Abstract
Premise of the study:
To study pollination networks in a changing environment, we need accurate, high-throughput methods. Previous studies have shown that more highly resolved networks can be constructed by studying pollen loads taken from bees, relative to field observations. DNA metabarcoding potentially allows for faster and finer-scale taxonomic resolution of pollen compared to traditional approaches (e.g., light microscopy), but has not been applied to pollination networks.
Methods:
We sampled pollen from 38 bee species collected in Florida from sites differing in forest management. We isolated DNA from pollen mixtures and sequenced rbcL and ITS2 gene regions from all mixtures in a single run on the Illumina MiSeq platform. We identified species from sequence data using comprehensive rbcL and ITS2 databases.
Results:
We successfully built a proof-of-concept quantitative pollination network using pollen metabarcoding.
Discussion:
Our work underscores that pollen metabarcoding is not quantitative but that quantitative networks can be constructed based on the number of interacting individuals. Due to the frequency of contamination and false positive reads, isolation and PCR negative controls should be used in every reaction. DNA metabarcoding has advantages in efficiency and resolution over microscopic identification of pollen, and we expect that it will have broad utility for future studies of plant–pollinator interactions.
Keywords: DNA metabarcoding, ITS, palynology, plant–pollinator interactions, pollination networks, rbcL
Mutualistic networks, the full set of interactions between mutually beneficial species in an ecosystem, have been recognized as a key driver of the creation and maintenance of biodiversity (Thompson, 1994). Mutualistic networks include plant–seed disperser networks and plant–pollinator networks and have been found to have a consistent “nested” structure, where more specialist species interact with a subset of the species interacting with the more generalist species, across many habitat types, plant and disperser/pollinator taxa, and geographic areas (Bascompte et al., 2003). This structure may help make these networks robust to perturbations (Memmott et al., 2004; Allesina and Tang, 2012; Staniczenko et al., 2013), although the relationship between mutualistic network structure and stability is a subject of contentious debate (Allesina and Tang, 2012; James et al., 2012; Staniczenko et al., 2013). It is important to better understand the structure and dynamics of pollination networks, particularly empirical networks (as opposed to models), to prevent their degradation or collapse (e.g., Lever et al., 2014) due to anthropogenic stressors (Potts et al., 2010; Ferreira et al., 2013).
Pollination networks are typically characterized by plant-focused visitation observations where pollinator visits to a single plant individual or to a defined area are observed and the pollinators are captured for finer-scale taxonomic classification. This method works well but is relatively inefficient because observers must wait for pollinators to arrive, limiting the number of interactions that can be characterized. Alternatively, pollination interaction networks focus on pollinators rather than on plants, given that each individual pollinator carries pollen grains that yield a record of the plants that a pollinator has visited. Bosch et al. (2009) were the first to try this approach, using visual microscopic identification of pollen, and compared it explicitly with plant-focused visitation surveys. They found that networks constructed from pollen carriage data were better characterized than those from plant visitation data; pollen carriage identified more interactions in the network, leading to higher connectance and fewer specialist plants and pollinators. Furthermore, better characterization allowed for the identification of clear phenological modules in the network that were not apparent from the plant visitation data. This approach was subsequently used by Burkle et al. (2013) to compare historical to contemporary networks, given that pollen carried by insect specimens is often maintained in insect collections.
While network characterization with pollen carriage clearly has advantages, it has not been widely used, partly because the identification of pollen via visual microscopy has a number of drawbacks. It requires highly specialized expertise, is time-consuming, and is often of low taxonomic resolution (in many or most cases pollen grains can only be identified to family or genus) (Bell et al., 2016). In addition, pollen is often only identified from a subsample of the available grains in a given pollen load, and thus may fail to detect rarer taxa (Bosch et al., 2009; Bell et al., 2016). Among other issues, this could potentially make networks appear to be less connected and less nested than they truly are, given that rare plant and pollinator species tend to interact with common, generalist species of the other group (e.g., Blüthgen, 2010).
A potential alternative methodology for the construction of pollen-carriage-based pollination networks involves the DNA barcoding of pollen (Keller et al., 2015; Sickel et al., 2015; Bell et al., 2016). DNA barcoding is the use of small regions of DNA that have a “barcoding gap,” defined as low intraspecific variation but high interspecific variation, to identify the species composition of a sample (Hebert et al., 2003). DNA metabarcoding is based on pollen-carriage-based methods but identifies multiple species in mixed-species samples using high-throughput sequencing. Pollen DNA metabarcoding offers a number of potential advantages over visual identification of pollen, including much greater speed; more comprehensive characterization of all grains in a sample; use of methods and equipment that are standard in any basic molecular biology laboratory (as opposed to the highly specialized knowledge needed to identify pollen visually); and higher taxonomic resolution, with 70–90% or more of plant taxa identified to the species level (CBOL Plant Working Group, 2009; Chen et al., 2010). This method, however, is still relatively new and, although the potential use in pollination networks has been discussed (Pornon et al., 2016), pollen DNA barcoding has not been widely used to characterize pollination networks.
Here we develop and describe basic methodology for pollination network characterization via DNA metabarcoding of pollen carriage. A key development is altering standard methods for often very small pollen loads carried by insects by maximizing the quantity of DNA extraction added to the PCR reaction and increasing the number of PCR cycles. We used pollinators sampled from a study of managed forests in Florida, USA, to show proof-of-concept of this method. Using DNA metabarcoding, we successfully identified mixed-species pollen loads collected from bees and used this information to construct a pollinator network. We compared our results to past research on pollinator network construction and have highlighted several areas for future work.
MATERIALS AND METHODS
Bee sampling and identification
Our aim in this work was to show proof-of-concept of using DNA metabarcoding to construct a pollination network, rather than to exhaustively document a particular network. The specimens we used consist only of bees, and thus do not represent a taxonomically complete pollination network from the flower-visitor perspective. The bee specimens we used came from a larger project investigating the impacts of biofuel production on bee communities across the southeastern USA. Here, we sampled pollen from bees collected in Florida from late April to early July 2014. We used bees from 13 field sites representing seven land use types (Appendix 1). We sampled bees via aerial netting along 200 × 2-m transects. We conducted aerial net sampling between the hours of 10:00 a.m. and 11:00 a.m. We collected each bee specimen into its own separate ethyl acetate kill jar to prevent pollen cross-contamination between specimens. We identified pinned bee specimens using Michener (2007) and Discover Life keys (http://www.discoverlife.org). A complete list of bee specimens is presented in Appendix S1 (53.5KB, docx) .
Pollen collection from bees
We extracted pollen from the bees by placing them each in a separate microcentrifuge tube containing water and a small amount (approximately 2 g/L) of liquid dish soap and vortexing the tube repeatedly until no pollen visibly remained on the bee. We examined bees under a stereomicroscope to ensure that the bulk of visible pollen was removed, and continued this process until any pollen remaining on the bee was considered negligible. We removed the bee (to be pinned and identified later) and then centrifuged the microcentrifuge tube containing the suspended pollen until the supernatant was clear or a definite pellet had formed at the bottom of the tube. We removed the supernatant and stored the pollen pellet at −20°C.
DNA isolations
We isolated the DNA from the majority of the pollen samples in April–July 2016 using the Macherey-Nagel NucleoSpin Food kit (Macherey-Nagel, Düren, North Rhine-Westphalia, Germany). A minority of pollen samples (marked with an asterisk in Appendix S1 (53.5KB, docx) ) were isolated in February–March 2015 using the MP Biomedicals FastDNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, California, USA). We used the Machery-Nagel kit for the majority of samples, because we found it to have higher yields. The subset of samples isolated with the MP Biomedicals kit was representative of all bee species, habitat types, and localities, so the change in DNA isolation method is unlikely to bias our results. When isolating DNA with the Macherey-Nagel NucleoSpin Food kit, we followed the “isolation of genomic DNA from honey or pollen” supplementary protocol designed to isolate genomic DNA from 10 g honey or a small pellet of pollen. The homogenization step was conducted in a Mini-BeadBeater-96 (BioSpec Products, Bartlesville, Oklahoma, USA) for 2 min, following the addition of Buffer CF from the NucleoSpin Food kit (Macherey-Nagel). To prevent tube breakage, we added the buffer prior to homogenization, rather than after, which increased the volume of liquid in the tubes during the homogenization step. Equivalent changes were also made to the protocol from the MP Biomedicals FastDNA SPIN Kit for Soil. We included “isolation negative” controls in each batch using water in place of a pollen sample.
PCR and sequencing
We used the dual-indexing pollen DNA metabarcoding strategy of Sickel et al. (2015), which is based on a method developed for the mixed-amplicon sequencing of bacterial 16S rRNA (Kozich et al., 2013). In addition to the ITS2 marker used by Sickel et al. (2015), we also included one of the standard DNA barcoding markers, part of the rbcL gene region of the chloroplast genome (CBOL Plant Working Group, 2009). For ITS2 amplification, we used the primers of Sickel et al. (2015). For rbcL, we used existing universal rbcL primers, rbcL2 (Palmieri et al., 2009) and rbcLaR (Kress and Erickson, 2007), which bind near the 5′ end of the rbcL gene and the middle of the rbcL gene, respectively. All primers were appended with MiSeq-specific adapters and Illumina index sequences (Kozich et al., 2013; Sickel et al., 2015). The forward primers included the index sequences SA501–SA508 and SB501–SB508, while the reverse primers included the index sequences SA701–SA712 and SB701–706, allowing for up to 288 possible primer combinations.
PCR was conducted as a duplex, combining both gene regions into a single PCR reaction. The PCR reactions contained 12.5 μL of KAPA HiFi ReadyMix (KAPA Biosystems, Boston, Massachusetts, USA), 0.25 μL of each primer (20 μM), and 11.5 μL of the template DNA (up to 20 ng/μL) for a total volume of 25 μL per reaction. To increase the chance of detecting all species in the mixture, each DNA extraction was included in three PCR reactions that were amplified separately, i.e., the reaction contents described above were divided between three PCR tubes. PCR cycles included an initial period of heat activation for 3 min at 95°C; followed by 35 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C; followed by a final extension of 10 min at 72°C and then held at 10°C. To investigate the effects of increased PCR cycles, which may be necessary to amplify small pollen samples, we performed additional reactions with 25 or 40 PCR cycles for a subset of three samples, each divided into three reactions as described above, covering a range of DNA concentrations (FL796 with a concentration of 2 ng/μL, FL765 with 17.6 ng/μL, and FL179 with 33.9 ng/μL). Reactions that were previously divided into three were recombined, and the success of the PCR reactions was confirmed using agarose gel electrophoresis. We included a PCR negative control for each batch of PCR, in addition to the “isolation negative” control described in the previous section. The PCR products were purified using Agencourt AMPure XP magnetic beads (Beckman Coulter, Danvers, Massachusetts, USA). At the Georgia Genomics Facility (Athens, Georgia, USA), DNA was quantified for each recombined and purified reaction, then an appropriate amount of each of these reactions was added to a pooled mixture so that the DNA quantities coming from each reaction were equal (except the negative controls from which the maximum volume of any pooled PCR product was added). The pooled mixture was sequenced at the Georgia Genomics Facility in a single flow cell on a 600-cycle run of the Illumina MiSeq instrument (Illumina, San Diego, California, USA). Sequence data have been deposited in the National Center for Biotechnology Information (NCBI; project number PRJNA344894).
Bioinformatics pipeline
We used the bioinformatics pipeline of Sickel et al. (2015) for species identification. This pipeline includes steps for joining forward and reverse reads using QIIME version 1.8.0 (Caporaso et al., 2010), removing low-quality reads with USEARCH version 8.0.1477 (Edgar, 2010), and taxonomic classification using Ribosomal Database Project (RDP) Classifier (Wang et al., 2007), separately for each marker. For ITS2 taxonomic classifications, we used the previously trained database for RDP Classifier described in Sickel et al. (2015). For taxonomic classifications using rbcL, we used the database described in Bell et al. (2017), currently available at https://github.com/KarenBell/rbcL-dual-index-metabarcoding. We used our negative controls to set threshold values for sequence removals; any taxonomic classifications recorded from fewer reads than the maximum read number we obtained from a negative control were considered to be either sequencing errors or sample contamination occurring in the DNA isolation, PCR, or sequencing stages, and were removed from further analysis. This may have the unintended consequence of removing any rare species from a particular pollen sample, favoring the species that an individual bee has been more frequently visiting. However, we decided that this was preferable to unintentionally including species that the bee had not actually visited.
Network construction and analysis
We constructed a pollination network by matching bee specimens to the plant taxa present in their pollen loads (from the bioinformatics pipeline) and pooling interactions within plant and bee taxa for the ITS2-based taxonomic classification to generate a quantitative network. This approach of using presence-absence data at the level of the individual pollen load and pooling data among individuals to build quantitative networks is also the typical approach with networks based on visual identification of pollen loads (e.g., Bosch et al., 2009). We did not use the combined rbcL and ITS2 classifications for this analysis because they did not give identical species-level results, and there is no straightforward method for combining the two separate data sets (although such conjoining is an interesting area for future research). We chose to use the ITS2 data, as this marker has greater coverage in its reference database and its taxonomic resolution is higher. We plotted the subsequent network and calculated a series of standard bipartite network metrics using the “bipartite” package (Dormann et al., 2009) for the R statistical programming language (R Core Team, 2016). We assessed the statistical significance of nestedness (measured as temperature), weighted NODF (nestedness metric based on overlap and decreasing fill), and H2′ using permutation tests with 9999 null models generated by randomly swapping matrix values, keeping both connectance and marginal row and column sums equivalent to those in the observed empirical network (Vázquez and Aizen, 2003).
RESULTS
Pollen DNA barcoding
Species identification
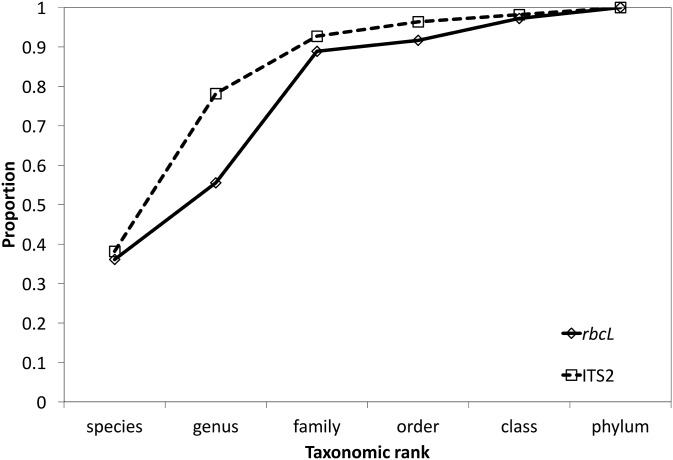
After joining and filtering raw data from Illumina MiSeq, we obtained 4,786,075 total sequencing reads. Of these, analysis using RDP Classifier identified 3,174,999 (66.3%) as ITS2 and 1,565,373 (32.7%) as rbcL, leaving 45,703 (1.0%) unable to be classified with either marker. With ITS2, most identifications were to the level of species (40% of taxa) or genus (38% of taxa), whereas with rbcL, only 55% of taxa were identified to genus or higher, with many more family-level identifications (33%; Fig. 1; Appendices S2 (914.3KB, xlsx) , S3 (1.4MB, xlsx) ). Overall, the most commonly identified taxon from ITS2 sequencing reads was a family-level identification of Cleomaceae (24% of total reads). Other common identifications were to the family Fabaceae, the genus Hypericum L. (Hypericaceae), the genus Ilex L. (Aquifoliaceae), and the genus Callicarpa L. (Lamiaceae). The most commonly identified plant taxon from rbcL data was a family-level identification of Fabaceae (13% of total reads). Other common identifications were to the species Haemodorum laxum R. Br. (Haemodoraceae), the family Lamiaceae, the genus Polanisia Raf. (Cleomaceae), and the genus Ilex (Aquifoliaceae). We did not identify any species in our samples that were solely dependent on wind pollination.
Fig. 1.
Proportions of classified taxa (i.e., those identified at least to phylum) identifiable to each taxonomic rank from DNA metabarcoding with rbcL and ITS2.
False positives
Negative control samples did not amplify enough PCR product to visualize using agarose gel electrophoresis. Sequencing of negative control samples yielded between 21 and 936 sequences identified as rbcL, between 42 and 1124 ITS2 sequences, and two to 99 sequences unable to be classified with either marker (Appendix S4 (22.1KB, docx) ). On average, negative controls had 1.6% of the total number of reads in an average sample. Most of these sequencing reads were a subset of the frequently recorded species in pollen samples, and were likely from small quantities of cross-contamination (Appendices S5 (27.3KB, docx) , S6 (30.4KB, docx) ). The exceptions were Fagopyrum Mill. ITS2 sequences and Populus L. rbcL sequences, which were present in negative controls, but rarely occurred in pollen samples. Neither of these species were present in any samples above the false positive threshold, so they were not included in any analyses. A small number of taxa were identified from a greater number of sequencing reads in negative controls than in samples and may have been the result of contamination from airborne pollen or DNA within the laboratory. For rbcL these were Picea abies (L.) H. Karst, Chamaecrista fasciculata (Michx.) Greene, Richardia scabra L., Populus deltoides W. Bartram ex Marshall, Poa pratensis L., Passiflora serratodigitata L., Teucrium scorodonia L., Vitex trifolia L., and Prunus davidiana Carrière; genus-level identifications to Populus, Prunus L., and Fagopyrum; family-level identifications to Rutaceae and Salicaceae; and order-level identification to Gentianales. For ITS2 these were Broussonetia papyrifera (L.) Vent., Alnus incana (L.) Moench, Poa trivialis L., Morus alba L., Allium ramosum L., Zea mays L., Amaranthus hybridus L., Fagopyrum esculentum Moench, and Populus alba L.; genus-level identifications to Fagopyrum, Musa L., Amaranthus L., Allium L., Alnus Mill., and Broussonetia L’Hér. ex Vent.; and family-level identifications to Polygonaceae and Amaranthaceae.
Effect of PCR cycles on species identifications
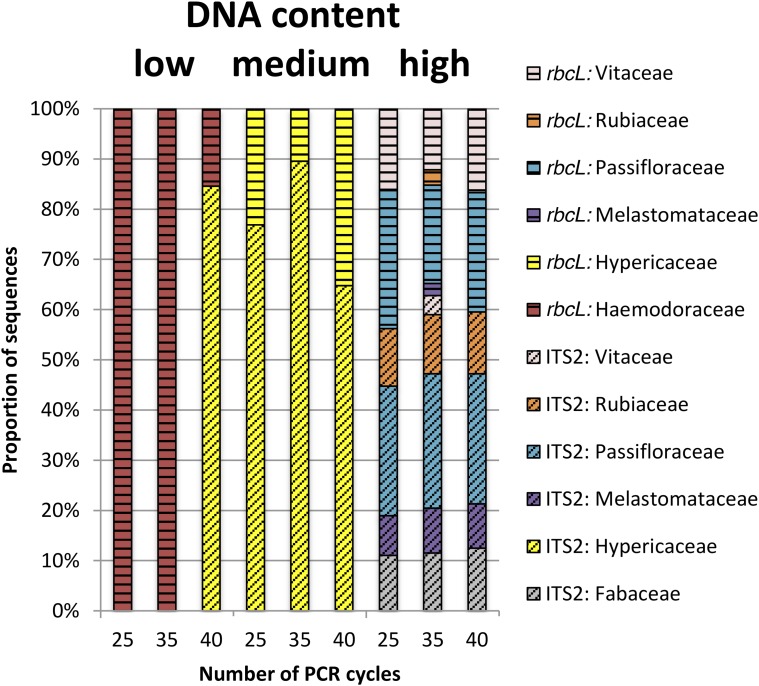
The impact of changing the number of PCR cycles on species-level identifications, proportions of species, and genetic markers was variable but small for most samples (Fig. 2). We found little effect of altering the number of PCR cycles for the sample with the highest DNA concentration (FL179). Increasing the number of PCR cycles for the sample having an intermediate DNA concentration (FL765) resulted in a higher proportion of ITS2 sequences compared to higher proportions of rbcL sequences detected when fewer PCR cycles were employed. However, for this sample, we only identified one family (Hypericaceae), so we cannot comment on the effects of the number of PCR cycles on inferred species composition or their relative proportions. By contrast, for the sample with the lowest DNA concentration (FL796), we detected only rbcL sequences of Haemodoraceae when the number of PCR cycles was 25 or 35, but when the number of cycles was increased to 40 we also detected ITS2 sequences of Hypericaceae.
Fig. 2.
Proportion of sequencing reads for family-level taxonomic identifications from samples FL179 (high DNA content), FL765 (moderate DNA content), and FL796 (low DNA content), with varying numbers of PCR cycles. Sequencing reads from rbcL are horizontally striped, sequencing reads from ITS2 are diagonally striped, and different families are represented in different colors.
Time and cost
For analysis of 90 pollen samples (+ six negative controls for isolation and PCR; combined into one 96-well plate) with rbcL and ITS2 in a duplex reaction, reagents cost approximately US$700 (excluding Illumina reagents), Illumina cost US$2300 (assuming PCR purification, DNA quantification, pooling, and sequencing are all outsourced to a sequencing facility), and approximately 42 h of labor were required (32 h for DNA isolation; 9 h for PCR; and 1 h for bioinformatics/pipeline loading, assuming that all required software has previously been installed). The minimum turnaround time from pollen samples to molecular identification following our protocol is 2 weeks, assuming two people working no more than 8-h days, five days a week; two PCR thermocyclers; overnight shipping to (or onsite availability of) the sequencing facility; no wait or delay at the sequencing facility; and a reasonably fast computer cluster for use in the bioinformatics analyses. This turnaround time increases with a single person, a single thermocycler, wait times at the sequencing facility, and use of a standard desktop computer for the bioinformatics analyses. Without labor, the cost is approximately US$30 per sample. Some savings could be achieved by using alternative reagents, although typically at the cost of additional labor. Further cost savings could be achieved by including more than 96 samples per Illumina flow cell. In the current study, we included 248 samples and 35 negative controls for two markers, and Sickel et al. (2015) used 384 samples with a single marker (ITS2). The practical limitations for multiplexing have not been assessed, but as the number of samples increases, the probability of detection of low-count pollen types decreases.
Pollinator network structure
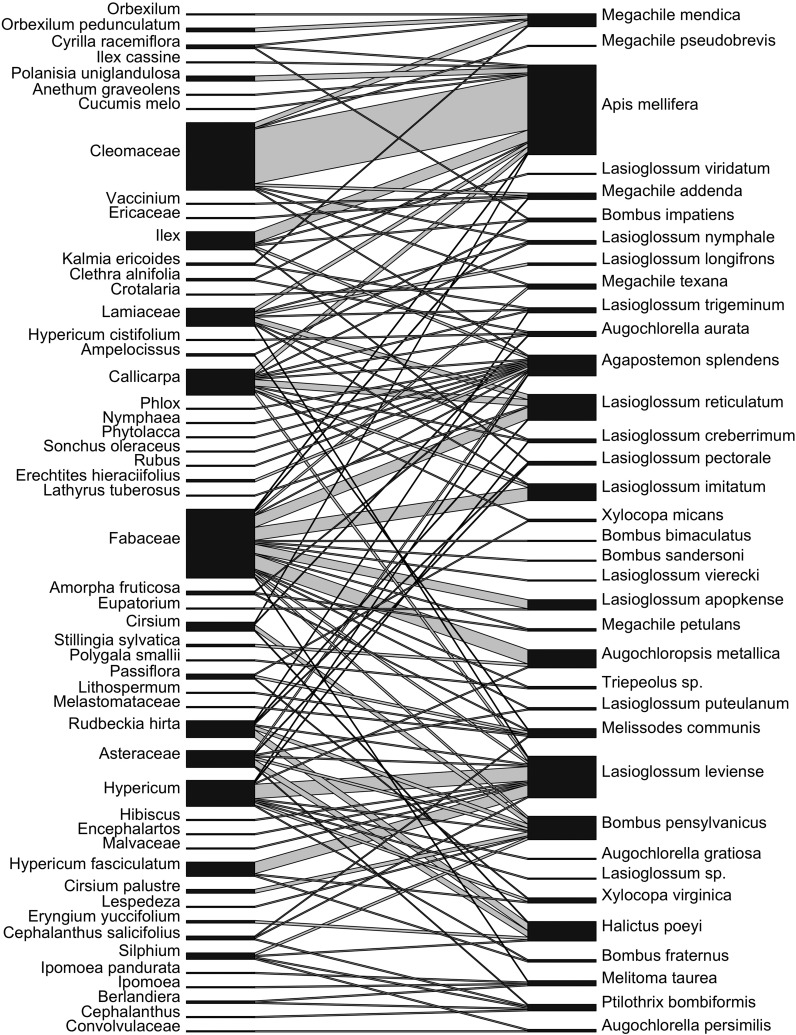
Because our aim was proof-of-concept of using DNA metabarcoding of pollen to construct pollination networks, we removed pollen only from bees and not from a taxonomically comprehensive set of flower visitors. The network metrics we report on should thus be considered to arise from one network module or subset, rather than an entire pollination network per se. From our Illumina sequence reads, we obtained a bipartite network with 37 pollinators and 51 plant taxa based on ITS2 taxonomic classification using RDP Classifier (Fig. 3). We report on network metrics in Table 1. Network connectance (realized proportion of possible links) was 0.069. Network “temperature” (a measure of nestedness) was 4.97, which was not statistically significant via permutation tests (P = 0.133). Weighted nestedness was 0.555, which is quite low compared to 25 empirical networks with a mean of 0.853 ± 0.047 SE (Bascompte et al., 2003). Weighted NODF was 7.372, which was statistically significant via permutation tests (P = 0.0001). H2′, a weighted measure of specialization, was 0.459 and statistically significant based on permutation tests (P = 0.000).
Fig. 3.
Bipartite pollination network of 37 bee specimens (top nodes) to the 51 plant taxa present in their pollen loads based on ITS2 taxonomic classification (bottom nodes). Interactions were pooled within plant and bee taxa. Links between plants and pollinators are represented with lines whose width is proportional to the number of interactions while the width of the nodes represents total abundance of that taxon across all of its interactions.
Table 1.
Summary metrics for the bipartite plant–pollinator network. Network analysis was completed in R with the package bipartite. Summary indices and values were calculated with the function networklevel. Detailed explanations of these indices are available in the literature (e.g., Dormann et al., 2009).
| Networklevel index or value | Value |
| Connectance | 0.069 |
| Web asymmetry | −0.146 |
| Links per species | 1.494 |
| No. of compartments | 1 |
| Compartment diversity | NA |
| Cluster coefficient | 0.059 |
| Nestedness | 4.968 |
| Weighted nestedness | 0.555 |
| Weighted NODF | 7.372 |
| Interaction strength asymmetry | −0.032 |
| Specialization asymmetry | 0.029 |
| Linkage density | 4.697 |
| Weighted connectance | 0.053 |
| Fisher alpha | 97.066 |
| Shannon diversity | 4.269 |
| Interaction evenness | 0.564 |
| Alatalo interaction evenness | 0.406 |
| H2′ | 0.459 |
| No. of pollinators | 37 |
| No. of plants | 51 |
| Mean no. of shared partners for pollinators | 0.512 |
| Mean no. of shared partners for plants | 0.263 |
| Cluster coefficient for pollinators | 0.143 |
| Cluster coefficient for plants | 0.206 |
| Weighted cluster coefficient for pollinators | 0.243 |
| Weighted cluster coefficient for plants | 0.287 |
| Niche overlap for pollinators | 0.156 |
| Niche overlap for plants | 0.083 |
| Togetherness for pollinators | 0.046 |
| Togetherness for plants | 0.021 |
| C-score for pollinators | 0.702 |
| C-score for plants | 0.817 |
| V ratio for pollinators | 3.053 |
| V ratio for plants | 4.317 |
| Discrepancy for pollinators | 88 |
| Discrepancy for plants | 83 |
| Extinction slope for pollinators | 2.421 |
| Extinction slope for plants | 1.575 |
| Robustness for pollinators | 0.701 |
| Robustness for plants | 0.612 |
| Functional complementarity for pollinators | 189.673 |
| Functional complementarity for plants | 198.561 |
| Partner diversity for pollinators | 1.37 |
| Partner diversity for plants | 1.289 |
| Generality for pollinators | 4.545 |
| Vulnerability for plants | 4.849 |
Note: NA = not available; NODF = nestedness metric based on overlap and decreasing fill.
DISCUSSION
We successfully identified mixed-species pollen loads collected from bees using DNA metabarcoding to show proof-of-concept for constructing quantitative pollination networks with molecular methods. Our work highlights several issues of particular interest, including areas for future work in many of them: comparison with traditional microscopic methods of pollen identification; variation in PCR cycle number; false positives; quantitation in DNA metabarcoding (i.e., relating sequence read numbers to pollen grain proportions); and differences in barcoding markers (rbcL vs. ITS2). As with all network studies, it is important to design the fieldwork to ensure successful pollinator network construction. Concerns such as which pollinator species to collect, whether to collect pollen from the pollen sac or the pollinator body, what time of day to collect, study site delimitation, and collecting methods are important, and are likely to depend on what questions are being addressed by the study. These are all worthwhile considerations, but are not limited to DNA metabarcoding, as they are also common considerations for network studies based on morphological identifications.
There may be situations where visual identification of pollen is more appropriate. This could be the case for small-scale studies, where only a small number of pollen grains need to be identified, eliminating any advantage of efficiency with DNA barcoding. If strict quantification of pollen grains on individual bees is required, then visual identification may be necessary. Overall though, our results suggest the strong utility of DNA metabarcoding as a method for constructing pollination networks via pollen carriage, and we expect that it will be increasingly used in pollination network studies in the future.
Comparison with microscopic pollen identification
DNA metabarcoding has a number of clear advantages compared with microscopic pollen identification. It is beyond the scope of this paper to quantitatively assess taxonomic resolution of the two methods, but such resolution is often limited to the family or genus level with microscopic identification (Rahl, 2008; Salmaki et al., 2008; Khansari et al., 2012). We identified 36% and 38% of taxa to species level with rbcL and ITS2, respectively. This level of success is remarkably high, given that currently only around 9% of known plant species are represented in the rbcL database, and 16% are represented in the ITS2 database (Bell et al., 2017). Previous studies have shown that, depending on the flora and the barcoding marker employed, 70–90% or more of taxa can be identified to the species level with barcoding when the barcode sequence for the species is known (Burgess et al., 2011; Hollingsworth et al., 2011). Certain lineages are likely to be better diagnosed with both DNA barcoding and morphological identification. Taxonomic resolution is also dependent on the presence of the species in databases for both DNA metabarcoding and visual identification. Certain geographic regions and plant groups are likely to be understudied, leading to a lack of representation in either DNA barcode or pollen morphology databases, or both. The latter should be considered to be temporary shortcomings of both DNA metabarcoding and visual pollen identification. Metabarcoding has a potentially faster turnaround time—around 12 days in a best-case scenario for 90 samples—which can be substantially longer in many contexts for microscopic identification. In terms of time allocation, microscopic identification involves essentially 100% hands-on time, whereas in metabarcoding, much of the time is hands-off (e.g., wait times during PCR, Illumina sequencing, and bioinformatics processing), during which researchers can work on other tasks. Metabarcoding also has the advantage that all pollen grains in a sample can be included in processing, in contrast to microscopic identification, in which often only a subset of pollen grains are identified due to the time costs involved (e.g., Bosch et al., 2009). Both DNA metabarcoding and microscopic pollen identification involve intensive training and learning, but the molecular methods involved in DNA metabarcoding are standard (DNA isolation, PCR) and are known by perhaps tens of thousands of people in the United States alone, whereas pollen identification training is highly specialized and known by perhaps a few dozen people in the United States. Furthermore, additional training is needed for each flora or geographic area from which pollen is to be identified using standard morphological techniques. Perhaps the major disadvantage of DNA metabarcoding is cost—approximately US$3000 with roughly 42 h of additional labor costs for a 96-well plate (90 samples plus negative controls). While this is a nontrivial cost, microscopic pollen identification likely costs more in some (and perhaps many) contexts depending on the cost of paying a trained palynologist, as well as factors affecting microscopic identification speed including individual variation in palynologists, diversity of plant community, training time for new plant communities, etc. One clear economic advantage of metabarcoding is the much higher predictability of time and sample costs relative to microscopic identification.
PCR cycles
Frequently pollen loads from bees, particularly those with small body size, contain only a small number of pollen grains, which can, in turn, lead to DNA templates of relatively low concentration following standard DNA isolation techniques. This may be particularly apparent if samples have been stored for many years before analysis, although we found no difference in the ability to amplify DNA from pollen stored for 2 yr at −80°C wet, dried, or in ethanol, or at room temperature dried or in ethanol (K.L.B., E.K.D., and B.B., unpublished data). Other types of pollinating insects may carry even smaller pollen loads, and to date these have not been tested with DNA metabarcoding. If pollen loads are very small, it may be necessary to combine pollen pellets from multiple individuals of the same pollinator species. However, this would prevent quantification based on the number of interacting individuals, and the data would need to be treated as presence-absence, unless it could be demonstrated that the read proportions were representative of pollen grain proportions. To obtain similar numbers of reads across samples, the relative quantity of DNA from these samples needs to be standardized. Although it may be plausible to increase the volume of DNA isolate added to the PCR reaction or increase the volume of PCR product added to the pooled sequencing reaction, these approaches are often limited by the total volume of the PCR or sequencing reaction, respectively. An alternative approach is to increase the number of PCR cycles to obtain more PCR product, although this also involves a set of potential problems. First, increasing the number of PCR cycles will increase the number of PCR products that are the result of contamination and PCR artifacts (Pinto and Raskin, 2012; Taberlet et al., 2012). One pollen-focused DNA metabarcoding study (Valentini et al., 2009) recommended using no more than 35 PCR cycles for these reasons. Based on our results, however, this did not appear to be a problem in our study; an increase in PCR cycles did not increase the number of reads allocated to species that were more common in negative controls, nor did it increase the number of reads that were unable to be classified. This may be due to our relatively conservative contamination threshold. A second potential problem is that the relative proportions of reads allocated to different species or different markers may change with the number of PCR cycles, due to the nonlinear nature of amplification. We found little effect of PCR cycle number on the proportions of sequencing reads in our sample that had the highest DNA concentration (FL179). For samples of moderate and low DNA concentration (FL765 and FL796), however, the number of PCR cycles did affect both the proportion of sequences assigned to different species (where multiple species were identified) and the proportion assigned to different markers. Notably, for the sample having the lowest DNA concentration (FL796), with 25 or 35 PCR cycles we detected only rbcL sequences of Haemodoraceae, but when the number of cycles was increased to 40, we also detected ITS2 sequences of Hypericaceae. This suggests either that Haemodoraceae is more readily amplified with rbcL primers than Hypericaceae is with ITS2 primers, or that Hypericaceae was present at a much lower quantity than Haemodoraceae, or that the copy number of Haemodoraceae rbcL is greater than the copy number of Hypericaceae ITS2. If the two loci were amplified separately, it is possible that both species would have been detected at a lower PCR cycle. Few other studies have explicitly tested the effect of the number of PCR cycles on species identification or quantification in a mixed-amplicon genetic species-identification analysis. A study of bacterial communities in the human gut found little effect on species identification and quantification when they increased the number of PCR cycles from 20 to 30 (Wu et al., 2010). However, they used 100 ng of DNA in their PCR reactions, a quantity that may not always be achievable for pollen DNA barcoding. While our results may simply be the result of the stochastic nature of PCR (and this could potentially be corrected by running more than triplicate PCR reactions for samples of low DNA concentration), we do not fully understand the mechanistic drivers of these results. Our results underscore that further experimentation with variation in PCR cycle number is certainly warranted, particularly for samples with low DNA concentrations.
False positives
False positives, due to contamination, are likely to be present in any pollen barcoding sample, and we strongly encourage the use of negative controls for both DNA isolation and for PCR. We also recommend sequencing these negative controls, even if no band is visible on a gel, to help estimate sample contamination. We suggest that negative controls be added to the flow cell at a volume no lower than the maximum volume added for any sample. Under these conditions, sequence reads from samples occurring at lower frequency than the highest-frequency reads from negative controls should be removed from subsequent analysis. In our study, this corresponded to around 5% of rbcL reads and 15% of ITS2 reads for most samples. This is probably overly conservative considering that on average the negative controls contained 1.6% of the number of reads present in an average sample. This conservatism may have had the unintended effect of removing rare interactions, leaving only the species visited most frequently by the individual bee in its current foraging trip. In studies where it is necessary to document very rare interactions, this may be a problem. In such situations, the number of contaminating sequences, and hence the number of species removed from analysis, could be reduced with conditions such as separate pre- and post-PCR rooms, laminar flow pre-PCR areas, and filtering of ambient air. We suspect that exclusion of very rare samples will have a negligible effect on most pollination network studies. Finally, this problem is not unique to DNA metabarcoding as most studies focused on visual identification of pollen use some arbitrary cut-off to decide which visually identified grains are legitimate vs. contamination.
Amplification of PCR products in our negative controls was too low to be visualized on a gel, but provided a few sequencing reads on the Illumina MiSeq. Sequencing of negative control samples yielded between 21 and 936 sequences identified as rbcL and between 42 and 1124 ITS2 sequences (containing between seven and 34 species). Two to 99 sequences were unable to be classified with either marker when analyzed with RDP Classifier (Appendix S4 (22.1KB, docx) ). Occasionally, the negative controls comprised a large number of sequences from the same species that was likely a contaminant from the laboratory (e.g., Fagopyrum, Populus), but the sequences of these species were never obtained from pollen samples above the threshold at which they would be included in analyses (Appendices S5 (27.3KB, docx) , S6 (30.4KB, docx) ). Most high-throughput sequencing studies do not include negative controls at the sequencing stage, but instead select an arbitrary limit below which a taxon is not included in the analysis, as it is expected to be either contamination, PCR artifact, or sequencing error (e.g., Cornman et al., 2015; Hawkins et al., 2015; Pornon et al., 2016). We found that including these samples allowed us to assess the impact of contamination, while using only a small proportion of the total sequencing reads provided by an Illumina MiSeq flow cell.
By contrast, other false positives may be the result of misidentifications from the bioinformatics pipeline. These false positives are more difficult to recognize, but in some cases very unlikely matches were returned. For example, we identified a large number of rbcL sequencing reads from 28 bee specimens from four sites, mostly in old, managed forest, long leaf pine reference sites (see Appendix 1 for details of habitat types), as Haemodorum laxum, a species that is endemic to southwestern Australia (FloraBase; https://florabase.dpaw.wa.gov.au/). While it is possible that this species may have been grown ornamentally (it appears, for example, as an incomplete record on the website of the U.S. National Gardening Association: https://www.garden.org), it is more likely, especially given their presence in less disturbed habitat types, that these sequences belong to another species of Haemodoraceae or a related family that is not present in the reference database. There are two species of Haemodoraceae recorded from North America, Lachnanthes caroliniana (Lam.) Dandy and Lophiola aurea Ker Gawl. (Flora of North America Editorial Committee, 1993). Both of these species occur in low, wet areas in savannas and pine forests in eastern North America, and could be expected to occur at our study sites. Neither of these species is in the rbcL database, and the family Haemodoraceae is not represented in the ITS2 database. This result points to the need for work that explicitly assesses false positives in DNA metabarcoding using known pollen samples.
Quantitation with pollen metabarcoding
One of the ultimate goals of pollen DNA metabarcoding is to record not only which species are interacting, but also which plant species are most frequently visited by each pollinator species. We quantified these interactions based on the number of interacting individuals, that is, the frequency at which a plant species was detected qualitatively in the mixed pollen load of a bee. Our work did not explicitly assess the quantitative efficacy of DNA metabarcoding, or how well the proportion of sequence reads matched the proportion of pollen grains in a sample. Other researchers have attempted to quantify the composition of mixed-species pollen loads carried by pollinators, and although most have found some correlation between the number of sequence reads and number of visually identified pollen grains (Pornon et al., 2016), most have found that this correlation is only moderate and that some species are found in vastly different proportions in different data sets (Keller et al., 2015; Kraaijeveld et al., 2015; Richardson et al., 2015). This may be due to biases among species in DNA isolation efficiency, pollen durability during storage, PCR efficiency, and copy number of ITS2 and rbcL (Bell et al., 2016). Other areas of bias are also common to visual pollen identification, including biases in the amount of pollen a plant produces, and the effect of grooming. This result pertains very strongly to networks, as quantitative networks—those that include information on visitation intensity—are often more informative than binary networks that include only presence-absence of a link (Blüthgen, 2010). For example, quantitative networks allow for the weighting of interactions to visitation intensity and decrease sensitivity to sampling effort, which in turn can lead to less biased estimates of network structure and topology (Blüthgen et al., 2008). We caution potential end-users of DNA metabarcoding for network construction to not use sequencing read proportions as if they were a precise quantification of visitation intensity. Rather, quantification can be achieved through recording the frequency of these presence-absence interactions. In addition to avoiding biases in the DNA metabarcoding data, this method of quantification also avoids biases in pollen output of different plant species, which may also be a limitation of pollinator networks based on microscopic pollen identification.
Relative success with ITS2 and rbcL
One clear outcome of our work is that the two barcoding markers employed in this study, rbcL and ITS2, did not give identical species-level results. This result is not surprising given that the markers have different taxonomic coverage in the DNA library to which our sequencing results were compared (38,409 plant species for rbcL and nearly twice as many—72,325—for ITS2 were present in our reference databases) and potentially different inherent levels of taxonomic resolution (up to ∼92% for ITS2 [Chen et al., 2010] vs. ∼72% for rbcL [CBOL Plant Working Group, 2009]). There could also be differences in the relative coverage of the two markers in our study flora in northern Florida. A detailed examination of three samples found that different species-level results obtained from rbcL and ITS2 were not due to different species-level identifications of the same pollen grains. Instead, the differences could be explained by differences in resolution due to a lack of species representation in the two databases, different levels of taxonomic resolution for the two markers, or differences in amplification success for the two markers. For example, although the species-level match was likely incorrectly identified with rbcL, Haemodoraceae was entirely undetected using ITS2 because the entire family is not represented in the database. Samples where we detected only Haemodoraceae with rbcL had no ITS2 identifications below the level of phylum. Problems associated with different amplification success for two markers could be reduced by amplifying the markers in separate reactions. This would make the results more robust, but would increase the costs. In our study, we obtained more reads for ITS2 than rbcL, which could also explain some of the differences in species-level resolution. Further research may be required to optimize the duplex PCR and compare results to separate amplifications. Optimizing the duplex PCR may have the added benefit of reducing potential primer-dimer issues, and hence increasing the number of reads that can be identified as rbcL or ITS2. Even without these minor improvements, using both markers certainly has the potential to increase taxonomic resolution. Combining markers could have even greater future potential if sequence reads from the same taxon can be quantitatively matched after development of quantitative correction factors as mentioned in the previous section.
In certain ecosystems, particularly those containing many closely related plant species, ITS2 and rbcL may be insufficient to identify all species present. In this case, other markers may be used. The most commonly used DNA barcoding markers include rbcL, matK, ITS2, trnL, and trnH-psbA. The relative merits of these markers have been discussed elsewhere (Fazekas et al., 2008; CBOL Plant Working Group, 2009; Ford et al., 2009; Hollingsworth et al., 2009, 2011; Chen et al., 2010; Shokralla et al., 2012), including in the context of pollen DNA metabarcoding (Bell et al., 2016). Using markers other than ITS2 and rbcL may also be useful when there are more existing data from other markers for species in the ecosystem. For example, in our study, including matK as a third marker may have identified Haemodoraceae to species level, because both North American species of Haemodoraceae have matK sequence data (https://www.ncbi.nlm.nih.gov, accessed 21 September 2016).
Conclusions
In this study, we have demonstrated the feasibility of using pollen DNA metabarcoding in the construction of pollinator networks, but have also highlighted some caveats and cautionary recommendations. We underscore that DNA metabarcoding is not quantitative in that sequence read proportions cannot be interpreted to reflect proportions of pollen grains in a sample. We therefore recommend either that any metrics calculated on networks constructed from barcoding should be based on binary, presence-absence networks, or that quantitative metrics (inferring interaction intensity) should be based on the frequency of these presence-absence data. Gene sequence reference libraries for barcode matching are not comprehensive in most geographic locations, and some taxonomic groups may not be represented even globally in such libraries. Usually this will lead to lower taxonomic resolution, but it may also lead to false positive identifications if a closely related species is present in the database. Results should be examined critically for identifications that are likely erroneous, and in some cases alternative matches could be inferred from knowledge of the local flora. Alternatively, researchers may wish to sequence key species from the study system and add these to the database before analysis. This can be quite easily achieved using tissue from any part of the plant, not necessarily pollen. The use of two or more barcoding markers can potentially improve taxonomic resolution, but there are no automated methods available at this time to reconcile identifications from multiple markers. Thus, multiple markers used in the same sample must be considered individually and will typically involve judgment calls on the part of the researcher. Contamination is likely to be a recurrent issue given the ubiquity of pollen and its durability in the environment, and we suggest that both DNA isolation and PCR negative controls be included in every batch of samples to allow for identification of contaminants and for setting thresholds of rare-sequence removals.
Despite these minor caveats, DNA metabarcoding offers tremendous advantages over visual microscopic identifications of pollen, in terms of taxonomic resolution and consistency of turnaround time. High-throughput methods for pollen identification are critically needed given global pollinator declines and a lack of knowledge of how structural changes in pollination networks affect their resilience to perturbations in response to ongoing anthropogenic environmental change. Although still in its infancy, DNA metabarcoding technology can be used today for construction of pollination networks and, with potential methodological refinements, holds tremendous promise for transforming their empirical study in the future.
Supplementary Material
Appendix 1.
Description of different management conditions at the study site.
| Management condition | Description of site |
| Longleaf pine (reference) | Longleaf pine trees greater than 30 yr in age with low degree of disturbance and management |
| Clear cut, debris left | All trees removed recently, woody debris remain at site |
| Clear cut, residues removed | All trees removed recently, no woody debris remain at site |
| Unthinned, managed | 12–15 yr of growth for slash or loblolly pine, no trees selectively removed during growth |
| Thinned, managed | 12–15 yr of growth for slash or loblolly pine, trees selectively removed during growth |
| Young, managed | Slash or loblolly pine stand 8–12 yr in age |
| Old, managed | Slash or loblolly stand more than 15 yr in age |
LITERATURE CITED
- Allesina S., Tang S. 2012. Stability criteria for complex ecosystems. Nature 483: 205–208. [DOI] [PubMed] [Google Scholar]
- Bascompte J., Jordano P., Melian C. J., Olesen J. M. 2003. The nested assembly of plant-animal mutualistic networks. Proceedings of the National Academy of Sciences, USA 100: 9383–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell K. L., de Vere N., Keller A., Richardson R. T., Gous A., Burgess K. S., Brosi B. J. 2016. Pollen DNA barcoding: Current applications and future prospects. Genome 59: 629–640. [DOI] [PubMed] [Google Scholar]
- Bell K. L., Loeffler V. M., Brosi B. J. 2017. An rbcL reference library to aid in the identification of plant species mixtures by DNA metabarcoding. Applications in Plant Sciences 5: 1600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüthgen N. 2010. Why network analysis is often disconnected from community ecology: A critique and an ecologist’s guide. Basic and Applied Ecology 11: 185–195. [Google Scholar]
- Blüthgen N., Fründ J., Vázquez D., Menzel F. 2008. What do interaction network metrics tell us about specialization and biological traits? Ecology 89: 3387–3399. [DOI] [PubMed] [Google Scholar]
- Bosch J., Martín González A. M., Rodrigo A., Navarro D. 2009. Plant-pollinator networks: Adding the pollinator’s perspective. Ecology Letters 12: 409–419. [DOI] [PubMed] [Google Scholar]
- Burgess K. S., Fazekas A. J., Kesanakurti P. R., Graham S. W., Husband B. C., Newmaster S. G., Percy D. M., et al. 2011. Discriminating plant species in a local temperate flora using the rbcL + matK DNA barcode. Methods in Ecology and Evolution 2: 333–340. [Google Scholar]
- Burkle L. A., Marlin J. C., Knight T. M., Klein A. M., Ollerton J., Winfree R., Tarrant S., et al. 2013. Plant-pollinator interactions over 120 years: Loss of species, co-occurrence, and function. Science 339: 1611–1615. [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CBOL Plant Working Group. 2009. A DNA barcode for land plants. Proceedings of the National Academy of Sciences, USA 106: 12794–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Yao H., Han J., Liu C., Song J., Shi L., Zhu Y., et al. 2010. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 5: e8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman R. S., Otto C. R. V., Iwanowicz D., Pettis J. S. 2015. Taxonomic characterization of honey bee (Apis mellifera) pollen foraging based on non-overlapping paired-end sequencing of nuclear ribosomal loci. PLoS ONE 10: e0145365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann C. F., Fruend J., Bluethgen N., Gruber B. 2009. Indices, graphs and null models: Analyzing bipartite ecological networks. Open Ecology Journal 2: 7–24. [Google Scholar]
- Edgar R. C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics (Oxford, England) 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Fazekas A. J., Burgess K. S., Kesanakurti P. R., Graham S. W., Newmaster S. G., Husband B. C., Percy D. M., et al. 2008. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 3: e2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira P. A., Boscolo D., Viana B. F. 2013. What do we know about the effects of landscape changes on plant–pollinator interaction networks? Ecological Indicators 31: 35–40. [Google Scholar]
- Flora of North America Editorial Committee [eds.]. 1993 onward. Flora of North America North of Mexico (online version), 18+ vols. Website http://www.efloras.org/flora_page.aspx?flora_id=1 [accessed 15 May 2017].
- Ford C. S., Ayres K. L., Toomey N., Haider N., van Alphen Stahl J., Kelly L. J., Wikström N., et al. 2009. Selection of candidate coding DNA barcoding regions for use on land plants. Botanical Journal of the Linnean Society 159: 1–11. [Google Scholar]
- Hawkins J., de Vere N., Griffith A., Ford C. R., Allainguillaume J., Hegarty M. J., Baillie L., Adams-Groom B. 2015. Using DNA metabarcoding to identify the floral composition of honey: A new tool for investigating honey bee foraging preferences. PLoS ONE 10: e0134735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P. D., Cywinska A., Ball S. L., DeWaard J. R. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London. Series B, Biological Sciences 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth M. L., Clark A. A., Forrest L. L., Richardson J., Pennington R. T., Long D. G., Cowan R. S., et al. 2009. Selecting barcoding loci for plants: Evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Molecular Ecology Resources 9: 439–457. [DOI] [PubMed] [Google Scholar]
- Hollingsworth P. M., Graham S. W., Little D. P. 2011. Choosing and using a plant DNA barcode. PLoS ONE 6: e19254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A., Pitchford J. W., Plank M. J. 2012. Disentangling nestedness from models of ecological complexity. Nature 487: 227–230. [DOI] [PubMed] [Google Scholar]
- Keller A., Danner N., Grimmer G., Ankenbrand M., von der Ohe K., von der Ohe W., Rost S., et al. 2015. Evaluating multiplexed next-generation sequencing as a method in palynology for mixed pollen samples. Plant Biology 17: 558–566. [DOI] [PubMed] [Google Scholar]
- Khansari E., Zarre S., Alizadeh K., Attar F., Aghabeigi F., Salmaki Y. 2012. Pollen morphology of Campanula (Campanulaceae) and allied genera in Iran with special focus on its systematic implication. Flora—Morphology, Distribution, Functional Ecology of Plants 207: 203–211. [Google Scholar]
- Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina Sequencing Platform. Applied and Environmental Microbiology 79: 5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraaijeveld K., de Weger L. A., Ventayol García M., Buermans H., Frank J., Hiemstra P. S., den Dunnen J. T. 2015. Efficient and sensitive identification and quantification of airborne pollen using next-generation DNA sequencing. Molecular Ecology Resources 15: 8–16. [DOI] [PubMed] [Google Scholar]
- Kress W. J., Erickson D. L. 2007. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2: e508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever J. J., van Nes E. H., Scheffer M., Bascompte J. 2014. The sudden collapse of pollinator communities. Ecology Letters 17: 350–359. [DOI] [PubMed] [Google Scholar]
- Memmott J., Waser N. M., Price M. V. 2004. Tolerance of pollination networks to species extinctions. Proceedings of the Royal Society of London. Series B, Biological Sciences 271: 2605–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michener C. 2007. The bees of the world. Johns Hopkins University Press, Baltimore, Maryland, USA. [Google Scholar]
- Palmieri L., Bozza E., Giongo L. 2009. Soft fruit traceability in food matrices using real-time PCR. Nutrients 1: 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto A. J., Raskin L. 2012. PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS ONE 7: e43093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornon A., Escaravage N., Burrus M., Holota H., Khimoun A., Mariette J., Pellizzari C., et al. 2016. Using metabarcoding to reveal and quantify plant-pollinator interactions. Scientific Reports 6: 27282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts S. G., Biesmeijer J. C., Kremen C., Neumann P., Schweiger O., Kunin W. E. 2010. Global pollinator declines: Trends, impacts and drivers. Trends in Ecology & Evolution 25: 345–353. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2016. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Website http://www.R-project.org/ [accessed 11 May 2017].
- Rahl M. 2008. Microscopic identification and purity determination of pollen grains. Methods in Molecular Medicine 138: 263–269. [DOI] [PubMed] [Google Scholar]
- Richardson R. T., Lin C.-H., Quijia J. O., Riusech N. S., Goodell K., Johnson R. M. 2015. Rank-based characterization of pollen assemblages collected by honey bees using a multi-locus metabarcoding approach. Applications in Plant Sciences 3: 1500043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmaki Y., Jamzad Z., Zarre S., Bräuchler C. 2008. Pollen morphology of Stachys (Lamiaceae) in Iran and its systematic implication. Flora—Morphology, Distribution, Functional Ecology of Plants 203: 627–639. [Google Scholar]
- Shokralla S., Spall J. L., Gibson J. F., Hajibabaei M. 2012. Next-generation sequencing technologies for environmental DNA research. Molecular Ecology 21: 1794–1805. [DOI] [PubMed] [Google Scholar]
- Sickel W., Ankenbrand M. J., Grimmer G., Holzschuh A., Härtel S., Lanzen J., Steffan-Dewenter I., Keller A. 2015. Increased efficiency in identifying mixed pollen samples by meta-barcoding with a dual-indexing approach. BMC Ecology 15: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staniczenko P. P. A., Kopp J. C., Allesina S. 2013. The ghost of nestedness in ecological networks. Nature Communications 4: 1391–1396. [DOI] [PubMed] [Google Scholar]
- Taberlet P., Coissac E., Hajibabaei M., Rieseberg L. H. 2012. Environmental DNA. Molecular Ecology 21: 1789–1793. [DOI] [PubMed] [Google Scholar]
- Thompson J. N. 1994. The coevolutionary process. University of Chicago Press, Chicago, Illinois, USA. [Google Scholar]
- Valentini A., Miquel C., Nawaz M. A., Bellemain E., Coissac E., Pompanon F., Gielly L., et al. 2009. New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: The trnL approach. Molecular Ecology Resources 9: 51–60. [DOI] [PubMed] [Google Scholar]
- Vázquez D. P., Aizen M. A. 2003. Null model analyses of specialization in plant–pollinator interactions. Ecology 84: 2493–2501. [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. D., Lewis J. D., Hoffmann C., Chen Y.-Y., Knight R., Bittinger K., Hwang J., et al. 2010. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiology 10: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.