Abstract
Antisense oligonucleotides (AONs) hold promise for therapeutic splice-switching correction in many genetic diseases. However, despite advances in AON chemistry and design, systemic use of AONs is limited due to poor tissue uptake and sufficient therapeutic efficacy is still difficult to achieve. A novel class of AONs made of tricyclo-DNA (tcDNA) is considered very promising for the treatment of Duchenne muscular dystrophy (DMD), a neuromuscular disease typically caused by frameshifting deletions or nonsense mutations in the gene-encoding dystrophin and characterized by progressive muscle weakness, cardiomyopathy, and respiratory failure in addition to cognitive impairment. Herein, we report the efficacy and toxicology profile of a 13-mer tcDNA in mdx mice. We show that systemic delivery of 13-mer tcDNA allows restoration of dystrophin in skeletal muscles and to a lower extent in the brain, leading to muscle function improvement and correction of behavioral features linked to the emotional/cognitive deficiency. More importantly, tcDNA treatment was generally limited to minimal glomerular changes and few cell necroses in proximal tubules, with only slight variation in serum and urinary kidney toxicity biomarker levels. These results demonstrate an encouraging safety profile for tcDNA, albeit typical of phosphorothiate AONs, and confirm its therapeutic potential for the systemic treatment of DMD patients.
Keywords: antisense oligonucleotides, Duchenne muscular dystrophy, preclinical, splice switching, tcDNA-AONs
Introduction
Duchenne muscular dystrophy (DMD) is an X-linked recessive disorder characterized by progressive muscle degeneration, loss of walking ability, decline of respiratory and cardiac functions, and early death.1, 2, 3 DMD is the most common and severe form of muscular dystrophy, with an incidence of about 1 in 3,500 newborn males.3 Various mutations in the DMD gene disrupt the open reading frame (ORF) and lead to the absence of functional dystrophin protein.4, 5 Becker muscular dystrophy (BMD) is also caused by mutations in the dystrophin gene, but is characterized by less severe symptoms, slower disease progression, and less severe muscle wasting than DMD.6 The cause for the difference between DMD and BMD is, in most cases, the conservation of the reading frame despite the mutation, which results in a truncated but partly functional dystrophin.
There is no effective therapy to stop the progression of the disease, although several promising experimental strategies are currently under investigation. Exon skipping using antisense oligonucleotides (AONs) to convert out-of-frame DMD mutations into in-frame deletions, thereby restoring expression of a truncated but functional dystrophin, is currently one of the most attractive approaches for the treatment of DMD. AON-based drugs have already entered clinical trials and demonstrated promising results.4, 5, 7, 8 However, although dystrophin could be restored at low levels using these synthetic naked oligonucleotide compounds in DMD patients,4, 9 these studies have failed to show a marked clinical benefit, and although the US Food and Drug Administration (FDA) has surprisingly granted accelerated approval to Eteplirsen (a phosphoroamidate morpholino oligomer [PMO-AO] targeting exon 51), additional clinical trials have been requested to confirm the drug’s clinical benefit, which has not yet been demonstrated.9, 10, 11, 12 Therefore, there is still a critical need to develop efficient AONs able to restore the expression of dystrophin in all relevant tissues and cells without jeopardizing the safety of patients. The novel class of AONs made of tricyclo-DNA (tcDNA), which displays unique pharmacological properties and unprecedented uptake in many tissues after systemic administration, is considered very promising for the treatment of DMD.13, 14 We have previously demonstrated the functional correction and neurobehavioral improvement in muscular dystrophic mice after treatment with a 15-mer splice-switching tcDNA and shown the superiority of this oligonucleotide analog compared to other naked chemistries (namely PMO and 2′OMe AONs).13 tcDNA is a conformationally constrained nucleotide analog that deviates from natural DNA by the presence of three additional carbon atoms between C5′ and C3′, to which a cyclopropane unit is fused for further enhancement of structural rigidity.15, 16 Fully modified as well as gapmer tcDNAs in the length range of 11–20 nt have been shown to produce potent antisense effects.17, 18 A further crucial feature of tcDNA chemistry is that it displays higher RNA-binding properties than 2′OMe and PMO,17 thus permitting use of shorter AONs. Importantly, this offers the advantage of reducing the mass of synthetic nucleotides administered while keeping a biological effect, which could reduce toxicity induced by accumulation of AONs. To investigate this possibility, we aimed to study the therapeutic potential of a short 13-mer tcDNA-AON that is fully modified and phosphorothiated (PS).
The majority of preclinical studies focus on assessing the efficacy of AONs in target tissues and improving delivery, and evaluation of toxicity is often neglected, even though this is particularly important when developing new generations of AONs or different drug delivery systems (DDSs). This can lead to subsequent failure of a new drug in further toxicological studies, as it happened with peptide conjugated PMO (PPMO) targeting the human dystrophin exon 50 (AVI-5038), which was found to cause mild tubular degeneration in the kidneys of cynomolgus monkeys.19 This example highlighted the fact that toxicity threshold varies between species. However, numerous specific and early biomarkers of toxicity can now be evaluated in mice (treated with high doses of AONs) to predict toxicity in pre-clinical development.20 These delivery challenges were recognized by experts from the COST Action BM1207 “Networking towards clinical implementation of antisense-mediated exon skipping for rare diseases”21 (http://exonskipping.eu), and it is becoming clear that these toxicological challenges should be addressed in the very early stages of new AON development to ensure the clinical translation of these studies.22 Because tcDNA chemistry has never been used in the clinic, as opposed to other naked chemistries, which have been evaluated for other applications before DMD, we believe it is of importance to evaluate its toxicological profile in mice before undertaking expensive and lengthy reglementary toxicological studies.
We therefore investigated here the therapeutic benefit and toxicology profile of a 13-mer tcDNA delivered intravenously (i.v.) in a mouse model of DMD (mdx mouse). We first show the efficiency of this short tcDNA, which allowed a restoration of dystrophin expression in skeletal muscles and to a lesser extent in the brain, leading to muscle function improvement and correction of behavioral features linked to the emotional/cognitive deficiency associated with the lack of dystrophin. Importantly, for the preclinical development of such a novel AON and considering how critical the early evaluation of the toxicity profile is in drug development, we also evaluated the toxicological profile of tcDNA-AONs after 12 weeks of treatment at a dose of 200 mg/kg/week using early and specific urinary kidney injury biomarkers (KIBs). We show that although tcDNA treatment was well-tolerated, with only minimal glomerular changes and few cell necroses in proximal tubules, some variation in serum and urinary biomarkers can be detected. Our results therefore demonstrate the efficacy of tcDNA treatment, with an overall encouraging safety profile, but highlight the well-known issue of PS-AON accumulation and associated kidney toxicity.
Results
Efficacy of 13-mer tcDNA-AON
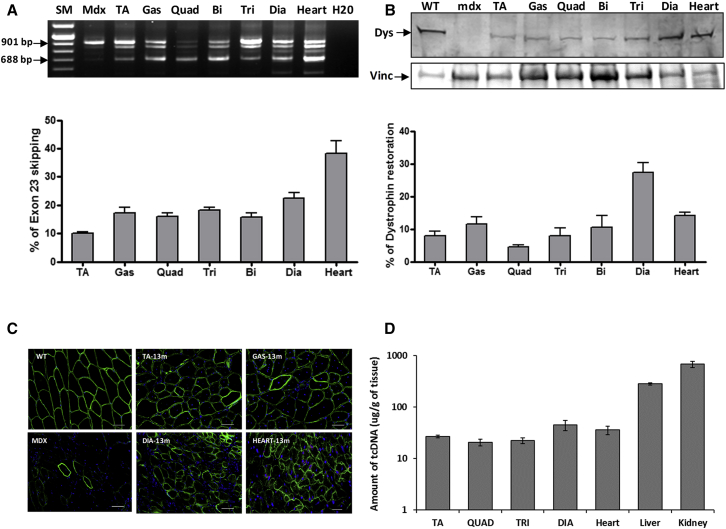
To evaluate the effectiveness of a 13-mer tcDNA, we designed a shorter tcDNA of 13 nt targeting the 5′ donor splice site on DMD intron 23 based on previously optimized and published sequences.13, 23 We first confirmed the efficacy of this 13-mer tcDNA-AON following intramuscular injections in the tibialis anterior (TA) muscle of adult mdx mice, which carry a nonsense mutation in exon 23 of the DMD gene24 (Figure S1). We then evaluated repeated systemic administrations of tcDNA-AONs, in which mice were treated intravenously with 200 mg/kg/week of tcDNA for a period of 12 weeks and tissues were analyzed 2 weeks after the final injection. RT-PCR results revealed a dystrophin transcript lacking exon 23 in all muscle samples from tcDNA-treated mice (Figure 1A), and quantification by qRT-PCR confirmed efficient exon 23 skipping in all examined tissues, reaching levels of 38% in the cardiac muscle (Figure 1A). We evaluated dystrophin restoration by western blot and detected the amount of dystrophin protein, ranging from 5% to 30%, in the different tissue samples, with particularly high levels in the diaphragm of tcDNA-treated mice (27%) compared to wild-type (WT) mice (Figure 1B). In order to visualize the localization of the restored dystrophin protein, we performed immunostainings and showed a correct expression and localization of dystrophin protein at the sarcolemma level of muscle fibers in the different skeletal muscle samples after tcDNA treatment (Figure 1C). tcDNA-AON amounts were also quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and detected in all tested tissues, with particularly high levels in the liver and kidneys, as expected for PS-AON (Figure 1D). These results showed, for the first time, the effectiveness of a shorter AON, 13-mer tcDNA, in exon-skipping approaches for DMD, with great restoration of dystrophin protein.
Figure 1.
13-mer tcDNA Treatment Induces Exon Skipping and Dystrophin Rescue
(A) Detection of exon-23-skipped dystrophin mRNA in mdx muscles after systemic delivery of tcDNA 13-mer at 200 mg/kg/week. The lower 688-bp fragment corresponding to the exon-23-skipped mRNA is detected by nested RT-PCR (blots). Intermediate-sized bands reflecting the formation of heteroduplex between the native and exon-23-skipped products can also be observed, as previously described.45 SM, size marker. Exon-23-skipped mRNA was quantified by TaqMan assay and expressed as the percentage of total dystrophin mRNA (shown in graphs below gels). n = 4 mice per group; error bars are mean ± SEM. (B) Western blot showing dystrophin (Dys) expression in different muscles from treated mice at 200 mg/kg/week compared to mdx and WT control mice. 100 μg of total protein were loaded for all samples and 25 μg for WT. Vinculin (Vinc) was used as internal control. The quantification of dystrophin has been done using a standard curve with 0%, 5%, 10%, and 20% of corresponding WT tissues. All western blots shown are representative of four treated mice. Below: diagrams depict quantification of dystrophin expressed as a percentage of WT expression using a standard curve of WT tissues after normalization with total protein loading. n = 4 mice per treated group; error bars are mean ± SEM. (C) Dystrophin immunostaining on transverse sections from mdx-treated muscles. Nuclei, blue (DAPI); dystrophin, green. Scale bar, 50 μm. (D) Quantification of tcDNA-13-mer amounts in various tissues by LC-MS/MS.
Functional Rescue
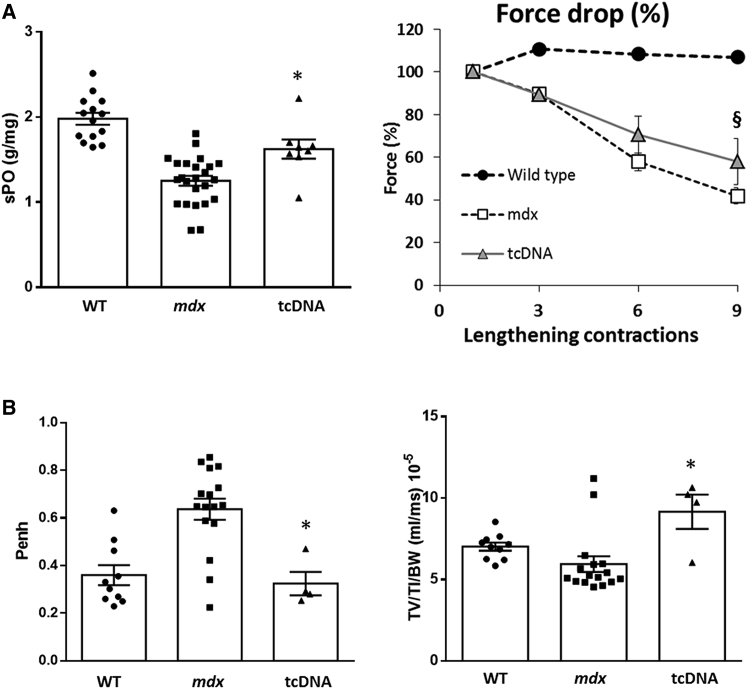
The functional properties were investigated by measuring the maximal specific force and resistance to eccentric contraction-induced skeletal muscle injury, which is an indicator of the structural integrity of the muscle fibers on semi-isolated TA muscles (Figure 2A, left panel). The TA muscles from the control mdx mice showed a decrease of 35% in the maximal specific force compared to the WT muscle. In contrast, treatment with the 13-mer tcDNA displaying 8% of restored dystrophin significantly improved the maximal specific force compare to the control mdx mice. Moreover, TA muscles from control mdx mice were unable to sustain tetanic tension, falling to 42% of their initial force after nine eccentric contractions. In contrast, TA muscles from tcDNA-treated mice maintained 58% of their force following the eccentric contractions, which correlated with the observed level of exon skipping and protein restoration of TA muscles following treatment (Figure 2A, right panel).
Figure 2.
Systemic Delivery of 13-mer tcDNA Improves the mdx Phenotype
(A) TA muscles of mdx mice treated with 200 mg/kg/week of tcDNA were analyzed for their maximal specific force (sP0) (left) and percentage of force drop following a series of three to nine eccentric contractions (right). n = 4 per group; error bars are mean ± SEM. (B) Respiratory function in mdx mice treated with 200 mg/kg of tcDNA for 12 weeks (n = 4 per group) compared to mdx control (n = 16) and WT mice (n = 10). penh and TV/TI/BW are shown. Error bars are mean ± SEM. *p < 0.05 and §p = 0.06 compared to mdx control mice (Mann-Whitney U tests).
Respiratory failure being one of the major causes of death in DMD patients,25 and because an improvement of this vital parameter is required for better quality of life, we explored respiratory function in mdx mice following 13-mer tcDNA treatment. Here, we confirmed, by measuring the respiratory function in mdx mice by whole-body plethysmography, that mdx control mice showed abnormalities for a number of respiratory parameters compared to age-matched WT mice (Figure 2B). We found that 13-mer tcDNA-treated mdx mice, which expressed 27% of rescued dystrophin protein in the diaphragm compared to WT controls, showed a significant improvement of respiratory function, as reflected by the enhanced pause (penh) and ratio of mean inspiratory flow expressed by tidal volume (TV) to inspiratory time (TI) normalized to body weight (TV/TI/BW), underlying the correlation between dystrophin restoration and amelioration of respiratory function (Figure 2B).
CNS Effect
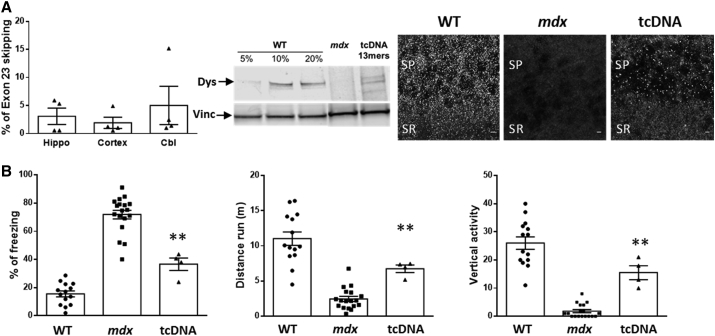
Dystrophin is expressed not only in muscles but also in central inhibitory synapses in the cerebellum, hippocampus, amygdala, and cerebral cortex, where it contributes to the clustering of synaptic γ-aminobutyric acid ([GABA]A) receptors.26, 27 In a previous study, we demonstrated for the first time that tcDNA chemistry was able to cross the blood-brain barrier and that systemic delivery of tcDNA targeting exon 23 of the mdx mouse promoted rescue of dystrophin expression in the brain.13 Here, we confirm this unique property and show, by qRT-PCR, even slightly higher levels of exon skipping in the hippocampus, cortex, and cerebellum of 13-mer tcDNA-treated mice compared to the previously reported levels using a 15-mer tcDNA-AON. We also observed restoration of dystrophin expression by western blot and confirmed the right localization of the restored dystrophin in stratum pyramidale (SP) and proximal stratum radiatum (SR) of CA1 hippocampus by immunostaining (Figure 3A).
Figure 3.
tcDNA Effect on the CNS
(A) Quantification of exon 23 skipping by qRT-PCR hippocampus (Hippo), cortex, and cerebellum (Cbl) following i.v. injection of 200 mg/kg/week of tcDNA for 12 weeks (left panel). Detection of restored dystrophin by western blot in cerebellum from tcDNA-treated mdx mice compared with WT and untreated mdx mice. 50 μg of total protein were loaded for all samples, with amounts of WT tissues ranging from 5% to 20% for the WT control (middle panel). Quantification using the standard of WT tissues reveals 5.3% ± 1.1% of dystrophin restoration. Dystrophin immunostaining in WT (left), PBS-injected control (middle) mdx mice, and tcDNA-treated mice (200 mg/kg/week, right) in the SP and proximal SR of the CA1 hippocampus. Scale bar, 20 μm. (B) Restraint-induced unconditioned fear responses expressed as a percentage of freezing time and horizontal (i.e., distance run in 5 min) and vertical activity. **p < 0.01 compared to mdx controls (Mann-Whitney U tests); n = 4 per treated group; error bars are mean ± SEM.
Brain defects due to dystrophin loss are associated with deficits in cognitive and executive functions, communication skills, and social behavior in DMD patients.28 The loss of dystrophin in the mdx mouse model of DMD has been associated with cognitive and emotional alterations, and an enhanced defensive behavior in response to stress has been reported.29 In order to evaluate a cognitive/emotional benefit following treatment, we measured the duration of tonic immobility (freezing) that resulted from a restraint-induced fear response in mdx mice. The mice were observed for a period of 5 min following an acute stress and, as expected, control mdx mice spent about 70% of the time freezing in contrast to only 20% for WT mice. Correlating with detected exon skipping and rescued dystrophin expression in the brain following 12 weeks of treatment with 13-mer tcDNA, we measured a significant improvement of the mdx emotional phenotype, reflected by the decreased freezing time, increased distance traveled, and vertical activity of the 13-mer-treated mice during the 5-min testing period (Figure 3B).
General Tolerability and Safety Profile
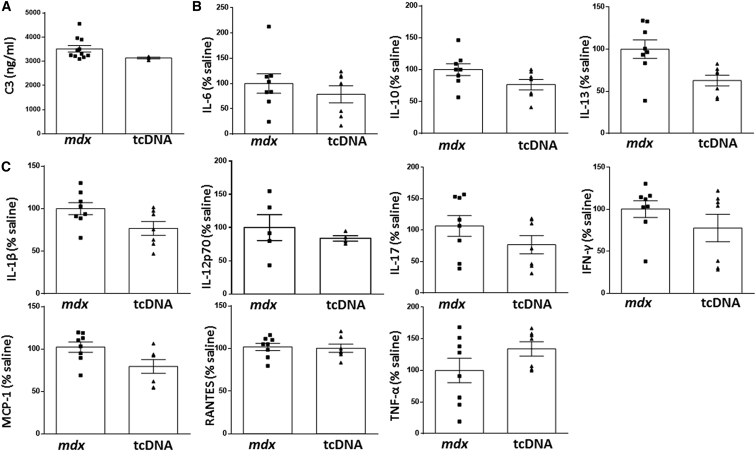
Safety pharmacological evaluation is a crucial aspect in drug development and needs to be considered relatively early for every new molecule with therapeutic purposes. In this regard, we carried out a detailed toxicity evaluation in mdx mice treated with 200 mg/kg/week of 13-mer tcDNA for 12 weeks. PS AONs are well-recognized for their immunostimulatory effects30 by activating the alternative pathway of complement among others.31, 32 Subsequently, proinflammatory cytokines are released, resulting in enhancement of innate immune responses.
In order to analyze the acute toxicologic effects of 13-mer tcDNA in mice, both complement activation and cytokine levels were analyzed (Figures 4A and 4B). The results showed that C3 protein levels, a complement protein with a central role in complement activation, remained unchanged after 13-mer tcDNA injection (Figure 4A). Complement activation was further investigated in human serum in vitro. Although positive control Zymosan significantly activates the complement pathway, tcDNA showed no effect, confirming the results obtained in vivo (Figure S2). Similarly, proinflamatory cytokines interleukin-1b (IL-1b), IL-12p70, IL-17, interferon-gamma (IFN-γ), monocyte chemoatractant protein (MCP)1, Rantes, and tumor necrosis factor alpha (TNF-α) as well as those with anti-inflammatory roles (IL-6, IL-10, and IL-13) showed no significant differences in mice injected with 13-mer tcDNA compared to mdx control mice (Figure 4B). Additional adverse and toxic effects of antisense molecules may include liver or renal injury, especially in repeated-dose studies, considering the amount of AONs that ends up in the liver and kidney (Figure 1D). In particular, the increased conferred stability of PS linkage may lead to tcDNA-PS accumulation within lysosomes from tubular renal cells and slower elimination by the urinary system. On the contrary, it is accepted that hepatotoxicity is unrelated to accumulation but to its interaction with hepatic proteins.20, 33
Figure 4.
Toxicology Profile following Intravenous Injection of tcDNA in mdx Mice
(A) C3 Complement activation analysis was carried out in mdx serum collected 1 hr after injection with 13-mer tcDNA (n = 3) or vehicle (PBS, n = 11). (B and C) Anti-inflammatory (B) and proinflammatory (C) cytokine levels were evaluated by multiplex inmunoassays using Luminex technology in mdx serum collected 1 hr after injection with 13-mer tcDNA (n = 7) or vehicle (PBS, n = 8). Data are mean ± SEM (p < 0.05).
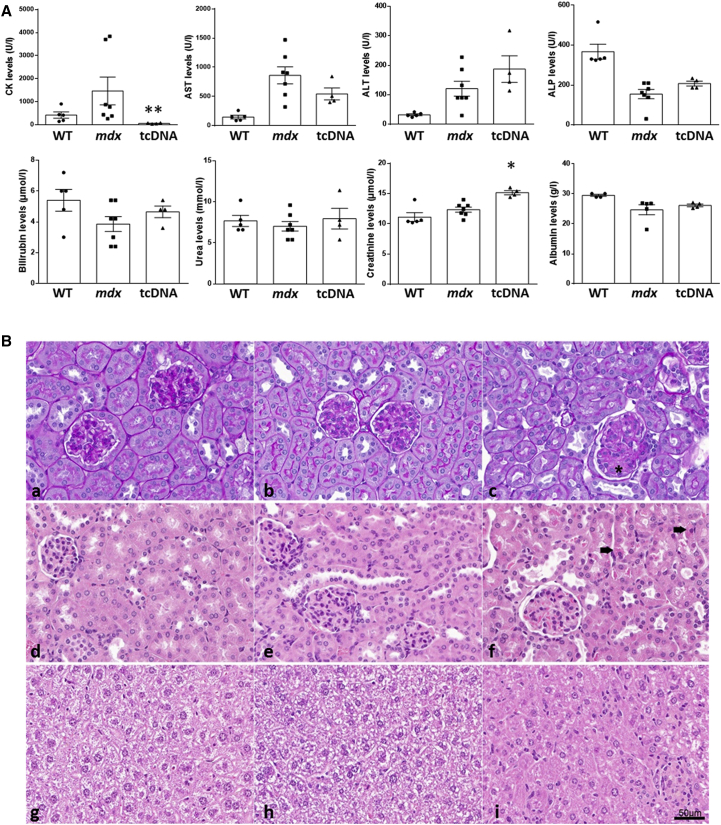
In this regard, we analyzed the serum levels of various general biomarkers in mice following 12 weeks of treatment with 200 mg/kg/week of 13-mer tcDNA. The serum creatine kinase (CK) level, a marker for muscle injury, was largely reduced in mice treated with 13-mer tcDNA, which confirmed the efficacy of the treatment to restore dystrophin and improve the dystrophic pathology of mdx mice (Figure 5A). We next quantified serum levels of bilirubin, alkaline phosphatase (ALP), and transaminases (ALT and AST) and observed no significant difference compared to mdx control mice (Figure 5A). These results confirm previous work, in which hepatic toxicities have been primarily related to locked nucleic acid (LNA) modifications rather than PS modifications.34
Figure 5.
Serum Biochemistry and Histopathological Profile of Liver and Kidneys in tcDNA-Treated mdx Mice
(A) Serum CK, AST, ALT, ALP, bilirubin, urea, creatinine, and albumin levels were measured at the end of the treatment in WT, mdx controls, and mdx mice treated with 200 mg/kg/week of 13-mer tcDNA for 12 weeks. n = 4 per treated group; error bars are mean ± SEM; *p < 0.05. (B) Histopathological profile of kidney and liver following tcDNA treatment. (a–c) PAS staining of WT (a) and control mdx (b) mouse kidney with negative staining of glomeruli. (c) In mice treated with 200 mg/kg/week of 13-mer tcDNA for 12 weeks, a slight increased mesangial matrix can be observed in the glomeruli, as shown with an asterisk. (d and e) H&E staining of WT (d) and control mdx (e) mouse kidney with normal glomeruli. (f) Few single cell necroses are observed within the proximal convoluted tubules in the tcDNA-treated mouse kidney (black arrow). (g and h) H&E staining of WT (g) and control mdx (h) mouse liver. (i) H&E staining of tcDNA-treated mouse liver reveals slightly increased inflammatory infiltrates.
First-line biomarkers to evaluate kidney function include serum urea, albumin, and creatinine because their abnormal values may indicate kidney dysfunction. Our results showed that both serum urea and albumin remained unchanged in tcDNA-treated mdx mice, whereas serum creatinine showed slightly increased values after 12 weeks of treatment when compared to WT and mdx control mice. This has already been observed with other AONs, such as 2′OMePS-modified AONs13 and may reflect an accumulation of the AON within lysosomes of the kidney proximal tubule (Figure 5A). Additionally, we explored the histopathological profile of kidney and liver in tcDNA-treated mdx in order to determine and evaluate the potential toxic response within the tissues. Histopathological findings were limited to minimal glomerular changes, characterized by slightly increased mesangial matrix in only very few glomeruli. Few single cell necroses were detected within the proximal convoluted tubules (Figure 5B). These findings could be the very early signs of a mild nephrotoxicity, characteristic of PS-AON accumulation in the kidney at this high dosing regimen. In livers from tcDNA-treated mice, increased inflammatory infiltrates could be detected without any necrotic hepatocytes, suggesting a minimal to moderate inflammation (Figure 5B).
Evaluation of Kidney Toxicity: Urinalysis and Genomic Biomarkers
In order to further study the potential renal toxicity that AON administration may induce due to its accumulation in proximal tubules, several described KIBs have been studied.35 Prior to evaluating the potential renal damage induced by tcDNA treatment, we first assessed the basal renal status in WT and mdx mice as well as their age-related progression. Although no significant differences were found in total urine protein or creatinine levels between WT and mdx mice, urine albumin was slightly increased in the latter (Table 1). Untreated mdx mice showed evidence of early renal injury (6 weeks old), which was further manifested by significantly increased expression of B2-microglobulin (B2M), renin, neutrophil gelatinase-associated lipocalin (NGAL), and osteopontin (OPN). With age, additional KIBs, such as cystatin C, were also found to be significantly elevated in mdx mice compared to their age-matched WT counterparts, whereas others were normalized (B2M, IFN-γ-induced protein 10 [IP10], and OPN) (Figure S3). Our results indicate that mdx mice develop early renal injury independently from AON treatment, which confirms previously published results.36
Table 1.
Urinary Levels of Albumin, Creatinine, and Total Protein in WT and mdx Mice Treated or Not with 13-mer tcDNA during 7 and 12 Weeks, Respectively
| Total Protein (μg/μL) | Albumin (μg/mL) | Creatinine (mg/dL) | |
|---|---|---|---|
| WT (n = 5) | 9.55 ± 2.15 | 29.85 ± 5.8 | 59.89 ± 7.54 |
| mdx (n = 4) | 9.72 ± 1.56 | 58.81 ± 17.38 | 68.18 ± 4.15 |
| tcDNA 7 weeks (n = 4) | 7.02 ± 0.49 | 178.4 ± 90.10 | 73.02 ± 7.01 |
| tcDNA 12 weeks (n = 4) | 7.73 ± 1.46 | 112.2 ± 15.95 | 70.19 ± 14.80 |
| p value (WT versus mdx) | >0.99 | 0.68 | >0.99 |
| p value (mdx versus tcDNA 7 weeks) | >0.99 | 0.59 | >0.99 |
| p value (mdx versus tcDNA 12 weeks) | >0.99 | 0.28 | >0.99 |
Data are expressed as mean ± SEM.
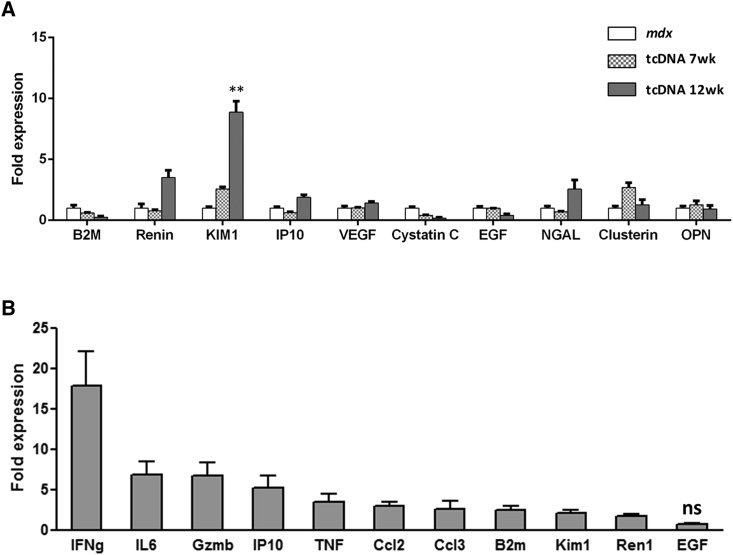
Treatment with tcDNA for 12 weeks did not affect total urine protein content or creatinine levels (Table 1). In addition, mdx mice treated for 7 or 12 weeks showed restored levels of B2M and cystatin C. However, kidney injury molecule 1 (KIM1) and renin were found to be further increased after 12 weeks of treatment (about 10- and 5-fold, respectively, compared to age-matched mdx controls) (Figure 6A; Table 2). Urinary albumin, although not statistically significant (p > 0.05), was also found to be elevated compared to untreated mice (Table 1). All other analyzed KIBs remained unchanged.
Figure 6.
Urinary KIB Levels after tcDNA Treatment
(A) KIBs were evaluated in urine collected from mdx mice controls and treated for a period of 7 or 12 weeks with tcDNA by multiplex assay using Luminex technology (n = 4 per group). Comparisons of statistical significance were assessed by Kruskal-Wallis followed by Dunn’s multiple comparison test (**p < 0.01). (B) KIB expression on the renal cortex of mice treated with 13-mer tcDNA was analyzed by qRT-PCR (n = 4 per group). The expression of all KIBs is significantly upregulated compared to age-matched mdx control (p < 0.0001), except for EGF (ns, not statistically significant). Normalized data are mean ± SEM.
Table 2.
KIB Urinary Biomarker Levels Normalized to Creatinine in mdx Mice Treated or Not with tcDNAs During 7 or 12 Weeks
| B2M (μg/mg) | Renin (μg/mg) | Kim1 (μg/mg) | IP10 (μg/mg) | VEGF (μg/mg) | Cystatin C (ng/mg) | EGF (μg/mg) | NGAL (ng/mg) | Clusterin (μg/mg) | OPN (ng/mg) | |
|---|---|---|---|---|---|---|---|---|---|---|
| mdx | 50.79 ± 11.83 | 2.465 ± 1.31 | 0.25 ± 0.04 | 2.28 ± 0.71 | 0.09 ± 0.02 | 827.33 ± 348.01 | 3.28 ± 0.28 | 729.05 ± 249.75 | 4.63 ± 0.53 | 1,360.4 ± 369.08 |
| tcDNA 7 weeks | 30.21 ± 2.52 | 1.87 ± 0.26 | 0.65 ± 0.05 | 1.42 ± 0.17 | 0.09 ± 0.01 | 335.87 ± 24.04 | 3.19 ± 0.02 | 513.64 ± 38.70 | 12.45 ± 1.83 | 1,712.39 ± 452.31 |
| tcDNA 12 weeks | 12.90 ± 5.70 | 8.58 ± 1.51 | 2.25 ± 0.23 | 4.31 ± 0.48 | 0.13 ± 0.01 | 138.41 ± 77.20 | 1.25 ± 0.38 | 1,865.94 ± 537.07 | 5.89 ± 1.94 | 1,258.42 ± 395.59 |
| p value (mdx versus week 7) | >0.99 | >0.99 | 0.472 | 0.866 | >0.99 | >0.99 | 0.231 | 0.867 | >0.99 | >0.99 |
| p value (mdx versus week 12) | 0.472 | <0.05 | <0.05 | 0.866 | 0.472 | 0.231 | 0.231 | 0.866 | >0.99 | >0.99 |
Data are expressed as mean ± SEM.
As previously published, the KIBs can also be measured by gene expression at the mRNA level on the mice kidney cortex,20 so we next confirmed these results by evaluating the gene expression of some KIBs on the mRNA level from tcDNA-treated mice and showed significant changes for several biomarkers (Figure 6B). Similar changes were previously reported with 2′OMePS-AON, confirming the expected effect of PS-AON accumulation in kidneys.
Discussion
Over the last decade, the prospect of successful AON-based splicing therapy for neuromuscular disorders has moved a step closer, particularly for DMD. Many of these promising therapies have now entered clinical trials and encouraging results have been obtained in most cases while also showing the limitations of currently used chemistries. The current generation of AONs has indeed failed to demonstrate clinical efficacy in DMD patients, largely due to a poor uptake and a few safety issues (for 2′OMePS at least) imposing a limited amount of AONs. Clinical studies in DMD face many other challenges that not only are common to trials in all rare diseases but are also specific to the Duchenne population, such as the number of patients with particular deletions amenable to exon skipping, the variation in clinical care across centers, or the lack of validated biomarkers.11, 37 Regarding the design and optimization of the AON itself, it has become clear that improving the delivery and safety profile are the major challenges, and international efforts are currently focusing on new generations of AONs or different drug delivery systems, such as various conjugations, to find the best clinical candidates in terms of efficiency and safety profile. Among these new promising chemistries, tcDNA has shown interesting properties and clear superiority compared to clinically tested AONs (2′OMe and PMO),13 which is partly due to its RNA-binding affinities, permitting use of AONs of decreased length. A previous study performed in a comparative cellular assay has already shown that a tcDNA of only 11 nt induced significant exon skipping.38 This crucial feature offers the advantage of reducing the mass of synthetic nucleotides administered while keeping a biological effect, thus reducing toxicity induced by the accumulation of AONs. In this study, we therefore focused on the efficacy and toxicology evaluation of a shorter tcDNA-AON of 13 nt targeting the mouse dystrophin exon 23 and demonstrate the therapeutic potential of tcDNA in the mdx mouse model following systemic treatment for 12 weeks. The 13-mer tcDNA was efficient in all tested skeletal muscles, albeit slightly less than the previously reported 15-mer tcDNA.13 Remarkably, in the heart, the 13-mer tcDNA induces particularly high levels of exon skipping and dystrophin restoration, suggesting a potential size advantage for uptake in the cardiac muscle.
Restoration of dystrophin expression significantly improved the mdx mouse phenotype and many functional improvements were observed, including gain in muscle-specific force and better respiratory function, which represents an indispensable requisite for a life-improving therapy, considering that respiratory failure is a major cause of death among patients with DMD.25
Exon skipping and dystrophin restoration were also detected in the brain following 13-mer tcDNA treatment, supporting the fact that tcDNAs are able to cross the blood-brain barrier. In the brain, dystrophin (Dp427) is found at the level of inhibitory GABAergic synapses in Purkinje cells of the cerebellum and in principal neurons of the hippocampus, cortex, and amygdala. Concurring with cognitive deficits and psychiatric symptoms observed in DMD, an enhanced defensive behavior in response to a threat has previously been reported in dystrophin-deficient mdx mice.29 Despite limited restoration of dystrophin observed in the brain, 13-mer tcDNA treatment was able to significantly restore certain behavioral aspects linked to the absence of dystrophin in the CNS, highlighting a great advantage of this chemistry for the systemic treatment of DMD. We are currently investigating these emotional/cognitive aspects in the mdx mouse model in more detail and assessing the effect of tcDNA local delivery to the specific structures of the brain (intraventricular, hippocampus, and cerebellum) in order to shed some light on the dystrophin localization/function in the CNS. The loss of full-length brain dystrophin in mdx mice has also been shown to impair their capacity of acquisition and long-term retention of cued and trace fear memories;39 thus, evaluating the effect of brain dystrophin rescue on long-term memory deficits following systemic and locoregional treatment of tcDNA may be particularly interesting to clarify the role of dystrophin in the brain.
Although the 13-mer tcDNA shows a great efficiency, similar to the previously reported 15-mer tcDNA, decreasing the sequence length makes it increasingly more challenging to identify a unique target site. Furthermore, increased binding energy has the capacity to not only increase potency against the intended target but also to “off-targets”, and the so-called hybridization-mediated off-target effects (OTEs) may become a more prevalent concern. However, because splicing modulation is highly dependent on target sequence position in the intended RNA target, effects on unintended off-target RNAs containing a perfect match are extremely unlikely with splice-switching AONs as opposed to gapmer AONs recruiting RNase H.30, 40 OTEs have not been noted with great abundance in preclinical or clinical studies to date, probably because not all off-target binding actually leads to functional effects on mRNA splicing. Not all sites on mRNA are accessible to an AON, nor are all off-target mRNAs in tissues that receive pharmacologic concentrations of an AON because they are differentially and temporally expressed in different tissues. Putative off-target interactions can be predicted using sequence alignment algorithms and pragmatic genomic screening strategies are in place in most companies developing AON-based drugs to delineate potential OTEs. The Oligonucleotide Safety Working group (OSWG) recommends assessment of OTEs for AONs during drug discovery and development, both computationally and experimentally,41 and such studies are currently ongoing for tcDNA candidates targeting human dystrophin exons. Although less relevant for future clinical evaluation, the 13-mer tcDNA used here and targeting the mouse exon 23 has been blasted and four potential targets have been identified (with a 12 nt/13 nt match) on mouse mRNA. However, expression of these targets was not modified after treatment with tcDNA (Figure S4).
Although these recommendations and screening are necessary, OTEs are generally not the main concern in the antisense field. Instead, the toxicities noted have been overwhelmingly associated with those expected by class. Each class of AON from first to third generation has stereotypic toxicity profiles, and these nuances are important for the toxicologic pathologist to be aware of.30 It is indeed becoming clear that these toxicological challenges should be addressed in the very early stages of new AON development to ensure the clinical translation of these studies.
Here, we evaluated for the first time the toxicology profile of a tcDNA-AON in a DMD mouse model and showed that high-dose tcDNA treatment (200 mg/kg/week for 12 weeks) was well-tolerated in all mice. Because AON can provoke an inflammatory reaction,36 we measured the levels of specific cytokines and chemokines following administration of tcDNA-AON and did not detect any acute pro-inflammatory response. We also evaluated the long-term toxicological profile within the tissues, and histopathological findings in tcDNA-treated animals were generally limited to minimal glomerular changes and few cell necroses in proximal tubules. Serum biochemistry analysis following 12 weeks of treatment did not reveal any significant increase in transaminases or bilirubin and only a slight variation in serum creatinine, which is typical of the PS-AON accumulation in kidneys. To investigate the toxicological profile of tcDNA more deeply, we measured some early biomarkers of renal toxicity at protein and genomic levels and detected some significant upregulation in several biomarkers. These results indicate some renal toxicity due to AON accumulation in kidney tubular cells and are in concordance with the duration and dose regimen of the AON treatment.20 Considering the potential to reach >200-fold in nephrotoxic models for KIM1 for example,42 the fold changes observed here can still be considered moderate and in the expected range for PS-AONs. Although these data confirm the renal toxicity associated with accumulation of PS-AONs in kidneys, no unexpected class-related toxicological issue emerged following tcDNA treatment.
Altogether, these results demonstrate a typical PS-AON safety profile for tcDNA, which enables dystrophin rescue in all tissues affected by the lack of dystrophin (including skeletal muscles, heart, and brain) following systemic administration. Importantly, this dystrophin rescue translated into functional improvements in a mouse model of DMD, especially of the respiratory system, and certain behavioral aspects. The properties of tcDNA make them particularly attractive for genetic diseases that are treatable by splice-switching approaches, and clinical evaluation of tcDNA for the treatment of DMD is currently being planned. However, the promise of tcDNA still crucially depends on how well it will be tolerated in humans and one should remain cautious until full reglementary toxicological studies are completed with the clinical candidate. We cannot exclude a possible toxicity that would be sequence specific because it has been previously reported that the LNAs induced unexpected, sequence-specific hepatotoxicity.34
Materials and Methods
Antisense Oligonucleotides and Animal Experiments
Animal procedures were performed in accordance with national and European legislation and approved by the French government (ministère de l’enseignement supérieur et de la recherche, Autorisation APAFiS #6518). mdx (C57BL/10ScSc-Dmdmdx/J) and C57BL/10 mice were bred in our animal facility at the Plateform 2Care, UFR des Sciences de la santé, Université de Versailles Saint Quentin, and were maintained in a standard 12-hr light/dark cycle with free access to food and water. Mice were weaned at weeks 4 to 5 postnatal, and two to five individuals were housed per cage.
The tcDNA-AON PS M23D 13-mer (5′-pCCTCGGCTTACCT-3′) targeting the donor splice site of exon 23 of the mouse dystrophin pre-mRNA used in this study was synthesized by SYNTHENA. Four 6- to 8-week-old mdx mice were injected intravenously in the retro-orbital sinus under general anesthesia using 1.5%–2% isoflurane once a week with the 13-mer tcDNA for a period of 12 weeks. An age-matched C57/BL10 (WT) group and mdx group receiving an equivalent volume of sterile saline were included as controls. 1 hr after the first injection, blood samples were collected from all mice to measure complement C3 and cytokine/chemokine levels. Additional blood samples were collected 1 week after the sixth injection (mid-treatment) and 1 week after the end of the treatment. 1 week after the last injection, behavioral tests were conducted and respiratory function was assessed. Mice were then placed in metabolic cages for urine collection over a period of 24 hr. Muscle function in situ was measured 2 weeks after the last injection as a final experiment before sacrifice. Muscles and tissues were then harvested and snap-frozen in liquid nitrogen-cooled isopentane and stored at −80°C before further analysis. Sample sizes and n values are indicated in each figure legend. Investigators were blinded for RNA and protein analysis, behavioral studies, muscle force, and respiratory function measurements.
Serum and Urine Analysis
Blood samples were collected from tail bleeds under general anesthesia. Analyses of serum CK, ALT, AST, ALP, bilirubin, creatinine, urea, and albumin levels were performed by the pathology laboratory at Mary Lyon Centre, Medical Research Council. Complement activation in mouse serum samples was measured by Microvue PS-C3 converter and SC5b-9 Plus kits (Quidel). For in vitro complement activation studies, tcDNA was incubated with normal pooled human serum (1:10) (Seralab) at 37°C for 45 min. Determination of complement activation was evaluated using the human SC5b-9 Plus kit (Quidel Co). 5 mg/mL Zymosan (Complement Technology) was used as positive control. Cytokine and chemokine levels in serum were analyzed by multiplex assays using Luminex technology. A Bio-Plex Pro Mouse Cytokine 10-Plex Immunoassay panel (Bio-Rad) was used to detect levels of IL-1β, IL-6, IL-10, IL-12p70, IL-13, IL-17, IFN-γ, MCP1, Rantes, and TNF-α according to the manufacturer’s instructions. Immunoassays were read using a Bio-Plex MAGPIX Multiplex reader, and the results were analyzed with Bio-Plex manager 6.1 software (Bio-Rad).
Urine was collected using metabolic cages over 24 hr directly in refrigerated tubes (4°C). Upon collection, urine was centrifuged at 10,000 × g for 10 min and supernatant was aliquoted and frozen at −80°C for further analysis. Urine creatinine was measured using the creatinine assay kit (R&D Systems) following the manufacturer’s instructions. Total protein in urine samples was measured as previously described.36 Briefly, proteins were precipitated from urine samples by adding 40 μL of dH2O and 200 μL of prechilled acetone to 10 μL of urine. Samples were then incubated at −20°C for 30 min, then centrifuged at 14,000 × g, 4°C, for 15 min. Pellets were resuspended in 40 μL of dH2O and protein concentration was measured using the Pierce BCA assay (Thermo Scientific). Albumin from urine samples was measured using the albumin ELISA kit (Bethyl Laboratories) following the manufacturer’s instructions. Acute kidney injury (AKI) biomarker levels were analyzed by multiplex assays using Luminex technology. The multiplex kidney injury panels (MKI1MAG-94K and MKI2MAG-94K, Merck Millipore) were used according to the manufacturer’s instructions to measure levels of B2M, renin, KIM1, IP10, vascular endothelial growth factor (VEGF), Cystatin C, epidermal growth factor (EGF), Lipocalin-2-NGAL, Clusterin, and OPN. The results were read using a Bio-Plex MAGPIX Multiplex reader and analyzed with the Bio-Plex manager 6.1 software (Bio-Rad).
RNA Analysis
Total RNA was isolated from intervening muscle sections collected during cryosection using TRIzol reagent according to the manufacturer’s instructions (Thermo Fisher Scientific). Aliquots of 500 ng of total RNA were used for RT-PCR analysis using the Access RT-PCR System (Promega) in a 50-μL reaction using the external primers Ex 20Fo (5′- CAGAATTCTGCCAATTGCTGAG-3′) and Ex 26Ro (5′-TTCTTCAGCTTGTGTCATCC- 3′). The cDNA synthesis was carried out at 45°C for 45 min, directly followed by the primary PCR of 30 cycles of 95°C (30 s), 55°C (1 min), and 72°C (2 min). 2 μL of these reactions were then re-amplified in nested PCRs by 22 cycles of 95°C (30 s), 55°C (1 min), and 72°C (2 min) using the internal primers Ex 20Fi (5′- CCCAGTCTACCACCCTATCAGAGC-3′) and Ex 26Ri (5′- CCTGCCTTTAAGGCTTCCTT-3′). PCR products were analyzed on 2% agarose gels. Exon 23 skipping was also measured by Taqman qRT-PCR, as previously described,13, 43 using Taqman assays that were designed against the exon 4-5 or exon 22-24 templates using the Custom Assay Design Tool (Life Technologies) (assay exon 4-5: forward: 5′-GGCACTGCGGGTCTTACA-3′; reverse: 5′-CATCCACTATGTCAGTGCTTCCTAT 3′; probe: 5′-TTCACTAAATCAACATTATTTTTC-3′; assay exon 22-24: forward: 5′- CTGAATATGAAATAATGGAGGAGAGACTCG-3′; reverse: 5′-CTTCAGCCATCCATTTCTGTAAGGT-3′; probe: 5′-ATGTGATTCTGTAATTTCC-3′). An inventoried 18S assay was utilized as an endogenous control (Life Technologies). 50 ng of cDNA was used as input per reaction, and all assays were carried out in triplicate. Assays were performed under fast cycling conditions on a Bio-Rad CFX96 Touch Real-Time PCR Detection System, and all data were analyzed using the comparative Ct method. For a given sample, the delta-Ct values of the exon 4-5 and exon 22-24 assays were used to calculate a relative abundance of total dystrophin and exon-23-skipped dystrophin mRNA, respectively. Exon 23 skipping was then expressed as a percentage against total dystrophin, as indicated by the exon 4-5 expression level.
Background levels of exon skipping detected in non-treated mdx controls (0.06% for TA, 0.12% for gastrocnemius, 0.20% for quadriceps, 0.09% for triceps brachialis, 0.004% for biceps brachialis, 0.01% for diaphragm, 0.004% for heart, 0.23% for cortex, 0.66% for hippocampus, and 0.27% for cerebellum) were deducted from the presented values.
Total RNA was also isolated from renal cortex samples as previously described,20 and qRT-PCR was performed on KIB genes IFN-γ, IL-6, Granzyme B (Gzmb), IP-10, TNF, chemokine ligand 2 (Ccl2), Ccl3, B2M, KIM1, Renin 1 (Ren1), and EGF and off-target genes Kremen1, Hopx, Ucn3, and Kif13a. After cDNA synthesis, real-time PCR was performed using the SYBR Green PCR Master Mix Protocol (Bio-Rad) in triplicate with a hotstart Taq polymerase. A 10-min denaturation step at 94°C was followed by 40 cycles of denaturation at 94°C for 10 s and annealing/extension at 60°C for 30 s. Primer PCR efficiency for each gene was determined using a standard dilution series (100–107 copies/μL), which subsequently enabled us to calculate the copy numbers from the Ct values. mRNA levels were normalized to GAPDH, and fold changes were calculated according to the -ΔΔCt method. The different primers sequences are described in the Supplemental Information (Table S1).
Western Blot and Immunohistochemistry Analysis
Protein extracts were obtained from pooled muscle sections treated with 125 mmol/L sucrose, 5 mmol Tris-HCl, pH 6.4, 6% of MOPS SDS Running buffer (Thermo Fisher Scientific), 10% SDS, 10% glycerol, and 5% β-mercaptoethanol. The samples were purified with the Compt-Able Protein Assay preparation reagent set (Thermo Fisher Scientific), and the total protein concentration was determined with the BCA Protein Assay Kit (Thermo Fisher Scientific). Samples were denatured at 95°C for 5 min, and 100 μg of protein were loaded onto Mini-Protean TGX stain-free gels 4%–15% (Bio-Rad) following the manufacturer’s instructions. Dystrophin protein was detected by probing the membrane with 1:50 dilutions of NCL-DYS1 primary monoclonal antibody (NCL-DYS1; Novocastra), and vinculin was detected as the internal control with the hVin-1 primary antibody (Sigma; 1:10,000), followed by incubation with a sheep anti-mouse secondary antibody (horseradish peroxidase conjugated; 1:15,000). Bands were visualized using the Odyssey CLx system (LI-COR Biosciences). The quantification has been done using the standard curve with 0%, 5%, 10%, and 20% of corresponding WT tissues and normalized to internal control (vinculin).
Sections of 10 μm were cut from at least two-thirds of the muscle length of the various tissues (TA, gastrocnemius, quadriceps femoris, biceps brachialis, triceps brachialis, diaphragm, and cardiac muscle) at 100-μm intervals. The intervening muscle sections were collected for RNA analysis and western blot analysis. The cryosections were examined for dystrophin expression using the rabbit polyclonal antibody dystrophin (dilution 1:500; cat. number RB-9024-P, Thermo Fisher Scientific), which was then detected by goat-anti-rabbit immunoglobulin Gs (IgGs) Alexa 488 (dilution 1:1000, Thermo Fisher Scientific).
Brain fresh-frozen cryosections of 30 μm were collected onto Superfrost+ slides, thawed for 2 min at room temperature (RT), fixed in acetone/methanol (1:1) for 5 min at −20°C, washed in PBS, incubated first in a blocking solution for 45 min (10% normal goat serum, 0.3% Triton X-100, and 1% BSA), then overnight at 4°C with a monoclonal anti-dystrophin primary antibody (DYS1 Leica; dilution: neat) and washed and incubated with secondary antibody Alexa 647 (1:400, 1 hr, RT). Controls prepared by omitting the primary antibody showed no specific staining. Images were taken at equivalent locations and exposure times using a laser scanning confocal microscope (Zeiss LSM 700, × 40 objective). Stacks of seven to eight images (1,024 × 1,024 pixels) spaced by 1 μm were recorded at a magnification of 156 nm/pixel.
Formalin-fixed kidneys and livers were paraffin-embedded, and 4-μm longitudinal and sagittal sections cut and stained with H&E or periodic acid shift (PAS) as previously described.44
Quantification of tcDNA by LC-MS/MS
Intervening sections were collected during cryosection of the frozen muscles and tissues and were homogenized to a concentration of 50 mg/mL in proteinase K buffer (100 mmol/L Tris-HCl, pH 8.5, 200 mmol/L NaCl, 5 mmol/L EDTA, and 0.2% SDS) containing 2 mg/mL of proteinase K (Invitrogen), followed by incubation for 2 hr (liver) or 4 hr (kidney, heart, and skeletal muscle) rotating at 55°C in a hybridization oven. Next, the samples were centrifuged for 15 min at maximum speed and the supernatant was used in the assay. Quantification of tcDNA was performed by LC-MS/MS analysis on a Dionex UltiMate 3000 RS HPLC system (Thermo Fisher Scientific) coupled to a Q Exactive Hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) as previously described.13
Muscle Function Analysis
Muscle function of mdx mice was evaluated by measuring TA muscle contraction in situ in response to nerve stimulation. Mice were anesthetized using pentobarbital (60 mg/kg intraperitoneally). Body temperature was maintained at 37°C using radiant heat. The knee and foot were fixed with pins and clamps, and the distal tendon of the muscle was attached to the lever arm of a servo-motor system (305B; Dual-Mode Lever; Aurora Scientific) using a silk ligature. The sciatic nerve was crushed proximally and stimulated distally by a bipolar silver electrode using supramaximal 0.1 ms duration square-wave pulses. We measured the absolute maximal isometric tetanic force (P0) generated during isometric contractions in response to electrical stimulation (frequency of 75–150 Hz, stimulation train of 500 ms). P0 was determined at L0 (length at which maximal tension was obtained during the tetanus). Absolute maximal isometric force was normalized to muscle mass as an estimate of specific maximal force (sP0), that is, specific force.
Fragility was estimated from the force decline resulting from lengthening contraction-induced injury. The sciatic nerve was stimulated for 700 ms (150 Hz stimulation frequency). A maximal isometric contraction of the TA muscle was initiated during the first 500 ms. Then, muscle lengthening (10% L0) at a velocity of 5.5 mm/s was imposed during the last 200 ms. All isometric contractions were made at an initial length L0. Nine lengthening contractions of the TA muscles were performed, each separated by a 60 s rest period. Maximal isometric force was measured 1 min after each lengthening contraction and expressed as a percentage of the initial maximal isometric force. As an indicator of active muscle stiffness, we measured the increase in force during the stretch of the first lengthening contraction. This force was expressed as a percentage of P0.
Respiratory Function
The respiratory function of mice was evaluated by whole-body plethysmography using an EMKA Technologies plethysmograph, as previously described,13 and essentially as recommended by TREAT-NMD (http://www.treat-nmd.eu/downloads/file/sops/dmd/MDX/DMD_M.2.2.002.pdf).
Briefly, unrestrained conscious mice were placed in calibrated animal chambers and the pressure difference between the reference and animal chambers was measured using a pressure transducer. Mice were allowed to acclimate in the chambers for 45 min at a stable temperature and humidity. Data were then collected every 5 s using the iox2 software (version 2.8.0.19; EMKA Technologies). TI was defined as the start of inspiration to the end of inspiration and the expiration time (TE) was defined as the start of expiration to the end of expiration. The relaxation time (RT) was defined as the time from the start of expiration to the time when 65% of the total expiratory pressure occurred. Pause and penh were defined and calculated by the following formulas: pause = (TE − RT)/RT and penh = (PEP/PIP) × pause, where PEP is peak expiratory pressure and PIP is peak inspiratory pressure. The value of each parameter was calculated from an average of 60 recordings of 5 s representing a total of 5 min. Inclusion criteria for each recording were >8 respiration events by 5 s and >80% of success rate as measured by the iox software.
Restraint-Induced Unconditioned Fear
Mice were handled firmly but gently using the scruff method, as for standard examination or intraperitoneal injection in laboratory mice. The mouse was restrained by a trained experimenter grasping the scruff and back skin between the thumb and index finger while securing the tail between the third and little fingers and tilting the animal upside down so that the ventral part of its body faced the experimenter. After 15 s, the mouse was released to a new cage (16 × 28 cm, with 12 cm high walls; illumination: ∼100 lx) and then video-tracked for 5 min using the ANY-maze software (Stoelting). All mice were tested between 10:00 am and 1:00 pm. Unconditioned fear responses induced by this acute stress were characterized by periods of tonic immobility (freezing) during the 5-min recording period in the novel cage. Complete immobilization of the mouse, except for respiration, was regarded as a freezing response. This was typically quantified as episodes of immobility lasting at least 1 s, with a 90% immobility sensitivity (10% body motion allowed). In all experiments, the percent time the mouse was freezing was calculated for group comparisons. Horizontal (i.e., the distance traveled) and vertical activity (number of ups) was also recorded. The investigator was blinded to the group allocations during the experiments.
Statistical Analysis
Data were analyzed by GraphPad Prism5 software and shown as the mean ± SEM. n refers to the number of mice per group. Comparisons of statistical significance were assessed by non-parametric Mann-Whitney U tests for the comparison of two groups or Kruskal-Wallis for the comparison of three or more groups, followed by Dunn’s multiple comparison test. Significant levels were set at *p < 0.05; **p < 0.01; and ***p < 0.001.
Author Contributions
G.G., K.R., L.E., F.Z., C.V., P.F., and A.G. designed and performed the laboratory experiments. G.G. performed muscle sectioning and RNA and protein analysis. K.R. and L.E. performed the toxicology analysis. P.F. performed histopathology on the liver and kidney. F.Z. and C.V. analyzed the CNS and performed the behavioral tests. K.R., L.E., C.L., L.G., and A.G. wrote the manuscript. A.G. and L.G. conceived the project, designed the experiments, supervised the entire study, and will serve as corresponding authors.
Conflicts of Interest
C.L. and L.G. are co-funders of SYNTHENA, which produces tricyclo-DNA oligomers.
Acknowledgments
We thank SYNTHENA, Berne, Switzerland, for providing the tcDNA-AONs used in this study. We thank the centre d’évaluation fonctionnelle of the University Pierre et marie Curie and Arnaud Ferry for the force measurements on mdx mice. We thank Stanislas Grassin Delyle and the MasSpecLab platform for the LC-MS/MS analysis. We would also like to thank Laurence Flette from the Pasteur Institute for her expertise in histopathology. This work was supported by the Agence nationale de la recherche (DYSther Project grant ANR-14-CE13-0037, Chair of Excellence HandiMedEx), the Institut National de la santé et la recherche médical (INSERM), the Association Monegasque contre les myopathies (AMM), and the Duchenne Parent project France (DPPF).
Footnotes
Supplemental Information includes four figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.06.013.
Contributor Information
Luis Garcia, Email: luis.garcia@uvsq.fr.
Aurélie Goyenvalle, Email: aurelie.goyenvalle@uvsq.fr.
Supplemental Information
References
- 1.Koenig M., Hoffman E.P., Bertelson C.J., Monaco A.P., Feener C., Kunkel L.M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 2.Muntoni F., Torelli S., Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 3.Emery A., Muntoni F. Oxford University Press; 2003. Duchenne Muscular Dystrophy. [Google Scholar]
- 4.Goemans N.M., Tulinius M., van den Akker J.T., Burm B.E., Ekhart P.F., Heuvelmans N., Holling T., Janson A.A., Platenburg G.J., Sipkens J.A. Systemic administration of PRO051 in Duchenne’s muscular dystrophy. N. Engl. J. Med. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 5.Cirak S., Arechavala-Gomeza V., Guglieri M., Feng L., Torelli S., Anthony K., Abbs S., Garralda M.E., Bourke J., Wells D.J. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mostacciuolo M.L., Lombardi A., Cambissa V., Danieli G.A., Angelini C. Population data on benign and severe forms of X-linked muscular dystrophy. Hum. Genet. 1987;75:217–220. doi: 10.1007/BF00281062. [DOI] [PubMed] [Google Scholar]
- 7.van Deutekom J.C., Janson A.A., Ginjaar I.B., Frankhuizen W.S., Aartsma-Rus A., Bremmer-Bout M., den Dunnen J.T., Koop K., van der Kooi A.J., Goemans N.M. Local dystrophin restoration with antisense oligonucleotide PRO051. N. Engl. J. Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 8.Kinali M., Arechavala-Gomeza V., Feng L., Cirak S., Hunt D., Adkin C., Guglieri M., Ashton E., Abbs S., Nihoyannopoulos P. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aartsma-Rus A., Krieg A.M. FDA approves eteplirsen for Duchenne muscular dystrophy: the next chapter in the eteplirsen saga. Nucleic Acid Ther. 2017;27:1–3. doi: 10.1089/nat.2016.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Q.-L., Cirak S., Partridge T. What can we learn from clinical trials of exon skipping for DMD? Mol. Ther. Nucleic Acids. 2014;3:e152. doi: 10.1038/mtna.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Straub V., Balabanov P., Bushby K., Ensini M., Goemans N., De Luca A., Pereda A., Hemmings R., Campion G., Kaye E. Stakeholder cooperation to overcome challenges in orphan medicine development: the example of Duchenne muscular dystrophy. Lancet Neurol. 2016;15:882–890. doi: 10.1016/S1474-4422(16)30035-7. [DOI] [PubMed] [Google Scholar]
- 12.Stein C.A. Eteplirsen approved for Duchenne muscular dystrophy: the FDA faces a difficult choice. Mol. Ther. 2016;24:1884–1885. doi: 10.1038/mt.2016.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goyenvalle A., Griffith G., Babbs A., El Andaloussi S., Ezzat K., Avril A., Dugovic B., Chaussenot R., Ferry A., Voit T. Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat. Med. 2015;21:270–275. doi: 10.1038/nm.3765. [DOI] [PubMed] [Google Scholar]
- 14.Goyenvalle A., Leumann C., Garcia L. Therapeutic potential of Tricyclo-DNA antisense oligonucleotides. J Neuromuscul Dis. 2016;3:157–167. doi: 10.3233/JND-160146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renneberg D., Leumann C.J. Watson-Crick base-pairing properties of tricyclo-DNA. J. Am. Chem. Soc. 2002;124:5993–6002. doi: 10.1021/ja025569+. [DOI] [PubMed] [Google Scholar]
- 16.Ittig D., Gerber A.-B., Leumann C.J. Position-dependent effects on stability in tricyclo-DNA modified oligonucleotide duplexes. Nucleic Acids Res. 2011;39:373–380. doi: 10.1093/nar/gkq733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renneberg D., Bouliong E., Reber U., Schümperli D., Leumann C.J. Antisense properties of tricyclo-DNA. Nucleic Acids Res. 2002;30:2751–2757. doi: 10.1093/nar/gkf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray S., Ittig D., Koller E., Berdeja A., Chappell A., Prakash T.P., Norrbom M., Swayze E.E., Leumann C.J., Seth P.P. TricycloDNA-modified oligo-2′-deoxyribonucleotides reduce scavenger receptor B1 mRNA in hepatic and extra-hepatic tissues--a comparative study of oligonucleotide length, design and chemistry. Nucleic Acids Res. 2012;40:6135–6143. doi: 10.1093/nar/gks273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moulton H.M., Moulton J.D. Morpholinos and their peptide conjugates: therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim. Biophys. Acta. 2010;1798:2296–2303. doi: 10.1016/j.bbamem.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Frazier K.S., Sobry C., Derr V., Adams M.J., Besten C.D., De Kimpe S., Francis I., Gales T.L., Haworth R., Maguire S.R. Species-specific inflammatory responses as a primary component for the development of glomerular lesions in mice and monkeys following chronic administration of a second-generation antisense oligonucleotide. Toxicol. Pathol. 2014;42:923–935. doi: 10.1177/0192623313505781. [DOI] [PubMed] [Google Scholar]
- 21.Aartsma-Rus A. Antisense-mediated exon skipping: networking to meet opportunities and to overcome challenges. Nucleic Acid Ther. 2014;24:1–3. doi: 10.1089/nat.2014.1500. [DOI] [PubMed] [Google Scholar]
- 22.Godfrey C., Desviat L.R., Smedsrød B., Piétri-Rouxel F., Denti M.A., Disterer P., Lorain S., Nogales-Gadea G., Sardone V., Anwar R. Delivery is key: lessons learnt from developing splice-switching antisense therapies. EMBO Mol. Med. 2017;9:545–557. doi: 10.15252/emmm.201607199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann C.J., Honeyman K., McClorey G., Fletcher S., Wilton S.D. Improved antisense oligonucleotide induced exon skipping in the mdx mouse model of muscular dystrophy. J. Gene Med. 2002;4:644–654. doi: 10.1002/jgm.295. [DOI] [PubMed] [Google Scholar]
- 24.Sicinski P., Geng Y., Ryder-Cook A.S., Barnard E.A., Darlison M.G., Barnard P.J. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 25.Kohler M., Clarenbach C.F., Böni L., Brack T., Russi E.W., Bloch K.E. Quality of life, physical disability, and respiratory impairment in Duchenne muscular dystrophy. Am. J. Respir. Crit. Care Med. 2005;172:1032–1036. doi: 10.1164/rccm.200503-322OC. [DOI] [PubMed] [Google Scholar]
- 26.Lidov H.G., Byers T.J., Kunkel L.M. The distribution of dystrophin in the murine central nervous system: an immunocytochemical study. Neuroscience. 1993;54:167–187. doi: 10.1016/0306-4522(93)90392-s. [DOI] [PubMed] [Google Scholar]
- 27.Knuesel I., Bornhauser B.C., Zuellig R.A., Heller F., Schaub M.C., Fritschy J.M. Differential expression of utrophin and dystrophin in CNS neurons: an in situ hybridization and immunohistochemical study. J. Comp. Neurol. 2000;422:594–611. doi: 10.1002/1096-9861(20000710)422:4<594::aid-cne8>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 28.Miranda R., Nagapin F., Bozon B., Laroche S., Aubin T., Vaillend C. Altered social behavior and ultrasonic communication in the dystrophin-deficient mdx mouse model of Duchenne muscular dystrophy. Mol. Autism. 2015;6:60. doi: 10.1186/s13229-015-0053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekiguchi M., Zushida K., Yoshida M., Maekawa M., Kamichi S., Yoshida M., Sahara Y., Yuasa S., Takeda S., Wada K. A deficit of brain dystrophin impairs specific amygdala GABAergic transmission and enhances defensive behaviour in mice. Brain. 2009;132:124–135. doi: 10.1093/brain/awn253. [DOI] [PubMed] [Google Scholar]
- 30.Frazier K.S. Antisense oligonucleotide therapies: the promise and the challenges from a toxicologic pathologist’s perspective. Toxicol. Pathol. 2015;43:78–89. doi: 10.1177/0192623314551840. [DOI] [PubMed] [Google Scholar]
- 31.Henry S.P., Beattie G., Yeh G., Chappel A., Giclas P., Mortari A., Jagels M.A., Kornbrust D.J., Levin A.A. Complement activation is responsible for acute toxicities in rhesus monkeys treated with a phosphorothioate oligodeoxynucleotide. Int. Immunopharmacol. 2002;2:1657–1666. doi: 10.1016/s1567-5769(02)00142-x. [DOI] [PubMed] [Google Scholar]
- 32.Agrawal S., Kandimalla E.R. Role of Toll-like receptors in antisense and siRNA [corrected] Nat. Biotechnol. 2004;22:1533–1537. doi: 10.1038/nbt1042. [DOI] [PubMed] [Google Scholar]
- 33.Dirin M., Winkler J. Influence of diverse chemical modifications on the ADME characteristics and toxicology of antisense oligonucleotides. Expert Opin. Biol. Ther. 2013;13:875–888. doi: 10.1517/14712598.2013.774366. [DOI] [PubMed] [Google Scholar]
- 34.Swayze E.E., Siwkowski A.M., Wancewicz E.V., Migawa M.T., Wyrzykiewicz T.K., Hung G., Monia B.P., Bennett C.F. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaidya V.S., Ferguson M.A., Bonventre J.V. Biomarkers of acute kidney injury. Annu. Rev. Pharmacol. Toxicol. 2008;48:463–493. doi: 10.1146/annurev.pharmtox.48.113006.094615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang A., Uaesoontrachoon K., Shaughnessy C., Das J.R., Rayavarapu S., Brown K.J., Ray P.E., Nagaraju K., van den Anker J.N., Hoffman E.P. The use of urinary and kidney SILAM proteomics to monitor kidney response to high dose morpholino oligonucleotides in the mdx mouse. Toxicol. Rep. 2015;2:838–849. doi: 10.1016/j.toxrep.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bladen C.L., Salgado D., Monges S., Foncuberta M.E., Kekou K., Kosma K., Dawkins H., Lamont L., Roy A.J., Chamova T. The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum. Mutat. 2015;36:395–402. doi: 10.1002/humu.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ittig D., Liu S., Renneberg D., Schümperli D., Leumann C.J. Nuclear antisense effects in cyclophilin A pre-mRNA splicing by oligonucleotides: a comparison of tricyclo-DNA with LNA. Nucleic Acids Res. 2004;32:346–353. doi: 10.1093/nar/gkh187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaussenot R., Edeline J.-M., Le Bec B., El Massioui N., Laroche S., Vaillend C. Cognitive dysfunction in the dystrophin-deficient mouse model of Duchenne muscular dystrophy: a reappraisal from sensory to executive processes. Neurobiol. Learn. Mem. 2015;124:111–122. doi: 10.1016/j.nlm.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 40.Kamola P.J., Kitson J.D.A., Turner G., Maratou K., Eriksson S., Panjwani A., Warnock L.C., Douillard Guilloux G.A., Moores K., Koppe E.L. In silico and in vitro evaluation of exonic and intronic off-target effects form a critical element of therapeutic ASO gapmer optimization. Nucleic Acids Res. 2015;43:8638–8650. doi: 10.1093/nar/gkv857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindow M., Vornlocher H.-P., Riley D., Kornbrust D.J., Burchard J., Whiteley L.O., Kamens J., Thompson J.D., Nochur S., Younis H. Assessing unintended hybridization-induced biological effects of oligonucleotides. Nat. Biotechnol. 2012;30:920–923. doi: 10.1038/nbt.2376. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Y., Vaidya V.S., Brown R.P., Zhang J., Rosenzweig B.A., Thompson K.L., Miller T.J., Bonventre J.V., Goering P.L. Comparison of kidney injury molecule-1 and other nephrotoxicity biomarkers in urine and kidney following acute exposure to gentamicin, mercury, and chromium. Toxicol. Sci. 2008;101:159–170. doi: 10.1093/toxsci/kfm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goyenvalle A., Babbs A., Wright J., Wilkins V., Powell D., Garcia L., Davies K.E. Rescue of severely affected dystrophin/utrophin-deficient mice through scAAV-U7snRNA-mediated exon skipping. Hum. Mol. Genet. 2012;21:2559–2571. doi: 10.1093/hmg/dds082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattison P.C., Soler-García A.A., Das J.R., Jerebtsova M., Perazzo S., Tang P., Ray P.E. Role of circulating fibroblast growth factor-2 in lipopolysaccharide-induced acute kidney injury in mice. Pediatr. Nephrol. 2012;27:469–483. doi: 10.1007/s00467-011-2001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aartsma-Rus A., Janson A.A.M., Kaman W.E., Bremmer-Bout M., den Dunnen J.T., Baas F., van Ommen G.J., van Deutekom J.C. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum. Mol. Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.