The insulin/insulin-like growth factor signaling (IIS) and target of rapamycin (TOR) pathways have long been thought to be involved in how organisms..
Keywords: multiparental populations, Drosophila melanogaster, insulin signaling, TOR signaling, life span, diet, eQTLs, Multiparent Advanced Generation Inter-Cross (MAGIC), MPP
Abstract
The nutritional environments that organisms experience are inherently variable, requiring tight coordination of how resources are allocated to different functions relative to the total amount of resources available. A growing body of evidence supports the hypothesis that key endocrine pathways play a fundamental role in this coordination. In particular, the insulin/insulin-like growth factor signaling (IIS) and target of rapamycin (TOR) pathways have been implicated in nutrition-dependent changes in metabolism and nutrient allocation. However, little is known about the genetic basis of standing variation in IIS/TOR or how diet-dependent changes in expression in this pathway influence phenotypes related to resource allocation. To characterize natural genetic variation in the IIS/TOR pathway, we used >250 recombinant inbred lines (RILs) derived from a multiparental mapping population, the Drosophila Synthetic Population Resource, to map transcript-level QTL of genes encoding 52 core IIS/TOR components in three different nutritional environments [dietary restriction (DR), control (C), and high sugar (HS)]. Nearly all genes, 87%, were significantly differentially expressed between diets, though not always in ways predicted by loss-of-function mutants. We identified cis (i.e., local) expression QTL (eQTL) for six genes, all of which are significant in multiple nutrient environments. Further, we identified trans (i.e., distant) eQTL for two genes, specific to a single nutrient environment. Our results are consistent with many small changes in the IIS/TOR pathways. A discriminant function analysis for the C and DR treatments identified a pattern of gene expression associated with the diet treatment. Mapping the composite discriminant function scores revealed a significant global eQTL within the DR diet. A correlation between the discriminant function scores and the median life span (r = 0.46) provides evidence that gene expression changes in response to diet are associated with longevity in these RILs.
GIVEN a finite amount of resources, a central requirement of all organisms is optimizing the allocation of those resources to competing anatomical structures and physiological functions, which necessitates the coordination of multiple organ systems within the nutritional environment. In eukaryotes, a commonly observed pattern is the extension of life span coupled with reduced reproduction in nutrient-limited environments (Sohal and Weindruch 1996; Browner et al. 2004; Hughes and Reynolds 2005; Kirkwood and Shanley 2005; Flatt and Schmidt 2009; Partridge et al. 2010; Piper et al. 2011), which has been hypothesized to result in part from an adaptive shift in resource allocation away from reproduction and toward somatic maintenance in low resource environments. This response is hypothesized to be adaptive in a selective regime of fluctuating resources (Neel 1962; Fischer et al. 2009; Wells 2009; Fischer et al. 2010). When food resources are scarce, individuals conserve resources and allocate to survival to ensure they will live until conditions improve. In good conditions, individuals allocate to reproduction. This hypothesis has been invoked many times to provide an evolutionary explanation for the extension of life span with dietary restriction (DR) that has been documented in a diversity of organisms (e.g., Hughes and Reynolds 2005; Kirkwood and Shanley 2005; Flatt and Schmidt 2009). Previous studies in Drosophila melanogaster have shown substantial genotype-by-diet interactions for phenotypes related to metabolism and resource allocation (Reed et al. 2010, 2014). This hypothesis leads to the central question: What are the genes that underlie the response of resource allocation to different nutritional environments?
The physiological mechanisms underlying the metabolism and allocation of nutrients have been well studied, and one of the key components is the insulin/insulin-like signaling/target of rapamycin (IIS/TOR) pathway (Teleman 2010; Nässel et al. 2015). This fundamental endocrine pathway shows remarkable conservation across animals. Orthologs of the human IIS/TOR pathway are present in rodents, fruit flies, and nematodes. Laboratory mutants of major genes in the IIS/TOR pathway specifically show very similar phenotypic effects across model organisms, suggesting its function is conserved across metazoa. Many studies in model organisms have demonstrated a role for the IIS/TOR pathway in growth, cellular and organismal metabolism, stress resistance, life span, and reproduction (Kenyon et al. 1993; Böhni et al. 1999; Clancy et al. 2001; Tatar et al. 2001; Bluher et al. 2003; Broughton et al. 2005; reviewed in: Garofalo 2002; Goberdhan and Wilson 2003; Giannakou and Partridge 2007; Kaletsky and Murphy 2010; Teleman 2010). For example, mutants lacking key components of the IIS/TOR pathway generally show reduced growth, increased life span, decreased fecundity, and increased stress resistance. Several lines of evidence suggest that the IIS/TOR pathway is also involved in the coordination of nutritional conditions with metabolism and growth: (1) insulin secretion is inhibited by nutrient deprivation (Geminard et al. 2009); (2) starvation mimics the effects of reduced IIS/TOR (Britton et al. 2002); and (3) overactivation of the IIS/TOR pathway can bypass the need for nutrients, causing growth in the absence of nutrients followed by death (Britton et al. 2002). These many studies and others support the hypothesis that the evolution of metabolism and allocation patterns ultimately results from the evolution of genes in the IIS/TOR pathway and other endocrine pathways (cf. Tatar et al. 2003; Flatt et al. 2005; Zera et al. 2007).
Most studies to date of the IIS/TOR pathway have followed a “one gene at a time” approach, altering one gene in isolation to characterize function. In this regard, they have been very successful at determining the pathway’s role in the regulation of metabolism and resource allocation and of the effects of altering each gene in isolation. It is largely due to these types of studies, which use techniques such as gene knockouts to experimentally produce large alterations to the expression of genes in the IIS/TOR pathway, that the IIS/TOR pathway has long been considered a prime candidate for the underlying mechanism of life-span extension under nutrient limitation. In stark contrast to the success of these studies in characterizing individual gene function, we know very little about the genetic basis of natural variation in the IIS/TOR pathway, how changes in the expression of many genes act in concert, and how diet-induced changes in IIS/TOR expression with diet relate to changes in potentially related phenotypes such as life span. While individuals experiencing DR and loss-of-function mutants in the IIS/TOR pathway show remarkably similar phenotypes, the evidence for a direct relationship between the IIS/TOR pathway and diet-induced life-span extension is mixed, with some studies supporting the relationship (e.g., Zid et al. 2009) and others not (Min et al. 2008; Whitaker et al. 2014; reviewed in Tatar et al. 2014). Thus, despite years of investigation on both the mechanisms of diet-induced life span and on the function of the IIS/TOR pathway, whether there is a connection between the two remains an unanswered question.
In this study, we use a multiparent population, the Drosophila Synthetic Population Resource (DSPR) (King et al. 2012a,b; Long et al. 2014; http://FlyRILs.org) to assay gene expression for ∼50 genes in the IIS/TOR pathway and median life span in multiple nutrition environments. We aim to answer three key questions:
How do IIS/TOR pathway genes respond to changes in diet?
What is the source of natural genetic variation in the response of IIS/TOR pathway genes to diet?
What is the relationship between IIS/TOR expression and life-span extension under DR?
Materials and Methods
Mapping population
To uncover the genetic basis of variation in gene expression within the IIS/TOR pathway, we used a large, multiparent population, the DSPR (King et al. 2012a,b; Long et al. 2014; http://FlyRILs.org). The DSPR is a panel of two sets of recombinant inbred lines (RILs) derived from two synthetic populations (pA and pB), each of which were created by crossing a different set of eight inbred founder lines for 50 generations, with one founder line shared between the populations. The DSPR founder lines have been fully sequenced, the RILs have been genotyped, and these data have informed a hidden Markov model that inferred the underlying founder haplotype at each position in each RIL. A detailed description of the DSPR and the associated data can be found in King et al. (2012a,b) and at http://FlyRILS.org. Raw resequencing data of the founder lines is deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under accession number SRA051316, and the RIL restriction-site-associated DNA (RAD) genotyping data are available under accession number SRA051306. The founder genotype assignments from the hidden Markov model are available as two data packages in R (http://FlyRILs.org/Tools/Tutorial) and are available from the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.r5v40).
Experimental diets
We used three experimental diets [DR, control (C), and high sugar (HS)] adapted from Bass et al. (2007) and Skorupa et al. 2008 (Supplemental Material, Table S1 in Supplementary Material R1). All experimental media was <2 weeks old and was stored at 4° to avoid using desiccated or degraded food. To avoid food degradation and larval interference, individuals were moved into vials with fresh media every 2–3 days.
IIS/TOR expression
Husbandry:
We employed a crossing design to avoid directly measuring inbred lines and potentially mapping variation associated with inbreeding depression. Briefly, we used 260 pB RILs crossed to a standard line. The F1 progeny from each cross (hereafter abbreviated RIX for recombinant inbred cross) were split among our three diet treatments with three replicate vials in each for a target total of 30 individuals per diet. This procedure is described in detail here. For each of 260 pB lines, we crossed pB females with the A4 founder line males (A4 was chosen as an arbitrary standard line) in six batches with each batch containing between 30 and 100 crosses. All expression measures were taken on the resulting female trans-heterozygous F1 individuals. Only female flies were used to limit the study to expression in a single sex (also assaying males would double the number of expression assays), and because females have more commonly been the focus of studies exploring the IIS/TOR pathway’s influence on life span and reproduction. We raised the parental lines (pB RILs and A4) and F1 larval offspring on a standard cornmeal-dextrose-yeast diet adapted from the Bloomington stock center (Table S1 in Supplementary Material R1). For each of the crosses, two virgin pB RIL females and two A4 males were placed into each of 9–10 vials (25 mm o.d. × 95 mm height), and were allowed to mate and lay eggs for 72 hr. We cleared all adult F1 flies that emerged before 11 days postoviposition (po) to control the age of the flies. Adult, mated female F1 individuals were collected and placed on experimental diets 14 days po. For each cross, 10 female progeny were placed in two to three replicate vials for each experimental diet (Figure S1 in Supplementary Material R1). Environmental conditions in the rearing chamber were held at 23°, 50% relative humidity, and a 24:0 light:dark cycle. Following 10 days on the diet treatment (24 days po), we immediately flash froze flies in liquid nitrogen to preserve gene expression levels, and placed the samples in a −80° freezer for storage. We began the process of flash freezing at approximately the same time of day for each batch and the order of flash freezing was haphazardly assigned to avoid any systematic bias in time of day. We pooled all replicates for a given cross together before performing expression assays. This strategy kept the number of samples assayed at a feasible level and maximized the number of lines we could assay while averaging over individual variability in expression. A small number of crosses failed to produce enough adults in one of the diets (if they failed in more than one diet, they were eliminated from the experiment). The final sample size for each diet was: DR, 249; C, 253; HS, 253).
Expression analysis:
We obtained RNA for whole flash frozen flies, aiming to assay global IIS/TOR expression. While it would be highly informative to obtain tissue-specific measures, this approach would have greatly increased the number of expression assays required. We first extracted RNA using a modified protocol based on the Life Technologies TRIzol RNA extraction protocol and then further purified the RNA via a clean-up step using a QAIGEN (Valencia, CA) RNeasy Plus Mini Kit. Between 20 and 30 flash-frozen flies (some flies were lost due to early deaths or escapees) were homogenized in a bead beater using stainless steel beads in 1 ml of TRIzol solution, RNA was then extracted and purified from the homogenate following the TRIzol extraction and QIAGEN protocols. Sample quality and concentration was evaluated using 2 μl of RNA extract on a Nanodrop 2000 system. The resulting RNA extracts were stored at −80° until they were reverse transcribed into complementary DNA (cDNA).
We converted RNA to cDNA using an Applied Biosystems (Foster City, CA) High Capacity Reverse Transcription Kit. We normalized the amount of starting RNA using the concentrations found on the Nanodrop system. The required volume to transfer 1.5 μg from each sample was distributed onto 96-well plates along with an appropriate amount of molecular grade double-distilled water to bring the total volume to 10 μl, and samples were subsequently reverse transcribed. These cDNA samples were stored at −80° to await quantitative PCR (qPCR).
We designed custom OpenArray 56 assay by 48 sample arrays with 56 Taqman Gene Expression Assays selected from the core components of the IIS/TOR pathway (Figure 1), and two housekeeping genes. One gene was included in error, ras, due to a misspecification when intending to select an assay for the similarly named Ras (also known as Ras85D). The selected gene assays were run on an Applied Biosystems QuantStudio 12K Flex Real-Time PCR System.
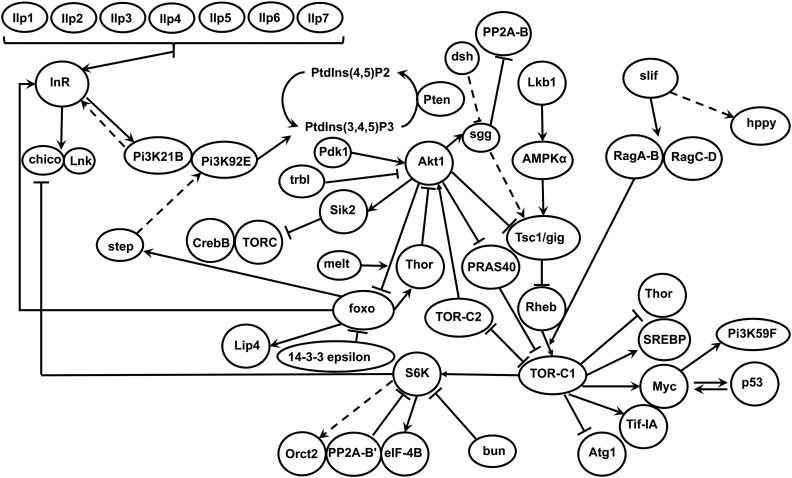
Figure 1.
Schematic of the assayed components of the IIS/TOR pathway and their proposed interactions in D. melanogaster. Arrows indicate activation and bar-ended lines indicate inhibition. Dashed lines indicate indirect or uncertain interactions. Mipp2, Tor, REPTOR, and rictor all form components of TOR-C1 and/or TOR-C2 and are not shown separately in the schematic. Adapted from Teleman (2010) and Nässel et al. 2015.
We ran our samples on a total of 20 arrays in five groups over a period of 1 month. In most cases, there was a single biological and a single technical replicate for each RIX. Nine RIXs were included in two batches and thus had two biological replicates. In addition, 14% of samples were run on a second array to fully use all arrays and thus had two technical replicates. Our raw and normalized expression measures (normalized by ΔCq only; see below for further preprocessing) have been deposited in NCBI’s Gene Expression Omnibus (GEO) (Edgar et al. 2002) and are accessible through GEO series accession number GSE93117 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93117).
Median life span
In a separate experiment, we measured life span for 80 pB RILs that were a subset of the original set of RILs used in crosses for the IIS/TOR expression assays. Small amounts of sample loss at various stages (e.g., cross failure, RNA extraction, OpenArray assay failure) led to less than complete overlap, with 64 RIL/RIX pairs with complete data for both gene expression and life span. We did not use the same crossing design as above, instead assaying the inbred RILs directly. While a crossing design is ideal, it is logistically more difficult to implement and it was not feasible at the time of the experiment. For each RIL, we set up 10 vials in which 6 females were mated to 4 males for 36 hr, after which adults were cleared. Early emerging flies were cleared 8 days po. At 10 days po, we transferred 30 females to each of two to three replicate vials in each of two diet treatments: C and DR (see Experimental diets). All larvae were reared on a standard cornmeal-dextrose-yeast diet. Flies were moved to new food every 2–3 days, during which we recorded the number of dead flies until all flies were dead. To calculate median life span per RIL per diet, we used the “Surv” and “survfit” functions in the survival package to fit a Kaplan–Meier survival curve and calculate median life span for each RIL in each diet treatment (Therneau and Grambsch 2000; Therneau 2015). Our raw daily mortality data are in Table S2 and our processed median life span data are in Table S3. The analysis procedure to calculate median life span given here can also be found in File S1.
Statistical analysis
All analyses described below were performed in R (version 3.3.1; R Core Team 2016).
Preprocessing and batch correction of gene expression:
Our expression data were normalized using the well-established ΔCq method (Livak and Schmittgen 2001) based on expression of the normalizing gene Ribosomal Protein L32 (RpL32). We initially chose two genes (RpL32 and Act42A) as potential normalizing genes that were invariant to treatment and sample quality based on preliminary qPCR experiments for a small set of samples (data not shown). We analyzed these genes again after we obtained all samples from the OpenArray. We confirmed that RpL32 did not show an effect of treatment (F2,913 = 0.635, P = 0.53) while Act42A did (F2,913 = 16.14, P < 0.001), thus we only used RpL32 as a normalizing gene. We also excluded two genes and three samples from our analysis that had a high assay failure rate (Ilp1 = 10% failure, Ilp4 = 40% failure).
Batch effects (both known and unknown) are a well-known issue in gene expression studies (Leek and Storey 2007; Leek et al. 2010, 2012; Mecham et al. 2010). There are several potential known sources of batch effects in our experiment. The sets of crosses were performed in batches, the RNA extraction was done in batches, and sets of arrays were run on different days. We note that samples from different treatments were deliberately distributed equally among these known sources to avoid confounding treatment with any batch. A principal component analysis revealed a strong effect distinguishing between the last batch of arrays run on the OpenArray and the initial four batches. Given this effect, we opted to selectively choose the technical replicate from the initial set in any case where we had two technical replicates in both sets. We used the “ComBat” function in the sva package to correct for both this known, strong batch effect and unknown batch effects (Leek et al. 2012, 2015). We included arrays in batches 1–4 vs. arrays in batch 5 as a known batch effect. The ComBat function detected two additional unknown/composite batch effects. After correcting for these batch effects, we performed parametric F-tests using the “f.pvalue” function to test for differential gene expression between the three diets. We then applied the false-discovery-rate (FDR) method (Benjamini and Hochberg 1995; Leek et al. 2012) to correct for the multiple tests performed and all P-values reported for differential expression are adjusted P-values for multiple tests. Following batch correction, we averaged over any biological or technical replicates. Both prior to and following batch correction, we performed quantile normalization on each gene to coerce normality (Pickrell et al. 2010), and all following analyses used this normalized data set. Our normalized, batch corrected gene expression measures are given in Table S4 and the preprocessing analysis as described above is also given in File S1.
Discriminant function analysis:
We performed a discriminant function analysis (DFA) to determine which genes best predict the diet treatment and collapse our 52 gene expression measures into composite variables that best account for diet-based differences (i.e., global changes in IIS/TOR expression in response to diet). We used the “lda” function in the MASS package in R (Venables and Ripley 2002) to fit the following model:
where Ti is the ith transcript abundance and βi is the ith effect estimate. Here, we report the results of a DFA considering only the DR and C treatments to facilitate a comparison with median life span and for the more straightforward interpretation of separating two vs. three groupings. Results for a DFA using all three diets were similar. Classification ability was nearly the same (72% for three diets vs. 77% for two diets), and the estimates of the standardized coefficients were highly correlated between the linear discriminant for two diets and the second linear discriminant for three diets (r = 0.91).
QTL mapping:
We performed QTL mapping for the following phenotypes: (1) gene expression for 52 genes measured in three diets, (2) the scores on the linear discriminant resulting from a DFA performed on our gene expression measures in two diets, and (3) median life span measured in two diets. We use the term “phenotype” generally hereafter to refer to the above-described measurements. For each of these phenotypes, we performed mapping within each diet treatment. For each RIL or RIX, we also calculated the difference in phenotype between environments and mapped the response to the environment.
To map QTL for each phenotype, we used Haley–Knott regression and regressed the eight additive founder haplotype probabilities on the phenotype of interest at positions regularly spaced across the genome in 10-kb intervals (Broman and Sen 2009; King et al. 2012a) by fitting the following model:
where yi is the phenotype (see descriptions of our sets of phenotypes above) of the ith RIL or RIX, pij is the probability the ith RIL or RIX has the jth haplotype at the locus, bij is the vector of effects for the jth haplotype, and ei is the vector of residuals. Mapping is performed in the same way in the RIXs (crossed to a standard) as in the inbred RILs, as all RIXs are crossed to the same standard line. The major difference is that an RIL will be homozygous for the founder genotype at a given position, while the RIX will have a single copy. To identify statistically significant QTL, we performed 1000 permutations of our data set to determine the FDR. Applying FDR procedures to genome scans is problematic due to dependencies among tests that arises from linkage (Chen and Storey 2006; Siegmund et al. 2011; Brzyski et al. 2017). To maintain the correlation structure of our gene expression measures, we permuted all expression phenotypes (both within and between diet measures), and our discriminant function scores together. We then calculated the average number of false cis (i.e., local) QTL and trans (i.e., distant) QTL across all phenotypes at different significance thresholds. To identify distinct peaks, we first identified all peak positions for a given genome scan. Then we removed any peaks that were within 2 cM of a higher peak. Peaks were considered cis if they were within ±5 Mb of the gene location. We then calculated the threshold that corresponded to an FDR (FDR = the number of false positives/the number of total positives; e.g., at a given threshold, FDR = the average number of significant cis-eQTL for the permuted data)/the number of significant cis-expression QTL (eQTL) for the observed data set) of 5 and 10% for cis- [5% FDR −log10(P) = 4.5; 10% FDR −log10(P) = 4.0] and trans-QTL [5% FDR −log10(P) = 5.8; 10% FDR −log10(P) = 5.4]. We used this same procedure separately for median life span, considering it an independent experiment. Here, no QTL were significant at an FDR <33% [−log10(P) = 3.67], precluding the calculation of FDR at 5 and 10% (no significant observed QTL translates to a zero in the numerator), thus we also report the threshold corresponding to a family-wise error rate of 5% [−log10(P) = 4.81; Churchill and Doerge 1994]. At these thresholds, with our sample sizes, we expect to overestimate the contribution of most QTL due to the Beavis effect (Utz and Melchinger 1994; Beavis 1998; Xu 2003; King and Long 2017). The lowest possible estimate for the percent variance explained by a QTL in our study is 11%, and all but the largest effect QTL will have a very large degree of uncertainty associated with this estimate. Thus, we do not report the percent variance explained by our peaks as it is largely uninformative in this case. Our QTL mapping results are in Table S5.
Data availability
All DSPR lines are available at http://FlyRILs.org/RequestFlies. All data are available centrally at http://FlyRILs.org/Data in addition to the public archives noted below. Raw resequencing data of the founder lines is deposited in the NCBI SRA (http://www.ncbi.nlm.nih.gov/sra) under accession number SRA051316, and the RIL RAD genotyping data are available under accession number SRA051306. The founder genotype assignments from the hidden Markov model are available as two data packages in R (http://FlyRILs.org/Tools/Tutorial) and are available from the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.r5v40). Our raw and normalized expression measures (normalized by ΔCq only; see below for further preprocessing) have been deposited in NCBI’s GEO (Edgar et al. 2002) and are accessible through GEO series accession number GSE93117 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93117). Our raw daily mortality data are in Table S2 and is available at from Zenodo: https://doi.org/10.5281/zenodo.322462. Our processed median life span data are in Table S3. Our normalized, batch-corrected gene expression measures are given in Table S4. All genome scan results are in Table S5.
Results
Differential expression in IIS/TOR genes
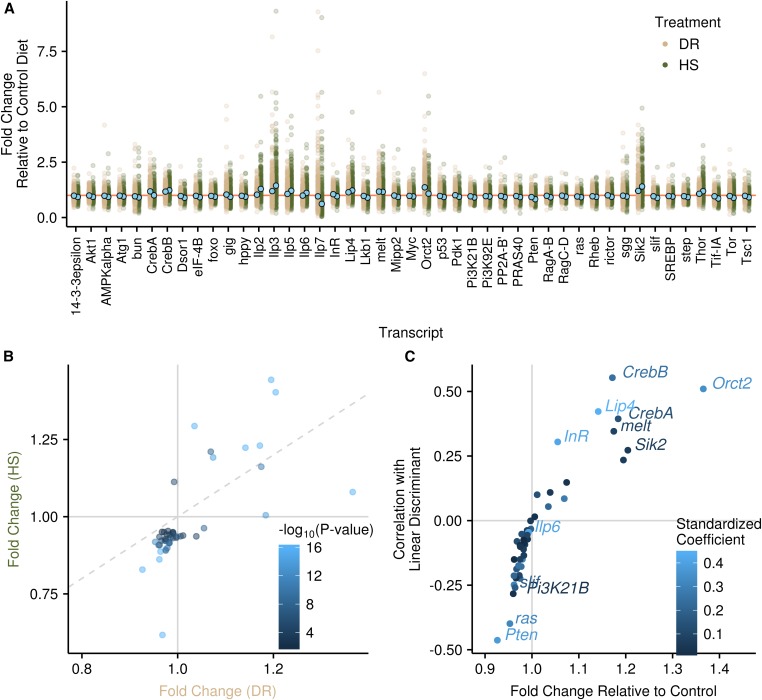
The majority of our genes (45 of 52 genes, 87%) were significantly differentially expressed between the three diets (Figure 2A), as would be expected for IIS/TOR genes in response to diet (see Introduction and references therein). Overall, the average expression change across diets was subtle, with no gene showing an average fold change greater than twofold between diets. There was a large amount of variability among RIXs in the change in expression across environments; however, we note that we did not replicate at the individual RIX level for most samples beyond pooling individuals, choosing instead to maximize the number of lines measured (thus replicating at the haplotype level) to optimize QTL mapping. Therefore, the variability at the RIX level should be interpreted with some caution, while haplotype means (see below) can be interpreted with increased confidence, as can average patterns of differential expression. Global patterns were similar in the DR and HS diets, with the HS diet typically showing greater differences (Figure 2B). In only a few cases did genes change in opposite directions in the HS vs. DR diet relative to the C diet (Figure 2B).
Figure 2.
Phenotypic patterns of gene expression across diets. (A) The fold change in the DR and HS diets relative to the C diet for each gene that was significantly differentially expressed. Each RIX is a single point on the plot. Means are shown with light blue points. The orange horizontal line denotes 1, i.e., no change relative to the control. One outlier point is not shown for Ilp7 in the DR treatment. (B) Comparison between the HS and DR fold change. Horizontal and vertical lines at 1 show when genes are over- or underexpressed relative to the C diet. Diagonal dashed line is the 1:1 line. Points in the quadrants above 1 for one diet and below 1 for the other are genes that trend in different directions in the DR vs. HS diet relative to C (top left and bottom right). Points falling above the 1:1 line in the top-right quadrant and below the 1:1 line in the bottom-left quadrant show a stronger effect in the HS diet than in the DR diet. Points are colored according to their significance. (C) The relationship between each gene’s fold change in the DR diet relative to the C diet and the correlation with the linear discriminant (i.e., loading). Points are colored according to the magnitude of the standardized coefficient from the linear discriminant analysis. Gene names are given for all potentially diagnostic genes (see Materials and Methods). Horizontal and vertical lines demark where genes are over- or underexpressed in the DR diet relative to the C diet.
The expression of individual genes in the different diets is visualized in Figure 2A with descriptions of major patterns of some key genes described here. The dIlps (Drosophila insulin-like peptides) showed some of the largest average differences with diet and showed high variability, either due to assay variation or large amounts of between-RIX variation. Compared to the C diet, expression changes followed the same pattern in the DR and HS diets, with the HS diet showing a greater change. Ilp2, Ilp3, and Ilp5 all showed increased expression (fold change relative to C diet: Ilp2: HS = 1.29, DR = 1.03, Padj < 0.001; Ilp3: HS = 1.44, DR = 1.20, Padj < 0.001; Ilp5: HS = 1.21, DR = 1.07, Padj < 0.001). Ilp6 showed similar expression in the DR and C diets and higher expression in the HS diet (fold change relative to C diet: HS = 1.11, DR = 0.99, Padj < 0.001). In contrast, relative to the C diet, Ilp7 showed lower expression in the HS diet, a pattern also found in the DR diet though with a much smaller difference (fold change relative to C diet: HS = 0.62, DR = 0.97, Padj < 0.001). Consistent with our observed change in llp7 expression, there is evidence for a specific role for Ilp7 in the regulation of oviposition on sucrose-containing diets (Yang et al. 2008), and HS diets have also been shown to reduce fecundity in adult flies (Skorupa et al. 2008).
Several additional key genes have been implicated or hypothesized to be involved in the transcriptional response to diet, including the insulin receptor (InR), the insulin receptor substrate (chico), the forkhead box type O transcription factor (foxo), and TOR. Surprisingly, chico was one of the few genes not significantly differentially expressed with diet. The difference in foxo expression was not large, though it was significant (fold change relative to C diet: HS = 0.94, DR = 0.98, Padj < 0.001). TOR showed the second-largest decrease relative to the C diet for the HS treatment, but showed only a subtle decrease in the DR treatment (fold change relative to C diet: HS = 0.89, DR = 0.97, Padj < 0.001). Very few genes showed contrasting patterns in the DR vs. HS diet but one of these was InR, which showed decreased expression in the HS diet and increased expression in the DR diet (fold change relative to C diet: HS = 0.96, DR = 1.05, Padj < 0.001).
Among the remaining genes, Sik2 was among the most highly expressed relative to the C diet for both the HS and DR treatments (fold change relative to C diet: HS = 1.40, DR = 1.20, Padj < 0.001) and Pten was among the most lowly expressed relative to the C diet (fold change relative to C diet: HS = 0.83, DR = 0.92, Padj < 0.001).
DFA
We performed a DFA for the DR and C diets to capture global patterns of expression in the pathway and to determine which genes best predict treatment, i.e., which genes are most diagnostic of the response to diet. DFA identifies the linear combination of predictor variables (here, genes) that maximizes the between-group variance and provides the optimal separation of groups (here, diets). We employed leave-one-out cross-validation to determine how accurately our set of gene expression measures could correctly classify samples into the C or DR diet. Overall, 77% of samples were classified correctly (74% for DR, 77% for C). Thus, while our set of genes does reasonably well, it is far from a perfect classifier, showing that expression measures of these IIS/TOR genes alone are not sufficient to definitively assign the diet treatment. Some of the variability in classification ability could stem from technical noise, and thus classification may be improved somewhat with increased technical replication.
We assessed the importance of individual genes to the linear discriminant in two ways: the standardized coefficients are the weights for each gene in the linear combination, and the loadings are the correlations of each gene with the linear discriminant (Figure 2C). We discuss potentially diagnostic genes here which meet one of two criteria: a standardized coefficient >0.4 or a loading >0.25 (i.e., explaining >6% of the variance). We note that loadings are full correlations (not partial or semipartial), and thus do not represent contributions independent of other genes, while the standardized coefficients represent the weighting of a given predictor variable after including all others. More genes are underexpressed in the DR diet relative to the C diet (31 vs. 14), but most show only a marginal effect and there are more diagnostic genes that are overexpressed (7 vs. 5). Among those underexpressed relative to the C diet are Pten (loading, −0.46; coefficient, −0.38), ras (loading, −0.40; coefficient, −0.32), slif (loading, −0.26; coefficient, −0.17), Pi3K21B (loading, −0.28; coefficient, −0.03), and Ilp6 (loading, −0.04; coefficient, −0.40). Diagnostic genes that are overexpressed relative to the C diet include Orct2 (loading, 0.51; coefficient, 0.34), CrebB (loading, 0.55; coefficient, 0.24), CrebA (loading, 0.39; coefficient, 0.14), Lip4 (loading, 0.42; coefficient, 0.45), melt (loading, 0.34; coefficient, 0.09), InR (loading, 0.30; coefficient, 0.42), and Sik2 (loading, 0.27; coefficient, 0.06). Pi3K21B, Sik2, and melt have high loadings showing they are predictive of diet on their own; however, their relatively low coefficients indicate more minor contributions once other genes are included, likely stemming from correlations with other genes. Of particular interest is Ilp6, which has a high coefficient but does not show a large fold change or large loading, indicating it is diagnostic only after variation is accounted for by other genes and underscoring the significant effect a gene might have in the absence of a large increase in abundance. One of the most diagnostic genes, ras, was not intentionally included in the array (see Materials and Methods) and there is no evidence it is involved in the IIS/TOR pathway, suggesting other genes outside the IIS/TOR pathway potentially show larger changes in expression with diet. We note that we also ran this analysis excluding the inadvertently measured gene, ras, and confirmed all results were qualitatively similar given it is not known to be a member of the IIS/TOR pathway.
The relationship between median life span and IIS/TOR expression
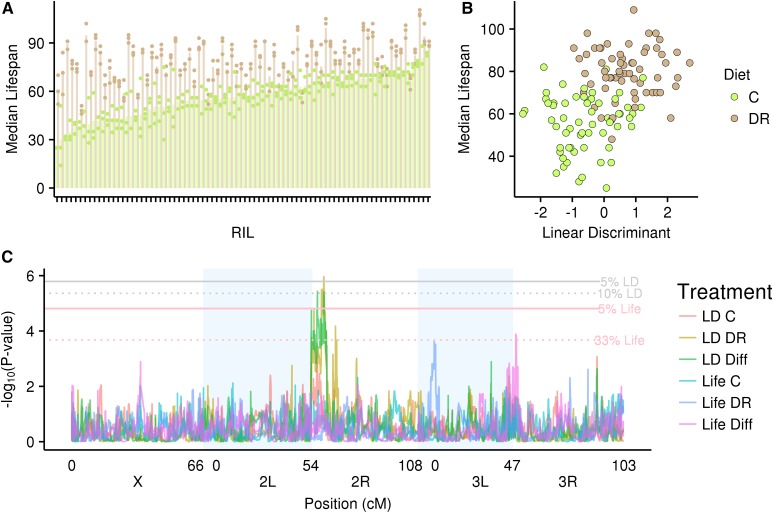
When pooled over treatments, RILs in the DR diet lived 37.5% longer at 50% survivorship compared to the C diet (77 vs. 56 days) (Figure 3A; Figure S2 in Supplementary Material R1 in File S2). There was a highly significant effect of diet (F1,318 = 1562.1, P < 0.001) and a highly significant diet-by-genotype interaction (F79,318 = 6.33, P < 0.001), indicating substantial variation among RILs in their response to diet. This variation is apparent both for median life span (Figure 3A) and for the age-specific survivorship curves of RILs on the DR vs. C diet (Figure S2 in Supplementary Material R1). Further, DR flies exhibited higher survivorship at all ages from about 20 days posteclosion onwards.
Figure 3.
(A) Median life span for each RIL in each diet. Each pair of bars represents the average median life span in each diet for each RIL. Each point is a single replicate. Different colors denote different diets. (B) Median life span vs. scores on the linear discriminant (LD; gene expression composite). Each point represents the mean for a single RIL/RIX. (C) Genome scans for median life span (1) within the C diet, (2) within the DR diet, and (3) between diets; and for the scores on the linear discriminant (1) within the C diet, (2) within the DR diet, and (3) between diets. Different colors denote the different genome scans. Horizontal lines denote the significance thresholds at different FDR values.
For 64 RILs/RIXs, we have both IIS/TOR expression measurements and median life span measurements. The correlation between these RIXs’ scores on the linear discriminant and the corresponding RILs’ median life span is high (r = 0.46; Figure 3B), producing a stronger signal than any one gene’s correlation with median life span (range of individual genes’ correlations with the linear discriminant: −0.27 to 0.21). To investigate the empirical significance of the correlation between median life span and the scores on the linear discriminant, we performed 10,000 permutations of our expression measures, permuting each set of 52 expression measures together to maintain the correlation structure. For each iteration, we then performed a DFA and computed the correlation between the scores of the linear discriminant and median life span (not permuted). The results of these permutations are shown in Figure S3 in Supplementary Material R1 and the associated empirical P-value is 0.001 (10 of 10,000 iterations resulted in correlations as high as or higher than our observed). This association indicates the cooccurrence of a holistic change in the expression of our set of genes (as indicated by the scores on the linear discriminant) and the dramatic change in life span between diets, but does not indicate direct causation. Additionally, the association between the linear discriminant and median life span does not hold within diet treatments (within DR: r = 0.01, P = 0.9; within C: r = 0.07, P = 0.6). However, we note that the range of variation in both median life span and the linear discriminant is reduced within diets, and with a single expression measurement per RIX per diet, technical noise may obscure the relationship within environments.
Quantitative trait loci
IIS/TOR gene expression:
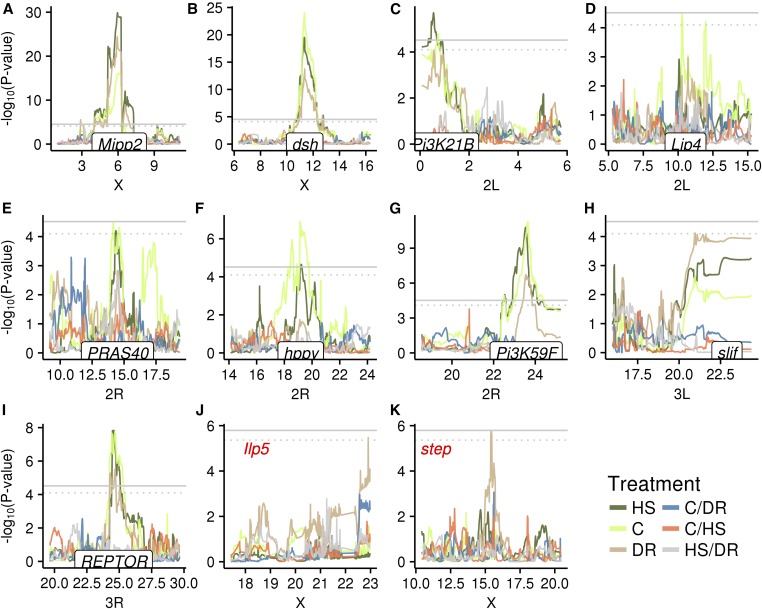
We identified cis (i.e., local) eQTL for six genes (dsh, hppy, Mipp2, Pi3K21B, Pi3K59F, and REPTOR) at an FDR of 5%, all of which are significant in multiple nutrient environments (Figure 4 and Table 1). An additional three genes (Lip4, PRAS40, and slif) had significant cis-eQTL at an FDR of 10%. While two of these additional cis peaks were only significant in a single diet, the trend in the other diets showed a similar pattern, suggesting strong concordance of gene expression in multiple diets (Figure 4, A–I). We also identified trans (i.e., distant) eQTL for two genes (Ilp5 and step) at an FDR of 10%, with both specific to a single nutrient environment (Figure 4, J and K, and Table 1). We did not map any eQTL, either cis or trans, for our measures of the difference in expression between environments at an FDR of 5 or 10%.
Figure 4.
Genome scans for gene expression measures within and between each diet at locations with significant QTL. Different colors denote different phenotypes. (A–I) cis-eQTLs show the location of the gene on the x-axis. (J–K) trans-eQTLs show the gene expression measure in red. Gray horizontal lines denote the significance threshold. Solid line, 5% FDR; dotted line, 10% FDR.
Table 1. Details of all identified QTL.
| Trait | Gene location | Treatment | Peak location | BCI | −log10(P-value) |
|---|---|---|---|---|---|
| Mipp2 | X: 6.08 | DR | X: 5.94 | 5.93–6.25 | 24.4 |
| Mipp2 | X: 6.08 | C | X: 6.15 | 5.91–5.95 | 16.1 |
| Mipp2 | X: 6.08 | HS | X: 5.94 | 5.91–6.24 | 29.9 |
| dsh | X: 11.35 | DR | X: 11.36 | 11.30–11.39 | 13.73 |
| dsh | X: 11.35 | C | X: 11.34 | 11.31–11.36 | 24.04 |
| dsh | X: 11.35 | HS | X: 11.34 | 11.28–11.36 | 19.53 |
| Pi3K21B | 2L: 0.30 | C | 2L: 0.75 | 0.07–0.94 | 4.80 |
| Pi3K21B | 2L: 0.30 | HS | 2L: 0.55 | 0.27–0.93 | 5.70 |
| Lip4 | 2L: 10.53 | C | 2L: 10.28 | 10.25–10.71 | 4.49a |
| hppy | 2R: 19.18 | C | 2R: 19.14 | 18.54–19.54 | 6.90 |
| hppy | 2R: 19.18 | HS | 2R: 19.19 | 18.90–20.22 | 4.65 |
| stepb | 2L: 21.74 | DR | X: 15.42 | 15.39–15.47 | 5.76a |
| PRAS40 | 2R: 14.48 | C | 2R: 14.15 | 13.98–14.78 | 4.53a |
| PRAS40 | 2R: 14.48 | HS | 2R: 14.36 | 14.27–14.61 | 4.20a |
| Pi3K59F | 2R: 23.56 | DR | 2R: 23.55 | 23.49–23.74 | 6.73 |
| Pi3K59F | 2R: 23.56 | C | 2R: 23.64 | 23.56–23.68 | 11.28 |
| Pi3K59F | 2R: 23.56 | HS | 2R: 23.54 | 23.47–23.60 | 10.79 |
| Ilp5b | 3L: 9.82 | DR | X: 22.91 | 22.60–22.98 | 5.47a |
| slif | 3L: 22.88 | DR | 3L: 20.98 | 3L 20.93–3R 3.46 | 4.74a |
| REPTOR | 3R: 24.56 | DR | 3R: 24.67 | 24.25–24.74 | 5.49 |
| REPTOR | 3R: 24.56 | C | 3R: 24.66 | 24.32–24.76 | 7.77 |
| REPTOR | 3R: 24.56 | HS | 3R: 24.60 | 24.46– 24.67 | 7.83 |
| Linear discriminant | — | DR | 2R: 9.41 | 6.12–9.63 | 5.96 |
| Linear discriminant | — | DR vs. C | 2R: 7.76 | 7.71–9.47 | 5.43c |
| Median life span | — | DR vs. C | 3R: 9.34 | 3L 17.35–3R 10.74 | 3.89c |
| Median life span | — | DR | 3L: 3.65 | 3.23–3.84 | 3.68c |
trans-eQTL (i.e., peaks at locations distant from gene location).
FDR = 10%.
FDR = 33%.
A powerful feature of the DSPR is the ability to estimate haplotype-specific effects at QTL peaks by estimating the mean phenotypic value for each set of RILs/RIXs harboring a given haplotype at the peak position. Here, and for all other QTL where we estimated haplotype means, we only considered haplotypes that were present in greater than three RIXs at a given position. For each gene with a significant eQTL, we estimated haplotype means within each diet and for the difference between each pair of diets at the peak position. When there were multiple significant peaks in different diets per gene, we chose the position with the highest −log10(P). Haplotype means were typically highly correlated for gene expression measured within each diet treatment (Table 2), particularly for strong cis-eQTLs, indicating the locus is influencing expression in a similar way in each diet. Correlations with between-diet gene expression estimates were generally lower, which was also reflected by the lack of colocalizing peaks for between-diet measures (Figure 4). We note that haplotype means correlations should be interpreted with caution as they consist of at most eight points when all haplotypes are represented at a position. Potential exceptions include suggestive signals for the HS vs. DR difference at the PRAS40 cis-eQTL, the C vs. DR difference at the Ilp5 trans-eQTL, and the C vs. DR difference at the step trans-eQTL (Figure 4, E and J).
Table 2. Correlations between haplotype means for each phenotype measured for a given gene with a significant eQTL.
| Gene ID | Within diet | Between diet | ||||
|---|---|---|---|---|---|---|
| DR | C | HS | DR vs. C | C vs. HS | DR vs. HS | |
| Mipp2 | * | 0.99 | 0.99 | 0.58 | 0.40 | 0.44 |
| dsh | 0.99 | * | 0.99 | 0.52 | 0.64 | 0.03 |
| Pi3K21B | * | 0.99 | 0.98 | 0.43 | 0.26 | 0.48 |
| Lip4 | 0.76 | * | 0.89 | 0.09 | 0.89 | 0.61 |
| PRAS40 | 0.24 | * | 0.52 | 0.62 | 0.53 | 0.64 |
| hppy | 0.58 | * | 0.73 | 0.42 | 0.76 | 0.31 |
| Pi3K59F | 0.93 | * | 0.81 | 0.50 | 0.33 | 0.15 |
| slif | 0.86 | 0.75 | * | 0.48 | 0.18 | 0.21 |
| REPTOR | * | 0.98 | 0.99 | 0.72 | 0.34 | 0.01 |
| Ilp5 | 0.99 | 0.93 | * | 0.82 | 0.74 | 0.78 |
| step | 0.64 | 0.82 | * | 0.61 | 0.69 | 0.89 |
For each gene, the * indicates the treatment with the highest peak location for a given gene. The correlations reported here for each gene are the correlations between the estimated haplotype means stemming from mapping expression within the * treatment and the estimated haplotype means stemming from mapping expression within and between (difference) all other treatments. All correlations include only haplotypes with at least three observations in our set of RIXs.
We searched the Bayesian credible intervals (BCIs) of our trans-eQTL to identify any obvious candidate genes that have evidence they are involved in IIS/TOR. We have no additional evidence for these candidate genes and are simply noting their presence in the intervals. The region associated with step contains just 12 genes. One of the genes located within the region, Shtd, has a known interaction with Diap1 (Tanaka-Matakatsu et al. 2007), which in turn has been shown to interact with Atg1 (Scott et al. 2007), foxo (Kanao et al. 2010), hppy (Resnik-Docampo and Celis 2011), and Myc (Levayer et al. 2015). There are 17 genes in the region associated with Ilp5. We found two genes, StnA and StnB, that have possible connections to the IIS/TOR network. Evidence suggests that both StnA and StnB are involved in synaptic transmission (Fergestad et al. 1999; Mohrmann et al. 2008). Stimson et al. (2001) indicated that StnA and StnB have associations with shi, a gene that is involved in numerous biological functions and interacts with Tor and slif (Hennig et al. 2006).
Linear discriminant:
We also mapped our composite linear discriminant and identified a single QTL within the DR diet at an FDR of 5%. This same QTL was also significant at an FDR of 10% for the difference in linear discriminant scores between diets (Figure 3C and Table 1). Given the relationship between the values within each diet and the calculated difference, it is not surprising to see correspondence between these measures. RIXs harboring the B2 haplotype at this position have higher scores on the linear discriminant on average, and show a larger difference with the control diet. RIXs harboring the B1 and B3 haplotypes have lower linear discriminant scores on average and show a smaller difference with the control diet, while the B6 and B8 haplotypes have intermediate values (Figure 5A). There is no obvious signal at this position among the genes identified as potentially diagnostic of diet, or for any other of our measured genes, either mapped within or between diets. One of our genes, RagC-D, is positioned within the peak interval (RagC-D location: 2R, 8.16 Mb; linear discriminant QTL BCI, 6.12–9.63 Mb). However, there is not strong evidence that RagC-D is primarily driving the signal at this QTL. There is not a significant cis-eQTL for RagC-D at this location, though there is a small signal at this position within the DR diet [peak −log10(P) = 2.5, peak position = 9.33 Mb]. Of our other measured genes, 12% have a peak this high or higher within this interval. Additionally, RagC-D did not show a large fold change between the DR and C diets (0.99), nor did it have a high loading (−0.07) or standardized coefficient (−0.04) associated with the linear discriminant.
Figure 5.
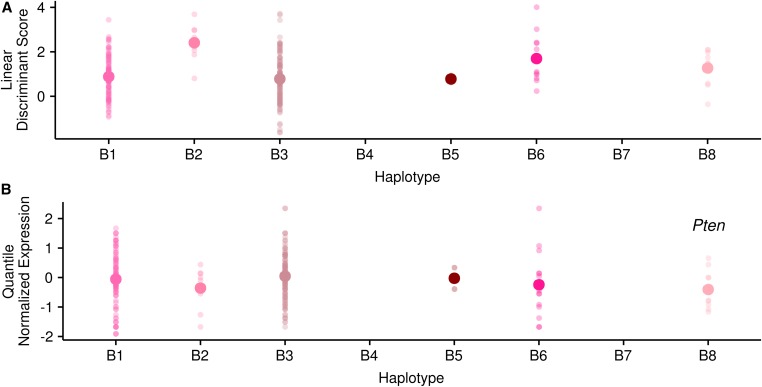
(A) Haplotype means for the linear discriminant at the linear discriminant QTL. (B) Haplotype means for Pten expression at the linear discriminant QTL. Each small point represents a single RIX. The single large point is the mean haplotype value.
Given that the linear discriminant is a composite of all our gene expression measures, we looked for corresponding patterns among haplotype means at the main DR peak for our set of genes within the DR diet. Overall, haplotype means for our gene expression measures in the DR treatment were highly correlated with the haplotype means for the linear discriminant at this position (average absolute value of correlations = 0.59). We also performed correlations between the haplotype means for the linear discriminant at the peak position and the haplotype means for our set of individual gene expression measures within the DR treatment at a set of 1000 randomly chosen positions to quantify the expected relationship between haplotype means stemming from chance alone. The mean average absolute value of correlations is lower (0.36) with 13% of positions greater than or equal to our observed value. The higher degree of concordance suggests a locus that influences the expression of many of our genes, which is perplexing in the face of the complete lack of QTL peaks among our genes in this region. For example, the haplotype means for Pten expression, one of our diagnostic genes, follow the same pattern as the haplotype means for the linear discriminant scores in the DR diet, however, the difference among the means is much less and thus there is not a significant peak at this position for Pten (Figure 5B). The linear discriminant peak was very wide (6.12–9.63 Mb), precluding a meaningful search for candidate genes in the interval in FlyBase.
Median life span:
No QTL were significant at an FDR of 5 or 10% for median life span; however, there were two peaks that were significant at an FDR of 33%, one within the DR treatment and one for the difference between treatments (Figure 3C and Table 1). To clarify the meaning of this threshold, for every set of genome scans for this set of three phenotypes (within C, within DR, and difference between), the expected number of false positives at this threshold is 0.65, and we identified 2. In our 1000 permutations, we identified two peaks 15% of the time. There was little correspondence between the median life span QTL map and the linear discriminant map (Figure 3C); there were no suggestive signals in the median life span QTL map at the linear discriminant peak and vice versa. The BCI for the peak for the difference in life span is quite wide (3L 17.35–3R 10.74 Mb) and spans the centromere. This effect is likely being driven by the presence of a second smaller peak on 3L. The 2-LOD drop is much narrower (3R 9.09–9.99 Mb). Several of our genes fall within the BCI for this peak (gig, tribbles, RagA-B, Rheb, slif, eIF-4B, and SREBP), but none fall within the 2-LOD drop. Of the genes in the BCI, only slif has a significant cis-eQTL, and slif also has a suggestive peak for the difference between DR and C in this region, which colocalizes with the life span peak [−log10(P) = 3.17, peak position = 10.20 Mb, BCI = 9.58–10.59 Mb]. slif was also among our diagnostic genes in our DFA and the correlation between the haplotype means for life span and slif at the life span peak position is moderate (r =0.48), and given the small sample size for life span and the differences in experimental design, we may not expect a high correspondence. None of our IIS/TOR genes are positioned in the peak identified within the DR diet, and no within DR eQTLs colocalize with this peak, even at a relaxed significance threshold. In addition, the peak for the difference between treatments overlaps with a QTL recently identified to affect life span in DSPR lines (Highfill et al. 2016) and a QTL identified using pooled sequencing of young and old flies in the DSPR B2 synthetic population (Burke et al. 2014). To assess if any additional genes occur in the narrower 2-LOD interval that have previously been linked with life span, we searched FlyBase (Attrill et al. 2016) for genes tagged with controlled vocabularies including “life span,” “aging,” “longevity,” and “lived.” We identified three genes under the QTL peak for the difference between the C and DR diet (Rel, pum, and Ras85D), and four genes under the DR peak (mir-282, Ide, sty, Strip, and PHGPx). Both Rel and pum were also identified as potential candidate genes in Highfill et al. (2016).
Discussion
Here, we have used a multiparent mapping panel, which allowed us to measure multiple related phenotypes in multiple environments on the same set of genotypes. Our study characterized natural patterns of diet-dependent changes in gene expression for the core components of the IIS/TOR pathway and took a multivariate approach to identify a novel locus that is associated with global expression changes. We linked these results to a second experiment using these same lines to explore the relationship between diet-induced life-span extension and the IIS/TOR pathway. Below, we discuss the relevance of our results to our three major questions.
How do IIS/TOR pathway genes respond to changes in diet?
Nearly all the genes we assayed showed differential expression between our diets. We note that different laboratories may use different diet recipes, temperatures, or photoperiods, all of which may influence IIS/TOR expression and complicate comparisons between this study and past or future studies. The changes in expression were generally small in magnitude, particularly between the DR and C diets. Somewhat unexpectedly, expression measures in the DR and HS diets trended in largely the same direction (Figure 2). One potential explanation for this effect is that expression is changing in response to the ratio of carbohydrate to protein. Relative to the C diet, both the DR diet and the HS diet have higher carbohydrate-to-protein ratios. Also consistent with this hypothesis is the fact that the HS carbohydrate-to-protein ratio is higher than the DR ratio, and expression changes tended to be greater in the HS diet. Additionally, Dobson et al. (2017) found HS diets early in adult life have the effect of inactivating foxo. Our results showing reduced expression of foxo and upregulation of many other IIS/TOR components are generally consistent with this result, particularly if the effect is general for diets with a high carbohydrate-to-protein ratio. Previous studies that have taken a “nutritional-geometry” approach support the idea that the ratio of nutrients is of critical importance (Lee et al. 2008; Behmer 2009; Piper et al. 2011; Tatar et al. 2014; Post and Tatar 2016).
In general, we observed small shifts in expression in many genes rather than a dramatic shift in a few key genes, which is not surprising for complex traits (Rockman 2012). We chose to measure overall expression changes using whole flies, with the implication being that we do not have tissue-specific expression information. If the changes in expression associated with diet are tissue specific with differing shifts in different tissues, our design would obscure this effect. In addition, some previous studies have found effects of IIS/TOR genes at the translational level rather than the transcriptional level (Zid et al. 2009), and it is possible there are post-transcriptional changes in IIS/TOR that are more dramatic than the changes we observed in transcription.
Many studies have shown that loss-of-function mutants in the insulin signaling pathway phenocopy the effects of DR (e.g., Kenyon et al. 1993; Clancy et al. 2001; Tatar et al. 2001), leading to the hypothesis that life-span extension is caused by the reduced insulin signaling associated with DR. However, direct evidence for the involvement of the IIS/TOR pathway in diet-dependent life span extension is still very unclear. Several studies have investigated whether IIS/TOR genes are required for the DR response and have found that while induced changes in expression in these genes often alter the response, no cases have been found where they eliminate the response (Min et al. 2008; Giannakou et al. 2008; Tatar 2011; Flatt 2014). We did not observe a strong signal indicative of reduced insulin signaling in the DR diet. For example, nearly all the Ilps had increased expression, with the exception of Ilp7. Previous studies have also found mixed evidence for reduced insulin signaling under DR. For example, Min et al. (2008) measured expression changes for the Ilps under control and DR conditions and found that only Ilp5 had increased expression. Post and Tatar (2016) measured expression of the Ilps and a few other key IIS genes in a nutritional geometry framework with many different diets and showed that each Ilp showed a unique response, with no trend toward a general reduction in expression on a low protein diet. In addition, the majority of studies use only a single genetic background to characterize expression changes, which may not apply generally across a population of genetically diverse individuals. Overall, there are relatively few studies characterizing typical changes in gene expression with diet among such individuals, leaving open the question of which genes show diet-dependent expression, the degree to which individuals vary in these changes, and what naturally occurring genetic variants influence these responses (Pletcher et al. 2002; Ding et al. 2014; Whitaker et al. 2014; Williams et al. 2015).
What is the source of natural genetic variation in the response of IIS/TOR pathway genes to diet?
Many previous studies of the genetic basis of gene expression have shown that cis (i.e., local) eQTL tend to have large effects on expression phenotypes (Gibson and Weir 2005; Gilad et al. 2008; Cookson et al. 2009; Ehrenreich et al. 2010; King et al. 2014). The majority of these studies have been done in a single environment and it is not known whether the response to the environment also tends to show strong cis-effects. There is some reason to expect that diet-dependent changes in the IIS/TOR pathway might be due largely to trans- (i.e., distant) rather than cis-eQTLs. The large effect loss-of-function genetic variants identified via classical genetic techniques described above are typically not segregating in natural populations, which is not surprising given the central role of the pathways involved (Van Voorhies et al. 2006). In addition, mapping studies and evolution experiments using natural populations have not independently identified these same genes as important contributors to natural genetic variation in traits such as life span (Remolina et al. 2012; Burke et al. 2014; Carnes et al. 2015). We identified primarily cis-eQTL mapped within diet treatments, confirming that at least some IIS/TOR components harbor strong cis-eQTLs. These cis-eQTLs showed very similar effects in each diet, and very little concordance with measures of the differences between environments (Figure 4 and Table 2), indicating they do not influence the response to the environment. Our two trans-eQTL were specific to a given diet and we hypothesize that the genetic basis of the response to the environment may stem largely from more subtle trans-effects. Unfortunately, these more subtle effects are more difficult to detect and we did not map any between diet eQTLs, making this a difficult hypothesis to confirm.
We used a multivariate approach, DFA, to take a more holistic approach to characterizing global changes in expression and successfully mapped a QTL influencing this composite expression measure (i.e., the linear discriminant; Figure 3C). Patterns among the haplotype means for individual genes at this peak support the hypothesis that this QTL influences the expression of multiple genes, though the effects on each individual gene are too small to be detected on their own.
What is the relationship between IIS/TOR expression and life-span extension under DR?
By comparing RIXs’ scores on the linear discriminant and their median life span, we were able to show an association between diet-dependent changes in IIS/TOR expression and life-span extension, though this association does not imply a direct relationship. At present we have no solid evidence beyond this association for a shared genetic basis between median life span and IIS/TOR expression. Our genome scans did not show colocalization of any QTL determining both phenotypes. However, we note that if there were shared QTL between these phenotypes, at our sample size and given the difference in experimental design, we should not be surprised by a lack of overlap, and thus this result should not be taken as evidence against a relationship (King and Long 2017). Recently, Paaby et al. (2014) demonstrated a link between a naturally occurring genetic variant in InR and several life history traits, including life span, though it is not known whether it is also related to the diet-dependent extension of life span. In addition, members of the IIS/TOR pathway show frequency changes with latitude, as do many life history traits (Kolaczkowski et al. 2011; Fabian et al. 2012). Future studies that take advantage of the naturally occurring genetic variation that exists in established mapping populations such as the DSPR to better characterize how gene expression and life span change in concert have the potential to uncover the mechanisms underlying diet-dependent life span extension.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.197780/-/DC1.
Acknowledgments
We thank Elizabeth Lo Presti, Vince Farinella, Anna Perinchery, and Maddie Taylor for help with fly rearing. Michelle Beckwith and Scott Reierstad provided technical assistance with the design and implementation of the OpenArray assays. Kristen Leach and David Braun aided with quantitative PCR assays. Anthony Long provided helpful advice about experimental design. This work was supported by National Institutes of Health grants F32 GM-099382 and R01 GM-117135 to E.G.K.
Footnotes
Communicating editor: S. F. Chenoweth
Literature Cited
- Attrill H., Falls K., Goodman J. L., Millburn G. H., Antonazzo G., et al. , 2016. FlyBase: establishing a gene group resource for Drosophila melanogaster. Nucleic Acids Res. 44: D786–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass T. M., Grandison R. C., Wong R., Martinez P., Partridge L., et al. , 2007. Optimization of dietary restriction protocols in Drosophila. J. Gerontol. A Biol. Sci. Med. Sci. 62: 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis W. D., 1998. QTL analyses: power, precision, and accuracy. Mol. Dissection Complex Traits 1998: 145–162. [Google Scholar]
- Behmer S. T., 2009. Insect herbivore nutrient regulation. Annu. Rev. Entomol. 54: 165–187. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B-Methodol. 57: 289–300. [Google Scholar]
- Bluher M., Kahn B., Kahn C., 2003. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299: 572–574. [DOI] [PubMed] [Google Scholar]
- Böhni R., Riesgo-Escovar J., Oldham S., Brogiolo W., Stocker H., et al. , 1999. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1–4. Cell 97: 865–875. [DOI] [PubMed] [Google Scholar]
- Britton J., Lockwood W., Li L., Cohen S., Edgar B., 2002. Drosophila’s insulin/P13-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2: 239–249. [DOI] [PubMed] [Google Scholar]
- Broman K. W., Sen S., 2009. A Guide to QTL Mapping with R/qtl. Springer, New York. [Google Scholar]
- Broughton S. J., Piper M. D. W., Ikeya T., Bass T. M., Jacobson J., et al. , 2005. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. USA 102: 3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browner W., Kahn A., Ziv E., Reiner A., Oshima J., et al. , 2004. The genetics of human longevity. Am. J. Med. 117: 851–860. [DOI] [PubMed] [Google Scholar]
- Brzyski D., Peterson C. B., Sobczyk P., Candès E. J., Bogdan M., et al. , 2017. Controlling the rate of GWAS false discoveries. Genetics 205: 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke M. K., King E. G., Shahrestani P., Rose M. R., Long A. D., 2014. Genome-wide association study of extreme longevity in Drosophila melanogaster. Genome Biol. Evol. 6: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes M. U., Campbell T., Huang W., Butler D. G., Carbone M. A., et al. , 2015. The genomic basis of postponed senescence in Drosophila melanogaster. PLoS One 10: e0138569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Storey J. D., 2006. Relaxed significance criteria for linkage analysis. Genetics 173: 2371–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy D. J., Gems D., Harshman L. G., Oldham S., Stocker H., et al. , 2001. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292: 104–106. [DOI] [PubMed] [Google Scholar]
- Cookson W., Liang L., Abecasis G. R., Moffatt M., Lathrop M., 2009. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 10: 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F., Gil M. P., Franklin M., Ferreira J., Tatar M., et al. , 2014. Transcriptional response to dietary restriction in Drosophila melanogaster. J. Insect Physiol. 69: 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A. J., Ezcurra M., Flanagan C. E., Summerfield A. C., Piper M. D. W., et al. , 2017. Nutritional programming of lifespan by FOXO inhibition on sugar-rich diets. Cell Rep. 18: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E., 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I. M., Gerke J. P., Kruglyak L., 2010. Genetic dissection of complex traits in yeast: insights from studies of gene expression and other phenotypes in the BYxRM cross. Cold Spring Harb. Symp. Quant. Biol. 74: 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian D. K., Kapun M., Nolte V., Kofler R., Schmidt P. S., et al. , 2012. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol. Ecol. 21: 4748–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergestad T., Davis W. S., Broadie K., 1999. The stoned proteins regulate synaptic vesicle recycling in the presynaptic terminal. J. Neurosci. 19: 5847–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer B., Taborsky B., Dieckmann U., 2009. Unexpected patterns of plastic energy allocation in stochastic environments. Am. Nat. 173: E108–E120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer B., Dieckmann U., Taborsky B., 2010. When to store energy in a stochastic environment. Evolution 65: 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T., 2014. Plasticity of lifespan: a reaction norm perspective. Proc. Nutr. Soc. 73: 532–542. [DOI] [PubMed] [Google Scholar]
- Flatt T., Schmidt P. S., 2009. Integrating evolutionary and molecular genetics of aging. Biochim. Biophys. Acta 1790: 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T., Tu M., Tatar M., 2005. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. BioEssays 27: 999–1010. [DOI] [PubMed] [Google Scholar]
- Garofalo R., 2002. Genetic analysis of insulin signaling in Drosophila. Trends Endocrinol. Metab. 13: 156–162. [DOI] [PubMed] [Google Scholar]
- Geminard C., Rulifson E. J., Leopold P., 2009. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 10: 199–207. [DOI] [PubMed] [Google Scholar]
- Giannakou M. E., Partridge L., 2007. Role of insulin-like signalling in Drosophila lifespan. Trends Biochem. Sci. 32: 180–188. [DOI] [PubMed] [Google Scholar]
- Giannakou M. E., Goss M., Partridge L., 2008. Role of dFOXO in lifespan extension by dietary restriction in Drosophila melanogaster: not required, but its activity modulates the response. Aging Cell 7: 187–198. [DOI] [PubMed] [Google Scholar]
- Gibson G., Weir B., 2005. The quantitative genetics of transcription. Trends Genet. 21: 616–623. [DOI] [PubMed] [Google Scholar]
- Gilad Y., Rifkin S. A., Pritchard J. K., 2008. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 24: 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goberdhan D., Wilson C., 2003. The functions of insulin signaling: size isn’t everything, even in Drosophila. Differentiation 71: 375–397. [DOI] [PubMed] [Google Scholar]
- Hennig K. M., Colombani J., Neufeld T. P., 2006. TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J. Cell Biol. 173: 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highfill C. A., Reeves G. A., Macdonald S. J., 2016. Genetic analysis of variation in lifespan using a multiparental advanced intercross Drosophila mapping population. BMC Genet. 17: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K., Reynolds R., 2005. Evolutionary and mechanistic theories of aging. Annu. Rev. Entomol. 50: 421–445. [DOI] [PubMed] [Google Scholar]
- Kaletsky R., Murphy C. T., 2010. The role of insulin/IGF-like signaling in C. elegans longevity and aging. Dis. Model. Mech. 3: 415–419. [DOI] [PubMed] [Google Scholar]
- Kanao T., Venderova K., Park D. S., Unterman T., Lu B., et al. , 2010. Activation of FoxO by LRRK2 induces expression of proapoptotic proteins and alters survival of postmitotic dopaminergic neuron in Drosophila. Hum. Mol. Genet. 19: 3747–3758. [DOI] [PubMed] [Google Scholar]
- Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R., 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- King, E. G., and A. D. Long, 2017 The Beavis effect in next-generation mapping panels in Drosophila melanogaster. G3 (Bethesda) 7: 1643–1652. [DOI] [PMC free article] [PubMed]
- King E. G., Macdonald S. J., Long A. D., 2012a Properties and power of the Drosophila synthetic population resource for the routine dissection of complex traits. Genetics 191: 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Merkes C. M., McNeil C. L., Hoofer S. R., Sen S., et al. , 2012b Genetic dissection of a model complex trait using the Drosophila synthetic population resource. Genome Res. 22: 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Sanderson B. J., McNeil C. L., Long A. D., Macdonald S. J., 2014. Genetic dissection of the Drosophila melanogaster female head transcriptome reveals widespread allelic heterogeneity. PLoS Genet. 10: e1004322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood T. B. L., Shanley D. P., 2005. Food restriction, evolution and ageing. Mech. Ageing Dev. 126: 1011–1016. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B., Kern A. D., Holloway A. K., Begun D. J., 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187: 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. P., Simpson S. J., Clissold F. J., Brooks R., Ballard J. W. O., et al. , 2008. Lifespan and reproduction in Drosophila: new insights from nutritional geometry. Proc. Natl. Acad. Sci. USA 105: 2498–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J. T., Storey J. D., 2007. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3: 1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J. T., Scharpf R. B., Bravo H. C., Simcha D., Langmead B., et al. , 2010. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 11: 733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J. T., Johnson W. E., Parker H. S., Jaffe A. E., Storey J. D., 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28: 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J. T., Johnson W. E., Parker H. S., Fertig E. J., Jaffe A. E., et al. , 2015. sva: Surrogate Variable Analysis. R package version 3.18.0. [Google Scholar]
- Levayer R., Hauert B., Moreno E., 2015. Cell mixing induced by myc is required for competitive tissue invasion and destruction. Nature 524: 476–480. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Long A. D., Macdonald S. J., King E. G., 2014. Dissecting complex traits using the Drosophila synthetic population resource. Trends Genet. 30: 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecham B. H., Nelson P. S., Storey J. D., 2010. Supervised normalization of microarrays. Bioinformatics 26: 1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min K.-J., Yamamoto R., Buch S., Pankratz M., Tatar M., 2008. Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell 7: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrmann R., Matthies H. J., Woodruff E., III, Broadie K., 2008. Stoned B mediates sorting of integral synaptic vesicle proteins. Neuroscience 153: 1048–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel D. R., Liu Y., Luo J., 2015. Insulin/IGF signaling and its regulation in Drosophila. Gen. Comp. Endocrinol. 221: 255–266. [DOI] [PubMed] [Google Scholar]
- Neel J., 1962. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am. J. Hum. Genet. 14: 353–362. [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., Bergland A. O., Behrman E. L., Schmidt P. S., 2014. A highly pleiotropic amino acid polymorphism in the Drosophila insulin receptor contributes to life-history adaptation. Evolution 68: 3395–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L., Alic N., Bjedov I., Piper M. D. W., 2010. Ageing in Drosophila: the role of the insulin/Igf and TOR signalling network. Exp. Gerontol. 46: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., Marioni J. C., Pai A. A., Degner J. F., Engelhardt B. E., et al. , 2010. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464: 768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M. D. W., Partridge L., Raubenheimer D., Simpson S. J., 2011. Dietary restriction and aging: a unifying perspective. Cell Metab. 14: 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher S., Macdonald S. J., Marguerie R., Certa U., Stearns S., et al. , 2002. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 12: 712–723. [DOI] [PubMed] [Google Scholar]
- Post S., Tatar M., 2016. Nutritional geometric profiles of insulin/IGF expression in Drosophila melanogaster. PLoS One 11: e0155628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2016. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: https://www.R-project.org/. [Google Scholar]
- Reed L. K., Williams S., Springston M., Brown J., Freeman K., et al. , 2010. Genotype-by-diet interactions drive metabolic phenotype variation in Drosophila melanogaster. Genetics 185: 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. K., Lee K., Zhang Z., Rashid L., Poe A., et al. , 2014. Systems genomics of metabolic phenotypes in wild-type Drosophila melanogaster. Genetics 197: 781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remolina S. C., Chang P. L., Leips J., Nuzhdin S. V., Hughes K. A., 2012. Genomic basis of aging and life-history evolution in Drosophila melanogaster. Evolution 66: 3390–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnik-Docampo M., de Celis J. F., 2011. MAP4K3 is a component of the TORC1 signalling complex that modulates cell growth and viability in Drosophila melanogaster. PLoS One 6: e14528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., 2012. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution 66: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. C., Juhász G., Neufeld T. P., 2007. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund D. O., Zhang N. R., Yakir B., 2011. False discovery rate for scanning statistics. Biometrika 98: 979–985. [Google Scholar]
- Skorupa D. A., Dervisefendic A., Zwiener J., Pletcher S. D., 2008. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell 7: 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal R., Weindruch R., 1996. Oxidative stress, caloric restriction, and aging. Science 273: 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimson D. T., Estes P. S., Rao S., Krishnan K. S., Kelly L. E., et al. , 2001. Drosophila stoned proteins regulate the rate and fidelity of synaptic vesicle internalization. J. Neurosci. 21: 3034–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M., Thomas B. J., Du W., 2007. Mutation of the Apc1 homologue shattered disrupts normal eye development by disrupting G1 cell cycle arrest and progression through mitosis. Dev. Biol. 309: 222–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M., 2011. The plate half-full: status of research on the mechanisms of dietary restriction in Drosophila melanogaster. Exp. Gerontol. 46: 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M., Kopelman A., Epstein D., Tu M., Yin C., et al. , 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292: 107–110. [DOI] [PubMed] [Google Scholar]
- Tatar M., Bartke A., Antebi A., 2003. The endocrine regulation of aging by insulin-like signals. Science 299: 1346–1351. [DOI] [PubMed] [Google Scholar]
- Tatar M., Post S., Yu K., 2014. Nutrient control of Drosophila longevity. Trends Endocrinol. Metab. 25: 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman A. A., 2010. Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem. J. 425: 13–26. [DOI] [PubMed] [Google Scholar]
- Therneau T. M., Grambsch P. M., 2000. Modeling Survival Data: Extending the Cox Model. Springer, New York. [Google Scholar]
- Therneau T., 2015. A Package for Survival Analysis in S. version 2.38. Available at: https://CRAN.R-project.org/package=survival. [Google Scholar]
- Utz H. F., Melchinger A. E., 1994. Comparision of different approaches to interval mapping of quantitative trait loci, pp. 195–203 in Biometrics in Plant Breeding: Applications of Molecular Markers, edited by van Ooijen J. W., Jansen J. European Association for Research on Plant Breeding, CPRO-DLO, Wageningen, The Netherlands. [Google Scholar]
- Van Voorhies W. A., Curtsinger J. W., Rose M. R., 2006. Do longevity mutants always show trade-offs? Exp. Gerontol. 41: 1055–1058. [DOI] [PubMed] [Google Scholar]
- Venables W. N., Ripley B. D., 2002. Modern Applied Statistics with S. Fourth Edition. Springer, New York. [Google Scholar]
- Wells J. C. K., 2009. Thrift: a guide to thrifty genes, thrifty phenotypes and thrifty norms. Int. J. Obes. 33: 1331–1338. [DOI] [PubMed] [Google Scholar]
- Whitaker R., Gil M. P., Ding F., Tatar M., Helfand S. L., et al. , 2014. Dietary switch reveals fast coordinated gene expression changes in Drosophila melanogaster. Aging 6: 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S., Dew-Budd K., Davis K., Anderson J., Bishop R., et al. , 2015. Metabolomic and gene expression profiles exhibit modular genetic and dietary structure linking metabolic syndrome phenotypes in Drosophila. G3 (Bethesda) 5: 2817–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., 2003. Theoretical basis of the Beavis effect. Genetics 165: 2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Belawat P., Hafen E., Jan L. Y., Jan Y.-N., 2008. Drosophila egg-laying site selection as a system to study simple decision-making processes. Science 319: 1679–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zera A. J., Harshman L. G., Williams T. D., 2007. Evolutionary endocrinology: the developing synthesis between endocrinology and evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 38: 793–817. [Google Scholar]
- Zid B. M., Rogers A. N., Katewa S. D., Vargas M. A., Kolipinski M. C., et al. , 2009. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139: 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All DSPR lines are available at http://FlyRILs.org/RequestFlies. All data are available centrally at http://FlyRILs.org/Data in addition to the public archives noted below. Raw resequencing data of the founder lines is deposited in the NCBI SRA (http://www.ncbi.nlm.nih.gov/sra) under accession number SRA051316, and the RIL RAD genotyping data are available under accession number SRA051306. The founder genotype assignments from the hidden Markov model are available as two data packages in R (http://FlyRILs.org/Tools/Tutorial) and are available from the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.r5v40). Our raw and normalized expression measures (normalized by ΔCq only; see below for further preprocessing) have been deposited in NCBI’s GEO (Edgar et al. 2002) and are accessible through GEO series accession number GSE93117 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93117). Our raw daily mortality data are in Table S2 and is available at from Zenodo: https://doi.org/10.5281/zenodo.322462. Our processed median life span data are in Table S3. Our normalized, batch-corrected gene expression measures are given in Table S4. All genome scan results are in Table S5.