Abstract
Epigenetic regulation of chromatin plays a critical role in controlling embryonic stem (ES) cell self-renewal and pluripotency. However, the roles of histone demethylases and activating histone modifications such as trimethylated histone 3 lysine 4 (H3K4me3) in transcriptional events such as RNA polymerase II (RNAPII) elongation and alternative splicing are largely unknown. In this study, we show that KDM5B, which demethylates H3K4me3, plays an integral role in regulating RNAPII occupancy, transcriptional initiation and elongation, and alternative splicing events in ES cells. Depletion of KDM5B leads to altered RNAPII promoter occupancy, and decreased RNAPII initiation and elongation rates at active genes and at genes marked with broad H3K4me3 domains. Moreover, our results demonstrate that spreading of H3K4me3 from promoter to gene body regions, which is mediated by depletion of KDM5B, modulates RNAPII elongation rates and RNA splicing in ES cells. We further show that KDM5B is enriched nearby alternatively spliced exons, and depletion of KDM5B leads to altered levels of H3K4 methylation in alternatively spliced exon regions, which is accompanied by differential expression of these alternatively splice exons. Altogether, our data indicate an epigenetic role for KDM5B in regulating RNAPII elongation and alternative splicing, which may support the diverse mRNA repertoire in ES cells.
INTRODUCTION
Gene expression in embryonic stem (ES) cells is regulated in part by packaging of DNA into chromatin, which is comprised of histones, DNA, RNA and associated proteins. Post-translational modification of histones, such as methylation, contributes to the regulation of chromatin structure (1) by influencing the activity of epigenetic modifiers and the transcriptional state of the underlying DNA sequence. Combined, these functions are important for controlling gene expression networks that promote self-renewal or differentiation.
Methylation of histone 3 lysine 4 (H3K4me3) is enriched at transcriptional start sites (TSS) of active genes (2–6), and is presumed to be a platform for RNA polymerase II (RNAPII) binding and target gene activation (7–9). Demethylation of H3K4me3 is facilitated by lysine demethylase 5 (KDM5) family members (10). KDM5 enzymes, such as KDM5B, which catalyzes H3K4 demethylation, have traditionally been presumed to be transcriptional repressors (11–13), although recent studies have described a more dynamic role for KDM5B in transcriptional regulation (14,15). KDM5B is important for normal embryonic development (16,17), ES cell differentiation (14,18,19) and is a barrier to the iPS cell reprogramming process (18). KDM5B has also been shown to focus H3K4 methylation near transcriptional regulatory elements such as promoters and enhancers by preventing H3K4 methylation from spreading to gene bodies and enhancer shores (14). Moreover, the redistribution of H3K4 methylation leads to dysregulated gene expression in KDM5B-depleted ES cells. While these findings demonstrate a critical role for KDM5B in regulating H3K4 methylation at promoters and gene body regions, it is unclear how the redistribution of H3K4 methylation from promoter to gene body regions impacts the regulation of the transcription cycle of RNAPII, including initiation and elongation events.
Therefore, to clarify the role for KDM5B in regulating RNAPII elongation, and splicing, a downstream co-transcriptional process that is affected by the rate of RNAPII elongation (20–24), we evaluated genome-wide changes in RNAPII distributions and RNA splicing in KDM5B-depleted ES cells. Our results show that depletion of KDM5B leads to decreased RNAPII promoter occupancy, which is correlated with decreased levels of H3K4me3 in promoter regions and decreased RNAPII initiation and elongation rates. In addition, we demonstrate that KDM5B is enriched nearby alternatively spliced exons (cassette exons), and depletion of KDM5B leads to altered levels of H3K4 methylation in alternatively spliced exon regions, which is accompanied by differential expression of these alternatively spliced exons. Together, our data implicate a novel role for KDM5B in regulating RNAPII elongation and alternative splicing in ES cells, thus contributing to the diverse mRNA repertoire in ES cells.
MATERIALS AND METHODS
ES cell culture
shLuc and shKdm5b (R1) ES cells were cultured as previously described with minor modifications (14,18). Briefly, R1 ES cells were cultured on irradiated mouse embryonic fibroblasts (MEFs) in Dulbecco's modified Eagle's medium/15% fetal bovine serum (FBS) media containing LIF (ESGRO) and 1 μg/ml puromycin at 37°C with 5% CO2. For chromatin immunoprecipitation (ChIP) experiments ES cells were cultured on gelatin-coated dishes in ES cell media containing 1.5 μM CHIR9901 (GSK3 inhibitor) for several passages to remove feeder cells. ES cells were passed by washing with phosphate buffered saline, and dissociating with trypsin using serological pipettes (sc-200279, sc-200281). For the rescue experiments, ES cells were transfected using lipofectamine 2000 with pCSH2-KDM5B-WT (wild-type) or pCSH2-KDM5B-mut (mutant; H499A) plasmids every 24 h for 72 h. Transfected ES cells were collected 48 h after the last transfection and subsequently used for downstream analyses (ChIP-PCR, western blotting).
ChIP-Seq analysis
ChIP-Seq experiments were performed as previously described with minor modifications (14,18,25,26). The mouse monoclonal RNAPII [8WG16] (ab817) and the rabbit polyclonal RNAPII Ser5P (ab5131) and RNAPII Ser2P (ab5095) antibodies were obtained from Abcam. The H3K4ac (07–539) antibody was also obtained from EMD Millipore. Briefly, 107–108 mouse ES cells (R1) were harvested and chemically crosslinked with 1% formaldehyde (Sigma) for 5–10 min at 37°C and subsequently sonicated. Sonicated cell extracts equivalent to 5 × 106 cells were used for ChIP assays. ChIP-enriched DNA was end-repaired using the End-It DNA End-Repair kit (Epicentre), followed by addition of a single A nucleotide and ligation of custom Illumina adapters. Polymerase chain reaction (PCR) was performed using Phusion High Fidelity PCR master mix. ChIP libraries were sequenced on Illumina HiSeq platforms according to the manufacture's protocol. Sequence reads were mapped to the mouse genome (mm9) using bowtie2 (27) with default settings. ChIP-Seq read enriched regions (peaks) were identified relative to control Input DNA using ‘Spatial Clustering for Identification of ChIP-Enriched Regions’ (SICER) software (28) with a window size setting of 200 bps, a gap setting of 400 bps, a false discovery rate (FDR) setting of 0.001. The SICER-compare function was used to compare multiple samples (FDR < 0.001, fold-change [FC] > 1.5). The RPBM measure (read per base per million reads) was used to quantify the density at genomic regions from ChIP-Seq datasets. At least two replicates were performed for the ChIP-Seq analyses. We have also applied the Kolmogorov–Smirnov test to obtain P-value statistics and compare densities at genomic regions.
Confirmation of ChIP binding using Q-PCR
Primers were designed for RNAPII ES cell ChIP-enriched genomic DNA regions using Primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/). Real-time Q-PCR was performed on non-amplified shLuc, shKdm5b and shKdm5b+mut RNAPII and Input ES cell ChIP DNA using an Applied Biosystems OneStepPlus machine.
RNA-Seq analysis
Poly-A mRNA was purified using a Dynabeads mRNA purification kit. Double-stranded cDNA was generated using a Super-Script double-stranded cDNA synthesis kit (Invitrogen). cDNA was subjected to library preparation as described above. RNA-Seq libraries were sequenced on an Illumina HiSeq platform according to the manufacturer's protocol.
The RPKM measure (read per kilobases of exon model per million reads) (29) was used to quantify the mRNA expression level of a gene from RNA-Seq data. Differentially expressed genes were identified using edgeR (FDR < 0.001; FC > 1.5) (30).
MISO (31) was used to analyze differential expression of cassette exons (upregulated, downregulated) between control and KDM5B-depleted ES cells using the following parameters (32): exons with the inclusion level change ΔΨ > 0.1 were downregulated while those with ΔΨ < −0.1 were upregulated. The Bayes factors calculated by MISO for differentially expressed exons were required to be >1.5. We extracted the database of cassette exons that is included in the MISO software package. The inclusion levels (Ψ) of alternatively spliced exons were estimated using the MISO algorithm with default parameters (filter_results = True and min_event_reads = 20).
Confirmation of alternative splicing using RT-PCR
Primers were designed for detecting alternatively spliced isoforms using PrimerSeq software (http://primerseq.sourceforge.net/) (33). RT-PCR was performed on non-amplified shLuc and shKdm5b ES cell cDNA.
RESULTS
Depletion of KDM5B leads to altered RNAPII occupancy at active genes
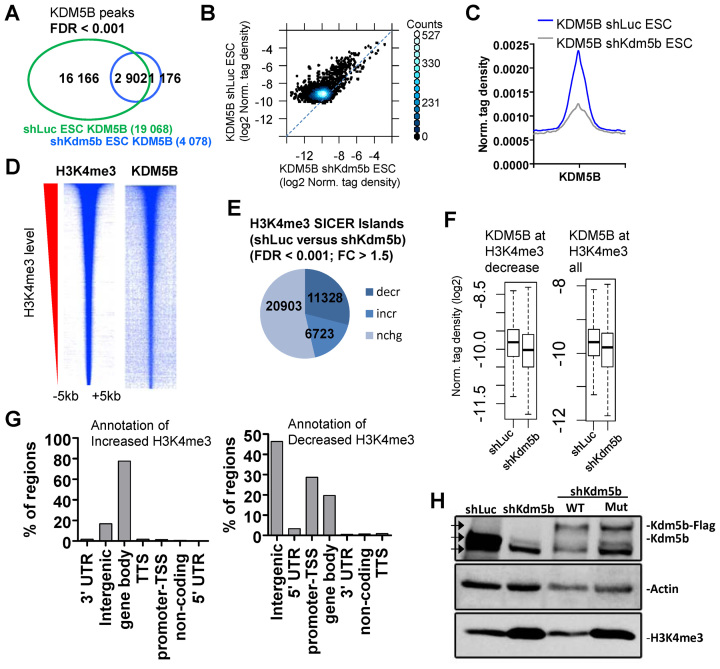
KDM5B was previously shown to be enriched near promoters and gene body regions (14), and depletion of KDM5B leads to decreased promoter H3K4 methylation, and increased gene body methylation (14). To investigate whether KDM5B binding sites are lost in KDM5B-depleted ES cells we performed ChIP-Seq. Results from these experiments show that >85% of KDM5B-binding sites (SICER islands; FDR < 0.001) are lost in KDM5B-knockdown ES cells (Figure 1A). Depletion of KDM5B binding in shKdm5b ES cells is also evident in a scatter plot (Figure 1B) and an average profile of KDM5B density (Figure 1C). We then investigated whether KDM5B levels are positively correlated with H3K4me3 levels. Heat maps demonstrate that genomic regions highly enriched with H3K4me3 are also bound by KDM5B (Figure 1D).
Figure 1.
KDM5B regulates H3K4me3 in ES cells. (A) Venn diagram showing overlap of KDM5B in control (shLuc) and KDM5B-depleted (shKdm5b) ES cells (See ‘Materials and Methods’ section for SICER-analysis; FDR < 0.001). (B) Scatter plot and (C) average profile of KDM5B densities in shLuc and shKdm5b ES cells. (D) Heat maps of KDM5B and H3K4me3 densities at H3K4me3 regions in ES cells. (E) Depletion of KDM5B leads to decreased H3K4me3 levels at promoters and increased levels in gene body regions (See ‘Materials and Methods’ section for SICER-analysis; fold-change > 1.5, FDR < 0.001). (F) Boxplots of KDM5B densities in shLuc and shKdm5b ES cells at H3K4me3 regions. (G) Annotation of H3K4me3 regions using HOMER (34) software. (H) Western blot of KDM5B and H3K4me3 in shLuc, shKdm5b, shKdm5b+ wild-type KDM5B (WT), shKdm5b+ mutant KDM5B (H499A; mut). Note the increased levels of H3K4me3 in shKdm5b and shKdm5b+mut ES cells relative to control (shLuc) and shKdm5b+WT ES cells. The three arrowheads in the KDM5B western blot represent the size of endogenous KDM5B (middle arrow), FLAG-tagged KDM5B (top arrow) and a non-specific band (bottom arrow).
To directly evaluate whether depletion affects the global distribution of H3K4me3 in shKdm5b ES cells (14,18) we used ChIP-Seq data (Figure 1E) (14). Spatial clustering for identification of SICER software (28) was used to define ChIP-enriched islands (FDR < 0.001; FC > 1.5). These results demonstrate that 32% of all H3K4me3 islands (see ‘Materials and Methods’ section; FC > 1.5, FDR < 0.001) decrease, while 19% of H3K4me3 islands increase in KDM5B-depleted ES cells. Moreover, KDM5B levels are reduced at regions with decreased H3K4me3 (Figure 1F). Annotation of differentially enriched H3K4me3 islands using HOMER software (34) revealed that decreased H3K4me3 islands mainly reside nearby promoter regions (28%) (±2 kb of TSS) or intergenic regions (46%), while H3K4me3 islands with increased levels in KDM5B-depleted ES cells are located in gene body regions (78%) but not promoter regions (1%) (Figure 1G).
To confirm the specificity of the shRNA sequences, and to determine whether alterations in H3K4me3 levels in KDM5B-depleted ES cells requires a KDM5B catalytically active JmjC domain, or KDM5B binding, we performed a rescue experiment by overexpressing an shRNA-resistant version of WT KDM5B or an shRNA-resistant enzymatically mutant version of KDM5B containing a point mutant in the first histidine residue of the Fe(II)-binding motif (H499A) (35). RNAi knockdown of KDM5B resulted in increased H3K4me3 (Figure 1H), which confirms our previous findings (18) and is in alignment with the known function of KDM5B as a H3K4 demethylase. Moreover, while H3K4me3 levels were similar between shKdm5b ES cells overexpressing WT KDM5B and control (shLuc) ES cells, H3K4me3 levels increased in shKdm5b ES cells overexpressing a mutant (mut) version of KDM5B relative to control ES cells (Figure 1H). These results confirm previous findings that the catalytic JmjC domain of KDM5B is important for regulating H3K4me3 levels.
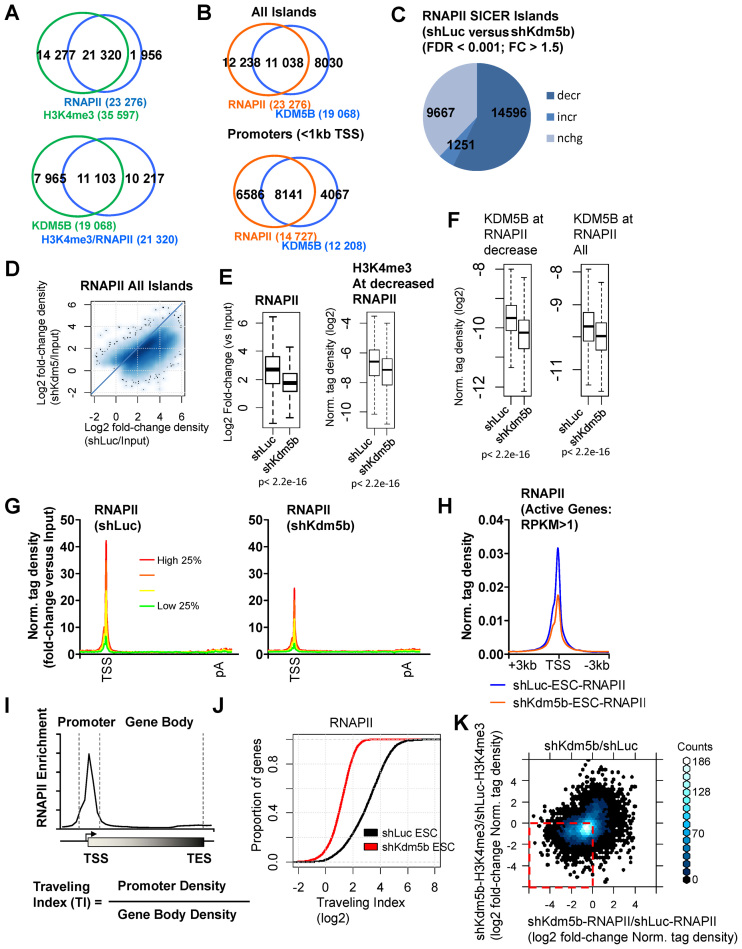
Because H3K4me3 is highly localized at promoter regions, we compared regions occupied by H3K4me3 and RNAPII, which is also localized at transcriptional start (TSS) regions and found that 92% of RNAPII bound regions contain H3K4me3 marks (Figure 2A, top). We also compared regions bound by KDM5B and regions co-occupied by H3K4me3/RNAPII, and found that RNAPII/H3K4me3 is enriched at 42% of KDM5B-bound regions (Figure 2A, bottom). In addition, RNAPII occupies 58% of all KDM5B-bound regions (Figure 2B, top), and co-binds 67% of KDM5B occupied promoter regions (Figure 2B, bottom).
Figure 2.
KDM5B regulates RNAPII occupancy at active genes in ES cells. (A) Venn diagrams showing overlap of RNAPII and H3K4me3 (left) and KDM5B and RNAPII/H3K4me3 at all SICER-defined islands. (B) Venn diagrams showing co-occupancy of KDM5B and RNAPII at all islands (top) or at promoter regions (bottom). (C) Change in the global distribution of RNAPII SICER-defined islands in shKdm5b ES cells relative to shLuc ES cells. (D) Scatter plots of RNAPII densities in shLuc and shKdm5b ES cells. x-axis: log2 fold-change density (shKdm5b/Input); y-axis: log2 fold-change density (shLuc/Input). (E) Boxplots of RNAPII densities in shLuc and shKdm5b ES cells. (F) Boxplots of KDM5B densities in shLuc and shKdm5b ES cells at RNAPII regions. (G) ChIP-Seq tag density of RNAPII binding normalized by input (log2 fold-change versuss Input) for all refseq genes sorted into quartiles based on their expression level in control (shLuc) ES cells. (H) ChIP-Seq tag density of RNAPII at TSS regions in shLuc and shKdm5b ES cells. (I) Schematic describing the calculation used to determine the traveling index (TI) at RNAPII marked genes in ES cells. The promoter bin is defined as a 1 kb window around the TSS of genes marked by RNAPII, while the transcribed region (gene body) is defined as the region extending to the TES. The TI is calculated from the ratio of the density of RNAPII in the promoter bin to the density of RNAPII in the gene body bin. (J) Empirical cumulative distribution for the TI of RNAPII across all genes for shLuc (black) and shKdm5b (red) ES cells. Y-axis shows the percentage of genes that exhibit a TI less than the value specified by the x-axis. A line shifted to the left means a systematic decrease in the TI. P-value for all <1E-5 (Kolmogorov–Smirnov test). Note the decreased TI for genes marked by RNAPII in shKdm5b ES cells. (K) Scatter plot of the ratio of relative tag densities of RNAPII and H3K4me3 in shKdm5b versus shLuc ES cells. x-axis: log2 fold-change (shKdm5b-RNAPII/shLuc-RNAPII); y-axis: log2 fold-change (shKdm5b-H3K4me3/shLuc-H3K4me3).
To investigate whether depletion of KDM5B alters the genome-wide distribution of RNAPII in KDM5B-depleted ES cells (14,18) we used ChIP-Seq. These results demonstrate that 57% of RNAPII islands had decreased levels in shKdm5b ES cells relative to control (shLuc) ES cells (FDR < 0.001, FC > 1.5) (Figure 2C). A scatter plot of densities of all RNAPII islands (Figure 2D) revealed that most RNAPII-occupied regions exhibit decreased levels in shKdm5b ES cells. Boxplots of densities at TSS to pA sites [poly (A)] also show global decreases in RNAPII levels in KDM5B-depleted ES cells (Figure 2E, left panel). Moreover, we observed decreased H3K4me3 levels at regions with decreased RNAPII binding (Figure 2E, right panel), and decreased KDM5B binding in KDM5B-depleted ES cells at RNAPII-bound regions (Figure 2F). Average profiles of densities at TSS to pA sites also show global decreases in RNAPII levels in KDM5B-depleted ES cells (Figure 2G).
Global decreases in RNAPII are also evident by evaluating average profiles of RNAPII densities around TSS regions in KDM5B-depleted ES cells (Figure 2H). To further characterize RNAPII binding in KDM5B-depleted ES cells we quantified the relative ratio of RNAPII in promoter to that in gene body regions, which we have termed the ‘traveling index’ (TI), which has also been called the traveling ratio (36) or pausing index (37) (Figure 2I). For genes where the rate of promoter clearance is similar to the rate of initiation, the TI is close to 1 (36,38), while genes with a TI greater than 1 exhibit promoter clearance of RNAPII at a rate lower than the initiation rate (38). It was previously found that 91% of genes in ES cells exhibit a RNAPII TI >2, demonstrating that RNAPII density is greater in proximal-promoter regions relative to gene body regions (38). Using this calculation, we observed a decrease in the TI for RNAPII in KDM5B-depleted ES cells (red) relative to control (shLuc) ES cells (black) (Figure 2J): We also observed 91% of genes with a RNAPII TI >2 in control ES cells, but only 58% of genes with a RNAPII TI >2 in KDM5B-depleted ES cells. To evaluate whether KDM5B binding influences the RNAPII TI we quantified the relative ratio of RNAPII in promoter to that in gene body regions for genes whose promoters are bound or unbound by KDM5B, respectively. Using this approach, we observed a greater decrease in the TI for RNAPII in KDM5B-depleted ES cells (red) relative to control (shLuc) ES cells (black) for genes whose promoters are bound by KDM5B relative to genes whose promoters are unbound by KDM5B (Supplementary Figure S1A and B).
To determine whether decreased RNAPII binding is due to decreased occupancy of KDM5B or altered H3K4 methylation levels we performed a rescue experiment by overexpressing an shRNA-resistant enzymatically mutant version of KDM5B (H499A) (35) in KDM5B-depleted ES cells, and subsequently evaluated RNAPII binding using ChIP-PCR. Our results confirm our ChIP-Seq findings, where we observed decreased RNAPII binding at several genomic regions in KDM5B-depleted ES cells relative to control (shLuc) ES cells (Supplementary Figure S1C). Moreover, we also observed decreased RNAPII occupancy in shKdm5b ES cells overexpressing mutant KDM5B (shKdm5b+mut), suggesting that decreased RNAPII occupancy is due to altered H3K4me3 levels and not decreased KDM5B binding (Supplementary Figure S1C).
These results suggest that depletion of KDM5B impacts occupancy of RNAPII at promoter regions and may suggest that KDM5B affects the rate of RNAPII elongation, including promoter clearance and intragenic speed of RNAPII. These results also demonstrate that a significant fraction of genes have a higher RNAPII density in gene body regions relative to promoter regions in KDM5B-depleted ES cells, which could be a result of cryptic RNAPII recruitment or initiation following spreading of H3K4 methylation from promoters to gene body regions in KDM5B-depleted ES cells.
We also found a correlation between decreased levels of RNAPII and H3K4me3 in KDM5B-depleted ES cells (Figure 2K), suggesting that a re-distribution of H3K4me3 from promoter regions to gene body regions may negatively impact RNAPII elongation.
KDM5B regulates RNAPII initiation in ES cells
RNAPII is phosphorylated on the C-terminal domain (CTD) of the large subunit during the transcription cycle. Following recruitment of RNAPII to the pre-initiation complex, which contains an unphosphorylated CTD, the CTD is phosphorylated at the serine five residue (Ser5) during initiation and the serine two residue (Ser2) during elongation (39). While the majority of RNAPII Ser5P peaks can be found nearby promoter regions, because RNAPII Ser5P can also be observed in gene body region (40), RNAPII Ser5P may also be involved in non-initiation transcriptional events.
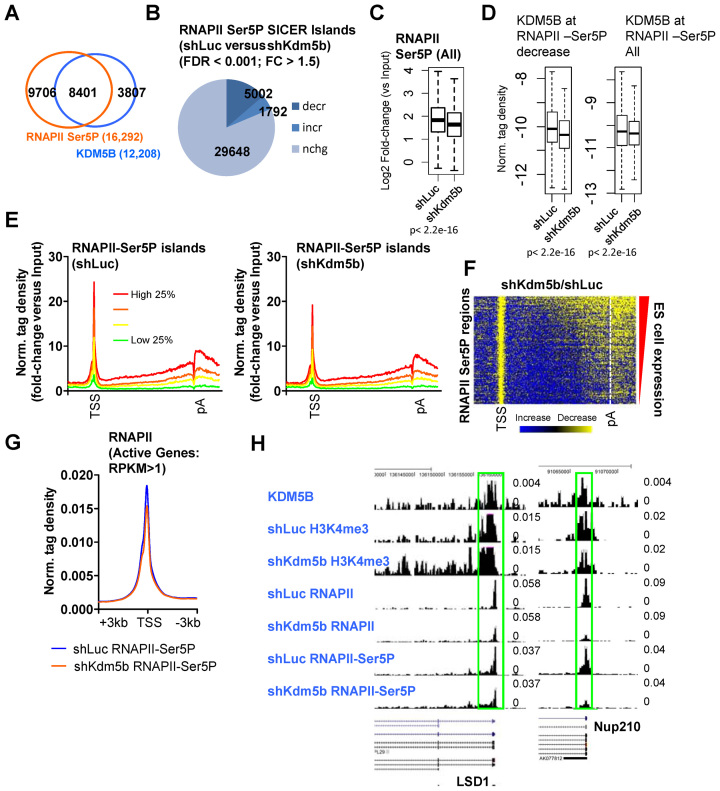
To investigate the role of KDM5B in RNAPII initiation we performed ChIP-Seq to evaluate RNAPII Ser5P levels in KDM5B-depleted ES cells. A comparison of KDM5B binding sites with RNAPII Ser5P bound regions revealed that >69% of KDM5B targets were enriched with RNAPII Ser5P at promoter regions (Figure 3A). By evaluating the global distribution of RNAPII Ser5P regions we observed a global decrease (Figure 3B and C), although a smaller number of RNAPII Ser5P occupied regions exhibited increased levels. Moreover, KDM5B binding levels decreased at RNAPII-Ser5P occupied regions in KDM5B-depleted ES cells relative to control ES cells (Figure 3D). Average profiles (Figure 3E) and heatmaps (Figure 3F) of densities at TSS to pA regions also revealed global decreases in RNAPII Ser5P levels in KDM5B-depleted ES cells.
Figure 3.
KDM5B regulates RNAPII initiation at active genes in ES cells. (A) Venn diagram showing overlap of RNAPII-Ser5P and KDM5B at promoter regions in ES cells. (B) Change in the global distribution of RNAPII-Ser5P in shKdm5b ES cells. (C) Boxplots of RNAPII-Ser5P densities in shLuc and shKdm5b ES cells. (D) Boxplots of KDM5B densities in shLuc and shKdm5b ES cells at RNAPII Ser5P regions. (E) Average profile of RNAPII-Ser5P binding normalized by input (log2 fold-change versus Input) of all refseq genes (TSS-TES) sorted into quartiles based on their expression level in ES cells. (F) Heat map of change in RNAPII-Ser5P in shKdm5b versus shLuc ES cells (yellow, decrease). (G) Average profile of RNAPII-Ser5P at TSS regions in shLuc and shKdm5b ES cells. (H) UCSC browser view of KDM5B, H3K4me3, RNAPII and RNAPII-Ser5P in shLuc and shKdm5b ES cells.
In addition, global decreases in RNAPII Ser5P are also observed by evaluating average profiles of RNAPII Ser5P densities around TSS regions (Figure 3G) in KDM5B-depleted ES cells. Because RNAPII levels decreased in KDM5B-depleted ES cells, to test whether KDM5B directly regulates RNAPII initiation or whether decreased RNAPII Ser5P levels are a result of decreased RNAPII binding in KDM5B-depleted ES cells, we evaluated RNAPII Ser5P levels by normalizing to total RNAPII. Results from these analyses demonstrate that RNAPII-Ser5P densities decrease in gene body regions of highly expressed genes in KDM5B-depleted ES cells relative to control (shLuc) ES cells (Supplementary Figure S2A). However, by normalizing to total RNAPII levels, we found that RNAPII-Ser5P promoter levels are similar between shLuc and shKdm5b ES cells (Supplementary Figure S2A). While these findings demonstrate that RNAPII-Ser5P levels decrease at active genes in KDM5B-depleted ES cells relative to control cells, because total RNAPII and initiated RNAPII (Ser5P) exhibit similar decreases in shKdm5b ES cells, it is likely that decreased RNAPII initiation (Ser5P) is a result of decreased RNAPII binding in KDM5B-depleted ES cells.
Overall, these results suggest that KDM5B regulates the rate-limiting step of initiation after RNAPII is recruited to promoters and subsequently modified on the CTD to Ser5P. It is possible that decreased initiation in KDM5B-depleted ES cells is due to spreading of H3K4 methylation from promoters to gene body regions. UCSC browser views also reveal decreased RNAPII and RNAPII Ser5P at promoters of active genes (Figure 3H). Overall, these results suggest that KDM5B positively regulates RNAPII initiation at promoter regions.
Depletion of KDM5B leads to dysregulated RNAPII elongation
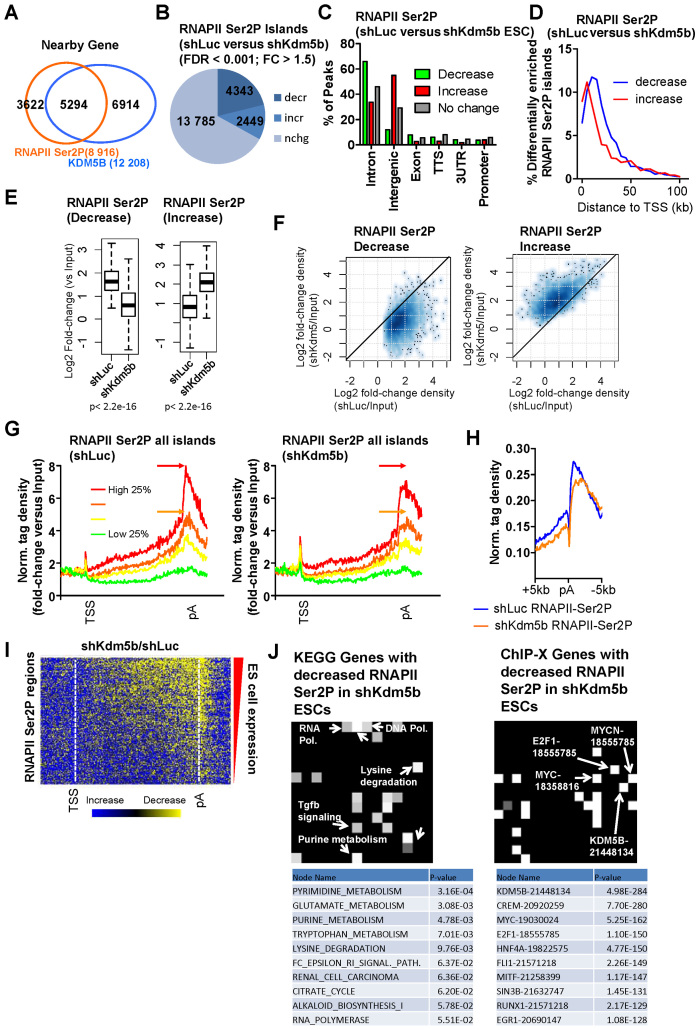
To evaluate the role for KDM5B in RNAPII elongation we interrogated RNAPII Ser2P occupancy in KDM5B-depleted ES cells using ChIP-Seq. A comparison of KDM5B binding sites with RNAPII Ser2P bound regions showed that >60% of RNAPII Ser2P enriched regions were co-occupied with KDM5B at genic regions (Figure 4A). An analysis of RNAPII Ser2P levels demonstrated a global decrease in KDM5B-depleted ES cells, although a subset displayed elevated levels (Figure 4B). Annotation of islands showed that RNAPII Ser2P levels decreased predominantly in introns (Figure 4C) while RNAPII Ser2P levels increased mainly in intergenic regions (Figure 4C). Moreover, islands with decreased RNAPII Ser2P levels were located further from TSS regions relative to islands with increased RNAPII Ser2P levels (Figure 4D), suggesting that RNAPII Ser2P elongation rates decrease as a function of distance from TSS regions in KDM5B-depleted ES cells. Combined, these results suggest a decreased rate of RNAPII elongation (RNAPII Ser2P), where decreased levels of RNAPII Ser2P were observed in gene body regions and increased RNAPII Ser2P levels were observed at a subset of regions closer to TSS regions. Although RNAPII Ser2P levels decreased at a subset of regions, it is possible that some of these increases reflect a shift in RNAPII Ser2P elongation closer to TSS regions (Figure 4D). Overall, these results suggest that elongation rates decrease globally in KDM5B-depleted ES cells.
Figure 4.
KDM5B regulates RNAPII elongation in ES cells. (A) Venn diagram showing overlap of RNAPII-Ser2P and KDM5B in gene body regions in ES cells. (B) Change in the global distribution of RNAPII-Ser2P in shKdm5b versus shLuc ES cells as defined by SICER analysis (see methods, fold-change > 1.5, FDR < 0.001). (C) Annotation of differentially enriched RNAPII-Ser2P regions shLuc and shKdm5b ES using HOMER software (34). (D) Histogram showing the distance of differentially enriched (increased or decreased) RNAPII Ser2P islands from TSS regions. Note that RNAPII Ser2P islands with decreased levels are located further from TSS regions relative to islands with increased levels. (E) Boxplots and (F) scatter plots of RNAPII-Ser2P densities in shLuc and shKdm5b ES cells. (G) Average profile of RNAPII-Ser2P binding normalized by input (log2 fold-change versus Input) at all refseq genes (TSS-TES) sorted into quartiles based on their expression level in control (shLuc) ES cells. The red and orange arrows denote the peak of RNAPII Ser2P in shLuc ES cells. (H) Average profile of RNAPII-Ser2P density at TES regions in shLuc and shKdm5b ES cells. RNAPII-Ser2P is known to be enriched at TES regions of genes. (I) Heat map of change in RNAPII-Ser2P in shKdm5b versus shLuc ES cells (yellow, decrease). (J) KEGG gene (left heatmap) and ChIP-X genes (right heatmap) evaluated using Network2Canvas demonstrates that metabolic and lysine degradation genes have decreased RNAPII-Ser2P in shKdm5b ES cells, and are bound by KDM5B. Each node (square) represents a gene list (shLuc versus shKdm5b bound genes bound by RNAPII-Ser2P associated with a gene-set library (KEGG or ChIP-X). The brightness (white) of each node is determined by its P-value.
Boxplots (Figure 4E) and scatter plots (Figure 4F) also revealed increased and decreased levels of RNAPII Ser2P in KDM5B-depleted ES cells. Dysregulated RNAPII Ser2P elongation is also evident by evaluating average profiles of RNAPII Ser2P densities around TSS-pA regions (Figure 4G) and TSS regions (Figure 4H). These analyses reveal decreased RNAPII Ser2P levels at TES regions in KDM5B-depleted ES cells (Figure 4G and H). Moreover, decreased RNAPII Ser2P levels are visible in a TSS-pA heatmap (Figure 4I).
Because RNAPII levels decreased in KDM5B-depleted ES cells, to test whether KDM5B directly regulates RNAPII elongation or whether decreased RNAPII Ser2P levels are a result of decreased RNAPII binding in KDM5B-depleted ES cells, we evaluated RNAPII Ser2P levels by normalizing to total RNAPII. Results from these analyses demonstrate that RNAPII-Ser2P densities decrease in gene body regions of highly expressed genes in KDM5B-depleted ES cells relative to control (shLuc) ES cells (Supplementary Figure S2B). These results suggest that RNAPII-Ser2P levels decrease at active genes in KDM5B-depleted ES cells relative to control cells. These findings implicate a role for KDM5B in regulating elongation after RNAPII is recruited to promoters and subsequently modified on the CTD to Ser2P. In addition, annotation of the RNAPII Ser2P differentially occupied regions revealed that the underlying genes encode transcripts involved in metabolic processes such as lysine degradation, and RNA polymerase (Figure 4J, left), and are bound by KDM5B (Figure 4J, right). In summary, these findings demonstrate a role for KDM5B in regulating RNAPII Ser2P elongation in ES cells.
Broad H3K4me3 domains are transcriptionally dysregulated in KDM5B-depleted ES cells
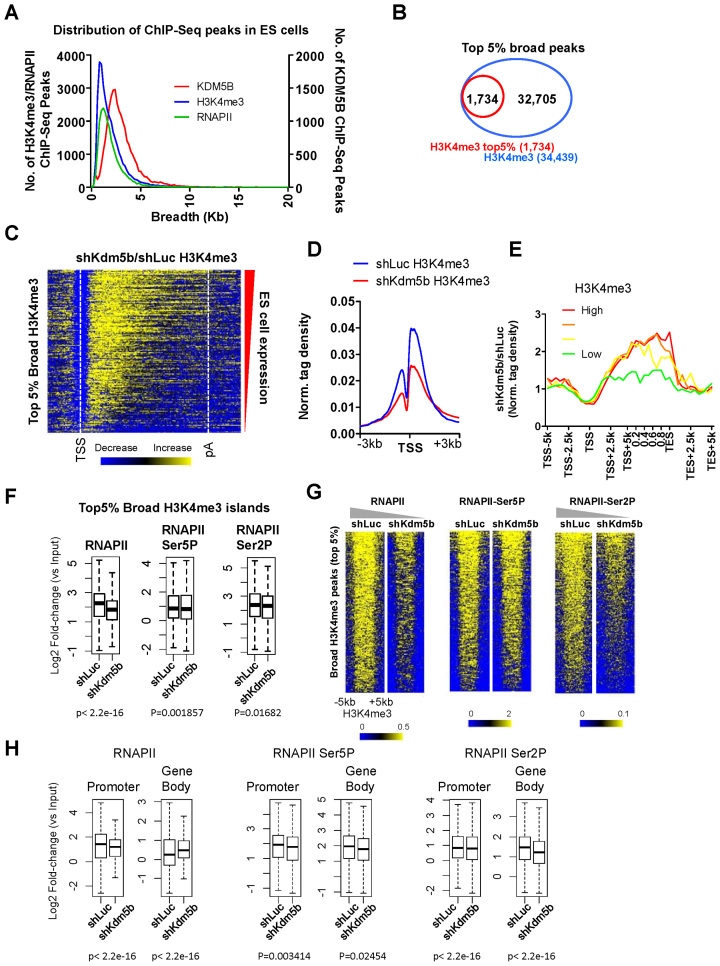
Broad H3K4me3 domains have been suggested to mark genes involved in cell identity (41), including pluripotency regulators. However, the role for broad H3K4me3 domains in transcriptional regulation, and how they are regulated by histone demethylases, is largely unknown. To investigate a potential role for H3K4me3 breadth in KDM5B-regulated transcriptional elongation, we first compared the breadth of H3K4me3, KDM5B and RNAPII in ES cells (Figure 5A). While the majority of KDM5B, H3K4me3 and RNAPII domains are 1–3 kb in length, domains broader than 35 and 45 kb were found for H3K4me3 and KDM5B, respectively, and the top 5% of broad H3K4me3 domains (Figure 5B) had an average length of ∼7kb. Because we previously found that depletion of KDM5B leads to decreased levels of H3K4 methylation at promoters and increased levels in gene body regions (14), we investigated whether regions of broad H3K4me3 exhibit a similar spreading of H3K4 methylation from promoter to gene body regions as was previously observed for all active genes (14). A heatmap (Figure 5C) and average profiles of H3K4me3 densities at TSS (Figure 5D) or TSS-pA regions (Figure 5E) revealed a marked decrease at promoter regions and an increase in gene body regions in KDM5B-depleted ES cells.
Figure 5.
KDM5B regulates RNAPII at genes with broad H3K4me3 domains. (A) Distribution of KDM5B, H3K4me3 and RNAPII SICER-defined ChIP-Seq peaks in ES cells. (B) Venn diagram showing the top 5% broadest H3K4me3 peaks in ES cells. (C) Heatmap of H3K4me3 ChIP-Seq tag density at TSS to pA regions of genes with the broadest (top 5%) H3K4me3 domains. (D) ChIP-Seq tag density of H3K4me3 around TSS regions of genes with the broadest (top 5%) H3K4me3 domains in shLuc and shKdm5b ES cells. (E) Correlation between changes in gene body histone methylation and expression level. Fold-change normalized tag density ratios (shKdm5b-ESC-H3K4me3/shLuc-ESC-H3K4me3) of H3K4me3 at genes with the broadest (top 5%) H3K4me3 domains, which were sorted into four groups based on their absolute expression level in control (shLuc) ES cells (red line, highest 25% expressed; green line, lowest 25% expressed). (F) Boxplots of RNAPII, RNAPII-Ser5P and RNAPII-Ser2P ChIP-Seq tag densities. log2 fold-change (shLuc-RNAPII/shLuc-Input); log2 fold-change (shKdm5b-RNAPII/shKdm5b-Input) at the broadest (top 5%) H3K4me3 domains in ES cells. (G) Heatmaps of RNAPII, RNAPII-Ser5P and RNAPII-Ser2P ChIP-Seq tag densities at the broadest (top 5%) H3K4me3 domains in ES cells. (H) Boxplots of densities of RNAPII, RNAPII-Ser5P and RNAPII-Ser2P ChIP-Seq tag densities at promoter and gene body regions of genes with the broadest (top 5%) H3K4me3 domains in ES cells.
To investigate whether transcriptional dynamics of genes marked by broad H3K4me3 domains are altered in KDM5B-depleted ES cells we evaluated the levels of RNAPII, RNAPII Ser5P and RNAPII Ser2P in shLuc and shKdm5b ES cells. Boxplots revealed that RNAPII, RNAPII Ser5P and RNAPII Ser2P levels decreased in KDM5B-depleted ES cells (Figure 5F), although the decrease of RNAPII was not as significant relative to all RNAPII marked regions (Figure 2E). Heatmaps also revealed decreased levels of RNAPII and RNAPII Ser2P in KDM5B-depleted ES cells (Figure 5G). While the decrease in H3K4me3 levels at promoter regions was similar between genes marked by broad H3K4me3 domains (Figure 5D) and all promoters (14), the increase in H3K4me3 in gene bodies was not as robust relative to genes marked with broad H3K4me3 domains (Figure 5E) (14). The slight differences in spreading of H3K4 methylation from promoters to gene bodies may be attributed to the breadth of H3K4me3 domains, which is significantly wider at these regions relative to all genes.
We also evaluated the relative levels of RNAPII, RNAPII Ser5P and RNAPII Ser2P in promoter and gene body regions that are at least 5 kb in length. While RNAPII, RNAPII Ser5P and RNAPII Ser2P levels decreased at promoters regions, and RNAPII Ser5P and RNAPII Ser2P levels decreased in gene body regions, RNAPII levels increased in gene body regions, suggesting that RNAPII may accumulate in gene bodies of regions with broad H3K4me3 domains (Figure 5H).
While our previous findings demonstrate that H3K4me3 domains are globally altered in KDM5B-depleted ES cells (14), it is unknown whether the sharpest H3K4me3 domains (top 5%) are altered in a similar way. As described above, the majority of H3K4me3 domains are 1–3 kb in length. In addition, the top 5% of sharp H3K4me3 domains (Supplementary Figure S3A) had an average length of 940 bp. An average profile (Supplementary Figure S3B) and boxplot (Supplementary Figure S3C) revealed that H3K4me3 levels decreased in KDM5B-depleted ES cells. We also found that regions of sharp H3K4me3 exhibit a similar spreading of H3K4 methylation from promoter to gene body regions as was previously observed for all active genes (14). Our results show that a heatmap (Supplementary Figure S3D) and an average profile (Supplementary Figure S3E) revealed a decrease at promoter regions and an increase in gene body regions in KDM5B-depleted ES cells.
In an analogous way as described above, to investigate whether transcriptional dynamics of genes marked by sharp H3K4me3 domains are altered in KDM5B-depleted ES cells we evaluated the levels of RNAPII, RNAPII Ser5P and RNAPII Ser2P in shLuc and shKdm5b ES cells. Boxplots showed that RNAPII, RNAPII Ser5P and RNAPII Ser2P levels decreased in KDM5B-depleted ES cells (Supplementary Figure S3F). Overall, these results demonstrate that depletion of KDM5B leads to decreased RNAPII, RNAPII Ser5P and RNAPII Ser2P at genes marked by broad or sharp H3K4me3 peaks.
KDM5B regulates splicing in ES cells
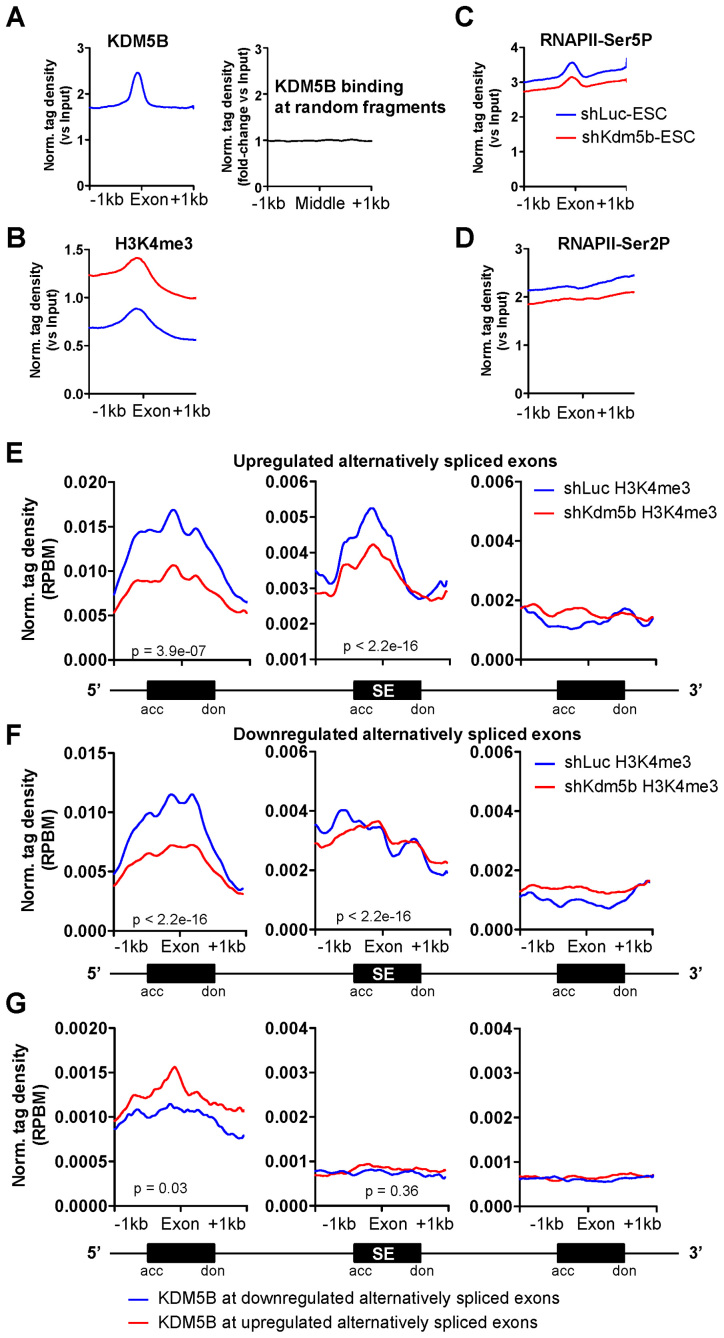
Co-transcriptional processes such as histone demethylation and splicing co-occur with the progression of RNAPII along gene bodies to facilitate elongation and to reset the underlying chromatin. In addition, histone modifications have important roles in controlling splice site choice (42–45), and H3K4me3 has been shown to be enriched at exon-intron junctions (46,47). These results suggest that spreading of H3K4 methylation from promoters to gene bodies, or depletion of KDM5B, may affect splicing. We also observed enrichment of KDM5B (Figure 6A), elevated H3K4me3 (Figure 6B), decreased RNAPII Ser5P (Figure 6C) and decreased RNAPII Ser2P (Figure 6D) in exon regions of gene bodies. Enrichment of KDM5B binding was markedly higher at exonic regions of gene bodies relative to random genomic regions of similar size and frequency (Figure 6A, right panel). These results implicate a potential role for KDM5B in regulating splicing.
Figure 6.
KDM5B is involved in regulating alternative splicing in ES cells. Average profiles of (A) KDM5B (left panel), (B) H3K4me3, (C) RNAPII-Ser5P and (D) RNAPII-Ser2P densities at exons (Norm. tag density versuss Input). Only exons located in gene body regions (>2 kb from TSS) were used for this analysis. The right panel in (A) shows KDM5B binding at random genomic sequences (Norm. tag density versus Input). An increase of H3K4me3 in gene body regions in KDM5B-depleted ES cells is consistent with previous findings. (E–G) Average profile of H3K4me3 densities at (E) upregulated or (F) downregulated alternatively spliced exon (cassette exons) regions and their flanking constitutively spliced exons in ES cells. Schematic of alternatively spliced exon and the flanking exons is shown below the graph. Densities were calculated at cassette exons from the two alternatively spliced exon groups (upregulated and downregulated) and levels of H3K4me3 are displayed relative to splice acceptor (acc) and donor (don) sites across cassette exons. (G) Average profile of KDM5B densities at upregulated or downregulated alternatively spliced exon regions and constitutively spliced flanking exons in ES cells. P-values for relative enrichment of H3K4me3 or KDM5B at differentially expressed alternatively spliced exons are shown.
To directly test whether KDM5B regulates splicing, we performed paired-end RNA-Seq to determine expression levels of annotated alternatively spliced exons and splicing patterns in control and KDM5B-depleted ES cells. Our analyses revealed that depletion of KDM5B leads to a significant upregulation of 275 alternatively spliced exons and downregulation of 302 alternatively spliced exons. We then examined the levels of H3K4me3 at the alternatively spliced exons and around the 5΄ and 3΄ neighboring exons (Figure 6E). These analyses revealed that upregulated alternatively spliced exons and their 5΄ neighboring exons had decreased levels of H3K4me3, while 3΄ exons had lower levels of H3K4me3 (Figure 6E), suggesting that H3K4me3 at alternatively spliced exons and upstream exons may regulate splicing. In addition, the 5΄ exons of downregulated alternatively spliced exons had decreased levels of H3K4me3, while 3΄ exons had lower levels of H3K4me3 (Figure 6F). We confirmed the differential expression of several alternatively spliced exons in shKdm5b ES cells using RT-PCR. These results reveal exon exclusion and inclusion relative to constitutively expressed 5΄ and 3΄ flanking exons in KDM5B-depleted ES cells (Supplementary Figure S4A). Interestingly, analysis of RNA-Seq data revealed that Pbrm1 is highly expressed in ES cells and during differentiation (18). Moreover, Pbrm1 has also been shown to be important for cardiac development (48). CD151 is also expressed during ES cell differentiation (18) and it is involved in diverse functions such as cell-cell adhesion, cell migration, platelet aggregation and angiogenesis (49).
Because we observed enrichment of KDM5B at all exons (Figure 6A), we evaluated levels of KDM5B at upregulated and downregulated alternatively spliced exons. These results revealed that 5΄ exons of upregulated alternatively spliced exons had elevated levels of KDM5B relative to downregulated alternatively spliced exons (Figure 6G). These analyses also show that KDM5B levels at 5΄ exons neighboring alternatively spliced exons are positively correlated with H3K4me3 levels. In this case, 5΄ exons of upregulated alternatively spliced exons have higher levels of KDM5B and H3K4me3 (shLuc) relative to downregulated alternatively spliced exons (Figure 6G). Overall, these results show KDM5B is enriched at 5΄ exons neighboring alternatively spliced exons, and depletion of KDM5B leads to decreased levels of H3K4me3 at alternatively spliced exons and their 5΄ neighboring exons, suggesting that KDM5B plays a role in regulating splicing in ES cells. Because depletion of KDM5B leads to decreased H3K4me3 at promoter regions, and we observed decreased H3K4me3 at 5΄ exons neighboring alternatively spliced exons, these results suggest that expression of exons near promoter regions are likely affected by depletion of KDM5B.
To test whether depletion of KDM5B affects splicing of exons near promoter regions we evaluated expression levels of exons in close proximity to TSS regions (5΄ exon within 1 kb of TSS). These results show that depletion of KDM5B leads to upregulation of 101 alternatively spliced exons and downregulation of 116 alternatively spliced exons. As described above, we also examined the levels of H3K4me3 at alternatively spliced exons and around the 5΄ and 3΄ neighboring exons. These results also show that upregulated alternatively spliced exons and their 5΄ neighboring exons (within 1 kb of TSS) had decreased levels of H3K4me3, while 3΄ exons had lower levels of H3K4me3 (Supplementary Figure S4B), further suggesting that H3K4me3 at alternatively spliced exons and upstream exons regulate splicing. We also found that the 5΄ exons (<1 kb of TSS) of downregulated alternatively spliced exons had decreased levels of H3K4me3, while 3΄ exons had lower levels of H3K4me3 (Supplementary Figure S4C). Moreover, KDM5B levels at 5΄ exons (<1 kb of TSS) neighboring alternatively spliced exons are positively correlated with H3K4me3 levels, and 5΄ exons of upregulated alternatively spliced exons have higher levels of KDM5B and H3K4me3 (shLuc) relative to downregulated alternatively spliced exons (Supplementary Figure S4D). Overall, these results show that depletion of KDM5B regulates splicing in ES cells.
While slow RNAPII elongation has been demonstrated to allow weak splice sites to be recognized, which can lead to higher inclusion of alternative exons (20), slow elongation can also cause exon skipping by promoting recruitment of negative splicing factors (22). Our findings, which demonstrate that slow elongation promotes exon inclusion or exclusion, is in alignment with two models that have been proposed to explain the regulation of alternative pre-mRNA splicing by RNAPII.
In addition, functional annotation showed that genes with differentially expressed alternatively spliced exons in KDM5B-depleted ES cells are developmentally regulated (Supplementary Figure S5A) and bound by KDM5B (Supplementary Figure S5A). We then compared these genes with differentially expressed alternatively spliced exons with global expression data from ES cells and embryoid body (EB) differentiated cells (50) using gene set enrichment analysis (51). This analysis shows that expression of genes with upregulated alternatively spliced exons is enriched in ES cells (Supplementary Figure S5B), while expression of genes with downregulated alternatively spliced exons is enriched in both ES cells and EB differentiated cells (Supplementary Figure S5B). Moreover, DAVID (52) gene ontology analysis revealed that genes with upregulated alternatively spliced exons are involved in metabolic (e.g. nucleotide) processes whereas genes with downregulated alternatively spliced exons are involved in RNA splicing, gene expression, RNA processing and chromatin modification (Supplementary Figure S5C), suggesting that a loss of KDM5B impacts ES cell function. Overall, these results demonstrate a role for KDM5B in regulating splicing in ES cells.
DISCUSSION
Although H3K4 methylation is widely known to be localized at active promoters (3), as more than 80% of genes with H3K4me3 marked promoters are actively transcribed (53), its role in transcription events such as elongation and alternative splicing are largely unknown. Evidence suggests that H3K4me3 is necessary for RNAPII recruitment to promoters, but not sufficient for elongation (54). Also, it was reported that breadth of H3K4me3 is associated with elevated transcription elongation (55), suggesting an epigenetically regulated transcriptional signature at active genes. It has also been suggested that exon length is correlated with activating histone modifications such as H3K4me3 and splicing events (56). Moreover, HDACs, which have been shown to interact with KDM5B (57), are also thought to regulate splicing (58), and H3K4 methyltransferase complex proteins have also been shown to regulate splicing (59).
In this study we found that KDM5B, an H3K4 demethylase, which is co-localized with RNAPII at promoters of active genes in ES cells, regulates RNAPII promoter occupancy, transcription initiation and elongation, and splicing in ES cells. Our findings also suggest that global decreases in promoter H3K4me3 levels leads to a dysregulated transcriptional cycle comprised of decreased RNAPII recruitment, decreased initiation, decreased elongation and altered splicing events.
There are several plausible mechanisms for globally decreased occupancy of RNAPII, RNAPII Ser5P and RNAPII Ser2P, and differential expression of alternatively spliced exons in KDM5B-depleted ES cells. First, decreased H3K4me3 levels at promoter regions caused by depletion of KDM5B may influence the rate of RNAPII recruitment to promoters, which subsequently affects RNAPII initiation, elongation and expression of alternatively spliced exons. Second, increased H3K4me3 levels in gene bodies of active genes caused by depletion of KDM5B (14) may lead to cryptic recruitment and initiation of RNAPII, which may influence the rate of RNAPII initiation, elongation and expression of alternatively spliced exons. Third, because KDM5B binds to gene body regions of active genes (14), and it has been suggested to interact with MRG15, an H3K36me3 interacting protein (60), a loss of KDM5B may negatively influence the rate of transcription elongation. Each of these scenarios would likely lead to decreased rates of elongation and more time for splicing machinery to recognize alternatively spliced exons in the final transcript. Our results support the model of transcription-coupled splicing, where decreased rates of elongating RNAPII Ser2P may lead to dysregulated splicing. Also, because KDM5B-depleted ES cells exhibit a decrease in the TI (Figure 2J), these results may suggest that KDM5B plays a role in regulating RNAPII pausing or occupancy of RNAPII at promoter regions. While promoter-proximal RNAPII levels decreased at active and non-productive genes in KDM5B-depleted ES cells, we also observed decreased levels of initiated RNAPII (Ser5P) and elongating RNAPII (Ser2P), which is thought to be essential for release of paused RNAPII and for productive elongation (61,62). Because depletion of KDM5B leads to decreased RNAPII Ser2P levels, and decreased pausing is associated with increased RNAPII Ser2P levels in gene bodies, while it is possible that KDM5B regulates pausing, our results suggest that depletion of KDM5B affects promoter occupancy rather than pausing. On the other hand, because depletion of KDM5B leads to decreased H3K4me3 levels in promoter regions and increased H3K4me3 in gene body regions, it is possible that increased release of paused RNAPII into gene bodies due to decreased levels of RNAPII and H3K4me3 in promoter regions is accompanied by slower elongation rates due to increased H3K4me3 in gene bodies. Moreover, it is likely that alterations in H3K4me3 are a result of depletion of KDM5B as opposed to differential expression of other H3K4 modifying enzymes, as previous work demonstrated that expression of Jarid family members is unaltered in KDM5B-depleted ES cells (18), and our current results demonstrate that expression of the H3K4 methyltransferases, Setd1a and Kmt2a/MLL1, is also unaltered in KDM5B-depleted ES cells (Supplementary Figure S6). Furthermore, it is likely that alterations in H3K4me3 or loss of KDM5B binding leads to altered RNAPII elongation in KDM5B-depleted ES cells as opposed to changes in histone acetylation due to depletion of KDM5B which could affect elongation rates. While H3K4ac has been shown to be enriched at active promoters and regulated by H3K4me3 (63), our results demonstrate that H3K4ac levels are similar between shLuc and shKdm5b ES cells. Average profiles (Supplementary Figure S7A and B), heat maps (Supplementary Figure S7C), scatter plots (Supplementary Figure S7D) and a UCSC genome browser view (Supplementary Figure S7E) show that H3K4ac levels are similar between shLuc and shKdm5b ES cells. Additionally, H3K4ac levels were similarly low in gene body regions of shLuc and shKdm5b ES cells (Supplementary Figure S7E and F). Western blots also confirmed similar levels of H3K4ac between shLuc and shKdm5b ES cells (Supplementary Figure S7G). Overall, our results demonstrate that KDM5B plays an important role in regulating RNAPII dynamics in ES cells.
Two models have been proposed to explain how RNAPII elongation dynamics regulate RNA splicing (64). The kinetic model suggests that the rate of RNAPII elongation regulates alternative splicing events (20). In this case, slow elongation generally favors exon inclusion, while an increased rate of elongation favors exon exclusion. The recruitment model suggests that splicing factors associating with transcriptional machinery or chromatin may influence splice choices. An integrative model has also been suggested to control splice choice, where similar alterations in elongation rates may lead to different splice choice (22). Our results are in agreement with the integrative model, where slow elongation leads to exon inclusion or exclusion.
It has been suggested that epigenetic information, such as H3K4me3, is predictive of mRNA production (54,65,66), RNAPII recruitment and elongation. While these previous analyses have evaluated static states of epigenetic data (e.g. histone modification levels in different cell types), rather than dynamic systems involving modulation of histone modifications, it is unclear whether co-localization of histone modifications with transcriptional machinery (e.g. RNAPII) at promoters regulates the transcriptional cycle. Because our results suggest that modulation of H3K4 methylation levels in promoter and gene body regions, by depleting KDM5B, influences RNAPII initiation and elongation, and splicing in ES cells, our study may to serve as a predictive platform for assessing RNAPII dynamics from histone modification information.
CONCLUSION
Together, these findings provide novel insights into the role for KDM5B and H3K4me3 in regulating RNAPII elongation and splicing in ES cells, where KDM5B-mediated focusing of H3K4 methylation near promoters to prevent the spread of these marks to surrounding regions maintains integrity of the transcription cycle.
ACCESSION NUMBER
The sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE94739.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr David Bentley for providing the pCSH2-KDM5B WT and mutant (H499A) plasmids, and Jiji Kurup for help with designing PCR primers. The DNA Sequencing Core facility of NHLBI assisted with this work. This work utilized the Wayne State University High Performance Computing Grid for computational resources (https://www.grid.wayne.edu/) and the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov).
Authors' contributions: B.L.K. conceived of the study, designed and carried out the experiments, analyzed the sequencing data and drafted the manuscript. R.H. helped with some of the experiments. All authors have read and approved the final version of this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wayne State University; Karmanos Cancer Institute; National Heart, Lung and Blood Institute [1K22HL126842-01A1 B.L.K.]. Funding for open access charge: Wayne State University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kouzarides T. Chromatin modifications and their function. Cell. 2007; 128:693–705. [DOI] [PubMed] [Google Scholar]
- 2. Ringrose L., Paro R.. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004; 38:413–443. [DOI] [PubMed] [Google Scholar]
- 3. Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K.. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837. [DOI] [PubMed] [Google Scholar]
- 4. Schubeler D., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J. et al. . The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004; 18:1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E. et al. . Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005; 122:517–527. [DOI] [PubMed] [Google Scholar]
- 6. Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B.. A high-resolution map of active promoters in the human genome. Nature. 2005; 436:876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sims R.J. 3rd, Nishioka K., Reinberg D.. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003; 19:629–639. [DOI] [PubMed] [Google Scholar]
- 8. Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T.. Active genes are tri-methylated at K4 of histone H3. Nature. 2002; 419:407–411. [DOI] [PubMed] [Google Scholar]
- 9. Schneider R., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T.. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004; 6:73–77. [DOI] [PubMed] [Google Scholar]
- 10. Mosammaparast N., Shi Y.. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 2010; 79:155–179. [DOI] [PubMed] [Google Scholar]
- 11. Lee M.G., Wynder C., Bochar D.A., Hakimi M.A., Cooch N., Shiekhattar R.. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell. Biol. 2006; 26:6395–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klose R.J., Zhang Y.. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007; 8:307–318. [DOI] [PubMed] [Google Scholar]
- 13. Christensen J., Agger K., Cloos P.A., Pasini D., Rose S., Sennels L., Rappsilber J., Hansen K.H., Salcini A.E., Helin K.. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007; 128:1063–1076. [DOI] [PubMed] [Google Scholar]
- 14. Kidder B.L., Hu G., Zhao K.. KDM5B focuses H3K4 methylation near promoters and enhancers during embryonic stem cell self-renewal and differentiation. Genome Biol. 2014; 15:R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamamoto S., Wu Z., Russnes H.G., Takagi S., Peluffo G., Vaske C., Zhao X., Moen Vollan H.K., Maruyama R., Ekram M.B. et al. . JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell. 2014; 25:762–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Albert M., Schmitz S.U., Kooistra S.M., Malatesta M., Morales Torres C., Rekling J.C., Johansen J.V., Abarrategui I., Helin K.. The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet. 2013; 9:e1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Catchpole S., Spencer-Dene B., Hall D., Santangelo S., Rosewell I., Guenatri M., Beatson R., Scibetta A.G., Burchell J.M., Taylor-Papadimitriou J.. PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int. J. Oncol. 2011; 38:1267–1277. [DOI] [PubMed] [Google Scholar]
- 18. Kidder B.L., Hu G., Yu Z.X., Liu C., Zhao K.. Extended self-renewal and accelerated reprogramming in the absence of Kdm5b. Mol. Cell. Biol. 2013; 33:4793–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmitz S.U., Albert M., Malatesta M., Morey L., Johansen J.V., Bak M., Tommerup N., Abarrategui I., Helin K.. Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J. 2011; 30:4586–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de la Mata M., Alonso C.R., Kadener S., Fededa J.P., Blaustein M., Pelisch F., Cramer P., Bentley D., Kornblihtt A.R.. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003; 12:525–532. [DOI] [PubMed] [Google Scholar]
- 21. Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S.. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011; 479:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dujardin G., Lafaille C., de la Mata M., Marasco L.E., Munoz M.J., Le Jossic-Corcos C., Corcos L., Kornblihtt A.R.. How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell. 2014; 54:683–690. [DOI] [PubMed] [Google Scholar]
- 23. Fong N., Kim H., Zhou Y., Ji X., Qiu J., Saldi T., Diener K., Jones K., Fu X.D., Bentley D.L.. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014; 28:2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shukla S., Oberdoerffer S.. Co-transcriptional regulation of alternative pre-mRNA splicing. Biochim. Biophys. Acta. 2012; 1819:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kidder B.L., Hu G., Zhao K.. ChIP-Seq: technical considerations for obtaining high-quality data. Nat. Immunol. 2011; 12:918–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kidder B.L., Zhao K.. Efficient library preparation for next-generation sequencing analysis of genome-wide epigenetic and transcriptional landscapes in embryonic stem cells. Methods Mol. Biol. 2014; 1150:3–20. [DOI] [PubMed] [Google Scholar]
- 27. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W.. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009; 25:1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B.. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008; 5:621–628. [DOI] [PubMed] [Google Scholar]
- 30. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katz Y., Wang E.T., Airoldi E.M., Burge C.B.. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010; 7:1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maunakea A.K., Chepelev I., Cui K., Zhao K.. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013; 23:1256–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tokheim C., Park J.W., Xing Y.. PrimerSeq: design and visualization of RT-PCR primers for alternative splicing using RNA-seq data. Genomics Proteomics Bioinformatics. 2014; 12:105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seward D.J., Cubberley G., Kim S., Schonewald M., Zhang L., Tripet B., Bentley D.L.. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat. Struct. Mol. Biol. 2007; 14:240–242. [DOI] [PubMed] [Google Scholar]
- 36. Reppas N.B., Wade J.T., Church G.M., Struhl K.. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell. 2006; 24:747–757. [DOI] [PubMed] [Google Scholar]
- 37. Zeitlinger J., Stark A., Kellis M., Hong J.W., Nechaev S., Adelman K., Levine M., Young R.A.. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007; 39:1512–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A.. c-Myc regulates transcriptional pause release. Cell. 2010; 141:432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fuda N.J., Ardehali M.B., Lis J.T.. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009; 461:186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nojima T., Gomes T., Grosso A.R., Kimura H., Dye M.J., Dhir S., Carmo-Fonseca M., Proudfoot N.J.. Mammalian NET-Seq reveals genome-wide nascent transcription coupled to RNA processing. Cell. 2015; 161:526–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benayoun B.A., Pollina E.A., Ucar D., Mahmoudi S., Karra K., Wong E.D., Devarajan K., Daugherty A.C., Kundaje A.B., Mancini E. et al. . H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014; 158:673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Luco R.F., Pan Q., Tominaga K., Blencowe B.J., Pereira-Smith O.M., Misteli T.. Regulation of alternative splicing by histone modifications. Science. 2010; 327:996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kolasinska-Zwierz P., Down T., Latorre I., Liu T., Liu X.S., Ahringer J.. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 2009; 41:376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schor I.E., Rascovan N., Pelisch F., Allo M., Kornblihtt A.R.. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:4325–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schor I.E., Fiszbein A., Petrillo E., Kornblihtt A.R.. Intragenic epigenetic changes modulate NCAM alternative splicing in neuronal differentiation. EMBO J. 2013; 32:2264–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Spies N., Nielsen C.B., Padgett R.A., Burge C.B.. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell. 2009; 36:245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou Y., Lu Y., Tian W.. Epigenetic features are significantly associated with alternative splicing. BMC Genomics. 2012; 13:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Z., Zhai W., Richardson J.A., Olson E.N., Meneses J.J., Firpo M.T., Kang C., Skarnes W.C., Tjian R.. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004; 18:3106–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wright M.D., Geary S.M., Fitter S., Moseley G.W., Lau L.M., Sheng K.C., Apostolopoulos V., Stanley E.G., Jackson D.E., Ashman L.K.. Characterization of mice lacking the tetraspanin superfamily member CD151. Mol. Cell. Biol. 2004; 24:5978–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R.. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006; 442:533–538. [DOI] [PubMed] [Google Scholar]
- 51. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dennis G. Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A.. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003; 4:P3. [PubMed] [Google Scholar]
- 53. Zhao X.D., Han X., Chew J.L., Liu J., Chiu K.P., Choo A., Orlov Y.L., Sung W.K., Shahab A., Kuznetsov V.A. et al. . Whole-genome mapping of histone h3 lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007; 1:286–298. [DOI] [PubMed] [Google Scholar]
- 54. Chen Y., Jorgensen M., Kolde R., Zhao X., Parker B., Valen E., Wen J., Sandelin A.. Prediction of RNA polymerase II recruitment, elongation and stalling from histone modification data. BMC Genomics. 2011; 12:544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen K., Chen Z., Wu D., Zhang L., Lin X., Su J., Rodriguez B., Xi Y., Xia Z., Chen X. et al. . Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat. Genet. 2015; 47:1149–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bieberstein N.I., Carrillo Oesterreich F., Straube K., Neugebauer K.M.. First exon length controls active chromatin signatures and transcription. Cell Rep. 2012; 2:62–68. [DOI] [PubMed] [Google Scholar]
- 57. Barrett A., Santangelo S., Tan K., Catchpole S., Roberts K., Spencer-Dene B., Hall D., Scibetta A., Burchell J., Verdin E. et al. . Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. Int. J. Cancer. 2007; 121:265–275. [DOI] [PubMed] [Google Scholar]
- 58. Hnilicova J., Hozeifi S., Duskova E., Icha J., Tomankova T., Stanek D.. Histone deacetylase activity modulates alternative splicing. PLoS One. 2011; 6:e16727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Teoh P.L., Sharrocks A.D.. WDR5, ASH2L, and RBBP5 control the efficiency of FOS transcript processing. Cell Mol. Biol. Lett. 2014; 19:215–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xie L., Pelz C., Wang W., Bashar A., Varlamova O., Shadle S., Impey S.. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011; 30:1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Eick D., Geyer M.. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 2013; 113:8456–8490. [DOI] [PubMed] [Google Scholar]
- 62. Ni Z., Saunders A., Fuda N.J., Yao J., Suarez J.R., Webb W.W., Lis J.T.. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol. Cell. Biol. 2008; 28:1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guillemette B., Drogaris P., Lin H.H., Armstrong H., Hiragami-Hamada K., Imhof A., Bonneil E., Thibault P., Verreault A., Festenstein R.J. et al. . H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 2011; 7:e1001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Luco R.F., Allo M., Schor I.E., Kornblihtt A.R., Misteli T.. Epigenetics in alternative pre-mRNA splicing. Cell. 2011; 144:16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Karlic R., Chung H.R., Lasserre J., Vlahovicek K., Vingron M.. Histone modification levels are predictive for gene expression. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:2926–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cheng C., Yan K.K., Yip K.Y., Rozowsky J., Alexander R., Shou C., Gerstein M.. A statistical framework for modeling gene expression using chromatin features and application to modENCODE datasets. Genome Biol. 2011; 12:R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.