in the early stages of PGC development.
Although spontaneous differentiation creates a vary rare population of germ cells in vitro, directed differentiation using defined conditions creates at least ten-fold more PGC like cells (PGCLCs) relative to differentiation as EBs, and these germ cells also correspond to PGCs in the early stage. There are three major approaches for directed differentiation of hESCs/hiPSCs (Table 1). The first approach uses pluripotent stem cells cultured under self-renewing conditions in a media called four-inhibitor (4i) on irradiated mouse embryonic fibroblasts (MEFs) (Irie et al., 2015). The 4i media contains human leukemia inhibitory factor (LIF), basic Fibroblast growth factor (bFGF), Transforming growth factor-β1 (TGFβ1), and the four inhibitors CHIR99021 (GSK3 inhibitor), PD0325901 (MEK inhibitor) SB203580 (MAPK inhibitor) and SP600125 (JNK inhibitor). Gene expression analysis using RNA-Seq comparing 4i cells to regular primed hESCs indicates that they are almost identical (>0.9 correlation).
Table 1.
Comparison of the directed differentiation approaches for generating PGCLCs in aggregates. In all medias, PGCLCs are induced in the aggregates and isolated by FACS. Only the critical media additives are included. See text for details
| PGCLC method | hESC/hiPSC self-renewal media | hESC/hiPSC self- renewal support/substrate | Pre-aggregate differentiation step | Aggregates containing PGCLCs |
|---|---|---|---|---|
| Irie (Irie et al., 2015) | LIF, bFGF, TGFβ1 CHIR99021 PD0325901 SB203580 SP600125 |
Irradiated MEFs | Not necessary | BMP4 LIF SCF EGF ROCK inhibitor |
| Sasaki (Sasaki et al., 2015) | StemFIT Commercial media | Laminin511 | CHIR99021 ACTIVIN A on Fibronectin for 42 hours | BMP4 LIF SCF EGF ROCK inhibitor |
| Sugawa (Sugawa et al., 2015) | Conventional MEF conditioned media containing bFGF/KSR | Matrigel | ACTIVIN A, BMP4, bFGF, ROCK inhibitor on Matrigel for 48 hours | BMP4 LIF ROCK inhibitor |
The second PGCLC induction approach starts with hiPSCs cultured under conventional primed feeder-free conditions in a media called “StemFit” on plates coated with the E8 fragment of recombinant LAMININ511 (Sasaki et al., 2015). StemFit is a commercial formula containing bFGF and other proprietary ingredients. When starting with hiPSCs cultured in StemFIT/LAMININ511, a two-step differentiation protocol is used. This is because the authors discovered a technical problem making 3D aggregates directly from StemFIT/LAMININ511 cultured cells, resulting in unacceptable amounts of cell death, and a low yield of PGCLCs. To overcome this, the authors introduced a 42-hour differentiation step where the hiPSCs are plated as single cells to create incipient mesoderm like cells (iMeLCs) (Sasaki et al., 2015). The iMeLC media includes a GSK3 inhibitor (same as 4i) as well as ACTIVIN A which uses the same receptor-regulated SMADs as TGFβ and ROCK inhibitor (ROCKi) (Table 1). The identity of iMeLCs is relatively undefined, with only 200 genes differentially expressed at >2-fold between the iPSCs and the iMeLCs.
Differentiating PGCLCs in aggregates from 4i cultured human pluripotent stem cells initially utilized a 48-hour pre-induction step on Vitronectin/Gelatin coated plates in media composed of bFGF and either TGFβ1 or ACTIVIN A. However subsequent experiments showed that the pre-induction step before aggregation was completely unnecessary (Irie et al., 2015). The aggregate media from 4i consists of LIF, Stem cell factor (SCF), Epidermal growth factor (EGF) and BMP4/BMP2 and ROCK inhibitor (Irie et al., 2015). Similarly, the media for generating aggregates from StemFIT through the iMeLC intermediate contains LIF, BMP4, SCF, EGF and ROCK inhibitor (Sasaki et al., 2015). The two formulations vary slightly in the concentrations of growth factors, however the percentage PGCLCs isolated from the aggregates on day 4 of differentiation by fluorescence activated cell sorting (FACS) are comparable between the two methods. Critically, the percentage PGCLCs acquired from StemFIT, ranges from around 20% to 60% (Sasaki et al., 2015), whereas in 4i, the percentage of PGCLCs ranges from 7% to 50% (Irie et al., 2015). The source of this variability in PGCLC percentage is unclear, and a comprehensive analysis of independently derived pluripotent stem cell lines will be required to determine if certain lines have higher or lower potential for PGCLC differentiation. Despite the variability in the percentage of PGCLCs, RNA-Seq data from the purified PGCLCs in both studies is highly correlated (Sasaki et al., 2015). Taken together the use of 4i/MEFs as a starting point for hESC culture verses StemFIT/LAMININ511 followed by iMeLC differentiation seems irrelevant, so long as the aggregates generated in PGCLC induction media are healthy, and the PGCLC population isolated by FACS is discreet and well separated.
The third approach involves starting with hESCs or hiPSCs cultured under conventional primed conditions on matrigel in the presence of MEF conditioned media (Sugawa et al., 2015). Similar to the approach of (Sasaki et al., 2015), PGCLCs are generated in aggregates after first creating a mesoderm-like cell intermediate in ACTIVIN A, BMP4, bFGF and ROCK inhibitor prior to aggregate formation. Unlike (Irie et al., 2015), the aggregate media where PGCLCs are induced only contains BMP4, LIF and ROCK inhibitor, while not containing EGC or SCF. PGCLCs generated in this media have reduced levels of PRDM14, and express transcripts consistent with PGCs that have an early PGC identity. The transcriptional program of PGCLCs created with this approach have not yet been compared to the PGCLCs created using 4i, or StemFIT cultured human pluripotent stem cells.
The transcription factor network for human germline specification
The above human pluripotent stem cell models do not reveal the origin of PGCs because the growth factors used to induce iMeLCs and PGCLCs are the same as those used to generate primitive streak (BMP4, ACTIVNA, GSK3i), mesendoderm (ACTIVINA) mesoderm (BMP4) and endoderm (ACTIVIN A, bFGF) from pluripotent stem cells (D’Amour et al., 2005; Davis et al., 2008; Oldershaw et al., 2010). However, one of the most notable discoveries in PGC specification using the 4i approach was that SOX17 is required upstream of PRDM1 to promote PGCLC fate (Irie et al., 2015). In contrast, in the mouse, SOX17 has no role in the differentiation of PGCs and instead is required for the differentiation of definitive gut endoderm (Kanai-Azuma et al., 2002). PRDM1 is required for both mouse and human PGCLC differentiation, however the mechanism by which PRDM1 functions in humans is still in dispute (Irie et al., 2015; Sasaki et al., 2015). Using CRISPR/Cas9 to delete PRDM1 in StemFit/LAMININ511, SOX17 and TFAP2C expression levels are unaffected in the Prdm1 mutant PGCLCs at day 2. Instead, PRDM1 mutant PGCs fail to up-regulate NANOS3, and by day 4, start to lose expression of Brachyury, and the primitive streak gene MIXL (Sasaki et al., 2015). This result is very different to the transcription factor network for PGC induction in mice (Figure 2). Similarly, PRDM14 has been depleted to around 50% of control levels with shRNAs during PGCLC differentiation, and this led to no effect on human PGCLC formation (Sugawa et al., 2015). Future studies using directed differentiation of PGCLCs should unveil the new transcription factor-network that has evolved to enable human fertility through the generation of an appropriate cohort of PGCs.
Future perspectives
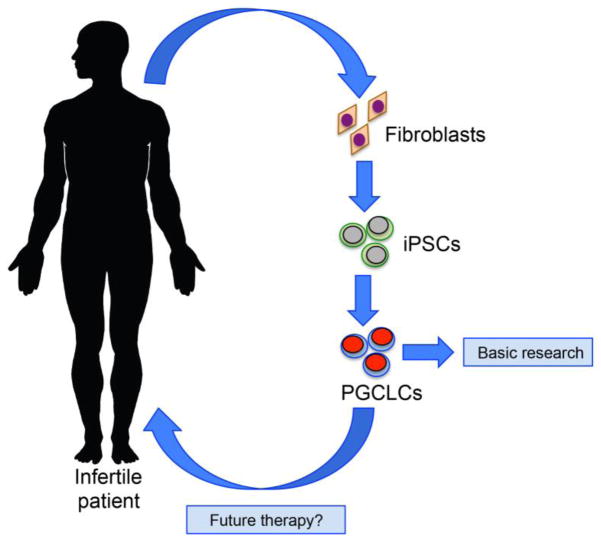
Human fetal tissue research and non human primate macaque models have been essential to the establishment of human PGCLC differentiation models to study mechanisms that contribute to early PGC allocation in the embryo. Together, these approaches have revealed that humans have evolved a new strategy for ensuring reproductive success. This new strategy is built in part on the mechanisms used in rodents, with notable exceptions, and is highly dissimilar to the strategy used by flies, worms, fish and frogs. Whether these molecular differences have made the human population more vulnerable to infertility is unclear at this stage. Finally, the hiPSC model will, for the first time, enable analysis of congenital diseases of the germline that can cause human infertility (Figure 3). Most notably, primary ovarian insufficiency where oocytes are lost in girls from a very young age, or human germ cell tumors, one of the most common cancers of adolescents and young adults which is speculated to originate in the PGC. Although germ cell tumors are highly treatable, survivors are at risk for morbidities on account of their treatment, and this young population would significantly benefit from a targeted therapy that could be revealed using PGCLCs differentiated from human pluripotent stem cells.
Figure 3. Using Human iPSCs to understand and treat human infertility.
Human iPSCs can be used to generate stem cells from infertile patients where the disease is speculated to originate from the germline. Today, differentiation of the hiPSCs into PGCLCs could be used as a model to understand fertility. In the future, hiPSC technology and the differentiation of PGCLCs could be used to restore fertility to individuals who lost their germline due to injury. For example, this therapy could be used in young adults who have become infertile after chemotherapy or radiation therapy to treat cancer when all other options have been exhausted.
Highlights.
Human PGC differentiation is distinct from rodents.
Human PGC differentiation in the embryo can be described as early, late and advanced.
Differentiation of PGCLCs from pluripotent stem cells creates early PGCs.
Acknowledgments
This work was supported by funds from the NIH grant P01HD075795 and R01HD079546 to ATC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aramaki S, Hayashi K, Kurimoto K, Ohta H, Yabuta Y, Iwanari H, Mochizuki Y, Hamakubo T, Kato Y, Shirahige K, et al. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev Cell. 2013;27:516–529. doi: 10.1016/j.devcel.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Beddington R, Rashbass P, Wilson V. Brachyury--a gene affecting mouse gastrulation and early organogenesis. Dev Suppl. 1992:157–165. [PubMed] [Google Scholar]
- Castrillon DH, Quade BJ, Wang TY, Quigley C, Crum CP. The human VASA gene is specifically expressed in the germ cell lineage. PNAS. 2000;97:9585–9590. doi: 10.1073/pnas.160274797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Surveys and Data Collection Systems from the Centers of Disease Control and Prevention: Key Statistics from the National Survey of Family Growth (2006–2010) 2012 [Google Scholar]
- Chiquoine A. The identification, origin and migration of the primordial germ cells in the mouse embryo. Anat Rec. 1954;118:135–146. doi: 10.1002/ar.1091180202. [DOI] [PubMed] [Google Scholar]
- Clark AT, Bodnar MS, Fox M, Rodriquez R, Abeyta M, Firpo M, Pera R. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum Mol Genet. 2004;13:727–739. doi: 10.1093/hmg/ddh088. [DOI] [PubMed] [Google Scholar]
- D’Amour K, Agulnick A, Eliazer S, Kelly S, Kroon E, Baetge E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Davis R, Ng E, Costa M, Mossman A, Sourris K, Elefanty A, Stanley E. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- Deglincerti A, Croft G, Pietila L, Zernicka-Goetz M, Siggia E, Brivanlou A. Self-organization of the in vitro attached human embryo. Nature. 2016;4:251–254. doi: 10.1038/nature17948. [DOI] [PubMed] [Google Scholar]
- Extavour C, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Felici M. In: Origin, Migration, and Proliferation of Human Primordial Germ Cells. oogenesis GA, Coticchio DF, De Santis L, editors. Springer; 2013. pp. 19–37. [Google Scholar]
- Gkountela S, Li Z, Vincent J, Zhang K, Chen A, Pellegrini M, Clark A. The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat Cell Biol. 2013;15:113–122. doi: 10.1038/ncb2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S, Zhang KX, Shafiq TA, Liao WW, Hargan-Calvopina J, Chen PY, Clark AT. DNA Demethylation Dynamics in the Human Prenatal Germline. Cell. 2015;161:1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, Yong J, Hu Y, Wang X, Wei Y, et al. The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Irie N, Walfred W, Tang M, MAS Germ cell specification and pluripotency in mammals: a perspective from early embryogenesis. Reproductive Medicine and Biology. 2014;13:203–215. doi: 10.1007/s12522-014-0184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie N, Weinberger L, Tang W, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna J, Surani M. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai-Azuma M, Kanai Y, Gad J, Tajima Y, Taya C, Kurohmaru M, Sanai Y, Yonekawa H, Yazaki K, Tam P, et al. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129:2367–2379. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yamaji M, Seki Y, Saitou M. Specification of the germ cell lineage in mice: a process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle. 2008 doi: 10.4161/cc.7.22.6979. Epub. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Dunn NR, Roelen BAJ, Zeinstra LM, Davis AM, Wright CVE, Korving JPWFM, Hogan BLM. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Development. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Hage WJ. Germline Development: Ciba Foundation Symposium. West Sussex, UK: John Wiley and Sons; 1994. Clonal analysis of the origin of primordial germ cells in the mouse; pp. 68–84. [DOI] [PubMed] [Google Scholar]
- Liu P. Requirement for wnt3 in vertebrate axis formation. Nature Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- Luckett W. Origin and differentiation of the yolk sac and extraembryonic mesoderm in presomite human and rhesus monkey embryos. Am J Anat. 1978;152:59–97. doi: 10.1002/aja.1001520106. [DOI] [PubMed] [Google Scholar]
- Magnusdottir E, Dietmann S, Murakami K, Gunesdogan U, Tang F, Bao S, Diamanti E, Lao K, Gottgens B, Azim Surani M. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat Cell Biol. 2013;15:905–915. doi: 10.1038/ncb2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KL. The Developing Human, Vol. 2. Vol. 1. Philadelphia: W.B. Saunders Company; 1977. [Google Scholar]
- Murakami K, Günesdogan U, Zylicz J, Tang W, Sengupta R, Kobayashi T, Kim S, Butler R, Dietmann S, Surani M. NANOG alone induces germ cells in primed epiblast in vitro by activation of enhancers. Nature. 2013;529:403–407. doi: 10.1038/nature16480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaki F, Hayashi K, Ohta H, Kurimoto K, Yabuta Y, Saitou M. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Okamoto I, Sasaki K, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Yamamoto T, Saitou M. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature. 2016 doi: 10.1038/nature19096. [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Oldershaw R, Baxter M, Lowe E, Bates N, Grady L, Soncin F, Brison D, Hardingham T. Directed differentiation of human embryonic stem cells toward chondrocytes. Nat Biotechnol. 2010;28:1187–1194. doi: 10.1038/nbt.1683. [DOI] [PubMed] [Google Scholar]
- Park TS, Galic Z, Conway AE, Lindgren A, van Handel BJ, Magnusson M, Richter L, Teitell MA, Mikkola HK, Lowry WE, et al. Derivation of primordial germ cells from human embryonic and induced pluripotent stem cells is significantly improved by coculture with human fetal gonadal cells. Stem Cells. 2009;27:783–795. doi: 10.1002/stem.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrett RM, Turnpenny L, Eckert JJ, O’Shea M, Sonne SB, Cameron IT, Wilson DI, Rajpert-De Meyts E, Hanley NA. The Early Human Germ Cell Lineage Does Not Express SOX2 During In Vivo Development or Upon In Vitro Culture. Biol Reprod. 2008 doi: 10.1095/biolreprod.107.066175. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Nakamura T, Okamoto I, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Shiraki N, Takakuwa T, et al. The germ cell fate of cynomolgus monkeys is specified in the nascent amnion. Dev Cell. 2016;39:169–185. doi: 10.1016/j.devcel.2016.09.007. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Yokobayashi S, Nakamura T, Okamoto I, Yabuta Y, Kurimoto K, Ohta H, Moritoki Y, Iwatani C, Tsuchiya H, et al. Robust In Vitro Induction of Human Germ Cell Fate from Pluripotent Stem Cells. Cell Stem Cell. 2015;17:178–194. doi: 10.1016/j.stem.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Shahbazi M, Jedrusik A, Vuoristo S, Recher G, Hupalowska A, Bolton V, Fogarty N, Campbell A, Devito L, Ilic D, et al. Self-organization of the human embryo in the absence of maternal tissues. Nat Cell Biol. 2016;18:700–708. doi: 10.1038/ncb3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber S, DeRosa M, Pineda J, Lenahan K, Grenia D, Gorman K, Gosden R. A series of monozygotic twins discordant for ovarian failure: ovary transplantation (cortical versus microvascular) and cryopreservation. Hum Reprod. 2008;23:1531–1537. doi: 10.1093/humrep/den032. [DOI] [PubMed] [Google Scholar]
- Sugawa F, Arauzo-Bravo M, Yoon J, Kim K, Aramaki S, Wu G, Stehling M, Psathaki O, Hubner K, Scholer H. Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. Embo J. 2015;34:1009–1024. doi: 10.15252/embj.201488049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Dietmann S, Irie N, Leitch H, Floros V, Bradshaw C, Hackett J, Chinnery P, Surani M. A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell. 2015;161:1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S, Dunn R, Sciammas R, Shapiro-Shalef M, Davis M, Calame K, Bikoff E, Robertson E. The zinc finger transcriptional represssor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- Weber S, Eckert D, Nettersheim D, Gillis A, Schäfer S, Kuckenberg P, Ehlermann J, Werling U, Biermann K, Looijenga L, et al. Critical function of AP-2 gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biology of Reproduction. 2009:214–223. doi: 10.1095/biolreprod.109.078717. [DOI] [PubMed] [Google Scholar]
- Witschi E. Migration of the germ cells of human embryos from the yolk sac to the primitive gonadal folds. Carnegie Inst Contrib Embryol. 1948;32:67–80. [Google Scholar]
- Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]