Abstract
The identification of immunoactive agents for clinical and mechanistic applications is a very active area of research. In this vein, analogues of the potent immunostimulant KRN 7000 with diverse cytokine profiles have attracted considerable attention. Herein, we report on the synthesis and activity for four new C-glycosides of KRN 7000, 11-phenylundecanoyl and 11-p-fluorophenylundecanoyl derivatives of C-KRN 7000, 2,3-bis-epi-C-KRN 7000 and the reverse amide of C-KRN 7000. In mice, compared to C-KRN 7000, 2,3-bis-epi-C-KRN 7000 stimulated higher release of the anti-inflammatory cytokine IL-4 and lower release of the inflammatory cytokines IFN-γ and IL-12. The phenyl terminated alkanoyl and reverse amide analogues were inactive. These data suggest that structure activity effects for KRN 7000 are not necessarily additive and their use in the design of new analogues will require an improved understanding of how subtle structural changes impact on cytokine activity.
Keywords: glycolipid, CD1d, NKT, cytokine, C-glycoside
Graphical Abstract

1. Introduction
The control of the immune system through the regulation of invariant natural killer T (iNKT) cells by glycolipids is an emerging approach to treating cancer and certain autoimmune disorders.1,2,3,4 To treat infections and tumors, a Th1 response is required, while a Th2 response is suited to the control of autoimmune diseases. However, an unbiased Th1/Th2 response is potentially problematic because it may lead to opposing effects against a particular disease condition.5 The potency of cytokine release may also present clinical hurdles because too high an upregulation of one pathway may induce iNKT cell hyporesponsiveness or anergy.6.7,8 Therefore, a major challenge for glycolipid-based therapies is the identification of glycolipid antigens that have well-defined Th-1/Th-2 profiles with measured potencies. The most well-studied glycolipid antigen is α-galactosylceramide (α-O-GalCer), also called KRN 7000, which has been tested in several mouse models of disease and in a limited number of human trials.9,10,11,12 However, clinical applications of KRN 7000 have been stymied; the consensus of the immunology community is that it is handicapped by its inability to induce a clear Th1/Th2 cytokine bias. In this context C- KRN 7000, the C-glycoside analog in which the glycoside oxygen is replaced with a “CH2”, emerged as a benchmark Th1 biased molecule in mice, inducing high levels of IFN-γ and IL-12 with negligible levels of IL-4. Subsequently C-KRN 7000 and other C-glycoside analogues have been studied in mouse models as a prevention agent for malaria, asthma, arthritis and as a vaccine adjuvant in tuberculosis and influenza. 13,14,15,16,17
The immunological effects of KRN 7000 are believed to involve its binding to CD1d on APCs, and presentation of the binary complex to TCRs on iNKT cells to form the bioactive ternary complex.18,19,20 This assemblage stimulates the production of cytokines. Against this backdrop, we were interested in the new C-glycosides 7–10. The activity of these new analogues together with X-ray data for known glycolipids may illuminate the molecular basis for cytokine regulation.21,22,23 The 11-phenyl- and 11-p-fluorophenyl-undecanoic derivatives 7 and 8 are interesting as the analogous O-glycosides are reported to show much higher Th-1 bias in against both mice and human iNKT cells compared to their hexacosanoic acid derivatives.24,25,26 Similarly, the nuanced cytokine profiles of C2–4 diastereomers of KRN 7000 and C-KRN 7000 relative to analogues with the natural stereochemistry, in particular the high Th-1 bias of 4-epiC-KRN7000, inspired 9.15,17,27 The reverse amide 10 is a probe for the binding of the amide residue.28,29,30,31,32
2. Results and Discussion
Synthesis
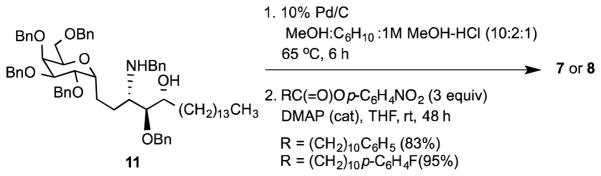
The phenyl terminated alkanoyl derivatives 7 and 8 were prepared from α-C-glycoside 11. The latter originated from α-C-allyl tetra-O-benzylgalactoside as previously described, and showed no evidence of the β-C-glycoside (Scheme 1). 33 Thus, transfer hydrogenation conditions on 11 was used for removal of the benzyl protecting groups because of the difficulty in cleavage of the N-benzyl under standard hydrogenolysis conditions.34 The crude product from this reaction was then treated with the p-nitrophenyl ester of the requisite fatty acid to give 7 and 8 in 83 and 95% yield respectively, over two steps. These materials were purified by silica gel chromatography with a gradient elution of 5–15 % methanol in chloroform, as previously described for related analogues of KRN 7000.16,17,32,35,36,37 The final compounds and their peracetylated derivatives were characterized by NMR and HRMS analysis. The synthesis of 2,3-bisepimer of C-KRN 7000, 9 and the reverse amide of C-KRN 7000, 10 has been reported as part of our earlier studies on C-glycosides.38
Scheme 1.
Synthesis of 11-phenylundecanoyl amides 7 and 8
Biological evaluation
The cytokine production levels induced by 7 – 10 were determined as reported.13,14 C-KRN 7000 2 and 3 were used as controls.14 Test compounds were administered to C57BL/6 mice by intraperitoneal injection. Sera were collected at 0, 6, 12 and 24 h after treatment for ELISA analysis of IFN-γ, IL-12 and IL-4 concentrations. In agreement with published data, 2 and 3 induced relatively high levels of Th1 cytokines IFN-γ and IL-12 compared to the Th2 cytokine IL-4. The C-KRN 7000 diastereomer 9 also induced higher IFN-γ than IL-4, but the level of IFN-γ production was lower, and that of IL-4 higher, compared to 2 and 3. Compared to 2 and 3, compound 9 induced a higher level of IL-4 than IL-12. The phenyl terminated alkanoyl and the reverse amide analogues 7, 8 and 10 did not induce any noticeable production of cytokines. (Figure 2).
Figure 2. Serum cytokine concentrations for mice administered with glycolipids.
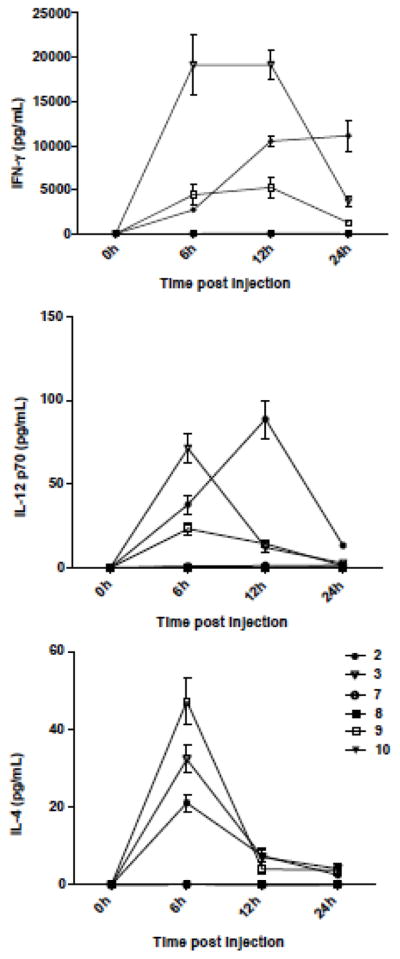
Representative data from one of three experiments. Error bars represent the standard deviation for triplicate wells. P < 0.05 as determined by the Student t-test with 2 and DMSO as controls.
The lower potency of IFN-γ induced by 9 compared to 2, together with the disparate activities of other diastereomers of 2, and analogous O-glycosides, suggest that cytokine activity is particularly sensitive to structural changes in the polar region of the ceramide residue.15,17,27 This is consistent with the fact that this region is intimately engaged in the ternary complex, and the notion that binding in the ternary complex impacts on cytokine potency and ratio.21,22,23,39 That compounds 7 and 8, the phenyl terminated undecanoyl analogues of 2 were inactive, whereas 2 was, is also noteworthy. In the O-glycoside series these phenyl terminated alkanoyl modifications have been shown to markedly increase the potency of cytokine production relative to their hexacosanoyl derivatives.24,25,26 Therefore, given the remote positions of these two loci, it was surprising that these fatty acid modifications deactivated rather than activated, the C-glycoside 2. This said, a similar loss in activity in a murine assay was observed when the hexacosanoic acid residue in C-glycosides 3 was replaced with an 8-phenyloctanoyl chain (cf 4), although against human iNKT cells, the expected increase in cytokine release was observed.14 Thus the cytokine activating effect of phenyl terminated alkanoyl chains may not be general for different classes of lipids, and for mice and human assays. Since the amide carbonyl in KRN 7000 is not believed to interact directly in the ternary complex, the loss of activity for the inverse amide 10 supports the notion that the amide NH acts as a H-bond donor and a precise location of the amide bond is critical for activity.40 In light of the observation that ester derivatives of KRN 7000 still retain some activity, the complete loss in activity of 10, suggests that the carbonyl residue may also be an important receptor contact. However, given our limited data, and the multi-factorial nature of cytokine stimulation, further speculation on the molecular basis for the observations on 7 – 10 is premature.
3. Conclusion
Four new C-glycosides of KRN 7000, 11-phenylundecanoyl and 11-p-fluorophenylundecanoyl derivatives of C-KRN7000, 2,3-bis-epi-C-KRN 7000 and the reverse amide of C-KRN 7000 were synthesized and their cytokine activity evaluated. In mice, compared to C-KRN 7000, 2,3-bis-epi-C-KRN7000 stimulated higher release of the anti-inflammatory cytokine IL-4 and lower release of the inflammatory cytokines IFN-γ and IL-12. The phenyl terminated alkanoyl and reverse amide analogues analogues were inactive. The activity of the 2,3-bis-epi-C-KRN 7000 was in line with the cytokine profile exhibited by diastereomeric O- and C- glycosides. The loss of activity in the reverse amide is consistent with structure activity hypotheses for the amide residue. However, given the high cytokine potency associated with phenyl terminated alkanoyl analogues of KRN 7000, the complete loss in activity for the 11-phenylundecanoyl C-glycosides was surprising. This unexpected loss reinforces the emerging idea that activity enhancements by changes at different loci of the parent O-glycoside are not necessarily additive. Thus the use of SAR data to design new analogs may require an improved understanding of how subtle structural changes impact on cytokine activity.
4. Experimental Section
4.1 General
Unless otherwise stated, all reactions were carried out under a nitrogen atmosphere in oven-dried glassware using standard syringe and septa technique. NMR spectra were obtained on 500 or 600 MHz instruments. Chemical shifts are relative to the deuterated solvent peak or the tetramethylsilane (TMS) peak at (δ 0.00) and are in parts per million (ppm). Assignments for selected nuclei were determined from 1H COSY experiments. Optical rotations [α]D are given in units of 10−1 degcm2g at 589 nm (sodium D-line). Thin layer chromatography (TLC) was done on 0.25 mm thick pre-coated silica gel HF254 aluminum sheets. Chromatograms were observed under UV (short and long wavelength) light, and/or were visualized by heating plates that were dipped in a solution of ammonium (VI) molybdate tetrahydrate (12.5 g) and cerium (IV) sulfate tetrahydrate (5.0 g) in 10% aqueous sulphuric acid (500 mL). Flash column chromatography (FCC) was performed using silica gel 60 (230–400 mesh) and employed a stepwise solvent polarity gradient, correlated with TLC mobility. Hexanes used for FCC had a boiling point in the 40–60 °C range.
4.2 C-KRN 7000 – 11-phenylundecanoyl amide 7
A mixture of 11 (85 mg, 0.083 mmol), 10% Pd/C (85 mg), cylclohexene (0.4 mL) and 1 N HCl (0.1 mL) in MeOH (2 mL), was heated at reflux for 6 h. The mixture was then cooled to rt, filtered over a pad of Celite and basic resin, and concentrated in vacuo to give a white solid (45 mg), which was used directly in the next step. A portion of this material (10 mg, 0.021 mmol) was taken up in THF (1 mL) and treated with p-nitrophenyl 1-phenylundecanoate (24 mg, 0.063 mmol) and DMAP (0.5 mg). The mixture was stirred at rt for 48 h, then concentrated in vacuo. The residue was taken up in MeOH (0.5 mL) and treated with 1M NaOMe in MeOH (0.5 mL) for 30 min. The solution was adjusted to pH 7 with Dowex 50WX8 resin, the mixture filtered over a Celite pad and the filtrate concentrated in vacuo. FCC of the residue using a gradient elution with 5–15% MeOH-CHCl3, afforded 7 as a white solid (10 mg, ca 83% over 2 steps): Rf =0.24 (15% MeOH-CHCl3); 1H NMR (500 MHz, C5D5N) δ 8.42 (d, J = 9.0 Hz, 1H, NH), 7.38–7.23 (m, 5H, ArH), 6.80–5.40 (bm, 6H, OH), 5.14 (m, 1H, H2), 4.74 (dd, J = 8.9, 5.5 Hz, 1H, H3’), 4.53 (m, 3H, H1’, H2’/4’, H6’), 4.38 (dd, J = 11.2, 4.6 Hz, 1H, H6’), 4.25 (m, 4H, H3, H4, H2’/4’, H5’), 2.72 (m, 1H, H1), 2.58 (m, 3H, Ha, CH2-11”), 2.45 (m, 2H, CH2-2”), 2.31 (m, 2H, Ha, H5), 2.22 (m, 1H, H1), 1.93 (m, 2H, H5, H6), 1.85 (m, 2H, CH2-3”), 1.70 (m, 1H, H6), 1.58 (m, 2H, CH2-10”), 1.40 – 1.10 (m, 34H), 0.87 (t, J = 6.8 Hz, 3H, CH3). 13C NMR (125 MHz, C5D5N) δ 173.9, 143.7, 129.3, 129.2, 126.5, 79.0, 77.5, 74.2, 73.1, 72.7, 71.1, 70.9, 63.2, 53.1, 37.4, 36.6, 34.9, 32.6, 32.3, 30.8, 30.7, 30.5, 30.4, 30.3, 30.2, 30.1, 30.0, 27.1, 27.0, 26.9, 23.4, 23.1, 14.7. HRMS (ESI, MNa+) m/z calc for C42H75NO8Na 744.5385, found 744.5388
4.3 Hexa-O-acetyl-C-KRN 7000 -11-phenylundecanoyl amide 7-OAc
Acetic anhydride (0.05 mL) was added to a solution of 7 (10 mg, 0.014 mmol) in a mixture of EtOAc (1 mL) and pyridine (0.05 mL). The reaction was stirred at rt for 16h, at which time the volatiles were removed in vacuo. FCC of the residue afforded 7-OAc (10 mg, 75%): 1H NMR (500 MHz, CDCl3) δ 7.26–7.13 (m, 5H, ArH), 5.71 (d, J = 9.7 Hz, 1H, NH), 5.38 (bs, 1H, H4’), 5.15 (m, 2H, H2’, 3’), 4.90 (m, 2H, H3, 4), 4.30 (dd, J = 9.0, 13.0 Hz, 1H, H6’), 4.17 (m, 1H, H2), 4.06 (m, 1H, H1’), 4.02 (m, 2H, H5’, 6’), 2.57 (t, J = 7.7 Hz, 2H, CH2-11”), 2.15 (t, J = 7.7 Hz, 2H, CH2-2”), 2.09 (s, 3H, Ac), 2.08 (s, 3H, Ac), 2.04 (s, 6H, Ac), 2.03 (s, 3H, Ac), 2.01 (s, 3H, Ac), 1.63 (m, 1H), 1.56 (m, 8H), 1.40 (m, 1H), 1.22 (m, 36H), 0.86 (t, J = 6.8 Hz, 3H, CH3). 13CNMR (125 MHz, CDCl3) δ 173.1, 171.3, 170.8, 170.2, 170.1, 170.0 (two peaks), 143.1, 128.6, 128.4, 125.8, 75.8, 72.9, 72.0, 68.7 (two peaks), 68.0, 67.6, 61.3, 48.7, 37.1, 36.2, 32.2, 31.8, 29.9, 29.8, 29.7, 29.6, 29.1, 27.0, 26.0, 25.7, 22.9, 22.7, 21.3, 21.1, 21.0, 20.9, 14.3. HRMS (ESI, MNa+) m/z calc for C54H87NO14Na 996.6019, found 996.6002.
4.4 C-KRN 7000-11-p-fluorophenyl-undecanoyl amide 8
A portion of the material from the hydrogenolysis of 11 (10 mg, 0.021 mmol) was taken up in THF (1 mL) and treated with p-nitrophenyl 4-fluorophenylundecanoate16 (25 mg, 0.063 mmol) and DMAP (0.5 mg). The reaction was processed as described for the preparation of 7. FCC of the residue afforded 8 as a white solid (12 mg, ca 88% over 2 steps): Rf = 0.26 (15% MeOH-CHCl3); 1H NMR (500 MHz, C5D5N) δ 8.46 (d, J = 9.0 Hz, 1H, NH), 7.20–7.11 (m, 4H, ArH), 5.15 (m, 1H, H2), 4.74 (dd, J = 8.9, 5.5 Hz, 1H, H3’), 4.52 (m, 3H, H1’, H2’/4’, H6’), 4.37 (dd, J = 11.2, 4.6 Hz, 1H, H6’), 4.24 (m, 4H, H2’/4’, H5’, H3, H4), 2.73 (m, 1H, H1), 2.60 (m, 1H, Ha), 2.51 (t, J = 7.9 Hz, 2H, CH2-11”), 2.48 (m, 2H, CH2-2”), 2.36 (m, 2H, Ha, H5), 2.24 (m, 1H, H1), 1.95 (m, 2H, H5, H6), 1.86 (m, 2H, CH2-3”), 1.70 (m, 1H, H6), 1.50 (m, 2H, CH2-10”) 1.45 – 1.10 (m, 34H), 0.88 (t, J = 6.7 Hz, 3H, CH3). 13C NMR (125 MHz, C5D5N) δ 173.4, 161.6 (d, JC-F = 240.5 Hz), 139.1 (d, JC-F = 2.8 Hz), 130.4 (d, JC-F = 7.4 Hz), 115.3 (d, JC-F = 20.8 Hz), 78.5, 77.1, 73.8, 73.1, 72.7, 72.2, 70.6, 70.4, 62.8, 52.7, 37.0, 35.2, 34.5, 32.2, 30.9, 30.4, 30.2, 30.1, 30.0, 29.9, 29.7, 29.6, 29.5, 26.6, 26.5, 23.0, 22.6, 14.3. HRMS (ESI, MNa+) m/z calc for C42H74FNO8Na 762.5291, found 762.5294.
4.5 Hexa-O-acetyl-C-KRN 7000-11-p-fluorophenylundecanoyl amide 8-OAc
Compound 8-OAc was prepared from 8, following the procedure used for 7-OAc. For 8-OAc: 1H NMR (500 MHz, CDCl3) δ 7.08 (m, dt, J = 2.0, 6.0 Hz, 2H, ArH), 6.92 (t, J = 6.0, 2H, ArH), 5.69 (d, J = 9.7 Hz, 1H, NH), 5.38 (bs, 1H, H4’), 5.14 (m, 2H, H2’, 3’), 4.89 (m, 2H, H3, 4), 4.30 (dd, J = 9.0, 13.0 Hz, 1H, H6’), 4.17 (m, 1H, H2), 4.06 (m, 1H, H1’), 4.00 (m, 2H, H5’, 6’), 2.53 (t, J = 7.7 Hz, 2H, CH2-11”), 2.15 (t, J = 7.7 Hz, 2H, CH2-2”), 2.08 (s, 3H, Ac), 2.07 (s, 3H, Ac), 2.04 (s, 6H, Ac), 2.03 (s, 3H, Ac), 2.01 (s, 3H, Ac), 1.74 (m, 1H), 1.56 (m, 8H), 1.40 (m, 1H), 1.22 (m, 36H), 0.85 (t, J = 6.8 Hz, 3H, CH3). 13CNMR (125 MHz, CDCl3) δ 173.1, 171.3, 170.8 (two peaks), 170.2, 170.1, 170.0, 161.6 (d, JC-F = 240.5 Hz), 139.1 (d, JC-F = 2.8 Hz), 129.9 (d, JC-F = 7.4 Hz), 115.1 (d, JC-F = 20.8 Hz), 75.7, 72.9, 72.0, 68.8, 68.7, 68.0, 67.5, 61.3, 48.7, 37.1, 35.3, 32.2, 31.8, 29.9, 29.8, 29.7, 29.6, 29.4, 29.1, 27.0, 26.0, 25.7, 22.9, 22.7, 21.3, 21.1, 21.0, 20.9, 14.3. HRMS (ESI, MNa+) m/z calc for C54H86FNO14Na 1014.5925, found 1014.5909.
4.6 Determination of serum cytokine concentration
6–8-week-old female C57BL/6 mice were purchased from the Jackson Laboratory and were maintained in animal facility of Weill Cornell Medical College. The cytokine production levels induced by glycolipids were determined as reported.13,14 The test compounds were taken up in DMSO (1 mg/mL) and warmed to 50–55 °C and sonicated. For each test compound 4 μL of the stock solution was diluted in 100 μL of 10 mM PBS buffer, warmed to 50–55 °C, sonicated again, and administered to C57BL/6 mice by intraperitoneal injection. Sera were collected at 0, 6, 12 and 24 h after glycolipid treatment for ELISA analysis of IFN-γ, IL-12 and IL-4 concentrations. IFN-γ production in serum was tested using enzyme-linked immunosorbent assay (ELISA, eBioscience). Serum concentrations of IL-12(p70) and IL-4 were measured using ELISA (BioLegend). Each compound was administered to two or three different mice in three independent experiments and the serum from each mice was tested in triplicate. Representative data for one of the three experiments are shown. The data were expressed as the mean ± standard deviation (SD) of triplicate wells. A p-value <0.05 was determined by the Student t-test with DMSO and 2 as controls.
Supplementary Material
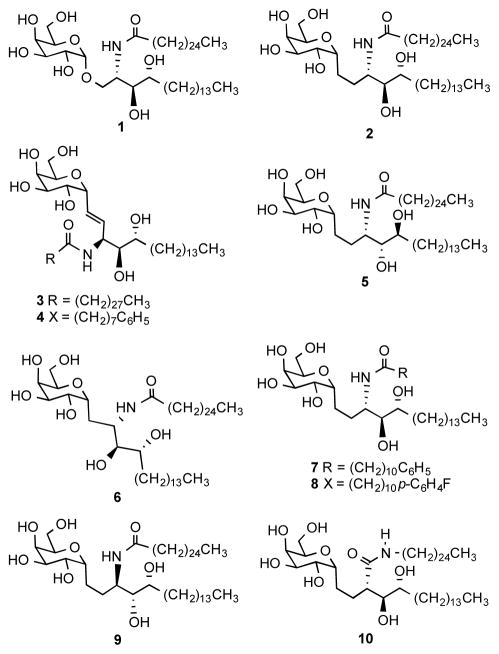
Figure 1.
Known and new analogues of KRN7000
C-glycosides of KRN 7000 with novel ceramide segments were prepared
The 2,3-bisepimer of C-KRN 7000 showed a lower Th-1 cytokine bias than C-KRN 7000
11-Phenyl undecanoic acid amides and the inverse amide of C-KRN 7000 were inactive
Structure activity effects for KRN 7000 are not necessarily additive
Acknowledgments
This work was supported by the National Science Foundation (CHE-1301330). A Research Centers in Minority Institutions Program grant from the National Institute of Health Disparities (MD007599) of the National Institutes of Health (NIH), which supports the infrastructure at Hunter College, and a Clinical Translational Science Center award (TR000457) from the NIH are also acknowledged.
Footnotes
NMR charts for compounds 7–10 can be found, in the online version, at
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Van Kaer L, Parekh VV, Wu L. Immunotherapy. 2011;3:59–75. doi: 10.2217/imt.10.85. [DOI] [PubMed] [Google Scholar]
- 2.Shissler SC, Bollino DR, Tiper IV, Bates JP, Derakhshandeh R, Webb TJ. Immunogenetics. 2016;68:623–628. doi: 10.1007/s00251-016-0928-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birkholz AM, Howell AR, Kronenberg M. J Biol Chem. 2015;290:20746–20746. doi: 10.1074/jbc.A115.647057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carreño LJ, Saavedra-Ávila NA, Porcelli SA. Clin Trans Immun. 2016;5:e69. doi: 10.1038/cti.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Garra A. Immunity. 1998;8:275–283. doi: 10.1016/s1074-7613(00)80533-6. [DOI] [PubMed] [Google Scholar]
- 6.Uldrich AP, Crowe NY, Kyparissoudis K, Pellicci DG, Zhan Y, Lew AM, Bouillet P, Strasser A, Smyth MJ, Godfrey DI. J Immunol. 2005;175:3092–3101. doi: 10.4049/jimmunol.175.5.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parekh VV, Wilson MT, Olivares-Villagómez D, Singh AK, Wu L, Wang C-R, Joyce S, Van Kaer L. J Clin Invest. 2005;115:2572–2583. doi: 10.1172/JCI24762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wingender G, Birkholz AM, Sag D, Farber E, Chitale S, Howell AR, Kronenberg M. J Immun. 2015;195:3838–3848. doi: 10.4049/jimmunol.1500203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banchet-Cadeddu A, Hénon E, Dauchez M, Renault JH, Monneaux F, Haudrechy A. Org Biomol Chem. 2011;9:3080–3104. doi: 10.1039/c0ob00975j. [DOI] [PubMed] [Google Scholar]
- 10.Tashiro T. Biosci Biotechnol Biochem. 2012;76:1055–1067. doi: 10.1271/bbb.120072. [DOI] [PubMed] [Google Scholar]
- 11.Laurent X, Bertin B, Renault N, Farce A, Speca S, Milhomme O, Millet R, Desreumaux P, Hénon E, Chavatte P. J Med Chem. 2014;57:5489–5508. doi: 10.1021/jm4010863. [DOI] [PubMed] [Google Scholar]
- 12.Marzabadi CH, Franck RW. Chem Eur J. 2016;22:1–16. [Google Scholar]
- 13.Schmieg J, Yang G, Franck RW, Tsuji M. J Exp Med. 2003;198:1631–1641. doi: 10.1084/jem.20031192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Li X, Chen G, Garcia-Navarro R, Franck R, Tsuji M. Immunol. 2008;127:216–225. doi: 10.1111/j.1365-2567.2008.02943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Li X, Shiratsuchi, Chen G, Dellabona P, Casorati G, Franck R, Tsuji M. J Immunol. 2009;183:4415–4421. doi: 10.4049/jimmunol.0901021. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Franck RW. C R Chimie. 2012;15:46–56. doi: 10.1016/j.crci.2011.05.006. and refs 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pu J, Franck RW. Tetrahedron. 2008;64:8618–8629. doi: 10.1016/j.tet.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Liu Z, Courtney AN, Metelitsa LS, Bittman R. ChemBioChem. 2012;13:1733–1737. doi: 10.1002/cbic.201200374. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu Z, Bittman R. Org Lett. 2012;14:620–623. doi: 10.1021/ol2032448. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Liu Z, Byun H-S, Bittman R. Org Lett. 2010;12:2974–2977. doi: 10.1021/ol1009976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang YJ, Hsuan YC, Lai ACY, Han YC, Hou DR. Org Lett. 2016;18:808–811. doi: 10.1021/acs.orglett.6b00090. [DOI] [PubMed] [Google Scholar]
- 18.Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Nature Rev Immun. 2012;12:845–857. doi: 10.1038/nri3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zajonc DM. Immunogenetics. 2016;68:561–576. doi: 10.1007/s00251-016-0931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Kaer L, Wu L, Joyce S. Trends Immun. 2016;37:738–754. doi: 10.1016/j.it.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aspeslagh S, Li Y, Yu ED, Pauwels N, Trappeniers M, Girardi E, Decruy T, Van Beneden K, Venken K, Drennan M, Leybaert L, Wang J, Franck RW, Van Calenbergh S, Zajonc DM, Elewaut D. EMBO J. 2011;30:2294–2305. doi: 10.1038/emboj.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel O, Cameron G, Pellicci DG, Liu Z, Byun HS, Beddoe T, McCluskey J, Franck RW, Castano AR, Harrak Y, Llebaria A, Bittman R, Porcelli SA, Godfrey DI, Rossjohn J. J Immunol. 2011;187:4705–4713. doi: 10.4049/jimmunol.1100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Nat Rev Immun. 2012;12:845–857. doi: 10.1038/nri3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Fujio M, Imamura M, Wu D, Vasan S, Wong CH, Ho DD, Tsuji M. Proc Natl Acad Sci USA. 2010;107:13010–13015. doi: 10.1073/pnas.1006662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu TN, Lin KH, Chang YJ, Huang JR, Cheng JY, Yu AL, Wong CH. Proc Natl Acad Sci USA. 2011;108:17275–17280. doi: 10.1073/pnas.1114255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu T-N, Lin K-H, Wu Y-T, Huang J-R, Hung J-T, Wu J-C, Chen C-Y, Chu K-C, Lin N-H, Yu AL, Wong C-H. ACS Chem Biol. 2016 doi: 10.1021/acschembio.6b00650. [DOI] [PubMed] [Google Scholar]
- 27.Park J-J, Lee JH, Seo K-C, Bricard G, Venkataswamy MM, Porcelli SA, Chung S-K. Bioorg Med Chem Lett. 2010;20:814–818. doi: 10.1016/j.bmcl.2009.12.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tashiro T, Hongo N, Nakagawa R, Seino K, Watarai H, Ishii Y, Taniguchi M, Mori K. Bioorg Med Chem. 2008;16:8896–8906. doi: 10.1016/j.bmc.2008.08.060. [DOI] [PubMed] [Google Scholar]
- 29.Shiozaki M, Tashiro T, Koshino H, Nakagawa R, Inoue S, Shigeura T, Watarai H, Taniguchi M, Mori K. Carbohydr Res. 2010;345:1663–1684. doi: 10.1016/j.carres.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Banchet-Cadeddu A, Martinez A, Guillarme S, Parietti V, Monneaux F, Hénon E, Renault JH, Nuzillard JM, Haudrechy A. Bioorg Med Chem Lett. 2011;21:2510–2514. doi: 10.1016/j.bmcl.2011.02.044. [DOI] [PubMed] [Google Scholar]
- 31.Tashiro T, Shigeura T, Watarai H, Taniguchi M, Mori K. Bioorg Med Chem. 2012;20:4540–4548. doi: 10.1016/j.bmc.2012.05.073. [DOI] [PubMed] [Google Scholar]
- 32.Wojno J, Jukes JP, Ghadbane H, Shepherd D, Besra GS, Cerundolo V, Cox LR. ACS Chem Biol. 2012;7:847–855. doi: 10.1021/cb2005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altiti AS, Mootoo DR. Org Lett. 2014;16:1466–1469. doi: 10.1021/ol5002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson AE, Johnstone RAW. Synthesis. 1976:685–687. [Google Scholar]
- 35.Tyznik AJ, Farber E, Girardi E, Birkholz A, Li Y, Chitale S, So R, Arora P, Khurana A, Wang J, Porcelli SA, Zajonc DM, Kronenberg M, Howell AR. Chem Biol. 2011;18:1620–1630. doi: 10.1016/j.chembiol.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trappeniers M, Van Beneden K, Decruy T, Hillaert U, Linclau B, Elewaut D, Van Calenbergh S. J Am Chem Soc. 2008;130:16468–16469. doi: 10.1021/ja8064182. [DOI] [PubMed] [Google Scholar]
- 37.Fujio M, Wu D, Garcia-Navarro R, Ho DD, Tsuji M, Wong CH. J Am Chem Soc. 2006;128:9022–9023. doi: 10.1021/ja062740z. [DOI] [PubMed] [Google Scholar]
- 38.Altiti AS, Bachan S, Mootoo DR. Org Lett. 2016;18:4654–4657. doi: 10.1021/acs.orglett.6b02284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu TN, Lin KH, Chang YJ, Huang JR, Cheng JY, Yu AL, Wong CH. PNAS. 2011;108:17275–17280. doi: 10.1073/pnas.1114255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hénon E, Dauchez M, Haudrechy A, Banchet A. Tetrahedron. 2008;64:9480–9489. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.