ABSTRACT
NMCP/CRWN (NUCLEAR MATRIX CONSTITUENT PROTEIN/CROWDED NUCLEI) is a major component of a protein fibrous meshwork (lamina-like structure) on the plant inner nuclear membrane. NMCP/CRWN contributes to regulating nuclear shape and nuclear functions. An NMCP/CRWN protein in Daucus carota (DcNMCP1) is localized to the nuclear periphery in interphase cells, and surrounds chromosomes in cells in metaphase and anaphase. The N-terminal region and the C-terminal region of DcNMCP1 are both necessary for localizing DcNMCP1 to the nuclear periphery. Here candidate interacting partners of the amino acid position 975–1053 of DcNMCP1 (T975–1053), which is present in the C-terminal region and contains a conserved sequence that plays a role in localizing DcNMCP1 to the nuclear periphery, are screened for. Arabidopsis thaliana nuclear proteins were subjected to far-Western blotting with GST-fused T975–1053 as a probe, and signals were detected at the positions corresponding to ∼70, ∼40, and ∼18 kDa. These ∼70, ∼40, and ∼18 kDa nuclear proteins were identified by mass spectrometry, and subjected to a yeast 2-hybrid (Y2H) analysis with T975–1053 as bait. In this analysis, the ∼40 kDa protein ARP7, which is a nuclear actin-related protein possibly involved in regulating chromatin structures, was confirmed to interact with T975–1053. Independently of the far-Western blotting, a Y2H screen was performed using T975–1053 as bait. Targeted Y2H assays confirmed that 3 proteins identified in the screen, MYB3, SINAT1, and BIM1, interact with T975–1053. These proteins might have roles in NMCP/CRWN protein-mediated biologic processes.
KEYWORDS: actin, chromatin, lamina-like structure, nuclear membrane, protein-protein interaction, plants, transcription factor
Introduction
Animal inner nuclear membrane (INM) is lined with a protein fibrous meshwork called the nuclear lamina,1,2 and plant INM is also lined with a lamina-like structure.3,4 The animal lamina consists of lamin proteins,1,2 while the plant lamina-like structure consists of NMCP/CRWN (NUCLEAR MATRIX CONSTITUENT PROTEIN/CROWDED NUCLEI) family proteins.5 The amino acid sequence similarities between lamin proteins and NMCP/CRWN proteins are low, thus they are thought to have different origins. However, they are predicted to have a similar secondary structure, which consists of the N-terminal head domain, the central coiled-coil domain, and the C-terminal tail domain.5 Lamin proteins are involved in regulating chromosome positioning in the nucleus,6-8 gene expression,9-12 nuclear mechanical properties, and nuclear shape.13 Some mutations in lamin proteins cause genetic diseases called laminopathies (for a review see ref. 14). NMCP/CRWN proteins also regulate nuclear shape,15,16,17 chromatin structure,17 and plant growth.15,17,18
The model plant Arabidopsis thaliana has 4 CRWN/NMCP proteins (CRWN1–4).15 CRWN1 physically interacts with 2 SUN (Sad1/UNC-84) proteins, SUN1 and SUN2.19 SUN proteins are present in the INM, and interact with WIP (WPP domain-interacting protein) outer nuclear membrane (ONM) proteins.20 WIP proteins interact with WIT (WIPP domain-interacting tail-anchored protein) ONM proteins,21 and WIT proteins interact with myosin XI-i, which would interact with cytosolic actin filaments.22 The complexes formed by the SUN, WIP, WIT, and myosin XI-i proteins are thought to act as the plant LINC (linker of nucleoskeleton and cytoskeleton) complexes, which transmit the force from the cytoplasmic actin filaments to the nuclear membrane, and thereby regulate nuclear shape. The NMCP/CRWN proteins may affect the structures and/or the functions of the LINC complexes via the interaction with the SUN proteins, and vice versa, although it has also been proposed that the NMCP/CRWN proteins and the LINC complexes have distinct roles in regulating nuclear shape.23 KAKU4, another regulator of nuclear shape, is localized to the nuclear periphery and interacts with CRWN1,24 but it is still unclear how the CRWN1-KAKU4 complex functions.
To localize an NMCP1/CRWN homolog in Daucus carota (DcNMCP1) to the nuclear periphery, its N-terminal region (position 1–141) and its C-terminal region (position 908–1053) are both necessary. The C-terminal region of DcNMCP1 contains the conserved sequence motif RYNLRRHK, and mutating it to either RYNLAAAA or RAAARRHK disrupts the nuclear periphery localization of DcNMCP1.25 At least 3 other D. carota NMCP/CRWN proteins (DcNMCP2–4) have been identified (Fig. 1). NMCP/CRWN proteins could be classified into 2 major clades.17 DcNMCP1, DcNMCP3, and DcNMCP4 belong to a clade that includes A. thaliana CRWN1–3, while DcNMCP2 belongs to the other clade with CRWN4. Among DcNMCP1–4, DcNMCP1 is the best characterized.
Figure 1.
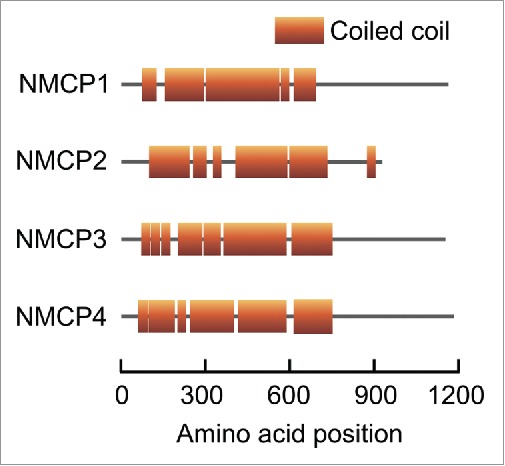
Schematic representation of DcNMCP1 (accession number: BAA20407.1), DcNMCP2 (BAI67718.1), DcNMCP3 (BAN14787.1), and DcNMCP4 (KX828842). Their predicted coiled-coil regions are shown as boxes.
Here, candidates for interacting partners of the C-terminal region of DcNMCP1 are screened for using far-Western blotting (FWB) and a yeast 2-hybrid (Y2H) system. The potential roles of the identified candidate interactors in regulating nuclear functions are also discussed.
Results
Far-Western blotting for detecting interacting partners of the partial tail domain of DcNMCP1
The partial tail domain corresponding to the amino acid position 975–1053 of DcNMCP1 (T975–1053) was expressed in Escherichia coli as glutathione S-transferase (GST)-fused protein, purified (Fig. S1), and used as the probe for FWB to detect T975–1053-binding nuclear proteins of A. thaliana suspension-cultured cells. The A. thaliana nuclear proteins were fractionated into hydrochloric acid (HCl)-soluble proteins and HCl-insoluble proteins. The HCl-soluble proteins should include histone,26 which is known to directly interact with a lamin protein in Drosophila melanogaster.27 The nuclear proteins were separated by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis), transferred to a membrane, and reacted with the probe and antibodies. In the FWB, signals were detected at the positions corresponding to ∼40, ∼38, ∼33, ∼19, ∼17 kDa for the HCl-soluble proteins, while at ∼70 and ∼40 kDa for the HCl-insoluble proteins (Fig. 2A). Similar banding patterns were observed when nuclear proteins were prepared using phenol extraction-based methods, which could reduce DNA contamination in protein samples (Fig. S2). When GST alone was used instead of the GST-T975–1053 fusion protein for the FWB, no clear signals were observed. When the blots were reacted with untagged T975–1053 before being reacted with GST-T975–1053 in the FWB, no clear signals were observed, either (Fig. 2B). These results suggest that the signals detected in the FWB are specific to the interaction between T975–1053 and other proteins. An in vitro pull-down assay was also performed using the same nuclear proteins and GST-T975–1053, but reproducibility of results was low (data not shown).
Figure 2.
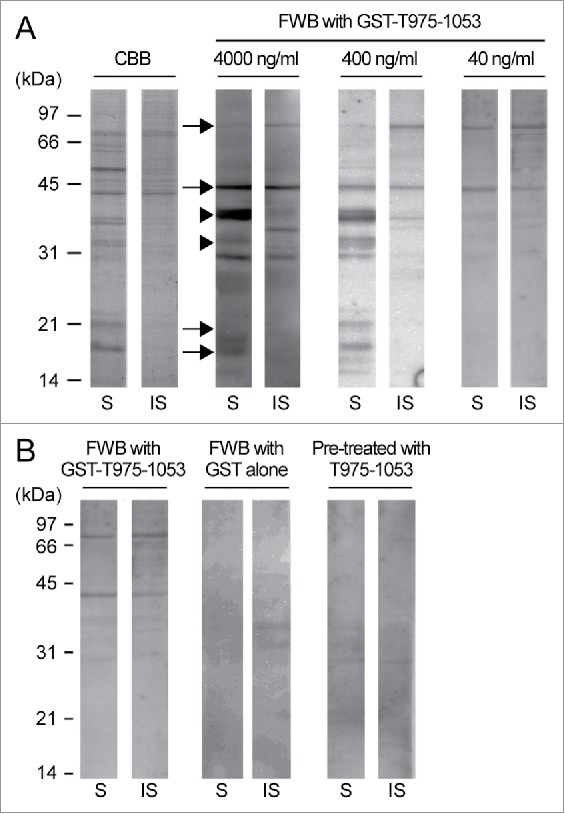
Far-Western blotting for detecting Arabidopsis thaliana nuclear proteins interacting with T975–1053. (A) HCl-soluble (S) and HCl-insoluble (IS) fractions of A. thaliana nuclear proteins were subjected to SDS-PAGE followed by either Coomassie brilliant blue staining (CBB) or far-Western blotting (FWB) with the indicated concentrations of the GST-T975–1053 probe. The positions of the ∼70-, ∼40-, ∼19-, and ∼17-kDa proteins are indicated by arrows, and the positions of the ∼38- and ∼33-kDa proteins are indicated by arrowheads. The experiments were repeated 3 times, and the representative result is presented. (B) The A. thaliana nuclear protein fractions were subjected to FWB with 40 ng/ml GST (for the middle 2 lanes) or 40 ng/ml GST-T975–1053 as the probe. For the right 2 lanes, the nuclear proteins on the membrane were reacted with untagged T975–1053, and then with GST-T975–1053. For the left 2 lanes, signals were detected as described in the panel A, and are shown as a control. The experiments were repeated 3 times, and the representative result is presented.
The signals of ∼70- and ∼40-kDa proteins were strong in the FWB even when a low concentration (40 ng/ml) of GST-T975–1053 was used (Fig. 2A, right panel), and the ∼19- and ∼17-kDa nuclear proteins should include histones. Such proteins detected in the FWB (Fig. 2A, arrows, and Fig. S2, arrowheads) were identified by the liquid chromatography-mass spectrometry (LC-MS). Some of the identified proteins were chosen for a Y2H analysis on the basis of their functional annotations and their coverage scores, which indicate what percentages of the full-length protein sequences were covered by the peptide sequences identified by the LC-MS (Table S1). In the targeted Y2H analysis, in summary, most of the tested proteins failed to enable yeast cells to grow on the selection medium when co-expressed with T975–1053, but the actin-related protein ARP7 (Arabidopsis Genome Initiative (AGI) code: AT3G60830) did (Fig. 2 and Table 1, upper 18 rows), supporting the idea that T975–1053 interacts with ARP7 in yeast.
Table 1.
The numbers of the colonies that could survive on DDO/X/A in the targeted Y2H analyses.
| 1AD-fused protein | 2BD-T975–1053 | Number of colonies assessed | Number of colonies surviving on DDO/X/A | 3P value in the chi-square test |
|---|---|---|---|---|
| ARP7 | + | 34 | 32 | < 10−10 |
| − | 31 | 2 | ||
| MED37F | + | 34 | 2 | 0.888 |
| − | 39 | 2 | ||
| PDI | + | 30 | 6 | 0.958 |
| − | 39 | 8 | ||
| HDT2 | + | 30 | 4 | 0.816 |
| − | 35 | 4 | ||
| RPS14A | + | 38 | 5 | 0.78 |
| − | 39 | 6 | ||
| HisH3.3 | + | 33 | 2 | 0.949 |
| − | 31 | 2 | ||
| HisH2AXa | + | 33 | 3 | 0.187 |
| − | 34 | 7 | ||
| HisH2B.11 | + | 31 | 5 | 0.42 |
| − | 33 | 8 | ||
| HAP1 | + | 33 | 2 | 0.348 |
| − | 31 | 4 | ||
| MYB3 | + | 35 | 31 | 0.00178 |
| − | 33 | 18 | ||
| BIM1 | + | 38 | 20 | < 10−4 |
| − | 34 | 2 | ||
| SINAT1 | + | 34 | 33 | < 10−10 |
| − | 31 | 3 |
AD: GAL4 activation domain.
BD: GAL4 DNA-binding domain. The presence of BD-fused T-975–1053 and its absence are indicated as + and −, respectively.
The chi-square test was performed for each AD-fused protein to examine whether the presence of BD-T975–1053 is independent of the percentages of the colonies that could survive on DDO/X/A.
Y2H screen for interacting partners of T975–1053
Independently of the FWB, a Y2H screen was performed using T975–1053 as bait and A. thaliana cDNA library. Approximately 200 thousand cDNA clones were screened, and 122 colonies survived on selection media. The majority of the plasmid inserts of these colonies were the genes encoding the MYB3 (AGI code: AT1G22640) and BIM1 (AT5G08130) transcription factors (TFs) and the putative E3 ubiquitin ligase SINAT1 (AT2G41980). The full-length coding sequences (CDSs) of MYB3, BIM1, and SINAT1 were recloned into the Y2H vector for a targeted Y2H analysis. In this analysis, MYB3, BIM1, and SINAT1 all enabled yeast cells when co-expressed with T975–1053 (Fig. 3 and Table 1, lower 6 rows), confirming that those proteins interact with T975–1053 in yeast.
Figure 3.
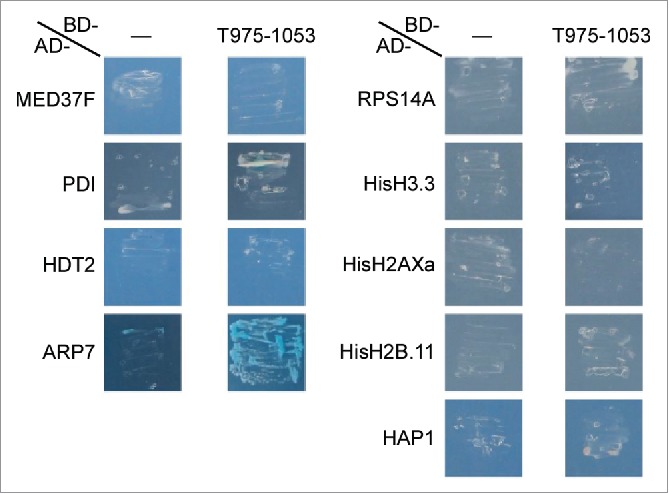
A targeted Y2H analysis using the proteins identified in the liquid chromatography-mass spectrometry (LC-MS). T975–1053 and one of the proteins identified in the LC-MS (see Table S1 for their information) were co-expressed as GAL4 DNA-binding domain (BD)-fused protein and GAL4 activation domain (AD)-fused protein, respectively, in Y2HGold Yeast Strain. For –, the BD alone was co-expressed with an AD-fused protein. The transformed cells were grown on the selection medium DDO/X/A, which contains X-α-gal and the Aureobasidin A antibiotic. More than 30 individual colonies obtained with the transformation were used for the analysis (see Table 1 for the exact numbers of the colonies used), and the representative results are presented.
Subcellular localization of the candidates for T975–1053-interacting partners
Full-length DcNMCP1 cannot be expressed as a GFP-fused protein in a transient expression system with Apium graveolens, but a DcNMCP1 variant that lacks the central coiled-coil domain can be, allowing to visualize positions where its tail domain is present.25 Such a GFP-fused DcNMCP1 variant (GFP-DcNMCP1HT) was co-expressed with mCherry-fused ARP7, MYB3, BIM1, or SINAT1 (ARP7-mCherry, MYB3-mCherry, BIM1-mCherry, or SINAT1-mCherry, respectively) to examine their subcellular localization. In agreement with the previous study,25 in all the cases studied, GFP-DcNMCP1HT was detected as either uniform or dotted signals at the nuclear periphery. Signals of ARP7-mCherry were detected in the nucleus and the cytosol, but in agreement with the previous finding that ARP7 is localized to the nucleus in interphase cells,28 the signal intensity of ARP7-mCherry was higher in the nucleus than in the cytosol (Fig. 4, top panels). Signals of MYB3-mCherry and BIM1-mCherry were also detected mainly in the nucleus (Fig. 4, second and third panels from the top). The signals of ARP7-mCherry, MYB3-mCherry, and BIM1-mCherry were not enriched in the nuclear periphery, but this does not rule out the possibility that these proteins could interact with the tail domain of DcNMCP1. Signals of SINAT1-mCherry was often detected as large dots (Fig. 4, fourth from the top), which may be protein aggregates, but they were detected at peripheral regions of the nucleus in some cells (Fig. 4, bottom panel). This difference in the localization of the SINAT1-mCherry signals might be due to the difference in their expression levels in the cells, although other factors might also be involved.
Figure 5.
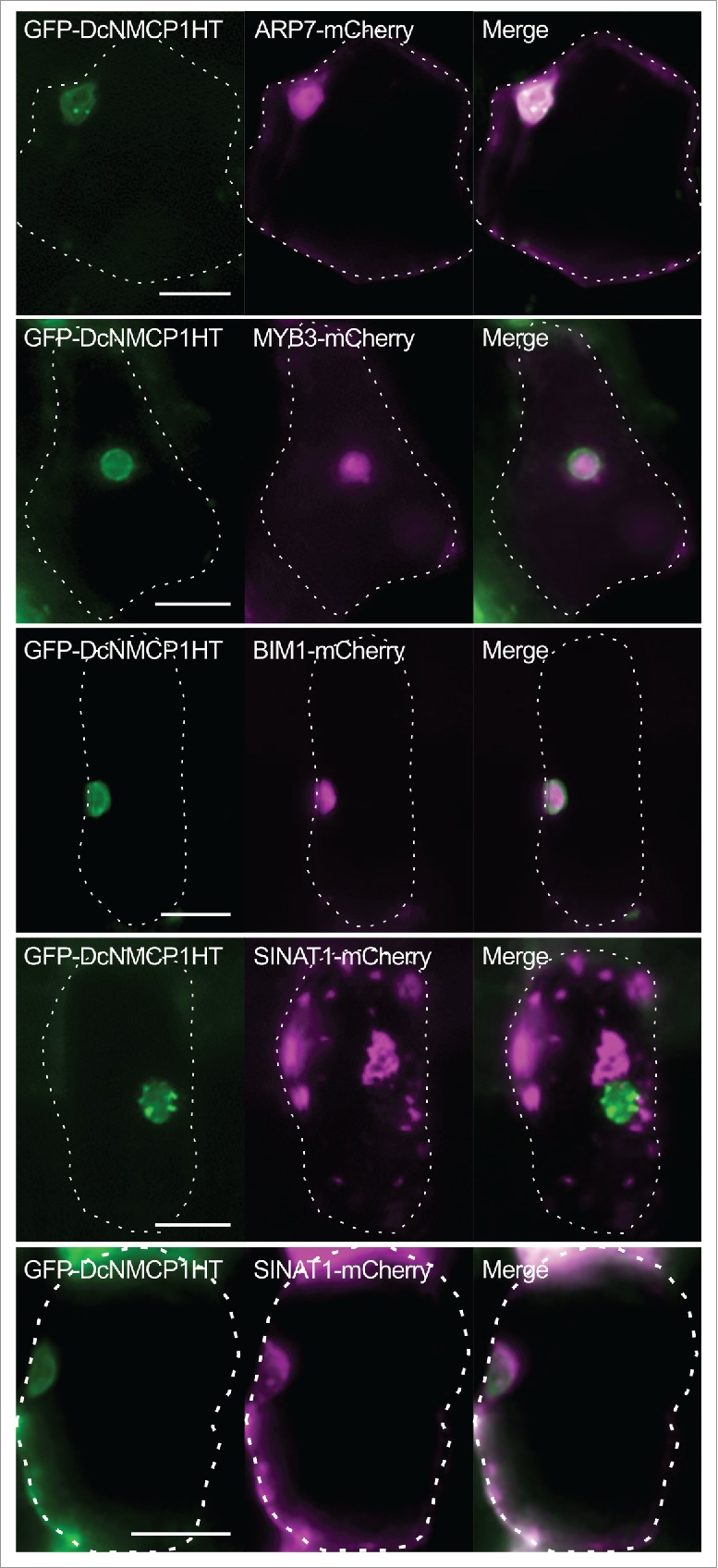
Subcellular localization of the possible interacting partners of the C-terminal region of DcNMCP1. A GFP-fused DcNMCP1 variant lacking the central coiled-coil domain (GFP-DcNMCP1HT) was co-expressed with mCherry-fused ARP7, MYB3, BIM1, or SINAT1 (ARP7-mCherry, MYB3-mCherry, BIM1-mCherry, or SINAT1-mCherry, respectively) in epidermal cells of Apium graveolens. For each combination of constructs, more than 10 transformed cells were observed, and representative results are shown. For SINAT1-mCherry, 2 patterns of signals, which might reflect different expression levels of SINAT1-mCherry in transformed cells, are shown (fourth and fifth panels from the top). Dotted lines indicate cell boundaries. Bars = 25 μm.
Figure 4.
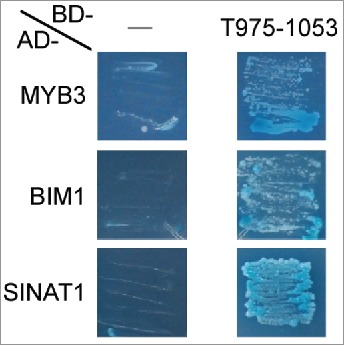
A targeted yeast 2-hybrid (Y2H) analysis using the proteins identified in the Y2H screen. T975–1053 and one of the proteins identified in the Y2H screen (MYB3, BIM1, or SINAT1) were co-expressed as GAL4 DNA-binding domain (BD)-fused protein and GAL4 activation domain (AD)-fused protein, respectively, in the Y2HGold yeast strain. For –, the BD alone was co-expressed with an AD-fused protein. The transformed cells were grown on the selection medium DDO/X/A, which contains X-α-gal and the Aureobasidin A antibiotic. More than 30 individual colonies obtained with the transformation were used for the analysis (see Table 1 for the exact numbers of the colonies used), and the representative results are presented.
Discussion
In this study, ARP7, MYB3, BIM1, and SINAT1 were identified as the candidate interactors of T975–1053. The idea that T975–1053 possibly interacts with ARP7 is consistent with a previous finding that the tail domain of human lamin A interacts with purified actin.29 ARP7 can be phylogenetically classified into a plant-specific clade in the actin-related protein family. ARP7 is localized in the cytosol in cells in metaphase and anaphase, and localized to the nucleus in cells in the interphase.28 During mitosis of Apium graveolens suspension-cultured cells, an NMCP/CRWN homolog, AgNMCP1, is associated to chromosomes in the metaphase, anaphase, and telophase, while it is localized to the nuclear periphery in the interphase and prophase.30 Thus, the nuclear localization of ARP7 is likely to depend on the formation of the nuclear membrane. It would be interesting to visualize the subcellular localization of both an NMCP1/CRWN protein and ARP7 at each phase of mitosis. Knockdown of ARP7 causes growth defects such as dwarfism and reduced fertility, and knockout of ARP7 causes embryonic lethality.31 Another nuclear actin-related protein, ARP6, is a component of the SWR1 complex, which catalyzes replacement of histone variants and thus regulates gene expression.32 It would be interesting to examine whether the regulation of chromatin structures by NMCP/CRWN proteins are mediated by nuclear actin-related proteins.
MYB3 is a MYB TF possibly involved in phenylpropanoid metabolism,33 and BIM1 is a basic helix-loop-helix (bHLH) TF involved in regulating the gene expression mediated by the phytohormone brassinosteroid,34 and SINAT1 is a putative E3 ubiquitin ligase functions of which have not been characterized. No TF or ubiquitin ligase subunit has been reported to interact with NMCP/CRWN proteins in plants. However, in animal cells, lamin A/C interacts with several TFs and DNA-binding proteins such as the c-Fos TF,35 the SREBP1 bHLH-leucine zipper TF,36 and the MYB telomere repeat-binding factor TRF2.37 Lamin A affects the ubiquitin-proteasome-dependent degradation of heterochromatin proteins, which promote heterochromatinization.38 These interactions are thought to facilitate the processes mediated by these proteins. A double knockout of CRWN1 and one of CRWN2–4 causes dwarfism, and further knockouts of them cause more severe dwarfism or lethality in A. thaliana.15,17 A double knockout of CRWN1 and CRWN3 stabilizes the ABI5 (ABSCISIC ACID INSENSITIVE 5) TF and increases sensitivity of plants to the stress-related phytohormone abscisic acid at the seed germination stage in A. thaliana.18 It would be interesting to examine whether NMCP/CRWN proteins regulate the subcellular localization, the protein stability, and/or the physiologic functions of MYB3, BIM1, and SINAT1. It would also be interesting to examine whether MYB3, BIM1, or SINAT1 regulates the physiologic functions of NMCP/CRWN proteins.
Materials and methods
Plant materials
Hypocotyls of D. carota and A. thaliana were incubated on the medium 1 and the medium 2 (Table S2), respectively, in the darkness at 25°C for approximately one month to obtain their calli. To develop suspension-cultured cells, the calli of D. carota and A. thaliana were liquid-cultured in the medium 1 and the medium 2, respectively, at 82 rpm at 25°C in the darkness. Every 7–10 days, 10 ml of the suspension cultures of D. carota and A. thaliana was transferred to 90 ml of the medium 1 and the medium 2, respectively, and cultured at 82 rpm at 25°C in the darkness to maintain these cells. These cells were used to prepare RNA and nuclear proteins.
RNA isolation and cDNA synthesis
Approximately 80 mg of the A. thaliana suspension-cultured cells was frozen in liquid nitrogen, ground with a motor and pestle to a fine powder, and resuspended in 1 ml TRIzol Reagent (Thermo Fisher Scientific, 15596026). The suspension was incubated for 5 min at room temperature, mixed with 200 μl chloroform, incubated for 2 min at room temperature, and centrifuged at 12000 × g for 15 min. The resulting upper aqueous phase was transferred to a new tube, mixed with 0.5 × volume of 2-propanol, incubated for 10 min at room temperature, and centrifuged at 12000 × g at 4°C for 10 min. The supernatant was removed and the pellet was washed with 750 μl of 75% (v/v) ethanol, dried, dissolved in 40 μl RNase-free water, and used as the RNA sample.
First strand cDNA was synthesized using Transcriptor First Strand cDNA Synthesis Kit (Roche, 04379012001). A 10 μl solution containing 1 μg of the total RNA and 12 μM Random Hexamer was incubated at 65 °C for 5 min, immediately put on ice, and mixed with 2 μl RNase-free water, 4 μl SCRIPT RT Buffer, 1 μl dNTP Mix, 1 μl of 100 mM DTT, 1 μl RNase Inhibitor, and 1 μl SCRIPT RT. The solution was incubated at 42°C for 10 min, 50°C for 40 min, 75°C for 15 min, and used as the cDNA sample.
Plasmid preparation
For FWB, the CDS of T975–1053 was amplified by PCR using the construct with the full-length DcNMCP1 in the pKD0330 background29 as the template and the primer pair shown in Table S3. The PCR products were digested by SalI, and inserted into the SalI site of the pGEX-6P-1 vector (GE Healthcare, 28954648), generating pGEX-6P-T975–1053.
For Y2H experiments, the same PCR products were digested by SalI and inserted into the SalI site of pGBKT7 DNA-BD Vector (Clontech, 630443), generating pGBK-T975–1053. To clone the other genes for the Y2H experiments, their full-length CDSs were amplified by PCR using the A. thaliana cDNA as template and the primer pairs shown in Table S3. The PCR products were digested and inserted into pGADT7 AD Vector (Clontech, 630442) using the restriction enzyme sites shown in Table S3. The pGAD vectors containing the full-length CDSs of MYB3, BIM1, and SINAT1 were extracted from the corresponding colonies identified in the Y2H screen (see the subsection “Y2H”), transformed into the E. coli strain DH5α, and purified from the transformed E. coli cells for further analyses.
To express GFP-DcNMCP1HT, a construct to express GFP-fused full-length DcNMCP125 was digested with BglII. The resulting larger DNA fragment was self-ligated, generating p35S-GFP-DcNMCP1HT. To express mCherry-fused proteins, the full-length CDSs of ARP7, MYB3, BIM1, and SINAT1 were amplified by PCR using the above pGAD vectors as templates and the primer pairs shown in Table S3. The PCR products were digested and inserted into pBS-35SMCS-mCherry39 using the restriction enzyme sites shown in Table S3.
Nuclear protein preparation
To prepare nuclear proteins of the D. carota and A. thaliana suspension-cultured cells, the cells were collected from 25 ml culture by leaving them without rotation, washed with the Man-MES-1/2N4 solution (0.35 M mannitol, 10 mM NaCl, 15 mM KCl, 1.5 mM MgCl2, 1 mM CaCl2, 20 mM 2-(N-morpholino)ethanesulfonic acid (MES)-KOH, pH 5.8), resuspended in 60 ml of the Man-MES-1/2N4 solution supplemented with 2% (w/v) CELLULASE “ONOZUKA” R-10 (Yakult Pharmaceutical Industry), 0.2% (w/v) MACEROZYME R-10 (Yakult Pharmaceutical Industry), 0.02% (w/v) Pectolyase Y-23 (Kyowa Kasei, 63–0501), and 2 mM 2-mercaptoethanol, and incubated at 25 °C for 4 h. The suspensions were centrifuged at 200 × g for 5 min, and the cells were resuspended in the Man-MES-1/2N4 solution. The centrifugation and resuspension were repeated twice, and the suspensions were centrifuged at 200 × g for 5 min. The cells were resuspended in 10 ml of the ice-cold homogenization buffer (20% (v/v) glycerol, 0.3 M sucrose, 1% (v/v) Triton X-100, 10 mM NaCl, 15 mM KCl, 1.5 mM MgCl2, 1 mM CaCl2, 2 mM 2-mercaptoethanol, 1× Protease Inhibitor Cocktail for General Use (Nacalai Tesque, 04080–24), 20 mM MES-KOH, pH 5.8), homogenized with a Potter glass homogenizer on ice, filtered through stainless steel mesh with 37 μm pores, and then through stainless steel mesh with 22 μm pores. The filtered suspensions were centrifuged at 2500 × g at 4°C for 15 min. The pellets were resuspended in 5 ml of the ice-cold homogenization buffer, and the suspensions were centrifuged at 2500 × g at 4°C for 15 min. The pellets were resuspended with Potter Teflon homogenizer in the ice-cold Man-MES-1/2N4 solution containing 25% (v/v) glycerol, and stored as the isolated nucleus sample at -40°C until used.
To extract proteins from the nuclei using HCl, the suspensions containing the isolated nuclei were mixed with an equal volume of ice-cold double-deionized water, and centrifuged at 2800 × g at 4°C for 10 min. The pellets were resuspended in 1.5 ml ice-cold double-deionized water. The suspensions were mixed with 1.5 ml of 0.8 M HCl, incubated at 4°C for 1 h with gentle shaking, and centrifuged at 9500 × g at 4°C for 15 min. The supernantants were transferred to a new tube. The pellets were resuspended in 3 ml of ice-cold 0.4 M HCl, and centrifuged 9500 × g at 4°C for 15 min. The supernatants were combined with those collected above. The resulting solutions were mixed with 36 ml ice-cold acetone, incubated at 4°C overnight, and centrifuged at 3600 × g at 4°C for 15 min. The pellets were dissolved in ice-cold 0.4 M HCl, and the solutions were dialyzed with Dialysis Membrane, Size 20, and double-deionized water. The resulting solutions were stored as the nuclear protein samples at −40°C until used. The HCl-insoluble pellets obtained above were dissolved in the denaturation solution (7 M urea, 3 mM EDTA, 6 mM 2-mercaptoethanol, 10 mM Tris-acetate, pH 7.6), and used for FWB.
Nuclear proteins were also prepared using SDS and phenol as described previously.40 Briefly, the solutions containing the isolated nuclei were centrifuged at 2000 × g at 4°C for 10 min. The pellets were washed twice with pre-chilled (-20 °C) 70% (v/v) ethanol, resuspended in the resuspension solution (2% (w/v) SDS, 0.1% (v/v) 2-mercaptoethanol, 20 mM EDTA, 20 mM Tris-HCl, pH 7.6), incubated at 100°C for 3 min, and incubated on ice for 1 min. The suspensions were mixed with an equal volume of phenol. The phenol phase was collected and dialyzed with Dialysis Membrane, Size 20, and the dialysis solution (0.1% (w/v) SDS, 0.1% (v/v) 2-mercaptoethanol, 10 mM Tris-HCl, pH 7.6) to obtain the nuclear protein samples. Nuclear protein samples were also prepared using a phenol- and guanidine isothiocyanate-based reagent, TriPure Isolation Reagent (Roche, 11667157001), according to the manufacturer's instructions. Protein concentrations in the protein samples were determined using the Pierce Coomassie Plus (Bradford) Protein Assay kit (Thermo Fisher Scientific, 23236).
FWB and LC-MS
BL21-Gold (DE3) Competent Cells (Agilent Technologies, 230132) were transformed with either pGEX-6P-1 or pGEX-6P-T975–1053, and liquid-cultured in 3 ml LBA (Luria-Bertani medium supplemented with 50 mg/l ampicillin) at 37°C for 4 h. The cultured cells were transferred to 90 ml LBA, further cultured at 37°C overnight, transferred to 360 ml LBA, and further cultured at 37°C for 1 h. Isopropyl β-D-1-thiogalactopyranoside was added to the culture at the 0.5 mM final concentration, and the cells were further cultured at 30°C for 5 h. The cells were collected by centrifugation at 9500 × g for 10 min, washed with PBS (phosphate-buffered saline: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1,76 mM KH2PO4, pH 7.4), resuspended in 10 ml cell lysis buffer (150 mM NaCl, 10 mM Tris-HCl, 0.2 mg/ml lysozyme, 1× Protease Inhibitor Cocktail for General Use, pH 8.0), incubated at room temperature for 10 min, mixed with 1 μl (250 units) of TurboNuclease (Accelagen, N0103P), and further incubated at room temperature for 20 min. The suspension was centrifuged at 33000 × g at 4°C for 10 min. The supernatant was transferred to a new tube, and ammonium sulfate was dissolved in it at the 0.56 g/ml final concentration. The solution was then incubated at 4°C overnight, and centrifuged at 33000 × g at 4°C for 10 min. The pellet was dissolved in PBS and used as the crude protein sample containing GST or GST-T975–1053.
GST or GST-T975–1053 in the crude sample was purified using Glutathione Sepharose 4B (GE Healthcare, 17075601) according to the manufacturer's instructions. Briefly, 10 ml of the crude protein solution was incubated with 1 ml of the Glutathione Sepharose 4B slurry equilibrated with PBS at room temperature for 1 h. The Glutathione Sepharose 4B resin was then washed with the washing solution (50 mM NaCl, 2 mM EDTA, 20 mM PIPES-KOH, pH 6.8), and incubated in 9 ml elution solution (20 mM reduced glutathione, 5 mM EDTA, 100 mM Tris-HCl, pH 8.0) to elute GST or GST-T975–1053. The eluent was dialyzed against PBS, and the resulting solution containing purified GST or GST-T975–1053 was stored as the probe solution at 4°C until used. The concentration of GST-T975–1053 in the solution was estimated to be 1.6 mg/ml on the basis of the Coomassie Briliant Blue (CBB) staining, where the signal intensity for GST-T975–1053 was compared with that of known amount of protein in a protein size marker (see the subsection “FWB”). To prepare untagged T975–1053, 1 ml of the solution containing purified GST-T975–1053 was mixed with 4 μl PreScission Protease (GE Healthcare, 27084301), and incubated at 25°C for 1 h. PreScission Protease and excised GST was removed from the solution using Glutathione Sepharose 4B according to the manufacturer's instructions. The resulting solution was dialyzed against PBS, and centrifuged at 16000 × g at 4°C for 10 min. The supernatant was used as the solution containing untagged T975–1053.
The nuclear protein samples were separated by SDS-PAGE (1.5 mg protein/lane) as described previously,41 transferred to Immun-Blot PVDF Membrane (Bio-Rad, 1620177) according to the manufacturer's instructions. The membrane was incubated at 30°C for 1 h in PBS containing 1% (w/v) Block Ace (DS Pharma Biomedical, UKB40) and 1% (w/v) bovine serum albumin, washed twice with PBST (PBS supplemented with 0.2% (v/v) Tween 20), incubated at 30°C for 3 h in PBST containing either GST or GST-T975–1053 as probe (probe concentrations are indicated in Fig. 1A), washed 3 times with PBST, incubated at 25°C for 3 h in PBST containing Anti-GST, Monoclonal Antibody, Peroxidase Conjugated (Wako, 011–21891), and washed 3 times with PBST. Signals were detected using the Chemi-Lumi One L chemiluminescent substrate (Nacalai Tesque, 07880), and the LumiVision Pro 400EX imager (AISIN). Images of FWB signals were processed using the Canvas X software (ACD Systems). To estimate the amounts and sizes of proteins, XL-Western Marker color plus (Apro Science, SP-2170) and SDS-PAGE Molecular Weight Standards, Low Range (Bio-Rad, 161–0304) were subjected to SDS-PAGE as well as the nuclear proteins. Proteins in the gel or on the membrane were stained with Quick-CBB PLUS (Wako, 178–00551).
For LC-MS, 1.5 mg of the A. thaliana nuclear proteins prepared with HCl (see the “Nuclear protein preparation” subsection) and SDS-PAGE Molecular Weight Standards, Low Range, were separated by SDS-PAGE, and stained with Quick-CBB PLUS. The gel blocks containing ∼70, ∼40, ∼19, ∼17 kDa protein bands were excised, and the proteins in these blocks were digested using In-Gel Tryptic Digestion Kit (Thermo Fisher Scientific, 89871) to elute the peptides from the gel. The peptides were analyzed with the LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) and the Proteome Discoverer 1.4 software (Thermo Fisher Scientific) provided by Instrumental Analysis Services of Global Facility Center at Hokkaido University.
Y2H experiments
For the Y2H screen, Y2HGold Yeast (Saccharomyces cerevisiae) Strain (Clontech, 630498) was transformed with pGBK-T975–1053 using the lithium acetate method.42 Yeast mating was performed using the transformed Y2HGold cells and Mate & Plate Library - Universal Arabidopsis (Normalized) (Clontech, 630487), which is a yeast cell library constructed from mRNA isolated from A. thaliana seedlings, leaves, stems, flowers, and siliques, according to the manufacturer's instructions. After the mating, the resulting cells were plated on the DDO/X/A selection medium (synthetic dextrose medium that lacks leucine and tryptophan and contains X-α-Gal (Clontech, 630407) and the Aureobasidin A antibiotic (Clontech, 630466)), and grown for 5 d at 30°C. Colonies that could grow on DDO/X/A were transferred to the more stringent medium QDO/X/A (DDO/X/A lacking adenine and histidine), and grown for 5 d at 30°C. Plasmid inserts in the colonies that could grow on QDO/X/A were amplified by yeast colony PCR using KOD FX (TOYOBO, KFX-101) and Matchmaker AD LD-Insert Screening Amplimer Set (Clontech, 630433), and sequenced with 3130 Genetic Analyzer (Applied Biosystems) at DNA Sequencing Facility of Research Faculty of Agriculture at Hokkaido University.
For targeted Y2H analyses, pGBK-T975–1053 and one of the pGADT7 constructs with genes of interest (see the subsection “Plasmid preparation”) were co-transformed into the Y2HGold strain with the lithium acetate method.42 Transformed cells were selected on the DDO medium (synthetic dextrose medium that lacks leucine and tryptophan), transferred to DDO/X/A, and grown at 30°C for 5 d to examine the reporter gene activation mediated by the interaction between T975–1053 and the proteins of interest. To obtain percentages of the colonies that survived on DDO/X/A, more than 30 individual colonies that grew on DDO were grown on DDO/X/A at 30°C for 5 d for each combination of constructs. Tiny or almost invisible colonies were regarded as non-surviving colonies, and the numbers of surviving colonies and non-surviving colonies were used for the chi-square test.
Expression of GFP- and mCherry-fused proteins
One of the mCherry constructs and p35S-GFP-DcNMCP1HT (see the “Plasmid preparation” subsection) (0.5 μg each) were mixed, and co-introduced into celery (Apium graveolens) epidermal cells using the Biolistic PDS-1000/He particle delivery system (Bio-Rad) as described previously.25 Cells were incubated for 24 h at room temperature after being transformed, and signals were observed using the BX50 epifluorescence microscope (Olympus) equipped with the ORCA-ER-1394 digital camera (Hamamatsu Photonics, Hamamatsu, Japan). The fluorescence mirror units U-MGFPHQ and U-MWIG2 (Olympus) were used to image GFP and mCherry, respectively. Images were processed with GIMP and Inkscape.
Accession numbers
Details regarding the sequences of the genes used in this study can be obtained with the following accession numbers (GenBank accession number for DcNMCP1 and AGI codes for A. thaliana genes): D64087 (DcNMCP1), AT5G42020 (MED37F), AT5G60640 (PDI), AT5G22650 (HDT2), AT3G60830 (ARP7), AT2G36160 (RPS14A), AT1G08880 (HisH2AXa), AT4G40030 (HisH3.3), AT5G59910 (HisH2B.11), AT1G02140 (HAP1), AT1G22640 (MYB3), AT5G08130 (BIM1), and AT2G41980 (SINAT1).
Supplementary Material
Abbreviations
- 2,4-D
2, 4-dichlorophenoxyacetic acid
- AGI
Arabidopsis Genome Initiative
- bHLH
basic helix-loop-helix
- CBB
Coomassie Briliant Blue
- CDS
coding sequence
- FWB
far-Western blotting
- GST
glutathione S-transferase
- INM
inner nuclear membrane
- LC-MS
liquid chromatography-mass spectrometry
- LINC
linker of nucleoskeleton and cytoskeleton
- MES
2-(N-morpholino)ethanesulfonic acid
- ONM
outer nuclear membrane
- SDS-PAGE
sodium dodecyl sulfate-PAGE
- TF
transcription factor
- Y2H
yeast 2-hybrid
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Masumi Yamagishi and his colleagues (Hokkaido University) for giving helpful advice and the DNA Sequencing Facility of Research Faculty of Agriculture at Hokkaido University for supporting DNA sequencing.
References
- [1].Goldman RD, Goldman AE, Green KJ, Jones JC, Jones SM, Yang HY. Intermediate filament networks: organization and possible functions of a diverse group of cytoskeletal elements. J Cell Sci Suppl 1986; 5:69-97; PMID:3308919 [DOI] [PubMed] [Google Scholar]
- [2].McKeon FD, Kirschner MW, Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature 1986; 319:463-8; PMID:3453101 [DOI] [PubMed] [Google Scholar]
- [3].Yu W, Moreno Díaz de la Espina S. The plant nucleoskeleton: ultrastructural organization and identification of NuMA homologues in the nuclear matrix and mitotic spindle of plant cells. Exp Cell Res 1999; 246:516-26; PMID:9925768 [DOI] [PubMed] [Google Scholar]
- [4].Masuda K, Takahashi S, Nomura K, Akimoto M, Inoue M. Residual structure and constituent proteins of the peripheral framework of the cell nucleus in somatic embryos from Daucus carota L. Planta 1993; 191:532-40. [Google Scholar]
- [5].Masuda K, Xu ZJ, Takahashi S, Ito A, Ono M, Nomura K, Inoue M. Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Exp Cell Res 1997; 232:173-81; PMID:9141634 [DOI] [PubMed] [Google Scholar]
- [6].Belmont AS, Zhai Y, Thilenius A. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol 1993; 123:1671-85; PMID:8276889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 1999; 145:1119-31; PMID:10366586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 2006; 38:1005-14; PMID:16878134 [DOI] [PubMed] [Google Scholar]
- [9].Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol 2008; 180:51-65; PMID:18195101; https://doi.org/ 10.1083/jcb.200706060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 2008; 452:243-7; PMID:18272965; https://doi.org/ 10.1038/nature06727 [DOI] [PubMed] [Google Scholar]
- [11].Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 2008; 4:e1000039; PMID:18369458; https://doi.org/ 10.1371/journal.pgen.1000039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dialynas G, Speese S, Budnik V, Geyer PK, Wallrath LL. The role of Drosophila Lamin C in muscle function and gene expression. Development 2010; 137:3067-77; PMID:20702563; https://doi.org/ 10.1242/dev.048231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol 1999; 147:913-20; PMID:10579712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Davidson PM, Lammerding J. Broken nuclei–lamins, nuclear mechanics, and disease. Trends Cell Biol 2014; 24:247-56; PMID:24309562; https://doi.org/ 10.1016/j.tcb.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell 2007; 19:2793-803; PMID:17873096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sakamoto Y, Takagi S. LITTLE NUCLEI 1 and 4 regulate nuclear morphology in Arabidopsis thaliana. Plant Cell Physiol 2013; 54:622-33; PMID:23396599; https://doi.org/ 10.1093/pcp/pct031 [DOI] [PubMed] [Google Scholar]
- [17].Wang H, Dittmer TA, Richards EJ. Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol 2013; 13:200; PMID:24308514; https://doi.org/ 10.1186/1471-2229-13-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhao W, Guan C, Feng J, Liang Y, Zhan N, Zuo J, Ren B. The Arabidopsis CROWDED NUCLEI genes regulate seed germination by modulating degradation of ABI5 protein. J Integr Plant Biol 2016; 58:669-678; PMID:26564029; https://doi.org/ 10.1111/jipb.12448 [DOI] [PubMed] [Google Scholar]
- [19].Graumann K. Evidence for LINC1-SUN associations at the plant nuclear periphery. PLoS One 2014; 9:e93406; PMID:24667841; https://doi.org/ 10.1371/journal.pone.0093406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhou X, Graumann K, Evans DE, Meier I. Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J Cell Biol 2012; 196:203-11; PMID:22270916; https://doi.org/ 10.1083/jcb.201108098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhao Q, Brkljacic J, Meier I. Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell 2008; 20:1639-51; PMID:18591351; https://doi.org/ 10.1105/tpc.108.059220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, Nishimura M, Hara-Nishimura I. Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr Biol 2013; 23:1776-81; PMID:23973298; https://doi.org/ 10.1016/j.cub.2013.07.035 [DOI] [PubMed] [Google Scholar]
- [23].Zhou X, Groves NR, Meier I. Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1. Nucleus 2015; 6:144-53; PMID:25759303; https://doi.org/ 10.1080/19491034.2014.1003512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Goto C, Tamura K, Fukao Y, Shimada T, Hara-Nishimura I. The Novel Nuclear Envelope Protein KAKU4 Modulates Nuclear Morphology in Arabidopsis. Plant Cell 2014; 26:2143-2155; PMID:24824484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kimura Y, Fujino K, Ogawa K, Masuda K. Localization of Daucus carota NMCP1 to the nuclear periphery: the role of the N-terminal region and an NLS-linked sequence motif, RYNLRR, in the tail domain. Front Plant Sci 2014; 5:62; PMID:24616728; https://doi.org/ 10.3389/fpls.2014.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273:5858-5868; PMID:9488723 [DOI] [PubMed] [Google Scholar]
- [27].Goldberg M, Harel A, Brandeis M, Rechsteiner T, Richmond TJ, Weiss AM, Gruenbaum Y. The tail domain of lamin Dm0 binds histones H2A and H2B. Proc Natl Acad Sci USA 1999; 96:2852-2857; PMID:10077600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kandasamy MK, McKinney EC, Meagher RB. Cell cycle-dependent association of Arabidopsis actin-related proteins AtARP4 and AtARP7 with the nucleus. Plant J 2003; 33:939-48; PMID:12609034 [DOI] [PubMed] [Google Scholar]
- [29].Simon DN, Zastrow MS, Wilson KL. Direct actin binding to A- and B-type lamin tails and actin filament bundling by the lamin A tail. Nucleus 2010; 1:264-72; PMID:21327074; https://doi.org/ 10.4161/nucl.1.3.11799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kimura Y, Kuroda C, Masuda K. Differential nuclear envelope assembly at the end of mitosis in suspension-cultured Apium graveolens cells. Chromosoma 2010; 119:195-204; PMID:19997923; https://doi.org/ 10.1007/s00412-009-0248-y [DOI] [PubMed] [Google Scholar]
- [31].Kandasamy MK, McKinney EC, Deal RB, Meagher RB. Arabidopsis ARP7 is an essential actin-related protein required for normal embryogenesis, plant architecture, and floral organ abscission. Plant Physiol 2005; 138:2019-32; PMID:16040647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Choi K, Park C, Lee J, Oh M, Noh B, Lee I. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 2007; 134:1931-41; PMID:17470967 [DOI] [PubMed] [Google Scholar]
- [33].Zhou M, Sun Z, Wang C, Zhang X, Tang Y, Zhu X, Shao J, Wu Y. Changing a conserved amino acid in R2R3-MYB transcription repressors results in cytoplasmic accumulation and abolishes their repressive activity in Arabidopsis. Plant J 2015; 84:395-403; PMID:26332741; https://doi.org/ 10.1111/tpj.13008 [DOI] [PubMed] [Google Scholar]
- [34].Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005; 120:249-59; PMID:15680330 [DOI] [PubMed] [Google Scholar]
- [35].Ivorra C, Kubicek M, González JM, Sanz-González SM, Alvarez-Barrientos A, O'Connor JE, Burke B, Andrés V. A mechanism of AP-1 suppression through interaction of c-Fos with lamin A/C. Genes Dev 2006; 20:307-20; PMID:16452503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Capanni C, Mattioli E, Columbaro M, Lucarelli E, Parnaik VK, Novelli G, Wehnert M, Cenni V, Maraldi NM, Squarzoni S, et al.. Altered pre-lamin A processing is a common mechanism leading to lipodystrophy. Hum Mol Genet 2005; 14:1489-502; PMID:15843404 [DOI] [PubMed] [Google Scholar]
- [37].Wood AM, Rendtlew Danielsen JM, Lucas CA, Rice EL, Scalzo D, Shimi T, Goldman RD, Smith ED, Le Beau MM, Kosak ST. TRF2 and lamin A/C interact to facilitate the functional organization of chromosome ends. Nat Commun 2014; 5:5467; PMID:25399868; https://doi.org/ 10.1038/ncomms6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chaturvedi P, Parnaik VK. Lamin A rod domain mutants target heterochromatin protein 1alpha and beta for proteasomal degradation by activation of F-box protein, FBXW10. PLoS One 2010; 5:e10620; PMID:20498703; https://doi.org/ 10.1371/journal.pone.0010620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tsugama D, Liu S, Takano T. A bZIP protein, VIP1, interacts with Arabidopsis heterotrimeric G protein β subunit, AGB1. Plant Physiol Biochem 2013; 71:240-246; PMID:23974356; https://doi.org/ 10.1016/j.plaphy.2013.07.024 [DOI] [PubMed] [Google Scholar]
- [40].LeStourgeon WM, Beyer AL. The rapid isolation, high-resolution electrophoretic characterization, and purification of nuclear proteins. Methods Cell Biol 1977; 16:387-406; PMID:886990 [PubMed] [Google Scholar]
- [41].Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227:680-5; PMID:5432063 [DOI] [PubMed] [Google Scholar]
- [42].Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 1992; 20:1425; PMID:1561104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


