Abstract
Ascertaining the molecular and physiological basis of domestication and breeding is an active area of research. Due to the current wide distribution of its wild ancestor, the wild boar, the pig (Sus scrofa) is an excellent model to study these processes, which occurred independently in East Asia and Europe ca. 9000 yr ago. Analyzing genome variability patterns in terms of metabolic pathways is attractive since it considers the impact of interrelated functions of genes, in contrast to genome-wide scans that treat genes or genome windows in isolation. To that end, we studied 40 wild boars and 123 domestic pig genomes from Asia and Europe when metabolic pathway was the unit of analysis. We computed statistical significance for differentiation (Fst) and linkage disequilibrium (nSL) statistics at the pathway level. In terms of Fst, we found 21 and 12 pathways significantly differentiated at a q-value < 0.05 in Asia and Europe, respectively; five were shared across continents. In Asia, we found six significant pathways related to behavior, which involved essential neurotransmitters like dopamine and serotonin. Several significant pathways were interrelated and shared a variable percentage of genes. There were 12 genes present in >10 significant pathways (in terms of Fst), comprising genes involved in the transduction of a large number of signals, like phospholipase PCLB1, which is expressed in the brain, or ITPR3, which has an important role in taste transduction. In terms of nSL, significant pathways were mainly related to reproductive performance (ovarian steroidogenesis), a similarly important target trait during domestication and modern animal breeding. Different levels of recombination cannot explain these results, since we found no correlation between Fst and recombination rate. However, we did find an increased ratio of deleterious mutations in domestic vs. wild populations, suggesting a relaxed functional constraint associated with the domestication and breeding processes. Purifying selection was, nevertheless, stronger in significantly differentiated pathways than in random pathways, mainly in Europe. We conclude that pathway analysis facilitates the biological interpretation of genome-wide studies. Notably, in the case of pig, behavior played an important role, among other physiological and developmental processes.
Keywords: behavior, domestication, pathway analysis, pig, genome sequence data
Plant and animal domestication were cornerstone events in mankind’s recent history (Diamond 2002). By ensuring a continuous and reliable supply of food, domestication allowed a steady increase in human population size that eventually resulted in the first urban societies, thereby facilitating the technological development that characterizes the human species. Although domestication has received considerable interest for many years from multiple disciplines, modern large-scale genomic technologies are shedding new light on a process where many unknowns still remain. This endeavor is largely facilitated in species, such as the pig, where a modern equivalent of the wild ancestor is available for comparison.
There is some ambiguity in defining what is domestication (Zeder 2015), given that many concurrent processes have occurred in the transition between wild specimens and the individuals bred by humans, and that domestication likely involved gradual discontinuities in gene flow between domestic and wild populations instead of a sudden stop (Frantz et al. 2015). Nevertheless, in animals, there are some shared characteristics among the major domestic species since they have been selected to meet human preferences; domestic animals have modified behavior, and distinctive growth and reproductive features compared to their wild ancestors. The genetic bases of these traits are clearly polygenic, as is evident for the numerous QTL that have been identified (www.animalgenome.org/cgi-bin/QTLdb/SS/index). This complicates the discovery of genes underlying their phenotypic variability because small effect sizes are difficult to detect.
Traditionally, studies looking for selective signals have analyzed individual SNPs or carried out a genomic scan in windows of contiguous SNPs of arbitrary size (e.g., Amaral et al. 2011; Burgos-Paz et al. 2013; Rubin et al. 2012). Since genes do not act in isolation but in concerted action with other genes, we argue, as other studies have done (Daub et al. 2013), that analyzing genomic variability patterns from a metabolic pathway point of view should facilitate the biological interpretation of the results. Compared to a genome window analysis, this approach could improve power when individual gene signals are weak. By adding up these individually weak signals in a pathway framework, a global significant statistic can eventually be obtained. Note that a pathway analysis differs from analyzing a posteriori a list of statistically significant genes using, by instance, gene ontology tools, since here we predefine a list of genes and we then study the collective behavior of genes of the whole list. The criticism by Pavlidis et al. (2012) is then less applicable when a prior hypothesis exists. For gene set approaches, see review in, e.g., Mooney et al. (2014).
Previous studies do show that taking into account how genes interact along metabolic pathways is enlightening. For instance, using a pathway approach, Daub et al. (2013) discovered that adaptation signals in humans, measured by increased differentiation, are enriched for pathogen resistance pathways. However, none of the genes were statistically outliers so this observation would probably have been overlooked had genes been analyzed individually. In cattle, Ha et al. (2015) identified several pathways associated with a number of key metabolites in dairy cows and Buitenhuis et al. (2014) revealed pathways associated with milk production. The main difference between those studies is the criterion that they used to merge individual signals, as numerous variants have been proposed [e.g., Wang et al. (2010)]. Here, we used Fisher’s statistics to combine several independent Fst values for each SNP into a gene P-value and, subsequently, those gene P-values were combined into a pathway P-value. We argue that combining signals from multiple SNPs should be less prone to false discoveries than taking the single most outlier signal for each gene.
Despite numerous studies in pig domestication [e.g., for a review see Ramos-Onsins et al. (2014)] so far, to our knowledge, pathway analysis has not been applied to improve our understanding of the domestication or breeding processes in this species. The fact that most phenotypic characteristics have a polygenic basis makes pathway analysis an attractive approach, provided that the pathway as a whole better explains the genetic basis of the trait than do individual genes. Here, we have used sequence data from 163 domestic and wild pigs to study how potentially selective processes associated with domestication and ensuing breeding have modeled the pig genome, when viewed from a pathway point of view. By using sequence instead of chips, we further avoid the issue of ascertainment bias, and provide a comprehensive, unbiased portray of nucleotide diversity. Note that a comparison between domestic and wild specimens necessarily confounds domestication and modern breeding signals, and truly disentangling genetic changes due to domestication from those caused by ensuing breeding requires ancient DNA studies at population scale, which is currently unrealistic despite some recent advances (Ramírez et al. 2014). Since we were predominantly interested in the shared signal left by domestication and breeding across breeds, we tried to minimize the specific breed effects. To this end, we combined genomes from several domestic breeds, sampling evenly the number of specimens per breed.
Materials and Methods
Pig samples
We analyzed a sample of 163 wild and domestic pig (Sus scrofa) genomes (Supplemental Material, Figure S1 and Table S1 in File S1). The 163 pigs were classified into Asian domestic pigs (ASDM, n = 60), Asian wild boars (ASWB, n = 20), European domestics (EUDM, n = 63), and European wild boar (EUWB, n = 20). ASDM represented 10 Chinese breeds (Meishan, Bamaxiang, Hetao, Laiwu, Luchuan, Minzhu, Sichuan, Tibetan, Wuzhishan, and Yunnan), which were chosen to represent the different geographic locations in China, six samples from each breed. ASWB comprised 10 boars from South China and 10 from North Asia (North China, Korea, and East Russia). EUDM were all from major breeds (Duroc, Landrace, Large White, and Pietrain), plus the local breeds Iberian, Mangalica, and the American miniature pig Yucatan, of Iberian descent (Burgos-Paz et al. 2013); 10 genomes per breed were chosen except for Mangalica and Iberian, where only five and eight were available, respectively. The 20 EUWBs were from Spain, France, The Netherlands, Switzerland, Italy, Greece, Tunisia, and the Near East. The domestic breeds used in this study are selected for a diversity of traits. For European breeds, meat content and growth are important targets, whereas Chinese breeds tend to be more prolific and fatter than their European counterparts.
Most of the sample sequences were available in public databases (Groenen et al. 2012; Rubin et al. 2012; Esteve-Codina et al. 2013; Molnár et al. 2014; Ai et al. 2015; Bianco et al. 2015a; Pérez-Enciso et al. 2016) and were downloaded from the short read archive (SRA, http://www.ncbi.nlm.nih.gov/sra). Two additional samples (Iberian pig IBGU1805 and a British Large White LWGB0348) were specifically sequenced for this study and have been submitted to the SRA (https://www.ncbi.nlm.nih.gov/sra; accessions SRX2787051 and SRX2788443 within study PRJNA255085). The VCF files containing both raw and imputed SNPs are available at https://bioinformatics.cragenomica.es/numgenomics (under the heading “data”).
NGS bioinformatics
We downloaded and mapped raw reads against the reference assembly (Sscrofa10.2, Groenen et al. 2012) using the BWA mem option (Li and Durbin 2009). We removed PCR duplicates using SAMtools rmdup v0.1.19 (Li et al. 2009) and realigned around indels with the GATK IndelRealigner tool (McKenna et al. 2010). We called genotypes with SAMtools mpileup and bcftools call v1.3.0 (Li et al. 2009) for each individual separately. To call a SNP, we set the minimum and maximum depths between 5 × and twice the average sample’s depth plus one, the minimum SNP quality was 10 in each sample, with the further requirements of minimum mapping quality and minimum base quality of 20. We also called the homozygous blocks, which are the parts of the sequence that are equal to the reference. Since SAMtools does not filter by default these homozygous blocks by depth, we filtered them fitting the same depth and quality requirements as for the SNP calling procedure using the “samtools depth” utility, BEDtools (Quinlan 2014), and custom scripts. In this way, both SNPs and homozygous blocks were filtered by the same criteria.
We then merged individual gVCF files into a multi-individual VCF file, with all the SNPs from the 163 samples. For this purpose, we followed a two-step approach, closely resembling that in Pérez-Enciso et al. (2016). In summary, we first generated a fasta file from the gVCF file for each individual and generated a multi-individual VCF file using the individual fasta file to identify whether a position is equal to the reference, polymorphic, or missing. An alternative approach would have been to call SNPs using all samples simultaneously, but this strategy has been shown to have less power (and similar type I errors) than the one followed here, because joint SNP calling is less sensitive to rare variants than individual calling (Nevado et al. 2014). Furthermore, Asian and European samples are highly divergent and multi-sample algorithms are optimized for single population analyses.
Once the multiple sample file was obtained, we discarded the singletons, SNPs in sex chromosomes, and the SNPs with >30% of missing data of the samples in each group (ASDM, ASWB, EUDM, and EUWB). If a given SNP was not called in at least ≥30% of samples in all groups, it was discarded from further analyses. Finally, we imputed the missing genotypes and inferred phases with Beagle 4.0 (Browning and Browning 2013). We annotated SNPs with Ensembl’s Variant Effect Predictor (McLaren et al. 2010). This tool also classifies nonsynonymous variants as tolerated or deleterious based on their SIFT scores (Sim et al. 2012), which predicts whether an amino acid substitution affects protein function. For each gene, we computed the ratio of deleterious vs. tolerated SNPs. These statistics were computed for each population (ASDM, ASWB, EUDM, and EUWB) separately. R (R Development Core Team 2014) was used to obtain a “heatmap” to represent Euclidean distances between samples’ genotypes.
Differentiation and disequilibrium metrics
Selection increases differentiation at positively selected loci between a control population and a population where the loci are beneficial, also causing an increase in linkage disequilibrium (LD) around selected haplotypes. These two well-known phenomena (e.g., Sabeti et al. 2006) can be captured by either Fst (allele frequency differentiation) or haplotype-based tests, such as nSL (Ferrer-Admetlla et al. 2014). Since the pig was independently domesticated in Asia and in Europe (Larson et al. 2005), we computed Fst (Weir–Cockerham estimate, Weir and Cockerham 1984) between wild and domestic populations in each continent, Asia and Europe, separately using VCFtools (Danecek et al. 2011). The nSL metrics are designed to detect the positive selection signal due to an increase in haplotype homozygosity; for this purpose, nSL measures the length of a segment of haplotype homozygosity in terms of number of mutations. We calculated the statistics with the program nSL (http://cteg.berkeley.edu/software.html) within the four different populations of interest (ASDM, ASWB, EUDM, and EUWB); the statistics were normalized according to derived allele frequency in 10 bins of size 0.10. The ancestral allele is needed for the nSL statistic and was inferred from a consensus outgroup allele, as explained in Bianco et al. (2015b). The consensus was obtained from several species: S. barbatus, S. cebifrons, S. verrucosus, S. celebensis, and African warthog (Phacochoerus africanus). The divergence between the different Sus species is ∼4.2 MYA, whereas that of Sus with warthog is ca. 10 MYA (Frantz et al. 2016). We removed those SNPs for which the ancestral allele could not be reliably identified or with less than two alleles. For each gene, we assessed the average recombination rate based in the linkage map by Tortereau et al. (2012). This map was based on four different F2 crosses between European and Chinese breeds; total autosomal length was ∼20 M. We obtained a smoothed recombination rate using the loess R package, to minimize the effect of gaps in the recombination map.
Pathway analysis
We downloaded the complete dataset with pig pathways and genes from NCBI Biosystems v.20160202 (Geer et al. 2010). The downloaded file contained 1789 pathways and 7157 genes. The median number of genes per pathway was 47 and ranged from 1 to 1519. The NCBI biosystems database contains records from different source databases, such as KEGG (http://www.genome.jp/kegg/, Kanehisa et al. 2008), REACTOME (http://www.reactome.org/, Matthews et al. 2009), or WikiPathways (http://www.wikipathways.org/, Pico et al. 2008), which are often redundant. For this reason, we filtered the pathways according to their size and redundancy in two steps. First, we removed pathways with <10 and >150 genes (150 corresponds to two SD in the distribution of number of genes per pathway); this was aimed at discarding pathways that were either not informative or too generic and complex. For instance, among the pathways with over 150 genes we find: metabolic pathways, gene expression, metabolism, hemostasis, immune system, and neuronal system. Second, for pathways sharing >50% of their genes, we selected the largest one.
We obtained an empirical P-value for Fst and nSL for each pathway following Dall’Olio et al. (2012). First, an empirical P-value for each SNP was obtained by ranking the statistics (Fst or nSL). Thus, a SNP with Fst (or nSL) ranked as the i-largest out of N SNPs, was assigned a P-value of i/N. Next, we obtained a gene P-value with Fisher’s statistics, which combines several independent P-values:
where S is the number of SNPs for the gene analyzed (i.e., those within the gene boundaries in the Ensembl database) and Pj each associated P-value; since x is distributed as a χ2 test with 2N d.f., we can obtain a combined P-value for the gene. In a second step, we repeated the same procedure by combining the P-values of each gene in the pathway to obtain a pathway P-value. The actual significance of this P-value is difficult to interpret since the null hypothesis is not clearly defined; therefore, we carried out permutations to determine significance. Since each pathway differs in number of genes, we carried out 1000 permutations for random gene sets of sizes 10–150 genes and differing by increments of 10 genes. In these permutations, dummy pathways were assembled using the P-values of randomly sampled genes, and the actual pathway P-value was compared with the null distribution obtained by permutation. To account for multiple testing, we used the q-value (Benjamini and Hochberg 1995), computed with R-package qvalue (Storey et al. 2015), to determine significant pathways using the P-values obtained by permutation.
Critically, Fisher’s statistics are based on the premise of independence between P-values, and this is not guaranteed with sequence data given the extreme disequilibrium between nearby SNPs. To avoid this, we pruned the SNP dataset by selecting those positions that minimized LD using the PLINK v.1.9 program (Chang et al. 2015), setting the variant inflation factor equal to two. With this approach, the P-value obtained from Fisher’s statistics was independent of the number of SNPs for each gene (Figure S2 in File S1). It should be mentioned that the nSL statistics were computed using all SNPs for which the ancestral allele could be determined, since nSL measures LD in the number of SNP units, but only the values for SNPs in equilibrium were retained to obtain the gene P-value, as for Fst metrics.
To investigate whether significance could be partly explained by Asian introgression in EUDM, we carried out a semisupervised ADMIXTURE (Alexander et al. 2009) analysis. We extracted SNPs from all genes pertaining to the given pathway and we ran ADMIXTURE with K = 2. We run a semisupervised analysis where all EUWB were assigned K = 1 and all Asian pigs were assigned K = 2, and we let the program compute the fraction of the EUDM genomes due to Asian origin. We did this for each significant pathway and for a random set of pathways with a similar number of genes.
Further, we built a coassociation network to visualize pathway relationships. Significant pathway-to-pathway connections were identified using the PCIT network inference algorithm (Reverter and Chan 2008). The PCIT algorithm is a soft-thresholding method that exploits the twin concepts of Partial Correlation and Mutual Information. In brief, it explores relationships between all possible triplets of nodes (i.e., pathways in our context), in an attempt to determine truly informative correlations between node pairs once the numerical influence of other nodes in the system has been accounted for. Clustering was based on 10 variables per pathway: the six pathway P-values for Fst (one value per continent) and nSL (one value per population) metrics, and nucleotide diversity in each of the four populations (ASWB, ASDM, EUWB, and EUDM), averaged for each gene in the pathway. We estimated Tajima’s nucleotide diversity (Tajima 1983, 1989) per gene per population with the methods developed by Ferretti et al. (2012), which account for missing data, using mstatspop software (S. Ramos-Onsins, unpublished data, available at http://bioinformatics.cragenomica.es/numgenomics/people/sebas/). We visualized the resulting network using Cytoscape (www.cytoscape.org, Shannon et al. 2003). In the visualization scheme, we mapped the pathways (nodes) to a series of attributes to help identify emerging properties. These included number of genes in the pathway, pathway source (KEGG or REACTOME), population with lowest Fst P-value (Asia or Europe), and pathway nucleotide variability.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results and Discussion
Genetics mirrors geography, to an extent
Out of the 163 genomes, we initially identified 71,458,035 autosomal SNPs. After quality filtering, removing the positions with >30% of missing data, and discarding singletons, these were reduced to 48,008,185 SNPs. Of those, 31,363,201 (65%) were annotated in dbSNP (https://www.ncbi.nlm.nih.gov/SNP), and the ancestral allele could be determined in 44,417,146 sites. By continent, Asia had a much larger number of private SNPs than Europe, 25,258,008 vs. 5,726,610, as expected from the fact that the species is of Asian origin and that European populations suffered a strong bottleneck, as has been observed in previous studies (Bianco et al. 2015b).
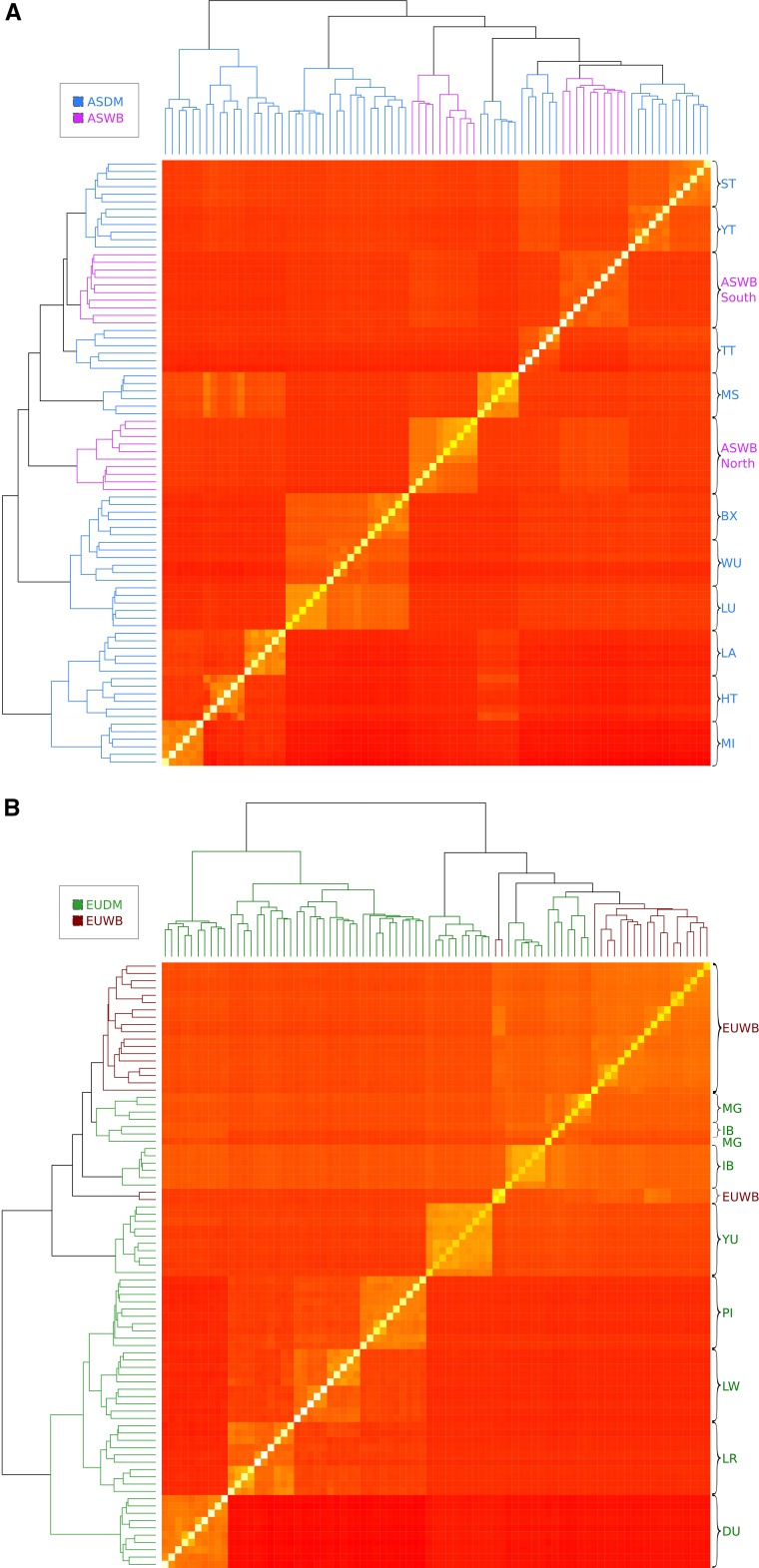
A heatmap of genetic distances between all samples and SNPs showed the well-known split between Asia and Europe (Figure S3 in File S1). The European heatmap (Figure 1A) shows that the main division is between wild boar and local breeds Iberian, Mangalica, and Yucatan vs. international pig breeds Pietrain, Landrace, Large White, and Duroc. Further, all EUWBs were clustered together except the two Near East wild boars, which were grouped in a separate branch. The Yucatan, a miniature pig developed in the USA starting with local Mexican pigs and that still retains an important percentage of ancestry from Iberian pigs (Burgos-Paz et al. 2013), formed a separate group but was closer to local pigs than to international breeds. Among those, Duroc was genetically more separate from the rest of the international pig breeds.
Figure 1.
(A) Heatmap of the European individuals using the molecular relationship matrix, computed using all available autosomal single nucleotide polymorphisms (SNPs). (B) Heatmap of the Asian pigs. In Europe, breed codes are DU, Duroc; IB, Iberian; LR, Landrace; LW, Large White; MG, Mangalitza; PI, Pietrain; and YU, Yucatan minipig. In Asia, breed codes are BX, Bamaxiang; HT, Hetao; LA, Laiwu; LU, Luchuan; MI, Minzhu; MS, Meishan; ST, Sichuan; TT, Tibet; WU, Wuzhishan; and YT, Yunnan. Colors are used to differentiate among the populations: ASDM (Asian domestic, blue), ASWB (Asian wild boar, purple), EUDM (European domestic, green), and EUWB (European wild boar, dark red).
The picture was somewhat more complex in Asia (Figure 1B), although pigs were also grouped by breed. In contrast to Europe, we observed a genetic split between North and South wild boars, in agreement with previous results (Ai et al. 2015). Nevertheless, this geographic pattern was not so evident among the domestic pigs, e.g., North Asian breeds (Laiwu, Hetao, and Minzhu), which are less separated from those from the South, compared to wild populations.
Pathway statistics
We retrieved 1789 pathways comprising 7157 genes from the NCBI database, which were reduced to a final set of 442 pathways with 5713 genes after filtering (Table S2 in File S1) by size (e.g., number of genes) and redundancy. Note that only 25% of pathways but 80% of genes were retained, showing the large redundancy in terms of genes across pathways. Most discarded pathways (676) were very small and contained <10 genes. The distribution of genes per pathway was highly leptokurtic (Figure S4 in File S1).
Differentiation metrics (Fst)
Differentiation (Fst) analysis indicates that allele frequency changes occurred in pathways associated with some important biological processes (Table 1). We found more significant pathways in Asia than in Europe; there were 21 pathways significantly differentiated at a q-value < 0.05 in Asia and 12 in Europe, involving a total of 1065 and 576 genes, respectively. Pathways were predominantly continent-specific, but five pathways were differentiated in both continents: integrin cell surface interactions, insulin secretion, pancreatic secretion, ABC transporters, and glutamatergic synapse. Our results are unlikely to be an artifact caused by differential recombination rate, as we found no correlation between Fst and recombination rate (Figure S5 in File S1). This contrasts with what has been observed in humans (Keinan and Reich 2010).
Table 1. Significant pathways (q-value < 0.05) obtained in the Fst analysis in Asia and/or Europe.
| Biological Process | Pathway Name | NCBI ID | Number of Genes | P-Valuea Asia | q-Value Asia | P-Valuea Europe | q-Value Europe |
|---|---|---|---|---|---|---|---|
| Behavior | Opioid signaling | 1337511 | 46 | 0.0005 | 0.010 | 0.1300 | 0.686 |
| Behavior | Glutamatergic synapse | 213816 | 75 | 0.0005 | 0.010 | 0.0005 | 0.018 |
| Behavior | Dopaminergic synapse | 469195 | 88 | 0.0005 | 0.010 | 0.0460 | 0.495 |
| Behavior | Serotonergic synapse | 525344 | 75 | 0.0005 | 0.010 | 0.1710 | 0.734 |
| Behavior | Long-term depression | 84497 | 41 | 0.0005 | 0.010 | 0.0230 | 0.442 |
| Behavior | Adrenergic signaling in cardiomyocytes | 908279 | 104 | 0.0005 | 0.010 | 0.0440 | 0.495 |
| Biological regulation | Renin secretion | 1223594 | 42 | 0.0140 | 0.245 | 0.0005 | 0.018 |
| Biological regulation | Phosphatidylinositol signaling system | 84464 | 70 | 0.0470 | 0.438 | 0.0005 | 0.018 |
| Cell communication | Cell–cell communication | 1336387 | 61 | 0.1820 | 0.570 | 0.0005 | 0.018 |
| Cell communication | Integrin cell surface interactions | 1337048 | 51 | 0.0005 | 0.010 | 0.0005 | 0.018 |
| Cellular process | Assembly of the primary cilium | 1336230 | 123 | 0.0005 | 0.010 | 0.0390 | 0.479 |
| Cellular process | Hippo signaling pathway | 749791 | 30 | 0.0005 | 0.010 | 0.1590 | 0.725 |
| Cellular process | Wnt signaling pathway | 84473 | 89 | 0.0005 | 0.010 | 0.2010 | 0.753 |
| Cellular process | Axon guidance | 84477 | 92 | 0.0005 | 0.010 | 0.0330 | 0.456 |
| DNA repair | DNA double-strand break repair | 1336358 | 106 | 0.0005 | 0.010 | 0.1100 | 0.685 |
| DNA repair | Nonhomologous end-joining | 92864 | 12 | 0.0005 | 0.010 | 0.0260 | 0.442 |
| Immune response | Complement cascade | 1336947 | 23 | 0.1350 | 0.535 | 0.0005 | 0.018 |
| Immune response | Fc-γ receptor (FCGR) dependent phagocytosis | 1336978 | 36 | 0.0005 | 0.010 | 0.0370 | 0.467 |
| Immune response | Chagas disease (American trypanosomiasis) | 147807 | 83 | 0.0005 | 0.010 | 0.5210 | 0.898 |
| Metabolic process | Glycosaminoglycan metabolism | 1336589 | 81 | 0.0005 | 0.010 | 0.2230 | 0.757 |
| Metabolic process | Phospholipase D signaling pathway | 1311111 | 104 | 0.1600 | 0.542 | 0.0005 | 0.018 |
| Metabolic process | G alpha (s) signaling events | 1337504 | 88 | 0.0005 | 0.010 | 0.2840 | 0.810 |
| Metabolic process | Pancreatic secretion | 169304 | 59 | 0.0005 | 0.010 | 0.0005 | 0.018 |
| Metabolic process | Insulin secretion | 777548 | 60 | 0.0005 | 0.010 | 0.0005 | 0.018 |
| Muscle contraction | Muscle contraction | 1337146 | 117 | 0.5250 | 0.856 | 0.0005 | 0.018 |
| Muscle contraction | Vascular smooth muscle contraction | 96246 | 85 | 0.0440 | 0.430 | 0.0005 | 0.018 |
| Regulation of transcription | Nuclear signaling by ERBB4 | 1337437 | 23 | 0.0005 | 0.010 | 0.1310 | 0.686 |
| Transport | ABC transporters | 84452 | 27 | 0.0005 | 0.010 | 0.0005 | 0.018 |
NCBI ID, National Center for Biotechnology Information database identifier.
P-value obtained from permutations.
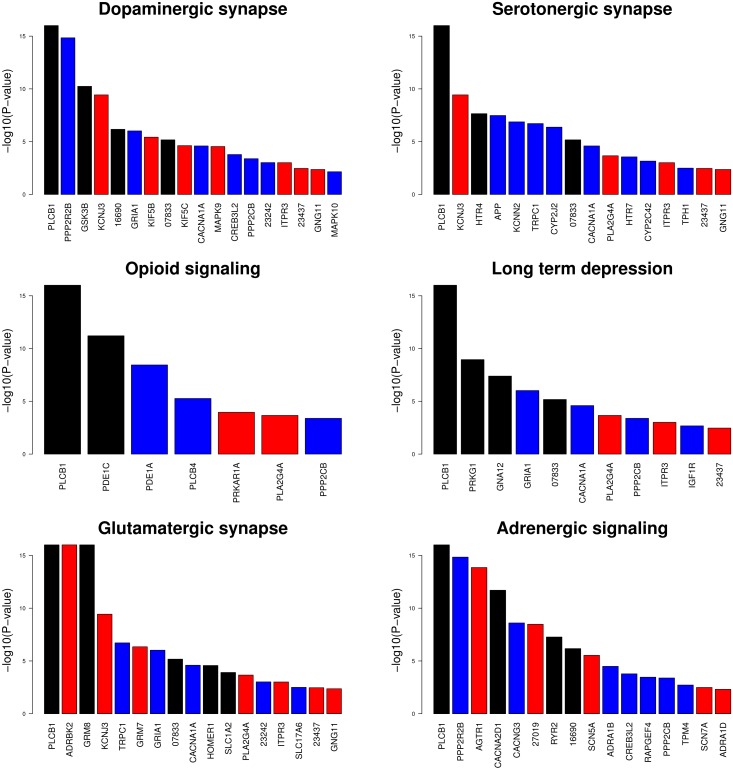
In Asia, we found six significant pathways related to behavior (serotonergic synapse, dopaminergic synapse, glutamatergic synapse, opioid signaling, long-term depression, and adrenergic signaling in cardiomyocytes). This is remarkable since it has long been recognized that domestication has affected behavior, yet the genetic basis for these changes has not been convincingly identified. The six pathways included a total of 264 genes, which codify for proteins involved in the metabolism of important neurotransmitters like serotonin, dopamine, and L-glutamate. Serotonin and dopamine are involved in aggression (serotonin) and reinforcement and reward (dopamine), whereas L-glutamate is the major excitatory neurotransmitter in the central nervous system. Clearly, aggression and reward must have played a role at least during the early stages of domestication and the genetic causes are likely shared between all domestic breeds. Pathway “adrenergic signaling in cardiomyocytes” involves several adrenaline receptors and calcium channels such as ryanodine receptor 2 (RYR2). While RYR2 is primarily expressed in cardiomyocytes, its isoform RYR1 is expressed in skeletal muscle and is well-known in pig genetics for being responsible for the pale, soft, and exudative syndrome (Fujii et al. 1991).
For the six behavior pathways, Figure 2 shows the P-values of all genes that were significant at the 1% nominal level either in Europe, Asia, or both. Although behavior pathways were mainly significant only in Asia (except glutamatergic synapse, Table 1), several individual genes were significant in both continents, foremost phospholipase C β 1 (PLCB1), which was significant in both continents and was present in all six behavior pathways. It can be suspected that these pathways were significant only because they contained the PLCB1 gene; however, PLCB1 was involved in a total of 35 pathways, and only 14 were significant (q-value < 0.05). Furthermore, we also computed the pathway P-value excluding PLCB1 and found only a modest decrease in significance (Table S3 in File S1). With our approach, it is unlikely then that a single gene is responsible for significance at the pathway level. Note that this is reasonable under a multi-cause mechanism but may prevent the researcher from identifying pathways where a single gene has main responsibility for the rate limiting effect on the whole pathway. In all, PLCB1 plays an important role in the intracellular transduction of many extracellular signals mediated by calcium. It cleaves the PIP2 molecule into IP3 and DAG. DAG, together with Ca2+ (its secretion is activated by ITPR3, also significant in Europe), activates PKC, which plays a central role in activating numerous functions such as transcription, the immune response, growth, learning, and smooth muscle contraction. This explains its presence in so many different pathways. Importantly, PLCB1 is expressed in select areas of the brain, including the cerebral cortex, hippocampus, amygdala, lateral septum, and olfactory bulb (Koh et al. 2007). In humans, deficiencies in this gene are associated with some kinds of epilepsy (Ngoh et al. 2014). Another interesting and significant gene in both continents was GSK3B (Glycogen synthase kinase-3), which is involved in energy metabolism, neuronal cell development, and body pattern formation.
Figure 2.
Gene P-values (−log10) of significant genes at the 1% nominal level in Europe (red bars), in Asia (blue bars,) or both continents (black bars) from the significant differentiated (Fst) pathways involved in behavior. When a gene was significant in both continents, the smallest P-value is plotted. Gene symbols are provided when available; otherwise numbers indicate ensembl ENSSSCG id.
The rest of the significantly differentiated pathways comprises those related to glucose metabolism (insulin and pancreatic secretion) and development (Wnt signaling, Hippo signaling, and axon guidance) in Asia, and recombination or muscle contraction in Europe. Hippo and Wnt signaling pathways are intimately related, and half of all significant genes were shared (Figure S6 in File S1). In contrast, vascular smooth muscle contraction and muscle contraction share only 16 significant genes out of 85 and 117 genes, respectively. Significant genes in the insulin pathway include PLCB1 and RYR2, as well as potassium and calcium channels that act on insulin granules and insulin transcription (KCNN1, KCNN2, and KCNMB1). The Hippo signaling pathway controls organ size, a fundamental target during domestication and modern breeding. Wnt signaling, in turn, is one of the most relevant and highly conserved signal transduction pathways, and it has a fundamental role in embryonic development. Hippo and Wnt are tightly interconnected signaling cascades, although their mechanisms differ; Hippo is mainly sensitive to cell density, whereas Wnt responds to concentrations of specific proteins (Irvine 2012). Interestingly, there is also a direct relationship between the Wnt and insulin pathways, as Wnt signaling increases cells’ insulin sensitivity. The three most significant genes in the Wnt pathway were PLCB1 (shared with other pathways, see Figure 2 and Figure S5 in File S1), inversin, which contains calmodulin domains and is involved in renal development, and GSK3B, also involved in body pattern formation. GSK3B is also a negative regulator of glucose hormone control.
It is finally worth mentioning two significant pathways involved in recombination, “DNA double strand break repair” and “nonhomologous end-joining” (Figure S7 in File S1, which also shows the rest of the significant Fst pathways). The issue of the effect of recombination on domestication has been debated for a long time in the literature. Theoretical models have predicted that domestication should increase recombination, as rapid selection indirectly favors an increased recombination rate such that the Hill–Robertson effect is less limiting for response, and this prediction has been confirmed using chiasma data from the literature (Ross-Ibarra 2004). In a classic paper, Ollivier (1995) also showed that the wild boar linkage map was ∼33% shorter than domestic pig maps. Nevertheless, other recent studies in sheep, goat, and dogs have ruled out changes in recombination rates compared to their wild ancestors (Munoz-Fuentes et al. 2015). Therefore, the significant differentiation found here in this pathway may not be paralleled with changes in recombination rate caused by domestication.
LD metrics
We repeated the same statistical procedure as for Fst with the nSL statistics, which measures LD instead of differentiation, and that is especially powerful for identifying soft sweeps (Ferrer-Admetlla et al. 2014). Each of the four populations, ASDM, ASWB, EUDM, and EUWB, were analyzed separately. Overall, there were fewer significant pathways at a q-value < 0.05 with nSL than with Fst (Table 1 vs. Table 2). In particular, we did not find a significant value in either EUWBs nor in ASWBs, perhaps because there were fewer wild than domestic pigs. In comparison to Fst, concordance between continents with nSL was very high in domestic pigs, as we found the same six out of seven significant pathways in both Asia and Europe. The only exception was pathway “Inflammatory mediator regulation of TRP channels” involved in the immune response, which was significant only in Asia. A potential matter of concern with pathway analysis is its definition. As an example, we found that arachidonic acid metabolism pathways annotated by KEGG (NCBI id 84417) and REACTOME (NCBI id 1336691) contained 24 shared genes out of a total of 47 and 39, respectively. Nevertheless, the significant genes (P-value < 0.01) of both pathways were the same. As noted by Mooney et al. (2014), different databases may contain different genes to represent the same biological process; this is a warning of the fact that pathway definition is not an unambiguous concept, and different criteria can legitimately be used to define a given biological process.
Table 2. Significant pathways (q-value < 0.05) obtained in the nSL analysis for the four populations, according to continent Europe (EU)/Asia (AS) and domestic (DM)/wild (WB) status.
| Biological Process | Pathway Name | NCBI ID | Number of Genes | P-Valuea ASDM | q-Value ASDM | P-Valuea ASWB | q-Value ASWB | P-Valuea EUDM | q-Value EUDM | P-Valuea EUWB | q-Value EUWB |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Behavior/reproduction | Ovarian steroidogenesis | 791446 | 30 | 5e−04 | 0.032 | 0.012 | 1 | 5e−04 | 0.037 | 0.524 | 1 |
| Biological regulation | Biological oxidations | 1336802 | 120 | 5e−04 | 0.032 | 0.148 | 1 | 5e−04 | 0.037 | 1 | 1 |
| Hormone synthesis | Steroid hormone biosynthesis | 84375 | 32 | 5e−04 | 0.032 | 0.029 | 1 | 5e−04 | 0.037 | 0.457 | 1 |
| Immune response | Inflammatory mediator regulation of TRP channels | 948291 | 73 | 5e−04 | 0.032 | 0.259 | 1 | 0.120 | 1.000 | 1 | 1 |
| Lipid metabolic process | Arachidonic acid metabolism | 1336691 | 39 | 5e−04 | 0.032 | 5e−04 | 0.110 | 5e−04 | 0.037 | 0.998 | 1 |
| Lipid metabolic process | Arachidonic acid metabolism | 84417 | 47 | 5e−04 | 0.032 | 0.041 | 1 | 5e−04 | 0.037 | 0.977 | 1 |
| Lipid metabolic process | Linoleic acid metabolism | 84418 | 24 | 5e−04 | 0.032 | 5e−04 | 0.110 | 5e−04 | 0.037 | 0.755 | 1 |
NCBI ID, National Center for Biotechnology Information database identifier; ASDM, Asian domestic; ASWB, Asian wild boar; EUDM, European domestic; EUWB, European wild boar.
P-value obtained from permutations.
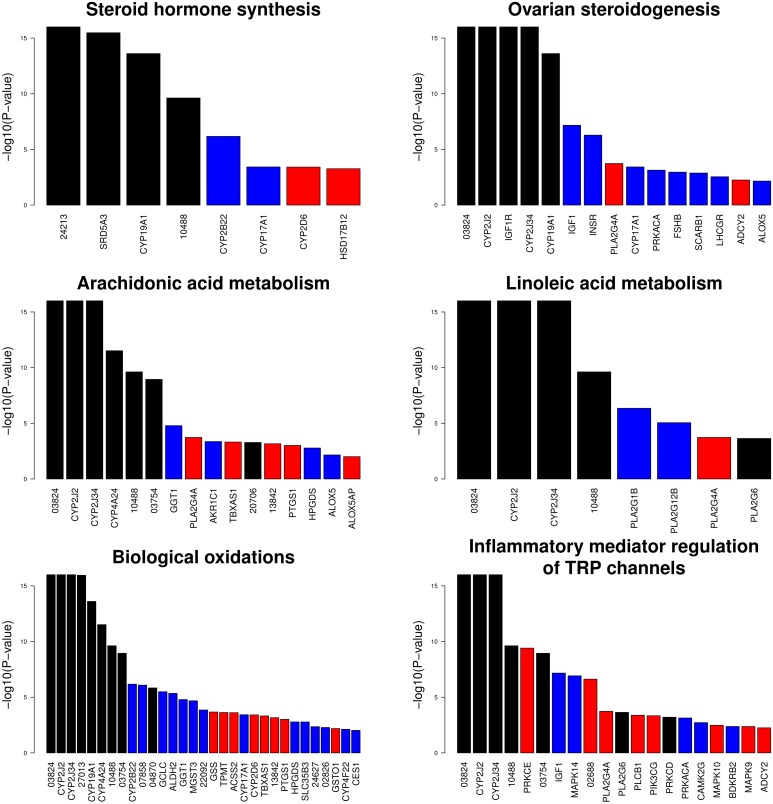
Two of the significant pathways are directly linked to reproductive performance (“Ovarian steroidogenesis” and “Steroid hormone biosynthesis”). Importantly, we also identified ovarian steroidogenesis in our previous work (Pérez-Enciso et al. 2016) in a much smaller study on domestication merging ASDM and EUDM vs. ASWB and EUWBs, and where a completely different analytical approach was employed. The remaining significant pathways were related to lipid metabolism, in particular to linoleic and arachidonic metabolism. Importantly, some of the final products of linoleic metabolism are THF-diols, which are converted into prostaglandins and are involved in the sexual behavior of males and the ovarian cycle in females. Therefore, most pathways identified with nSL are interrelated and linked to reproduction.
Among the most significant genes in ovarian steroidogenesis, there appears the uncharacterized gene ENSSSCG00000003824. This gene seems to be orthologous to UGT2B (UDP glycosyltransferase), a cluster of genes involved in the glucuronidation of estrogens. Figure 3 shows the significant gene P-values for the significant nSL pathways. It is interesting to note that a more coherent signal across continents emerged with nSL than with Fst metrics, since the most significant genes with nSL are shared between continents (e.g., compare Figure 2 and Figure 3).
Figure 3.
Significant genes at the 1% nominal level either in Europe, Asia or both, present in the significant pathways obtained from the nSL (linkage disequilibrium) analysis. Gene symbols are provided when available; otherwise numbers indicate ensembl ENSSSCG id.
Pathways are interrelated
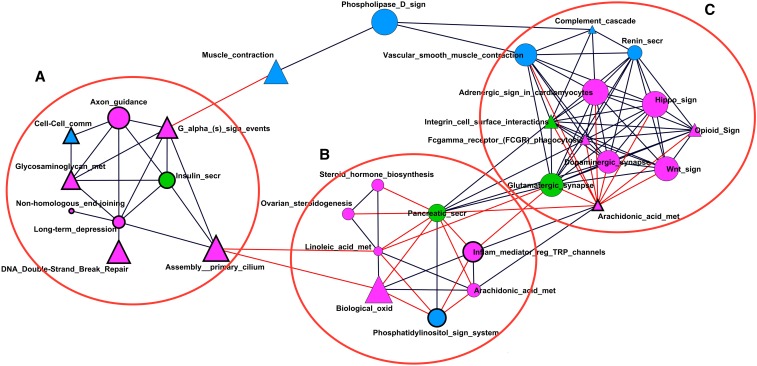
Much as genes do not act in isolation, neither do pathways. To represent this, a coassociation network was built with all 35 significant pathways, either with Fst or nSL analysis (Table 1 and Table 2 merged). The metrics used for the clustering contained the pathway Fst and nSL P-values together with nucleotide variabilities (see Materials and Methods). The entire network of pathways contained 83 negative and 129 positive connections. Note that the interpretation of a “positive” or “negative” connection is not straightforward, as is often the case in multivariate methods. The sign would indicate that domestication and/or breeding has exerted similar or opposite effects on the variables used to build the network, conditional on the fact that pathways are significant in at least one analysis. The three most connected pathways, each with 19 connections, were arachidonic acid metabolism (NCBI id 1336691) with 45 genes, glutamatergic synapse with 122 genes, and dopaminergic synapse with 119 genes. When the minimum correlation was set to 0.80 in absolute value (Figure 4), four pathways were not sufficiently connected (nuclear signaling by ERBB4, Chagas disease, serotonergic synapse, and ABC transporters). In turn, three clusters of highly interconnected pathways are immediately apparently in the network visualization of Figure 4. Cluster A contains nine pathways with higher than average nucleotide diversity that show strong positive connections between them. Prominent in this cluster is the axon guidance pathway with 119 genes. Its connections with cell–cell communication, G α signaling events, and assembly of the primary cilium via the insulin secretion pathways suggest that this cluster mainly involves extracellular guidance such as growth and hormonal regulation, helping axons reach their targets (Dickson 2003).
Figure 4.
Coassociation network among the 31 interconnected significant pathways. Each node represents a pathway that is connected by an edge if partial correlation with another pathway is significant and >0.8 (in absolute value). Node size is proportional to number of genes in the pathways. Node shapes represent pathway source: triangles for REACTOME and circles for KEGG. Colors indicate the population with lowest Fst (statistical significance for differentiation) P-value: pink for Asia, blue for Europe, and green for equal significance. Node line width to pathway variability: thin and thick lines for pathways with variability below and above average, respectively. Black and red edges represent positive and negative correlations between pathways, respectively. The three main pathway clusters are identified with letters A–C.
Most of the corresponding pathways of the other two clusters are negatively related between them. Cluster C is composed of processes related with the sympathetic nervous system, which is activated in response to stress by neurotransmitters such as dopamine (dopaminergic synapse) and glucocorticoids, which induce glutamate release (glutamatergic synapse, Popoli et al. 2011). Several other processes in cluster C are activated in response to the activation of the sympathetic nervous system, for instance increased heart contraction (adrenergic signaling in cardiomyocytes), blood vessels constriction in some parts of the body (vascular smooth muscle contraction and renin secretion), blood vessel dilatation in muscle and muscle contraction (muscle contraction), and energy obtainment by lipids and carbohydrate degradation (pancreatic secretion). These processes are negatively connected with pathways in cluster B, which contains hormone-controlled processes related to reproduction (steroid hormone biosynthesis, ovarian steroidogenesis, linoleic acid metabolism, and arachidonic acid metabolism), and that are inhibited in stress events, like the pathway “Inflammatory mediator regulation of TRP channels.”
Finally, some genes also appeared repeatedly across pathways, which may indicate a central role in some biochemical routes. The list of genes present in at least 10 of the significant pathways is in Table 3. Most of these genes are enzymes involved in general processes, such as phospholipases PLCB1 and PLCB3 involved in the transduction of many signals; PLA2G4A and PLA2G4B, which release arachidonic acid; kinases (PRKCA, PRKCG, PRKACA, and MAPK1) involved in development; and adenylate cyclases (ADCY2, ADCY3, and ADCY4), which are part of the signal transduction of G proteins, e.g., affecting dopamine. In addition, we also found the receptor ITPR3, which has an important role in taste transduction and is involved in the activation process of PKC that, as explained above, acts on several processes. Some taste receptors have been shown to be affected by domestication (da Silva et al. 2014).
Table 3. Genes present in 10 or more significant pathways (Fst).
| Gene Symbol | Ensembl Gene ID | Pathways | Significant Pathways | P-Value (Fst Asia) | P-Value (Fst Europe) | Genomic Position |
|---|---|---|---|---|---|---|
| PLCB3 | ENSSSCG00000013034 | 35 | 15 | 0.128 | 0.741 | 2: 6911684–6927124 |
| PLCB1 | ENSSSCG00000007056 | 35 | 15 | 1e−16 | 5e−04 | 17: 19691509–19860912 |
| PRKCA | ENSSSCG00000017268 | 46 | 13 | 0.248 | 0.887 | 12: 13502757–13602445 |
| PRKACA | ENSSSCG00000013771 | 47 | 13 | 0.830 | 0.583 | 2: 65350514–65371241 |
| PLA2G4A | ENSSSCG00000023351 | 21 | 11 | 0.388 | 2e−04 | 9: 140460880–140623439 |
| PRKCG | ENSSSCG00000003256 | 36 | 11 | 0.985 | 0.157 | 6: 52982004–53001741 |
| ITPR3 | ENSSSCG00000001518 | 27 | 11 | 0.938 | 9e−04 | 7: 34443056–34510838 |
| ENSSSCG00000000175 | ENSSSCG00000000175 | 35 | 11 | 0.579 | 0.385 | 5: 15129052–15145294 |
| ENSSSCG00000023437 | ENSSSCG00000023437 | 25 | 10 | 0.435 | 0.003 | 13: 67554829–67604679 |
| ADCY2 | ENSSSCG00000017101 | 32 | 10 | 0.024 | 0.249 | 16: 80358753–80624210 |
| MAPK1 | ENSSSCG00000010081 | 74 | 10 | 0.696 | 0.633 | 14: 53590167–53614842 |
| ADCY3 | ENSSSCG00000008578 | 32 | 10 | 0.999 | 0.994 | 3: 121107128–121201171 |
| ENSSSCG00000007833 | ENSSSCG00000007833 | 39 | 10 | 7e−06 | 0.003 | 3: 23052200–23174810 |
| ADCY4 | ENSSSCG00000001988 | 32 | 10 | 0.999 | 0.463 | 7: 80227590–80243075 |
ID, identifier; Fst, statistical significance for differentiation.
Impact of deleterious mutations rate
We found 123,571 synonymous and 138,121 nonsynonymous SNPs, of which 75,486 were predicted by the VEP tool (McLaren et al. 2010) to be tolerated and 62,635 to be deleterious. In order to investigate whether the significant pathways and their genes have a larger proportion of deleterious vs. tolerated variants than the rest of pathways, we classified the SNPs in three groups: (i) SNPs in nonsignificant genes of nonsignificant pathways; (ii) SNPs in nonsignificant genes (P-value > 0.01) from significant pathways; and (iii) SNPs in significant genes (P-value < 0.01) of significant pathways. Table 4 shows the count of predicted deleterious and tolerated SNPs by group according to continent and domestic/wild status populations. In all four populations, there was a systematic trend of decreasing deleterious/tolerated rate with SNPs in significant genes of significant pathways compared to SNPs from non-significant pathways. The χ2 test was significant in Europe and when all populations were jointly considered (P < 0.01) but not in Asia. These results can be interpreted as an increased functional constraint (lower ratio of deleterious mutations) in significant genes from significant pathways rather than in genes from nonsignificant pathways.
Table 4. Deleterious and tolerated SNPs grouped according to Europe (EU)/Asia (AS) continent and domestic (DM)/wild (WB) status.
| Continent | Population | SNP Typea | Nonsignificant Genes of Nonsignificant Pathways | Significant Genes of Nonsignificant Pathways | Nonsignificant Genes of Significant Pathways | Significant Genes of Significant Pathways | P-Valueb |
|---|---|---|---|---|---|---|---|
| Asia | ASDM | Tolerated | 8534 | 1,195 | 2,625 | 1,176 | 0.756 |
| Deleterious | 6,825 | 954 | 2,144 | 927 | |||
| λASDM | 0.800 | 0.798 | 0.817 | 0.788 | |||
| ASWB | Tolerated | 6,245 | 843 | 2,029 | 636 | 0.033 | |
| Deleterious | 4,432 | 587 | 1,320 | 391 | |||
| λASWB | 0.710 | 0.696 | 0.651 | 0.615 | |||
| ASDM/ASWB | λASDM/λASWB | 1.127 | 1.147 | 1.255 | 1.282 | ||
| Europe | EUDM | Tolerated | 5,191 | 716 | 1,675 | 620 | 0.023 |
| Deleterious | 3,429 | 483 | 1,200 | 349 | |||
| λEUDM | 0.661 | 0.675 | 0.716 | 0.563 | |||
| EUWB | Tolerated | 2,654 | 416 | 893 | 346 | 0.001 | |
| Deleterious | 1,153 | 198 | 382 | 103 | |||
| λEUWB | 0.434 | 0.476 | 0.428 | 0.298 | |||
| EUDM/EUWB | λEUDM/λEUWB | 1.523 | 1.418 | 1.675 | 1.891 | ||
| Total | Total | Tolerated | 22,624 | 3,170 | 7,222 | 2,778 | 0.003 |
| Deleterious | 15,839 | 2,222 | 5,046 | 1,770 | |||
| λTotal | 0.700 | 0.701 | 0.699 | 0.637 |
SNP, single nucleotide polymorphism; ASDM, Asian domestic; ASWB, Asian wild boar; EUDM, European domestic; EUWB, European wild boar.
λ corresponds to the ratio of deleterious vs. tolerated SNPs.
P-value obtained from χ2 test of the 2 × 2 table containing nonoverlapping SNP sets (nonsignificant genes from nonsignificant pathways vs. significant genes from significant pathways).
Previous studies suggest that domestication has resulted in an increased accumulation of deleterious mutations (Cruz et al. 2008; Renaut and Rieseberg 2015; Pérez-Enciso et al. 2016). In agreement with this, we observed larger ratio of deleterious vs. tolerated mutations in domestics than in wild boars (λASDM/λASWB and λEUDM/λEUWB in Table 4). Interestingly, these ratios are higher in significant genes than in random gene SNPs, and also higher in Europe than in Asia. Therefore, these data suggest that potential purifying selection is weaker/less effective in Europe than in Asia, likely because of the well-known low effective population size of old world pigs (Groenen et al. 2012). Besides, even if within-population purifying selection was stronger in significant genes, it was comparatively weaker in domestic than in wild populations.
General discussion
We report a functional analysis of pig domestication and breeding using a large complete sequence dataset that consisted of 40 wild boars and 123 domestic pig genomes from Asia (mainly China) and Europe. Rather than a standard exploratory genome-wide analysis, we focused on an analysis where the unit of study was the pathway. Genes do not function in isolation but coordinately, and thus metabolic pathways provide a reasonable scaffold to accommodate this fact (e.g., Daub et al. 2013). In a previous study, we observed that the “heritability” of domestic status varied according to pathway and that differences were not due to the number of genes in the pathway, suggesting that pathway can be a meaningful analysis unit (Pérez-Enciso et al. 2016). In fact, one of the main advantages of this approach is that it provides a direct biological interpretation of the analyses, although an independent source of information may be required to conclude which tissue and developmental stage the perturbed pathway may act in. In contrast, standard window-based genome-wide scans may pinpoint regions devoid of annotations or where the functional relation between significant windows is unknown. We assessed two metrics, differentiation (Fst) and disequilibrium (nSL), and although some of the pathways were connected (Figure 4), we found little concordance between the two analyses. Lack of agreement between differentiation and disequilibrium statistics have been reported previously (e.g., Chen et al. 2016; Dall’Olio et al. 2012), and this is likely because of the different timing and persistence of effects caused by selection (Sabeti et al. 2006). In particular, since disequilibrium erodes rapidly, our analysis suggests that reproductive changes (Table 2) are among the most recent ones, whereas others such as development and behavior (Table 1) were earlier targets of domestication and/or breeding. This is coherent with current knowledge, as behavioral changes must have occurred in earlier stages of domestication, as exemplified by the important experiment for tameness in foxes (Kukekova et al. 2012), whereas an emphasis on increasing reproductive performance is a more recent target of modern breeding.
Our approach has limitations as well. Foremost, many genes are not assigned to any pathway. In the NCBI Biosystems database v.160202 used here, 7157 genes out of a total of 21,691 annotated genes (Ensembl genes v. 83) were assigned to at least one pathway. After filtering, we further restricted the analysis to 442 pathways containing 5713 genes; these correspond to a total of 220 Mb or ∼8.5% of the whole genome. Another issue is redundancy and the definition of a pathway itself, since there are several databases (KEGG, REACTOME, and Interactome, etc.) that contain lists of functionally related genes. Definitions of the same pathways can actually be quite different between databases (Mooney et al. 2014), as we observed here with the arachidonic metabolism pathways (Table 2). Here, as in Daub et al. (2013), we decided to initially consider all available pathways from the NCBI biosystems database, although we set a maximum redundancy between pathways of 50%. However, in contrast to Daub et al. (2013) who considered the most significant SNP from each gene and removed all gene redundancy between pathways, here we combined all SNPs (after pruning for LD) from a given gene into a single statistics using Fisher’s method, and we allowed a 50% gene redundancy. It is not evident which method is best, but it seems that our approach is more conservative since outlier Fst will be smoothed out unless a general trend across SNPs in the whole gene is maintained. In turn, allowing for gene redundancy allows us to keep the original gene set instead of pathway pruning.
The history of domestication and domestic breeds is quite complex. In addition to multiple independent domestication events, as occurred in Asia and in Europe in the case of the pig, local adaptive processes have occurred since different breeds have been selected for different traits. Therefore, it is not surprising that previous works (e.g., Amaral et al. 2011) reported that most of selective signals were breed-specific, although our work demonstrates that shared domestication and breeding signals across breeds can still be detected. These signals are numerous and none of them are strong enough to explain the whole process. Once more, the polygenic model prevails. We further show that Asian and European domestication/breeding processes have both distinctive and shared pathways, and that multiple processes have been involved, such as an increase in disequilibrium and in differentiation. Differentiation metrics (Fst) revealed a larger number of signals than disequilibrium (Table 1 vs. Table 2), but this may be due to the experimental design: we analyzed several breeds jointly and disequilibrium is more rapidly eroded by demographic processes than differentiation (Sabeti et al. 2006). Nevertheless, we often found genes that were significant in both continents, such as several genes involved in behavior (PLCB1, GSK3B, and HTR4, Figure 2), while the pathways they belong to were significant in only one continent. Given that most European breeds have been admixed with Asian pigs (Groenen 2016), it is possible that these shared signals may actually be due to introgression. To verify this, we ran a semisupervised ADMIXTURE analysis on all significant (Table 1 and Table 2) and a set of random pathways (Figure S8 in File S1). The average Asian component in EUDM pigs across significant pathways was q = 0.11 (SD = 0.03), which is nearly identical to that observed in a random set of pathways (0.11, SD = 0.02). Similarly, we did not find differences in Asian component between shared significant pathways across continents (q = 0.105) and those that were continent-specific (q = 0.102). This suggests that Asian introgression is unlikely to have caused a shared signal between continents, which can be explained because the Asian signature in European breeds seems to be quite heterogeneous, i.e., due to different Asian origins (Bosse et al. 2014; Bianco et al. 2015a).
In contrast to previous works in humans, which reported an enrichment of pathways related to pathogen response in adaptation (Daub et al. 2013), we did not find a strong overrepresentation of immune system-related pathways. Only three related pathways were detected with Fst (complement cascade, Chagas disease, and FCGR phagocytosis, Table 1). This could be due to the fact that domestication was accompanied by stronger selection for traits other than disease resistance such as behavior, reproduction, or development. Another explanation is that these disease resistance signals were breed-specific and therefore remained undetected in this experimental design.
As in other studies (Cruz et al. 2008; Renaut and Rieseberg 2015; Pérez-Enciso et al. 2016), we found an increased accumulation of deleterious mutations in domestic animals. We systematically observed a higher proportion of deleterious variants in domestic groups compared to wild boars. This was observed for all genes, regardless of whether they were significant or not. On the other hand, a decreased accumulation of deleterious mutations was observed in significant genes from significant pathways, suggesting, as shown in Table 3, that these genes perform essential and central tasks in the physiology and development of the pig. Therefore, these genes seem to be under stronger functional constraint than randomly sampled genes.
Conclusions
We have studied the functional basis of domestication and breeding in the pig. This was possible because a modern equivalent of the wild ancestor is still available for study and was facilitated by the numerous sequences in the public domain. We show that these processes predominantly involved pathways related to behavior, especially in Asia, but also others like insulin, organ size development, recombination, and female reproduction. At least in part, these results can be explained by a relaxation of purifying selection associated with the domestication and/or breeding processes. Nevertheless, this purifying selection was stronger in genes and pathways that were significant using Fst than in random genes, likely because these genes play central roles and are highly functionally constrained. Negative selection was also stronger in Asia than in Europe, likely due to the larger effective size of the Asian population. In all probability, this analysis is conservative since we have focused on SNPs and genes that are consistently differentiated between all domestic breeds pooled together vs. wild boar. Focusing on a specific breed may have increased power but also could be prone to false discovery rate, identifying signals that are breed specific rather than domestic specific.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.042671/-/DC1.
Acknowledgments
We thank Sara Guirao-Rico, Marcel Amills, and Sebastián Ramos-Onsins for comments. Part of this work was developed while M.P.-E. was visiting CSIRO Agriculture at the St. Lucia campus (Brisbane, Australia), funded by a CSIRO McMaster Visiting Fellowship. J.L.-C. is the recipient of a Training Grant for Research Staff (FPI) grant, to achieve a PhD, from the Ministry of Economy and Science (MINECO, Spain). Work was also funded by grants AGL2013-41834-R and AGL2016-78709-R from MINECO to M.P.-E. We also acknowledge the support of MINECO for the “Centro de Excelencia Severo Ochoa 2016–2019” award SEV-2015-0533 and of the Centres de Recerca de Catalunya program (CERCA, Generalitat de Catalunya, Spain) to the Centre for Research in Agricultural Genomics.
Footnotes
Communicating editor: E. Akhunov
Literature Cited
- Ai H., Fang X., Yang B., Huang Z., Chen H., et al. , 2015. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat. Genet. 47: 217–225. [DOI] [PubMed] [Google Scholar]
- Alexander D. H., Novembre J., Lange K., 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19: 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral A. J., Ferretti L., Megens H.-J., Crooijmans R. P., Nie H., et al. , 2011. Genome-wide footprints of pig domestication and selection revealed through massive parallel sequencing of pooled DNA. PLoS One 6: e14782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57: 289–300. [Google Scholar]
- Bianco E., Soto H. W., Vargas L., Pérez-Enciso M., 2015a The chimerical genome of Isla del Coco feral pigs (Costa Rica), an isolated population since 1793 but with remarkable levels of diversity. Mol. Ecol. 24: 2364–2378. [DOI] [PubMed] [Google Scholar]
- Bianco E., Nevado B., Ramos-Onsins S. E., Pérez-Enciso M., 2015b A deep catalog of autosomal single nucleotide variation in the pig. PLoS One 10: e0118867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse M., Megens H.-J., Madsen O., Frantz L. A., Paudel Y., et al. , 2014. Untangling the hybrid nature of modern pig genomes: a mosaic derived from biogeographically distinct and highly divergent Sus scrofa populations. Mol. Ecol. 23: 4089–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning B. L., Browning S. R., 2013. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194: 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitenhuis B., Janss L. L. G., Poulsen N. A., Larsen L. B., Larsen M. K., et al. , 2014. Genome-wide association and biological pathway analysis for milk-fat composition in Danish Holstein and Danish Jersey cattle. BMC Genomics 15: 1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos-Paz W., Souza C. A., Megens H. J., Ramayo-Caldas Y., Melo M., et al. , 2013. Porcine colonization of the Americas: a 60k SNP story. Heredity (Edinb) 110: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M., et al. , 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Pan D., Ren H., Fu J., Li J., et al. , 2016. Identification of selective sweeps reveals divergent selection between Chinese Holstein and Simmental cattle populations. Genet. Sel. Evol. 48: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz F., Vilà C., Webster M. T., 2008. The legacy of domestication: accumulation of deleterious mutations in the dog genome. Mol. Biol. Evol. 25: 2331–2336. [DOI] [PubMed] [Google Scholar]
- da Silva E. C., de Jager N., Burgos-Paz W., Reverter A., Perez-Enciso M., Roura E., 2014. Characterization of the porcine nutrient and taste receptor gene repertoire in domestic and wild populations across the globe. BMC Genomics 15: 1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Olio G. M., Laayouni H., Luisi P., Sikora M., Montanucci L., et al. , 2012. Distribution of events of positive selection and population differentiation in a metabolic pathway: the case of asparagine N-glycosylation. BMC Evol. Biol. 12: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub J. T., Hofer T., Cutivet E., Dupanloup I., Quintana-Murci L., et al. , 2013. Evidence for polygenic adaptation to pathogens in the human genome. Mol. Biol. Evol. 30: 1544–1558. [DOI] [PubMed] [Google Scholar]
- Diamond J., 2002. Evolution, consequences and future of plant and animal domestication. Nature 418: 700–707. [DOI] [PubMed] [Google Scholar]
- Dickson B. J., 2003. Molecular mechanisms of axon guidance. Science 298: 1959–1964. [DOI] [PubMed] [Google Scholar]
- Esteve-Codina A., Paudel Y., Ferretti L., Raineri E., Megens H.-J., et al. , 2013. Dissecting structural and nucleotide genome-wide variation in inbred Iberian pigs. BMC Genomics 14: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A., Liang M., Korneliussen T., Nielsen R., 2014. On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol. Biol. Evol. 31: 1275–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti L., Raineri E., Ramos-Onsins S., 2012. Neutrality tests for sequences with missing data. Genetics 191: 1397–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz L., Meijaard E., Gongora J., Haile J., Groenen M. A. M., et al. , 2016. The evolution of suidae. Annu. Rev. Anim. Biosci. 4: 61–85. [DOI] [PubMed] [Google Scholar]
- Frantz L. A. F., Schraiber J. G., Madsen O., Megens H.-J., Cagan A., et al. , 2015. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat. Genet. 47: 1141–1148. [DOI] [PubMed] [Google Scholar]
- Fujii J., Otsu K., Zorzato F., de Leon S., Khanna V. K., et al. , 1991. Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science 253: 448–451. [DOI] [PubMed] [Google Scholar]
- Geer L. Y., Marchler-Bauer A., Geer R. C., Han L., He J., et al. , 2010. The NCBI BioSystems database. Nucleic Acids Res. 38: D492–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen M. A. M., 2016. A decade of pig genome sequencing: a window on pig domestication and evolution. Genet. Sel. Evol. 48: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen M. A. M., Archibald A. L., Uenishi H., Tuggle C. K., Takeuchi Y., et al. , 2012. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491: 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha N.-T., Gross J. J., van Dorland A., Tetens J., Thaller G., et al. , 2015. Gene-based mapping and pathway analysis of metabolic traits in dairy cows. PLoS One 10: e0122325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine K. D., 2012. Integration of intercellular signaling through the Hippo pathway. Semin. Cell Dev. Biol. 23: 812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., et al. , 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A., Reich D., 2010. Human population differentiation is strongly correlated with local recombination rate. PLoS Genet. 6: e1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh H.-Y., Kim D., Lee J., Lee S., Shin H.-S., 2007. Deficits in social behavior and sensorimotor gating in mice lacking phospholipase C beta 1. Genes Brain Behav. 7: 120–128. [DOI] [PubMed] [Google Scholar]
- Kukekova A. V., Temnykh S. V., Johnson J. L., Trut L. N., Acland G. M., 2012. Genetics of behavior in the silver fox. Mamm. Genome 23: 164–177. [DOI] [PubMed] [Google Scholar]
- Larson G., Dobney K., Albarella U., Fang M., Matisoo-Smith E., et al. , 2005. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 307: 1618–1621. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L., Gopinath G., Gillespie M., Caudy M., Croft D., et al. , 2009. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 37: 619–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., et al. , 2010. Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics 26: 2069–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár J., Nagy T., Stéger V., Tóth G., Marincs F., et al. , 2014. Genome sequencing and analysis of Mangalica, a fatty local pig of Hungary. BMC Genomics 15: 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney M. a., Nigg J. T., McWeeney S. K., Wilmot B., 2014. Functional and genomic context in pathway analysis of GWAS data. Trends Genet. 30: 390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Fuentes V., Marcet-Ortega M., Alkorta-Aranburu G., Forsberg C. L., Morrell J. M., et al. , 2015. Strong artificial selection in domestic mammals did not result in an increased recombination rate. Mol. Biol. Evol. 32: 510–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevado B., Ramos-Onsins S. E., Perez-Enciso M., 2014. Resequencing studies of nonmodel organisms using closely related reference genomes: optimal experimental designs and bioinformatics approaches for population genomics. Mol. Ecol. 23: 1764–1779. [DOI] [PubMed] [Google Scholar]
- Ngoh A., McTague A., Wentzensen I. M., Meyer E., Applegate C., et al. , 2014. Severe infantile epileptic encephalopathy due to mutations in PLCB1: expansion of the genotypic and phenotypic disease spectrum. Dev. Med. Child Neurol. 56: 1124–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollivier L., 1995. Genetic differences in recombination frequency in the pig (Sus scrofa). Genome 38: 1048–1051. [DOI] [PubMed] [Google Scholar]
- Pavlidis P., Jensen J. D., Stephan W., Stamatakis A., 2012. A critical assessment of storytelling: gene ontology categories and the importance of validating genomic scans. Mol. Biol. Evol. 29: 3237–3248. [DOI] [PubMed] [Google Scholar]
- Pérez-Enciso M., de los Campos G., Hudson N., Kijas J., Reverter A., 2016. The “heritability” of domestication and its functional partitioning in the pig. Heredity (Edinb) 118: 160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pico A. R., Kelder T., Van Iersel M. P., Hanspers K., Conklin B. R., et al. , 2008. WikiPathways: pathway editing for the people. PLoS Biol. 6: 1403–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoli M., Yan Z., McEwen B. S., Sanacora G., 2011. The stressed synapse: the impact of stress and glucocorticoids on glutamate transmission. Nat. Rev. Neurosci. 13: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., 2014. BEDTools: the swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics. 47: 11.12.1–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team , 2014. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Ramírez O., Burgos-Paz W., Casas E., Ballester M., Bianco E., et al. , 2014. Genome data from a sixteenth century pig illuminate modern breed relationships. Heredity (Edinb) 114: 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Onsins S. E., Burgos-Paz W., Manunza A., Amills M., 2014. Mining the pig genome to investigate the domestication process. Heredity (Edinb) 113: 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaut S., Rieseberg L. H., 2015. The accumulation of deleterious mutations as a consequence of domestication and improvement in sunflowers and other compositae crops. Mol. Biol. Evol. 32: 2273–2283. [DOI] [PubMed] [Google Scholar]
- Reverter A., Chan E. K. F., 2008. Combining partial correlation and an information theory approach to the reversed engineering of gene co-expression networks. Bioinformatics 24: 2491–2497. [DOI] [PubMed] [Google Scholar]
- Ross-Ibarra J., 2004. The evolution of recombination under domestication: a test of two hypotheses. Am. Nat. 163: 105–112. [DOI] [PubMed] [Google Scholar]
- Rubin C.-J., Megens H.-J., Martinez Barrio A., Maqbool K., Sayyab S., et al. , 2012. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA 109: 19529–19536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P. C., Schaffner S. F., Fry B., Lohmueller J., Varilly P., et al. , 2006. Positive natural selection in the human lineage. Science 312: 1614–1620. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., et al. , 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., et al. , 2012. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40: W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, J. D., A. J. Bass, A. Dabney, and D. Robinson, 2015 qvalue: Q-value Estimation for False Discovery Rate Control. Available at: http://github.com/jdstorey/qvalue. Accessed: June 1, 2016.
- Tajima F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortereau F., Servin B., Frantz L., Megens H.-J., Milan D., et al. , 2012. A high density recombination map of the pig reveals a correlation between sex-specific recombination and GC content. BMC Genomics 13: 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B. S., Cockerham C. C., 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H., 2010. Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet. 11: 843–854. [DOI] [PubMed] [Google Scholar]
- Zeder M. A., 2015. Core questions in domestication research. Proc. Natl. Acad. Sci. USA 112: 3191–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.