Abstract
Recognition and rejection of heterospecific male gametes occurs in a broad range of taxa, although the complexity of mechanisms underlying these components of postmating cryptic female choice is poorly understood. In plants, the arena for postmating interactions is the female reproductive tract (pistil), within which heterospecific pollen tube growth can be arrested via active molecular recognition and rejection. Unilateral incompatibility (UI) is one such postmating barrier in which pollen arrest occurs in only one direction of an interspecific cross. We investigated the genetic basis of pistil-side UI between Solanum species, with the specific goal of understanding the role and magnitude of epistasis between UI QTL. Using heterospecific introgression lines (ILs) between Solanum pennellii and S. lycopersicum, we assessed the individual and pairwise effects of three chromosomal regions (ui1.1, ui3.1, and ui12.1) previously associated with interspecific UI among Solanum species. Specifically, we generated double introgression (‘pyramided’) genotypes that combined ui12.1 with each of ui1.1 and ui3.1, and assessed the strength of UI pollen rejection in the pyramided lines, compared to single introgression genotypes. We found that none of the three QTL individually showed UI rejection phenotypes, but lines combining ui3.1 and ui12.1 showed significant pistil-side pollen rejection. Furthermore, double ILs (DILs) that combined different chromosomal regions overlapping ui3.1 differed significantly in their rate of UI, consistent with at least two genetic factors on chromosome three contributing quantitatively to interspecific pollen rejection. Together, our data indicate that loci on both chromosomes 3 and 12 are jointly required for the expression of UI between S. pennellii and S. lycopersicum, suggesting that coordinated molecular interactions among a relatively few loci underlie the expression of this postmating prezygotic barrier. In addition, in conjunction with previous data, at least one of these loci appears to also contribute to conspecific self-incompatibility (SI), consistent with a partially shared genetic basis between inter- and intraspecific mechanisms of postmating prezygotic female choice.
Keywords: epistasis, female choice, postmating isolation, speciation, unilateral incompatibility
Traits that underpin sexual recognition and rejection can be critical both for mate choice within species and for prezygotic isolating barriers between species. Such traits can contribute to premating interactions, including behavioral and chemical signals that indicate appropriate mating partners, or can act after mating but before fertilization, including interactions between gametes and/or between gametes and the female reproductive tract (Bernasconi et al. 2004). In the latter case, many sexual organisms—including plants—appear to exhibit “cryptic female choice,” in which genotype-specific interactions between female tissues and male gametes (in the case of plants, pollen tubes, or “gametophtyes”) determine the paternity of offspring following mating with one or more male genotypes (Alonzo et al. 2016). Similarly, cryptic female choice can influence the outcome of mating between species, when females can recognize and reject heterospecific male gametes. The specific mechanisms by which this choice is exercised, either within or between species, have been identified in some select systems (e.g., Price 1997; Manier et al. 2013; Castillo and Moyle 2014). However, much remains to be understood about the complexity and redundancy of these postmating prezygotic female traits, the specific loci that are necessary and sufficient for recognition and rejection, and the extent to which these mechanisms are shared between intraspecific and interspecific sexual interactions.
In flowering plants (angiosperms), recognition and rejection of pollen can occur at several stages after pollen is transferred (e.g., via wind or animal vectors) to the female receptive stigma (the receiving tissue for pollen deposition), including during pollen germination and pollen tube growth (via cell growth/elongation) down the female style (the reproductive tract that connects the stigma to the ovary). These “pollen–pistil” interactions (the pistil is composed of the stigma, style, and ovary) are roughly equivalent to postcopulatory interactions in animals, with the exception being that pollen tubes (male gametophytes) actively express a substantial fraction of their genome (Rutley and Twell 2015). Molecular mechanisms of pistil-mediated recognition and rejection of conspecific pollen tubes are arguably best understood in the context of genetic SI, whereby pollen from self or close relatives is recognized and rejected in the female style. Although there are several different types of genetic SI (Charlesworth et al. 2005), within Solanaceae SI is mediated by the “S-locus” that encodes at least two molecules responsible for the self-rejection mechanism, including an S-RNase (the female/stylar component) that recognizes one or more pollen-expressed F-box protein(s) in germinated pollen tubes and arrests pollen tube growth within the style. Moreover, Solanaceous SI is a “gametophytic” system in which pollen is rejected in styles when it bears an S-allele that is identical to an S-allele of the pistil (maternal) parent (McClure et al. 1989). Because individuals will always share S-alleles with themselves, this genetic system prevents self-fertilization when pollen is transferred within a flower or between flowers on the same individual. In addition to genes at the S-locus, other factors are also known to be required for SI function in Solanaceous species, including HT—a small asparagine-rich protein (McClure et al. 1999)—and other stylar glycoproteins (McClure et al. 2000; Cruz-Garcia et al. 2003; Hancock et al. 2003; de Graaf 1999) on the pistil-side, and pollen-side proteins including Cullins that are components of pollen protein complexes (Zhao et al. 2010; Li and Chetelat 2014; Hua and Kao 2006). SI genotypes can give rise to SC lineages when one or more of these molecular components has a loss-of-function mutation(s) (Charlesworth and Charlesworth 1979; Mable 2008; Stone 2002; Takayama and Isogai 2005; Tao and Iezzoni 2010; Covey et al. 2010).
Pistil-mediated pollen tube rejection can also act as an important species barrier among plant lineages, especially those that are otherwise weakly isolated by trait differences associated with premating (e.g., pollinator or flowering time) isolation. In comparison to SI pollen–pistil interactions, the molecular basis of recognition and rejection of heterospecific pollen is less well understood. Nonetheless, in several cases there is strong evidence that elements of this behavior are mechanistically associated with SI. Classical studies of interspecific pollen–pistil barriers in some species groups indicate that these are often observed between lineages that differ in the presence/absence of SI, such that SI species styles reject pollen from self-compatible (SC) species, but the reciprocal cross does not show stylar rejection (the so-called “SI × SC rule”; de Nettancourt 1977; Bedinger et al. 2011; Lewis and Crowe 1958; Murfett et al. 1996). Molecular and functional analysis of the resulting UI has confirmed that genetic components of SI can be shared in part with those of UI. For example, in some Nicotiana species, transforming nonrejecting SC genotypes with a functional S-RNase can be sufficient to confer the ability to reject pollen from other SC species (Hancock et al. 2003). Additional factors required for SI can also play a role in stylar (female)-side UI. For example, both S-RNase and HT protein are required for SI Nicotiana alata rejection of pollen from SC N. plumbaginifolia; normal S-RNase expression is individually insufficient (Hancock et al. 2003; McClure et al. 1999). Similarly, in Solanum, cotransformation of functional copies of both HT and S-RNase into the SC species Solanum lycopersicum is sufficient to confer the ability to reject pollen from other SC species (Tovar-Méndez et al. 2014).
While these observations clearly indicate that SI-associated molecular mechanisms can be sufficient to enable pistil-side pollen rejection between species, other data indicate that these mechanisms are more complex in nature. In particular, there is also evidence for S-RNase-independent UI mechanisms (Murfett et al. 1996), often from species pairs that exhibit UI, but do not follow the SI × SC rule. For example, in Solanum, some SC populations of S. pennellii and S. habrochaites lack S-RNase expression but continue to be competent to reject interspecific pollen from SC species (Covey et al. 2010; Baek et al. 2015; Chalivendra et al. 2013). Less directly, QTL mapping between Solanum species that both differ in SI status and show UI, have detected UI QTL that do not colocalize with the S-locus. In particular, an analysis between SI S. habrochaites and SC S. lycopersicum detected three UI QTL, only one of which was localized to the S-locus (Bernacchi and Tanksley 1997). A second study between SI S. pennellii and SC S. lycopersicum detected two UI QTL, neither of which was at the S-locus (Jewell 2016). Interestingly, the two non-S-locus QTL detected in these studies (ui3.1 and ui12.1) both localize to the same genomic regions on chromosomes 3 and 12, suggesting a common genetic basis for UI among different species. Moreover, in both studies, one of these QTL (ui12.1) colocalized with the known genomic location of HT, and in one case (Jewell 2016) the presence/absence of HT expression in mature styles was significantly associated with the phenotypic strength of UI.
Overall, these data suggest that there might be substantial overlap between molecular mechanisms of SI and UI—including both S-RNase-dependent or -independent mechanisms— and also the involvement of additional loci that have yet to be molecularly identified, and whose relationship to SI is unknown. As such, several important aspects of the genetics of pistil-side UI remain unclear, including the minimum number of factors sufficient to express S-RNase-independent UI, the degree of overlap with molecular loci underpinning S-RNase-dependent mechanisms, and therefore the level of redundancy between alternative mechanisms underlying these important postmating forms of female mate choice.
Here, our goal was to assess the specific role of three chromosomal regions in affecting pistil-side UI between species. Focusing on the UI QTL previously identified in two different Solanum species crosses—ui1.1, ui3.1, and ui12.1 (Bernacchi and Tanksley 1997; Jewell 2016)—our aim was to evaluate the individual and joint effects of these three unlinked chromosomal regions on the expression of UI. To do so, we used ILs in which single chromosomal regions from a donor species genotype (S. pennellii) have been introgressed into the genetic background of an otherwise isogenic recipient species (S. lycopersicum). Lines incorporating three different introgressed regions (from chromosomes 1, 3, and 12) were examined individually and in pairwise combinations; the latter DILs (Canady et al. 2006; sometimes called “pyramid lines”: Gur and Zamir 2004) were created via crosses among ILs (see Materials and Methods). Two criteria were used to evaluate evidence for epistasis between these target loci. First, we examined evidence for transmission ratio distortion (TRD) in the products of crosses between different ILs, to look for evidence that particular genotypes were over- or underrepresented. Second, we quantified the strength of pistil-side UI response phenotypes in the DIL lines and compared this to the same phenotype in single IL genotypes. This comparison allowed us to evaluate whether the quantitative effects of individual introgressions differs in the presence of a second introgressed locus, and to evaluate the minimum number of loci required to express pistil-side interspecific pollen rejection. We find evidence that loci on chromosomes 3 and 12 are simultaneously required to express UI; lines in which these loci are represented individually are unable to reject heterospecific pollen. One of these QTL, ui12.1, likely involves a known molecular contributor to SI (HT protein) thus further supporting the inference that factors associated with SI contribute to the expression of the UI phenotype. In addition, comparisons among DILs created using overlapping sections of ui3.1 suggest that this other QTL might be underpinned by at least two separate genetic factors that additively contribute to the genetic variation in the strength of UI.
Materials and Methods
Study system
The tomato clade (Solanum section Lycopersicon) contains 13 closely related species known to be separated by a range of incomplete pre- and postzygotic isolating barriers (Moyle 2008), including pollen–pistil incompatibility, and specifically UI (Bedinger et al. 2011; Covey et al. 2010). Baek et al. (2015) directly examined the strength of pollen tube rejection between all 13 species within the clade and found evidence that UI was strongest and most consistently observed in SI × SC species crosses. Among other genetic mapping resources in this group are several IL libraries in which chromosomal regions representing most or all of a donor species genome have been serially introgressed into the genetic background of a recipient species, usually the domesticated tomato S. lycopersicum (Bernacchi and Tanksley 1997; Eshed and Zamir 1995). For this study, we used lines drawn from a IL library previously developed between S. pennellii, a wild tomato species, and S. lycopersicum, where each line contains a marker delimited homozygous region of S. pennellii accession LA0716 introgressed into the genomic background of S. lycopersicum accession LA3475 (Eshed et al. 1992; Eshed and Zamir 1995, 1994). We used four different lines drawn from this library, based on whether these lines carried an S. pennellii introgression that encompassed the markers previously used to delimit the location of each UI QTL (Supplemental Material, Table S1 in File S1). IL1-1 overlaps the genomic location of the S-locus as well as the location of ui1.1, the UI locus previously mapped in an F2 population between S. lycopersicum and S. habrochaites (Bernacchi and Tanksley 1997). IL3-3 and IL3-4 contain S. pennellii introgression regions that overlap the previously mapped ui3.1 in Bernacchi and Tanksley (1997) and in Jewell (2016); together these introgressions span a broad genomic region (∼70 cM), with an overlapping region of ∼44 cM (Figure S1). IL12-3 overlaps the previously mapped ui12.1 in prior studies (Bernacchi and Tanksley 1997). The location of markers delimiting each QTL, and the upper and lower bounds of each S. pennellii introgression in the ILs, were cross-referenced against marker locations on the marker map in Eshed and Zamir (1994) and, when necessary, the tomato genome (at solgenomics.net; Table S1 in File S1).
Note that the known chromosomal location of HT protein falls within both IL12-3 and ui12.1. HT was duplicated in the ancestor of Solanum, resulting in two tandemly arrayed paralogs (HT-A and HT-B) at this chromosome 12 location (Covey et al. 2010). Moreover, both HT-A and HT-B are expressed in S. pennellii LA0716 but are nonfunctional in S. lycopersicum due to null mutations in both HT loci (Kondo et al. 2002; Covey et al. 2010). In contrast, the S-RNase protein at the S-locus is nonfunctional in both S. pennellii and S. lycopersicum genotypes in this experiment; S. pennellii is normally an SI species, but SI has recently been lost in this population due to loss of the S-RNase gene (Solyc01g055200) (Li and Chetelat 2015). Therefore, we included an IL spanning ui1.1 here in order to assess whether other genes contained within this chromosomal region also contribute to S-RNase-independent mechanisms of UI.
Construction of DILs
Seeds for our four target ILs were obtained from the Tomato Genetics Resource Center (tgrc.ucdavis.edu). To generate lines with two introgressed S. pennellii regions (DILs), crosses were performed pairwise between ILs (Table 1), and resulting heterozygous F1s were then selfed to generate F2 seeds (a “DIL population”) for genotyping and ultimately phenotyping. In this experiment, three different DIL combinations were generated in which we combined ui12.1 with ui1.1 and with the two alternative (overlapping) S. pennellii regions at ui3.1 (Table 1). We were unable to generate offspring from reciprocal crosses in all pairwise IL-IL combinations, as some of these did not produce seeds in one of the crossing directions despite numerous attempts, or failed to produce viable seed that was homozygous for target S. pennellii introgressions across each target QTL region. Patterns of marker representation and segregation distortion in F2 progeny from the crosses are discussed further below.
Table 1. Observed and expected (in parentheses) genotype frequencies in DIL populations generated from crossing our four target ILs.
| Genotype | |||
|---|---|---|---|
| Genotype at IL 12-3 | |||
| Genotype at IL 3-3 | LL | LP | PP |
| LL | 1 (1.63) | 1 (3.25) | 1 (1.63) |
| LP | 0 (3.25) | 2 (6.5) | 13 (3.25) + |
| PP | 1 (1.63) | 1 (3.25) | 6 (1.63) + |
| Genotype at IL 3-4 | |||
| Genotype at IL 12-3 | LL | LP | PP |
| LL | 14 (7.31) | 15 (14.63) | 11 (7.31) |
| LP | 18 (14.63) | 23 (29.25) | 23 (14.63) |
| PP | 4 (7.31) | 3 (14.63) − | 6 (7.31) |
| Genotype at IL 12-3 | |||
| Genotype at IL 1-1 | LL | LP | PP |
| LL | 9 (4.18) | 13 (8.375) | 7 (4.18) |
| LP | 10 (8.375) | 17 (16.75) | 5 (8.375) |
| PP | 2 (4.18) | 1 (8.375) − | 3 (4.18) |
The IL genotype used as the maternal parent in the original IL × IL cross is listed at the top of each cross. Genotypes that are over- or underrepresented are indicated by + or −, after Bonferroni correction. See Table S4 in File S1 for exact P-values after Bonferroni correction. IL, introgression line; LL, homozygous S. lycopersicum genotype; LP, heterozygous genotype; PP, homozygous S. pennellii genotype.
Genotyping and scoring individuals
Progeny were genotyped within each F2 “DIL population” in order to identify individuals that were homozygous for each S. pennellii introgression region and to describe patterns of marker TRD at these introgressions. These genotypes were used to identify individuals that were homozygous at both introgression regions, for further phenotypic assessment. Genotypes were also used to calculate overall genotype frequencies in segregating populations, to assess if there was evidence of non-Mendelian patterns of transmission that might be consistent with selection against certain genotypic combinations.
Genomic DNA was extracted using a modified CTAB protocol and Cleaved Amplified Polymorphic Sequence (CAPS) genotyping was used to characterize the allelic identity (S. lycopersicum: L or S. pennellii: P) using the target markers (solgenomics.net; Table S2 in File S1). CAPS markers are restriction fragment variants caused by single nucleotide polymorphisms or insertion/deletions, which create or abolish restriction enzyme recognition sites. Here we identified and genotyped markers that were designed to distinguish S. lycopersicum and S. pennellii alleles.
For each individual, DNA was amplified using PCR primers for each target marker and checked with gel electrophoresis (Table S3 in File S1). For each individual at each marker locus, a subsample of the amplicon was incubated with the relevant restriction endonucleases, and the digestion products were separated on 1.5% agarose gels, visualized with Ethidium Bromide staining, and imaged prior to manual scoring. Markers were chosen such that an allele from one parent would yield unique sized fragments (in bp) and the allele from the other parent would yield fragments of a different size (Table S2 in File S1). Thus, each F2 individual could be scored as homozygous for a parental allele (either S. lycopersicum or S. pennellii) at each marker, or as heterozygous in a case where the sample had the cleaved banding patterns representative of both parental alleles (Table S2 in File S1).
Each F2 individual was genotyped at 2–4 markers associated with the chromosomal region they were expected to carry. For each target introgression on chromosomes 1 and 12, three markers were selected to span the length of the S. pennellii introgressed region. At the chromosome 3 locus, five markers in total were used for genotyping: three were located in the region shared between IL3-3 and IL3-4, and one each was located in the region exclusive to either IL3-3 or IL3-4. Each individual was scored as a homozygous S. lycopersicum (LL), heterozygous (LP), or homozygous S. pennellii (PP) genotype for each chromosomal region based on these marker genotypes. During the generation of DILs, recombination events could occur within the introgression S. pennellii region within each line, so scoring LL, LP, and PP individuals at each locus required some additional criteria. In particular, individuals were classified into genotypic categories based on the identity (L vs. P) of the majority of markers scored within each target region. For example, on IL3-3, if three of the four markers were PP and one marker was heterozygous (LP), this individual was scored as double homozygous for S. pennellii (PP) at this chromosomal region. For IL12-3, if the three markers within the introgressed region disagreed, marker 74.00 was used as the tiebreaker as this marker is physically close to the genomic location of HT-A and HT-B. For each DIL population, genotype frequencies at both loci (e.g., chromosome 3 and 12) were determined by combining data from the two chromosomal regions to calculate observed two-locus genotype frequencies. Because of the number of markers used for genotyping, the size of the introgression regions, and the potential for recombination events between donor and recipient regions (in heterozygotes) during the generation of DILs, this procedure does not guarantee that the target gene(s) are maintained within the resulting DIL individuals, even if they carry S. pennellii alleles at each marker site. Nonetheless, we note that the procedure is conservative with respect to our inferences about UI; if the target locus/loci were lost from DIL individuals, we would fail to detect the relevant UI phenotype. Therefore, although we cannot exclude instances of lost alleles in one or more of our DIL individuals, our observation that some DIL genotype combinations do show significant rejection phenotypes (Results, below) is consistent with these lines retaining S. pennellii allele(s) for UI responses.
Quantifying the presence and speed of UI
To determine pollen tube growth phenotypes in each of our IL and DIL lines, and therefore the phenotypic expression of UI, we used an assay in which pollen is manually applied to a target stigma, allowed to germinate and grow in the style for 24 hr, and then the style is fixed and stained to visualize and measure the extent of pollen tube growth (Figure 1). Evidence of UI is demonstrated via rejection of pollen tubes in the style, quantified in terms of the proportional distance of pollen tube growth out of the total style length: a value that can vary between zero (very rapid rejection at the top of the style) to 1 (complete pollen tube growth down the entire length of the style).
Figure 1.
Representative images of the presence (A) and absence (B) of a UI pollen rejection response in the style. The white bar indicates a 1 mm scale. (A) UI cross, illustrated here with the phenotype observed in DIL 3-4 × 12-3, in which pollen rejection occurs approximately three-fourths down the length of the style. The arrowhead indicates where the majority of pollen tubes halt within the female reproductive tract (pistil) in this genotype. (B) Compatible cross in which pollen tubes successfully reach the ovary, illustrated with the phenotype observed for IL 3-4. DIL, double introgression line; UI, unilateral incompatibility.
For each assay, an unopened bud was emasculated (1 d prior to opening) by removing the entire anther cone using hand forceps. Hand pollination was performed the following day, using S. lycopersicum (accession LA3475) pollen. This accession is the S. lycopersicum genomic background in our ILs and DILs; its pollen is expected to be rejected in a style that has an S. pennellii-derived genotype sufficient to mount a UI response. At 24 hr postpollination, styles were collected into a 3:1 mixture of 95% EtOH:Glacial Acetic Acid in individual eppendorf tubes, and stored in the −20 freezer until imaging. For normal pollen tube growth down the complete length of the style, 24 hr is more than sufficient, except when a UI response has been mounted. Pollination protocols were identical for both our IL and DIL lines. We assayed three to five styles (technical replicates) per biological individual (Table 2).
Table 2. Proportion of pollen tube growth for each genotype with upper and lower 95% C.I.s.
| Genotype (Female × Male) | Mean ± SE | 95% C.I. Lwr | 95% C.I. Upr | # of Biological Replicates |
|---|---|---|---|---|
| IL 1-1 | 0.94 ± 0.02 | 0.865 | 1.014 | 3 |
| IL 3-3 | 0.94 ± 0.03 | 0.862 | 1.011 | 3 |
| IL 3-4 | 0.92 ± 0.04 | 0.868 | 1.002 | 4 |
| IL 12-3 | 0.98 ± 0.01 | 0.921 | 1.036 | 5 |
| DIL 1-1 × 12-3 | 0.85 ± 0.02 | 0.781 | 0.911 | 4 |
| DIL 12-3 × 3-3 | 0.51 ± 0.03 | 0.469 | 0.599 | 4 |
| DIL 3-4 × 12-3 | 0.76 ± 0.05 | 0.708 | 0.814 | 6 |
DIL genotypes are named so that the IL used as the maternal parent in the original IL × IL cross is specified first. Lwr, lower 95% C.I.; Upr, upper 95% C.I.; #, number; IL, introgression line; DIL, double IL.
To score pollen tube growth phenotypes, collected styles were placed in 5 M NaOH and allowed to soften for 20–24 hr. Following softening, styles were washed and stained using 200 µl of Aniline blue fluorochrome (Biosupplies Australia Pty Ltd) for 3.5 hr in the dark, as described previously (Covey et al. 2010; Bedinger et al. 2011; Jewell 2016). Styles were then imaged using an EVOS FL microscope with the DAPI emission filter. Stained pollen tubes fluoresce under these conditions, allowing us to differentiate style tissue from pollen tubes, and therefore determine the extent of pollen tube growth in each style. Because styles are generally too long to be captured in a single image, multiple images of the style were taken at 4 × magnification and then stitched together using the program AutoStitch (Brown and Lowe 2007). Images were visualized for measurement using ImageJ (Schneider et al. 2012). Measurements taken on each style included length of the style and the five longest pollen tubes. The average of these five pollen tubes was taken as our measurement of absolute distance traveled within this style, and UI was quantified as the proportional distance of pollen tube growth out of the total style length.
Statistical analyses
To determine whether our observed genotype frequencies significantly deviated from expected genotype frequencies in each “DIL population,” we calculated the expected proportion of each genotype and then performed a binomial test with a Bonferroni correction (Table 1). For each pairwise combination of loci, we used the upper and lower 95% C.I.s around the regression coefficient to verify that the differences for genotypic classes were significant. We then performed planned independent contrasts for two different types of comparisons. Specifically, we asked: (1) are DILs different from their parental ILs in terms of pollen tube growth and (2) are DILs different from each other? All analyses were run in RStudio version 0.99 (RStudio Team 2015).
Evaluating efficacy of observed UI
Finally, in cases where we saw a significant UI phenotype (i.e., were pollen growth was arrested in the style), we performed additional replicate crosses using S. lycopersicum LA3475 pollen, to evaluate the degree to which this style phenotype reflected an effective barrier to hybridization (i.e., to confirm that this rejection phenotype is associated with partial or complete inability to make fruits in the same cross). At least five crosses were performed per genotype as described above, but instead of collecting styles 24 hr post pollination, they were allowed to remain on the flower which was tagged and evaluated for a minimum period of 2 wk for evidence of fruit development (retention of the flower, followed by swelling and enlargement of the ovary). We also observed whether individuals were able to produce fruits following self-pollination, and confirmed that all genotypes in the experiment were self-fertile.
Data availability
ILs are available from the Tomato Genetics Resources Center (tgrc.ucdavis.edu). Conditions for PCR and marker information are supplied as supplemental material to the paper (Tables S2 and S3 in File S1).
Results
Departure from Mendelian segregation ratios
We detected significant TRD in all three DIL populations generated from crosses between different ILs, such that the frequency of genotypes generated between all crosses deviated from expected values [IL12-3 × IL3-3: X2 (8, N = 26) = 51.217, P = 2.38e−08; IL3-4 × IL12-3: X2 (8, N = 117) = 25.86, P = 0.001; and IL12-3 × IL1-1: X2 (8, N = 67) = 19.649, P = 0.001; Table 1]. This suggests that there are interactions among alleles at these loci that specifically affect the likelihood of transmission of different introgression combinations. For example, a strong deviation in our 12-3 × 3-3 DIL population is due to overrepresentation of homozygotes for S. pennellii alleles at both chromosomal regions, as well as overrepresentation of individuals that are heterozygous (LP) on chromosome 3 and homozygous (PP) on chromosome 12. This observed pattern of TRD is consistent with selection against S. lycopersicum alleles on a heterozygous F1 pistil (a genotype that has one allele at each of these two S. pennellii regions) (Figure 2). While some genotypes within the other DIL combinations deviate somewhat from the expected ratios (Table 1), only two additional comparisons survived Bonferroni correction: in the 3-4 × 12-3 DIL combination, we found evidence of underrepresentation of homozygotes for S. pennellii (PP) alleles at chromosome 12, specifically when heterozygous (LP) on chromosome 3. Similarly, in the 12-3 × 1-1 DIL population, genotypes homozygous for S. pennellii (PP) at chromosome 1 and heterozygous (LP) at chromosome 12 were significantly underrepresented.
Figure 2.
One model of gametophytic selection on different haploid pollen genotypes that could result in transmission ratio distortion among F2s. Two loci (ui3.1 and ui12.1) are designated by whether they have alleles from S. pennellii (i.e., 3P and 12P) or S. lycopersicum (i.e., 3L and 12L). Left side: when two ILs are crossed (3P3P 12L12L × 3L3L12P12P), the F1 is heterozygous at the two regions (3P 12L 3L12P). Selfing this F1 produces four different haploid pollen genotypes: 3L12L, 3L12P, 3P12L, and 3P12L. Right side: based on the genotype of our heterozygous F1 individual (3L3p 12L12P), preferential pollen use and stylar selection against specific pollen genotypes (e.g., 3L12L, 3L12P, and 3P12L) could generate deviations from expected Mendelian ratios. ILs, introgression lines; NILs, near-isogenic lines.
UI phenotypes are observed in pairwise genotype combinations of ui3.1 with ui12.1
We found that, while individual IL lines showed no significant UI response, several DIL genotype combinations exhibited significant UI (Figure S2). The proportion of pollen tube growth down the style in the ILs ranged from 0.93 to 0.98, with C.I.s that overlapped 1.00, consistent with pollen tubes that have grown the entire length of the style and reached the ovary (Figure 3 and Table 2). In addition, the 95% C.I.s on each IL mean overlap, consistent with no differences among ILs in their UI phenotype (Figure 3).
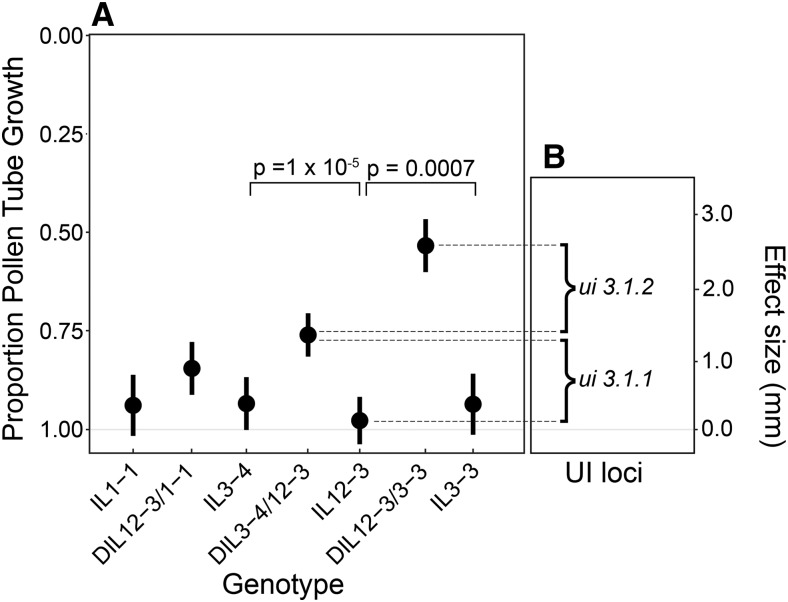
Figure 3.
Proportion of pollen tube growth by genotypic class. (A) Mean value with upper and lower 95% C.I.s for distance traveled through the length of the style for each genotypic class. The top of the graph (0) represents the stigma, where pollen is placed. The bottom of the graph (1.0) represents the ovary, which is at the base of the style. (B) The estimated effect size of the two loci (ui3.1.1 and ui3.1.2) inferred to underlie ui3.1 (see text). DIL, double introgression line; IL, introgression line; UI, unilateral incompatibility.
In comparison, mean pollen tube growth was generally reduced in the DIL genotypes and more highly variable between them (0.54–0.85; Table 2). Of these DIL combinations, two show evidence for significant reductions in proportional pollen tube growth, consistent with a quantitative UI response; both these DILs involve combinations of S. pennellii alleles on chromosomes 3 and 12 (i.e., IL3-3 or IL3-4 with IL12-3). Planned independent contrasts confirmed that DILs that combine S. pennellii alleles on chromosomes 3 and 12 were significantly different from their respective IL parental genotypes (DIL12-3 × 3-3: F1,6 = 15.21, P = 0.001; and DIL 3-4 × 12-3: F1,6 = 30.21, P = 1.60e−05). In comparison, the DIL combining S. pennellii alleles on chromosomes 1 and 12 was not significantly different to its parental ILs (F 1, 6 = 2.226, P = 0.150), and its 95% C.I.s overlapped with the parental IL genotypes, consistent with no significant interaction giving rise to a UI response in this DIL.
Patterns of pollen rejection suggest two loci underlie UI QTL on chromosome 3
In addition to displaying significant UI phenotypes, we also found that the quantitative strength of UI differed significantly between DILs with chromosome 3 S. pennellii alleles, depending upon which specific chromosome 3 introgression they contained (12-3 × 3-3 vs. 3-4 × 12-3: F 1, 6 = 12.835; P = 0.002). In particular, DIL individuals carrying the S. pennellii alleles from IL3-3 showed a more rapid UI response (pollen tube growth arrests half way down the style) in comparison to DIL individuals carrying the S. pennellii alleles from IL3-4 (Figure 3A). These alternative DILs carry chromosome 3 introgressions that overlap by 44 cM (Figure S1), as well as unique S. pennellii regions of 12 and 14 cM in IL3-3 and IL3-4, respectively, so the differences in UI phenotype between them could be explained by allelic differences within these unique regions. While there are several possible genetic interpretations of our observed patterns, the most parsimonious explanation requires only two loci at which S. pennellii alleles contribute to the quantitative expression of UI (in combination with ui12.1). The first locus (ui3.1.1) is contained within the chromosomal region shared between IL3-3 and IL3-4 and contributes an average effect of 1.42 mm to the quantitative strength of UI (see Figure 3B). In addition, the IL3-3 region contains a second locus which, in combination with ui3.1.1 and ui12.1, contributes an additional average effect of 1.13 mm to the quantitative strength of UI (see Figure 3B) when homozygous for S. pennellii alleles.
Finally, our inferences from pistil phenotypes about the relative strength of UI exhibited by DIL 3-4 × 12-3 and DIL 12-3 × 3-3 are borne out in additional replicate crosses (n = 5), where we evaluated the frequency with which each of these UI-associated DIL genotypes was able to set fruit. We found that DIL 12-3 × 3-3 did not set any fruit with S. lycopersicum pollen, consistent with our observed strong UI pistil phenotype. Results for DIL 3-4 × 12-3 were similar, except that two of the crosses set fruit; this is consistent with our observation that the UI rejection response acts later in this genotype, and it suggests a partially effective UI stylar barrier that reduces the frequency of fertilization with S. lycopersicum pollen. These data provide additional support for our inference that UI is acting as a partial or complete barrier to fertilization by heterospecific SC pollen, and that this barrier can be visualized in terms of pollen rejection phenotypes in the postpollinated style.
Discussion
While data suggest that there might be substantial overlap between molecular mechanisms of SI and UI, including both S-RNase-dependent or -independent mechanisms, several important aspects of the genetics of pistil-side UI remain unclear, including the minimum number of factors sufficient to express S-RNase-independent UI, the degree of overlap with molecular mechanisms underpinning S-RNase-dependent mechanisms, and therefore the level of redundancy between alternative mechanisms underlying these important postmating mechanisms of female mate choice. Here, our goals were to evaluate the minimum number of loci required to express pistil-side interspecific S-RNase-independent UI, and to evaluate the relative contribution of three different chromosomal regions to this phenotype. To do so, we examined the individual and combined (epistatic) effects of three candidate loci, pyramided as DILs, on the expression of pistil-side pollen rejection. We find evidence that factors on both chromosomes 3 and 12 are jointly required for the expression of S-RNase-independent UI, whereas these loci have no effect individually. In addition, we find some evidence consistent with gametophytic selection against certain genotypes, in the form of TRD. Together, these results suggest a strong role for the joint (epistatic) action of relatively few loci in determining the expression of pollen-pistil compatibility in this system.
Two loci are jointly required to express S-RNase-independent UI between species
Our findings clearly support a strong role for interactions between more than one molecular factor in the expression of S-RNase-independent UI. Specifically, we show that S. pennellii alleles from both chromosome 3 and 12 are necessary (and sufficient) for the expression of quantitative pistil-side UI against SC S. lycopersicum pollen. This confirms the general expectation that pollen recognition and rejection requires coordinated molecular interactions between several proteins, consistent with other kinds of molecular recognition and rejection mechanisms (McClure et al. 1999, 2000), but also suggests that the products of relatively few loci are sufficient to mount an S-RNase-independent UI response. Our study was designed based on previous mapping studies of UI in Solanum that identified main effect loci for UI on chromosomes 3, 12, and/or 1 (Bernacchi and Tanksley 1997). Other analyses have confirmed that ui1.1 is associated with the presence/absence of functional pistil-side S-RNase, at least between Solanum lineages in which one genotype is SI. The involvement of S-RNase for interspecific pollen rejection was first confirmed via when transforming SC Nicotiana species with functional S-RNase was shown to be sufficient to reconstitute UI in an otherwise non-UI SC species (Murfett et al. 1996). Nonetheless, because the S-locus is a large and genetically complex chromosomal region, additional loci within this region might also contribute to S-RNase-independent UI among genotypes that lack S-RNase but still show UI phenotypes. However, our results do not support the involvement in UI rejection of additional pistil-side loci at ui1.1, at least in this particular species pair; we detected no additional effect of S. pennellii alleles at ui1.1 between our two genotypes, both of which lack S-RNase function.
In contrast to ui1.1, our results confirm that S-RNase-independent UI is the joint product of S. pennellii alleles at chromosomes 3 and 12. Molecular analyses in other Solanum species pairs indicate that pistil-side HT protein contributes to the UI effect associated with ui12.1. HT protein has been shown to be required for pollen rejection from N. plumbaginifolia (Hancock et al. 2003). Within Solanum, expression of one of the tandemly duplicated HT proteins (HT-A) is phenotypically associated with the expression of UI in a number of species (Covey et al. 2010). In addition, a QTL analysis between S. lycopersicum and a different (SI) genotype of S. pennellii (Jewell 2016) indicated that the presence/absence of HT protein in styles was significantly associated with the strength of the UI response in a segregating F2 population. Although we do not have equivalently direct data on the molecular underpinnings of ui12.1 here, based on these other studies our working hypothesis is that the effect of IL12-3 on UI is partly or solely due to S. pennellii alleles at HT.
In comparison to ui1.1 and ui12.1, the molecular loci underpinning ui3.1 remain unknown, although Jewell (2016) identified several potential genes, and three especially strong candidates, for this locus by combining additional genomic and gene expression data (Pease et al. 2016) with the mapped location of ui3.1 in that study. Interestingly, our findings here suggest that ui3.1 is more complex than revealed in prior mapping experiments. QTL analysis has known limitations in terms of identifying number, location, and individual effects of loci, because detection depends on the heritability of the trait, the size of the segregating population, and the density of genetic markers (Mackay et al. 2009). Accordingly, the observation that single QTL can resolve into more than one underlying locus is not uncommon (Mackay et al. 2009), especially when the C.I.s on this locus are broad. In this case, ui3.1 was mapped to a region of ∼25 cM between S. pennellii and S. lycopersicum (Jewell 2016) and ∼10 cM between S. habrochaites and S. lycopersicum (Bernacchi and Tanksley 1997), both of which could easily harbor more than one contributing locus.
Given this, currently the most parsimonious inference from our observations is that (at least) two loci contribute additively to quantitative UI expression at ui3.1, one located in the genomic region overlapping between IL3-3 and IL3-4, and one in the region unique to IL3-3 (see Results and Figure 3B). Nonetheless, there are other alternative interpretations of our observations. For example, tomato chromosomes are known to be enriched for pericentromeric heterochromatin (Wang et al. 2006; Tanksley et al. 1992). Marker delineated breakpoints indicate that IL3-3 overlaps the centromeric region, and therefore contains centromeric heterochromatin from S. pennellii that matches the species origin of the rest of the introgression. In comparison, IL3-4 does not contain the S. pennellii centromeric region. Given this, it is possible that the phenotypic difference between IL3-3 and IL3-4 is instead due to position effects at ui3.1 that are associated with proximity to conspecific (IL3-3) vs. heterospecific (IL3-4) centromeric heterochromatin. Interestingly, this still implies that two loci are involved in the phenotypic patterns we observed at ui3.1, just that the second “locus” is the regulatory environment associated with species differences in the pericentromeric region. Regardless, in terms of our goals to evaluate the minimum number of loci required to express pistil-side S-RNase-independent UI, and evaluate the relative contribution of three different chromosomal regions to this phenotype, our data indicate that two to three loci at two genomic locations—on chromosomes 3 and 12—are jointly required and sufficient to express this important postmating interspecific barrier.
Source of TRD
We detected evidence of TRD during the generation of our DILs. One interpretation is that these distorted genotypes are due to gametophytic selection against particular haploid pollen genotypes in the F1 (doubly heterozygous) style in each case. Genetic differences among male gametophytes could result in either differential gametophytic selection or competition, both of which could potentially influence genotype frequencies in the next generation (Snow and Mazer 1988). For example, gametophytic selection could change the probability of fertilization based on genetic differences expressed in the male gametophyte, according to a simple model illustrated in Figure 2. In this case, pollen that carries both S. pennellii alleles for IL 3-3 and IL 12-3 has a growth or persistence advantage in the heterozygous F1 pistil whereas pollen carrying S. lycopersicum alleles is preferentially selected against (Figure 2). This model implies that S. pennellii-derived proteins in the style are able to differentially recognize and reject pollen that lacks S. pennellii alleles at ui3.1 and ui12.1. This is intriguing because these loci are expected to have pistil-side functions but are not necessarily expected to mediate pollen-side involvement in UI. In contrast, known loci that influence pollen-side expression of UI are located on chromosomes 1, 6, and 10 (Li and Chetelat 2010, 2015; Li et al. 2010; Chetelat and DeVerna 1991), although these observations do not exclude the potential involvement of additional loci on other chromosomes. We note also that dissimilar patterns of TRD are observed in DIL populations from the two other IL combinations, where some genotypes with S. pennellii alleles are significantly underrepresented (Results). However, in these later cases, we cannot exclude the possibility that pollen carrying heterospecific chromosomal segments is simply less viable or less competitive than “pure” S. lycopersicum pollen (i.e., that TRD is due to reduced hybrid male fertility in these genotypes). In addition, patterns of TRD can also be due to other complex causes, such as dysfunction (hybrid inviability) in both male and female gametes that results in TRD due to differential (sex independent) gamete survival (Koide et al. 2008) or to differential survival of postfertilization hybrid zygotes. Therefore, although gametophytic selection might explain part or all of the patterns of TRD we observed, other complex causes are also possible.
UI and reproductive isolation
The active recognition and rejection of heterospecific pollen tubes growing within the pistil is a form of postmating cryptic female choice against heterospecific mates. In the Solanaceae, UI is a particularly common reproductive isolating barrier, especially in genera that have both SI and SC species (Bedinger et al. 2011; Lewis and Crowe 1958). Dissecting the mechanisms that contribute to UI can therefore provide insight into the expression of this reproductive barrier among lineages. Here, we have shown that relatively few loci are sufficient to express UI. Moreover, these pistil-side mechanisms likely share genetic components with SI mechanisms within species. Previous work has shown that S-RNase necessary for SI is also a key pistil-side contributor to UI (Murfett et al. 1996), as is HT (McClure et al. 1999). Moreover, HT appears to contribute to both S-RNase-dependent and S-RNase-independent mechanisms of UI among some species (Covey et al. 2010; Tovar-Méndez et al. 2017). Given these dual functions, it is plausible that the molecular machinery for mounting a UI response is already present in the styles of SI species, and its phenotypic expression merely requires encountering heterospecific SC pollen. Nonetheless, the existence of several redundant mechanisms (i.e., S-RNase-independent and -dependent mechanisms) of UI suggests that this heterospecific barrier is not merely a pleiotropic by-product of stylar competence for SI. Indeed, it is possible that the molecular mechanism(s) underlying ui3.1 are not shared with SI recognition and rejection, consistent with independently selected and maintained mechanisms specifically for UI. Evaluating this possibility awaits further fine-mapping and functional characterization of this locus (or loci) in the future.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.041673/-/DC1.
Acknowledgments
The authors thank Ashley Huh, C. J. Jewell, and Christian King for data collection and assistance with experimental material, and the Indiana University Bloomington greenhouse staff. The authors would also like to thank Dean Castillo for advice on statistical analyses, and two anonymous reviewers for comments that greatly improved the manuscript. This research was funded by National Science Foundation grant MCB-1127059.
Footnotes
Communicating editor: D. Zamir
Literature Cited
- Alonzo S. H., Stiver K. A., Marsh-Rollo S. E., 2016. Ovarian fluid allows directional cryptic female choice despite external fertilization. Nat. Commun. 7: 12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek Y. S., Covey P. A., Petersen J. J., Chetelat R. T., McClure B., et al. , 2015. Testing the SI× SC rule: pollen–pistil interactions in interspecific crosses between members of the tomato clade (Solanum section Lycopersicon, Solanaceae). Am. J. Bot. 102(2): 302–311. [DOI] [PubMed] [Google Scholar]
- Bedinger P. A., Chetelat R. T., McClure B., Moyle L. C., Rose J. K., et al. , 2011. Interspecific reproductive barriers in the tomato clade: opportunities to decipher mechanisms of reproductive isolation. Sex. Plant Reprod. 24(3): 171–187. [DOI] [PubMed] [Google Scholar]
- Bernacchi D., Tanksley S. D., 1997. An interspecific backcross of Lycopersicon esculentum× L. hirsutum: linkage analysis and a QTL study of sexual compatibility factors and floral traits. Genetics 147(2): 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi G., Ashman T.-L., Birkhead T., Bishop J., Grossniklaus U., et al. , 2004. Evolutionary ecology of the prezygotic stage. Science 303(5660): 971–975. [DOI] [PubMed] [Google Scholar]
- Brown M., Lowe D. G., 2007. Automatic panoramic image stitching using invariant features. Int. J. Comput. Vis. 74(1): 59–73. [Google Scholar]
- Canady M. A., Ji Y., Chetelat R. T., 2006. Homeologous recombination in Solanum lycopersicoides introgression lines of cultivated tomato. Genetics 174(4): 1775–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo D. M., Moyle L. C., 2014. Intraspecific sperm competition genes enforce post-mating species barriers in Drosophila. Proc. Biol. Sci. 281: 20142050 The Royal Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalivendra S. C., Lopez-Casado G., Kumar A., Kassenbrock A. R., Royer S., et al. , 2013. Developmental onset of reproductive barriers and associated proteome changes in stigma/styles of Solanum pennellii. J. Exp. Bot. 64(1): 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., 1979. The evolution and breakdown of S-allele systems. Heredity 43(1): 41–55. [Google Scholar]
- Charlesworth D., Vekemans X., Castric V., Glemin S., 2005. Plant self-incompatibility system: a molecular evolutionary perspective. New Phytol. 168(1): 61–69. [DOI] [PubMed] [Google Scholar]
- Chetelat R., DeVerna J., 1991. Expression of unilateral incompatibility in pollen of Lycopersicon pennellii is determined by major loci on chromosomes 1, 6 and 10. Theor. Appl. Genet. 82(6): 704–712. [DOI] [PubMed] [Google Scholar]
- Covey P. A., Kondo K., Welch L., Frank E., Sianta S., et al. , 2010. Multiple features that distinguish unilateral incongruity and self-incompatibility in the tomato clade. Plant J. 64(3): 367–378. [DOI] [PubMed] [Google Scholar]
- Cruz-Garcia F., Hancock C. N., McClure B. A., 2003. S-RNase complexes and pollen rejection. J. Exp. Bot. 54(380): 123–130. [DOI] [PubMed] [Google Scholar]
- de Graaf, B. H. J., 1999 Pistil proline-rich proteins in Nicotiana tabacum. Ph.D. Thesis, Catholic University, Nijmegen, The Netherlands. [Google Scholar]
- de Nettancourt D., 1977. Incompatibility in Angiosperms, Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- Eshed Y., Zamir D., 1994. A genomic library of Lycopersicon pennellii in L. esculentum: a tool for fine mapping of genes. Euphytica 79(3): 175–179. [Google Scholar]
- Eshed Y., Zamir D., 1995. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141(3): 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y., Abu-Abied M., Saranga Y., Zamir D., 1992. Lycopersicon esculentum lines containing small overlapping introgressions from L. pennellii. Theor. Appl. Genet. 83(8): 1027–1034. [DOI] [PubMed] [Google Scholar]
- Gur A., Zamir D., 2004. Unused natural variation can lift yield barriers in plant breeding. PLoS Biol. 2(10): e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock C. N., Kondo K., Beecher B., McClure B. A., 2003. The S-locus and unilateral incompatibility. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358(1434): 1133–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z., Kao T.-h., 2006. Identification and characterization of components of a putative Petunia S-locus F-box–containing E3 ligase complex involved in S-RNase–based self-incompatibility. Plant Cell 18(10): 2531–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell, C. P., 2016 Genetics and Evolution of Reproductive Behavior in Solanaceae. Ph.D. Thesis, Indiana University, Bloomington. [Google Scholar]
- Koide Y., Onishi K., Nishimoto D., Baruah A. R., Kanazawa A., et al. , 2008. Sex‐independent transmission ratio distortion system responsible for reproductive barriers between Asian and African rice species. New Phytol. 179(3): 888–900. [DOI] [PubMed] [Google Scholar]
- Kondo K., Yamamoto M., Matton D. P., Sato T., Hirai M., et al. , 2002. Cultivated tomato has defects in both S‐RNase and HT genes required for stylar function of self‐incompatibility. Plant J. 29(5): 627–636. [DOI] [PubMed] [Google Scholar]
- Lewis D., Crowe L., 1958. Unilateral interspecific incompatibility in flowering plants. Heredity 12: 233–256. [Google Scholar]
- Li W., Chetelat R. T., 2010. A pollen factor linking inter-and intraspecific pollen rejection in tomato. Science 330(6012): 1827–1830. [DOI] [PubMed] [Google Scholar]
- Li W., Chetelat R. T., 2014. The role of a pollen-expressed cullin1 protein in gametophytic self-incompatibility in solanum. Genetics 196(2): 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Chetelat R. T., 2015. Unilateral incompatibility gene ui1. 1 encodes an S-locus F-box protein expressed in pollen of Solanum species. Proc. Natl. Acad. Sci. USA 112(14): 4417–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Royer S., Chetelat R. T., 2010. Fine mapping of ui6. 1, a gametophytic factor controlling pollen-side unilateral incompatibility in interspecific solanum hybrids. Genetics 185(3): 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable B. K., 2008. Genetic causes and consequences of the breakdown of self-incompatibility: case studies in the Brassicaceae. Genet. Res. 90(1): 47–60. [DOI] [PubMed] [Google Scholar]
- Mackay T. F., Stone E. A., Ayroles J. F., 2009. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10(8): 565–577. [DOI] [PubMed] [Google Scholar]
- Manier M. K., Lüpold S., Belote J. M., Starmer W. T., Berben K. S., et al. , 2013. Postcopulatory sexual selection generates speciation phenotypes in Drosophila. Curr. Biol. 23(19): 1853–1862. [DOI] [PubMed] [Google Scholar]
- McClure B. A., Haring V., Ebert P. R., Anderson M. A., Simpson R. J., et al. , 1989. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature 342(6252): 955–957. [DOI] [PubMed] [Google Scholar]
- McClure B., Mou B., Canevascini S., Bernatzky R., 1999. A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proc. Natl. Acad. Sci. USA 96: 13548–13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure B. A., Cruz-Garcia F., Beecher B., Sulaman W., 2000. Factors affecting inter- and intraspecific pollen rejection in Nicotiana. Ann. Bot. (Lond.) 85: 113–123. [Google Scholar]
- Moyle L. C., 2008. Ecological and evolutionary genomics in the wild tomatoes (Solanum sect. Lycopersicon). Evolution 62(12): 2995–3013. [DOI] [PubMed] [Google Scholar]
- Murfett J., Strabala T. J., Zurek D. M., Mou B., Beecher B., et al. , 1996. S RNase and interspecific pollen rejection in the genus Nicotiana: multiple pollen-rejection pathways contribute to unilateral incompatibility between self-incompatible and self-compatible species. Plant Cell 8(6): 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease J. B., Guerrero R. F., Sherman N. A., Hahn M. W., Moyle L. C., 2016. Molecular mechanisms of postmating prezygotic reproductive isolation uncovered by transcriptome analysis. Mol. Ecol. 25(11): 2592–2608. [DOI] [PubMed] [Google Scholar]
- Price C. S., 1997. Conspecific sperm precedence in Drosophila. Nature 388(6643): 663–666. [DOI] [PubMed] [Google Scholar]
- RStudio Team , 2015. RStudio: Integrated Development for R. RStudio, Inc., Boston, MA. [Google Scholar]
- Rutley N., Twell D., 2015. A decade of pollen transcriptomics. Plant Reprod. 28(2): 73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9(7): 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow A. A., Mazer S. J., 1988. Gametophytic selection in Raphanus raphanistrum: a test for heritable variation in pollen competitive ability. Evolution 42(5): 1065–1075. [DOI] [PubMed] [Google Scholar]
- Stone J. L., 2002. Molecular mechanisms underlying the breakdown of gametophytic self-incompatibility. Q. Rev. Biol. 77(1): 17–32. [DOI] [PubMed] [Google Scholar]
- Takayama S., Isogai A., 2005. Self-incompatibility in plants. Annu. Rev. Plant Biol. 56(1): 467–489. [DOI] [PubMed] [Google Scholar]
- Tanksley S., Ganal M., Prince J., De Vicente M., Bonierbale M., et al. , 1992. High density molecular linkage maps of the tomato and potato genomes. Genetics 132(4): 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R., Iezzoni A. F., 2010. The S-RNase-based gametophytic self-incompatibility system in Prunus exhibits distinct genetic and molecular features. Sci. Hortic. (Amsterdam) 124(4): 423–433. [Google Scholar]
- Tovar-Méndez A., Kumar A., Kondo K., Ashford A., Baek Y. S., et al. , 2014. Restoring pistil-side self-incompatibility factors recapitulates an interspecific reproductive barrier between tomato species. Plant J. 77: 727–736. [DOI] [PubMed] [Google Scholar]
- Tovar-Méndez A., Lu L., McClure B., 2017. HT proteins contribute to S‐RNase‐independent pollen rejection in Solanum. Plant J. 89: 718–729. [DOI] [PubMed] [Google Scholar]
- Wang Y., Tang X., Cheng Z., Mueller L., Giovannoni J., et al. , 2006. Euchromatin and pericentromeric heterochromatin: comparative composition in the tomato genome. Genetics 172(4): 2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Huang J., Zhao Z. H., Li Q., Sims T. L., et al. , 2010. The Skp1-like protein SSK1 is required for cross-pollen compatibility in S-RNase-based self-incompatibility. Plant J. 62(1): 52–63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ILs are available from the Tomato Genetics Resources Center (tgrc.ucdavis.edu). Conditions for PCR and marker information are supplied as supplemental material to the paper (Tables S2 and S3 in File S1).