ABSTRACT
The nuclear pore complex (NPC) comprises more than 30 nucleoporins (Nups). NPC mediates macromolecular trafficking between the nucleoplasm and the cytoplasm, but specific roles of individual Nups are poorly understood in higher plants. Here, we show that the novel nucleoporin unique to angiosperm plants (designated as Nup82) functions in a salicylic acid-dependent defense in a redundant manner with Nup136, which is a component of the nuclear basket in the NPC. Arabidopsis thaliana Nup82 had a similar amino acid sequence to the N-terminal half of Nup136 and a Nup82-GFP fusion was localized on the nuclear envelope. Immunoprecipitation and bimolecular fluorescence complementation analyses revealed that Nup82 interacts with the NPC components Nup136 and RAE1. The double knockout mutant nup82 nup136 showed severe growth defects, while the single knockout mutant nup82 did not, suggesting that Nup82 functions redundantly with Nup136. nup82 nup136 impaired benzothiadiazole (an analog of salicylic acid)-induced resistance to the virulent bacteria Pseudomonas syringae pv. tomato DC3000. Furthermore, transcriptome analysis of nup82 nup136 indicates that deficiency of Nup82 and Nup136 causes noticeable downregulation of immune-related genes. These results suggest that Nup82 and Nup136 are redundantly involved in transcriptional regulation of salicylic acid-responsive genes through nuclear transport of signaling molecules.
KEYWORDS: Arabidopsis thaliana, nuclear envelope, nucleoporin, Nup136/Nup1, Nup82, plant immunity, proteome, transcriptome
Introduction
The nuclear pore complex (NPC), which spans the outer and inner nuclear membranes, tightly regulates protein and RNA trafficking between the cytoplasm and the nucleus. Although recent works suggested that large ribonucleoprotein complexes could be exported from the nucleus by budding from the nuclear membrane,1 most of nucleocytoplasmic exchange occurs through the NPC. Ultrastructure analysis indicated that the overall structure of the NPC is conserved among eukaryotes.2-4 The NPC is a cylindrical channel comprising repetitive subunits that are organized with 8-fold radial symmetry around the central axis. Despite its large size (∼60 MDa in yeast and ∼120 MDa in mammals), the NPC comprises 30 species of proteins, termed nucleoporins (Nups) that are present in multiple copies.5,6 By combining a diverse set of immunoelectron microscopic, crystallographic and proteomics experiments, a detailed architectural map of the yeast NPC was calculated and determined.7,8 According to this model, nucleoporins can be subdivided into 5 classes: transmembrane ring, core scaffold (inner ring, outer ring and linker), cytoplasmic filaments, nuclear basket and central barrier.7-10 Transmembrane ring and core scaffold nucleoporins are thought to play important roles in the assembly of the NPC on the nuclear envelope, while other nucleoporins are responsible for regulation of trafficking, via their Phenylalanine-Glycine (FG) repeats, which can bind nuclear transport receptors. The nuclear basket is composed of 3 nucleoporins, including NUA, Nup146/Nup1 and Nup50 in higher plants.11,12 Interestingly, the nuclear basket plays multiple nuclear envelope-associated functions, such as RNA biogenesis, regulation of SUMO homeostasis, chromatin maintenance and the control of cell division.13
In addition to the transport system, the NPC provides attachment sites for the underlying chromatin, and regulates genome organization and gene expression by establishing distinct chromatin environments.14,15 An early electron microscopy study demonstrated that the NPC is surrounded by a decondensed chromatin domain, while the nuclear membrane is largely associated with heterochromatin.16 Nucleoporins in mammals and yeast interact with chromatin or transcriptional machineries to regulate gene expression.17,18 Recently, it was also shown that tethering reporter genes to the NPC significantly affected their expression level in Arabidopsis.19 These results suggested that mechanism of gene expression regulation by the NPC is well-conserved among eukaryotes.
In the past 10 years, genetics and proteomics have identified many plant nucleoporins and their physiologic roles.11,12 Importantly, plant nucleoporins mediate many biologic processes,20 including pathogen interactions,21-23 symbiosis24-26 hormone responses,27-30 abiotic stresses31-33 and the circadian clock.34 However, it is largely unknown how different nucleoporins in the NPC control the trafficking of cargo molecules for the signaling pathways. Previously, we identified Arabidopsis Nup136/Nup1, which has a plant specific sequence, using an interactive proteomics approach.35 Based on its structure, its dynamics on the nuclear envelope, and interactors, Nup136 was proposed as a functional homolog of mammalian Nup153, which is localized in the nuclear basket of the NPC.7,8 Nup136 deficiency resulted in pleiotropic phenotypes, such as early flowering, abnormal nuclear shape and lower fertility,35,36 suggesting the importance of Nup136 for plant growth. In this study, we isolated and characterized a novel nucleoporin, Nup82, whose sequences are similar to those of N-terminal half of Nup136. Our results showed that Nup82 is functionally redundant with Nup136 in mediating the salicylic acid-dependent immune response, which functions via changes in gene expression.
Results and discussion
Nup82 is a novel nucleoporin conserved in angiosperms
To identify novel nucleoporins from Arabidopsis, we searched protein databases with sequences of known plant nucleoporins. This analysis indicated that Arabidopsis thaliana contains a gene (At5g20200) that encodes a homologous protein to Nup136 (At3g10650), with 40% similarity in overall amino acid sequences. The At5g20200 gene was predicted to encode an 82-kDa protein whose sequences are homologous to those of N-terminal half of Nup136 (Fig. S1). We therefore named the protein as Nup82. In contrast to Nup136, Nup82 has no FG repeats, which are involved in nucleocytoplasmic transport, nor does it have any known functional domain. Homologs of Nup82 were found in other plant species, including Glycine max (dicot), Populus trichocarpa (dicot), Nelumbo nucifera (eudicot), Zea mays (monocot), Oryza sativa (monocot), and Amborella trichopoda (basal angiosperm), but not in Physcomitrella patens (moss), Selaginella moellendorffii (fern), or Picea abies (gymnosperm) (Fig. 1A). A phylogenetic tree with Nup82 and Nup136 members in plants clearly demonstrated that 2 nucleoporin ancestors existed before lineage-specific expansions (Fig. 1A). Therefore, Nup82 is a unique nucleoporin that might have been acquired during angiosperm evolution to provide the NPC with additional functions. To clarify the subcellular localization of Nup82, we expressed a Nup82–GFP fusion protein in Arabidopsis plants and protoplasts, stably and transiently, respectively. As observed for other GFP-fused nucleoporins,35 Nup82-GFP clearly labeled the nuclear periphery (Fig. 1B and C), which was similar to the NPC fluorescent signals obtained from other Arabidopsis and eukaryote nucleoporins.35,37 These results suggest that Nup82-GFP is localized in the NPC.
Figure 1.
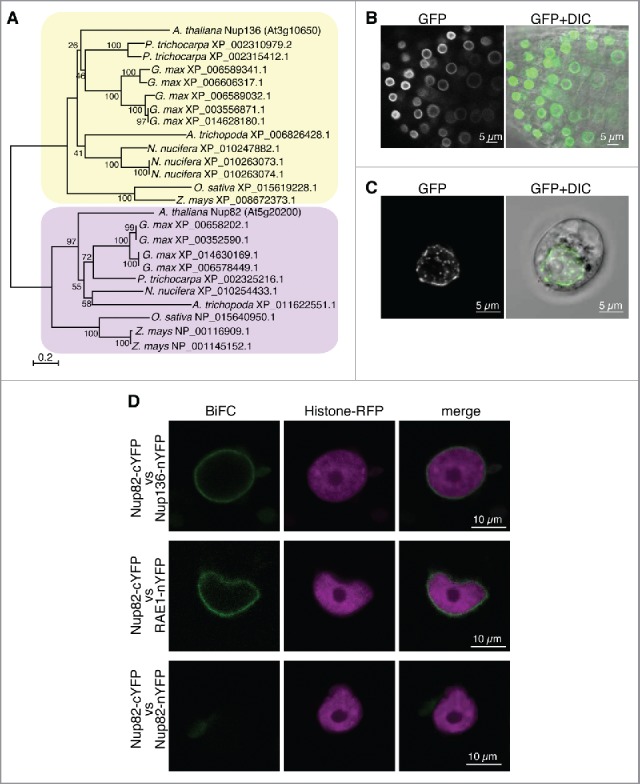
Nup82 is a nucleoporin that is unique to angiosperms and is localized on the nuclear envelope. (A) The phylogenetic tree of Nup82 (yellow box) and Nup136 (purple box) proteins from representative angiosperms (Amborella trichopoda), monocots (Oryza sativa and Zea mays), eudicots (Nelumbo nucifera), and dicots (Glycine max, Populus trichocarpa and Arabidopsis thaliana). The aligned amino acid sequences were assembled into a phylogenetic tree using the boot-strapped neighbor-joining algorithm48 in MEGA 6.06 with 1000 trials (http://www.megasoftware.net/). Bootstrap values are indicated as percentages of 1000 trials at their respective nodes. Labels at the branch tips indicate NCBI Reference Sequence Numbers or gene names. (B) Confocal fluorescence images of root tip cells from transgenic Arabidopsis stably expressing Nup82-GFP. (C) Confocal fluorescence images of protoplasts of Arabidopsis cultured cells transiently expressing Nup82-GFP. (D) Bimolecular fluorescence complementation (BiFC) assays in tobacco leaf epidermal cells. Histone-RFP was coexpressed as a marker for visualizing nuclei in the transformed cells. nYFP, N-terminal half of YFP. cYFP, C-terminal half of YFP.
We next determined whether Nup82 interacts with other nucleoporins in the NPC. An anti-GFP antibody was used to immunoprecipitate detergent-solubilized fractions from the transgenic Nup82–GFP plants. To identify the components of the immunoprecipitates, proteins were separated by SDS-PAGE, followed by a mass spectrometry analysis using an LTQ-Orbitrap instrument. The mass spectrometry analysis demonstrated that Nup82-GFP coimmunoprecipitated with 2 nucleoporins (RAE1 and Nup136) and one nucleocytoplasmic transport protein (Ran-binding protein1 domain-containingprotein) (Table 1 and Supplemental Table 1). RAE1 and Nup136 are components of outer-ring and nuclear basket of the NPC, respectively.11,12 Ran is an evolutionary conserved member of small GTPase and regulates nucleocytoplasmic transport.38 Bimolecular fluorescence complementation (BiFC) assays confirmed the interaction of Nup82 with Nup136 and RAE1 but not with Nup82 itself (Fig. 1D). These results indicate that Nup82 interacts with nucleoporins, forming the NPC in Arabidopsis. Previously, we found that Nup136–GFP, which dynamically associates and disassociates with the NPC, was also coimmunoprecipitated with only a few nucleoporins.35 These results of the coimmunoprecipitates with Nup82-GFP and Nup136-GFP suggest that Nup82 interacts directly with Nup136 in the basket of the NPC. We hypothesized that Nup82 has a function related to that of Nup136.
Table 1.
Identification of Arabidopsis nucleoporins and Ran-binding protein that interact with Nup82-GFP by mass spectrometry.
| AGI code | Assigned name | Peptide | Score |
|---|---|---|---|
| At5g20200 | Nup82 | 30 | 545 |
| At4g11790 | Ran-binding protein | 4 | 195 |
| At1g80670 | RAE1 | 1 | 31 |
| At3g10650 | Nup136 | 3 | 30 |
The Arabidopsis Genome Initiative (AGI) codes were obtained from the TAIR database (http://www.arabidopsis.org). Peptide represents the number of unique peptides matched by mass spectrometry. Scores were calculated using MASCOT (Matrix Science).
Nup82 and Nup136 function redundantly in plant growth
To determine the physiologic function of Arabidopsis Nup82, 2 T-DNA insertion mutant lines, nup82–1 and nup82–2, were isolated (Fig. 2A). Reverse transcription polymerase chain reaction (RT-PCR) confirmed that not full-length but truncated NUP82 transcripts were present in either allele (Fig. 2B and Fig. S2). However, the mutants grew normally and were indistinguishable from wild-type (WT) plants under our standard our growing condition (data not shown), suggesting that Arabidopsis has functionally redundant nucleoporins. We speculated that Nup136 shares a function with Nup82. We, therefore, produced double knockout mutants for Nup136 and Nup82, which share similar amino acid sequences (Fig. S1). At the seedling stage, the nup82 nup136 double mutants grew normally, compared with either the WT or the nup136 single mutant (Fig. 2C). However, after bolting, the nup82 nup136 double mutants showed a more severe phenotype, which included stronger dwarfism and lower fertility (Fig. 2C and D). We previously reported that Nup136 deficiency led was important in nuclear morphology.35 However, the nup82 mutation had no additive/synergistic effect on the nuclear morphology in the nup82 nup136 mutant (Fig. S3). These results suggest that Nup82 functions redundantly with Nup136 in plant growth but not in the nuclear morphology.
Figure 2.
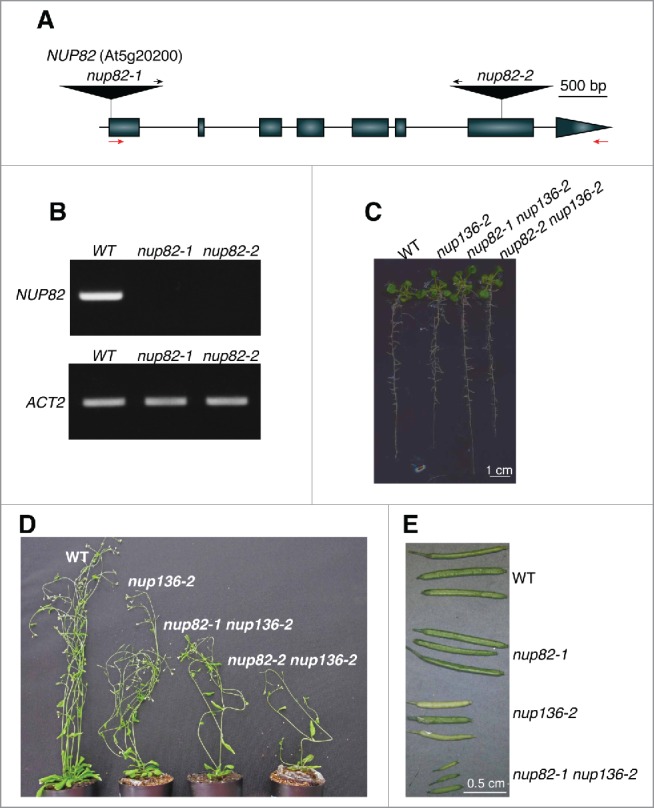
Isolation of nup82 nup136 double mutants. (A) A schematic representation of the NUP82 gene, which contains 8 exons. The positions of T-DNA insertions in nup82–1 and nup82–2 are shown. Closed boxes and solid lines indicate exons and introns, respectively. Black arrows indicate the orientation of the left border sequence. Red arrows indicate the primers used for RT-PCR in (B). (B) RT-PCR analysis of NUP82 and ACTIN2 (ACT2) transcripts in the wild-type (WT), nup82–1, and nup82–2. Amplification of NUP82 and ACT2 required 40 and 27 PCR cycles, respectively. These data are representative of 2 biologic replicates and 2 technical replicates. (C) Thirteen-day-old seedlings of wild-type (WT), nup136–2, nup136–2 nup82–1, and nup136–2 nup82–2 plants. (D) Five-week-old wild-type (WT), nup136–2, nup136–2 nup82–1, and nup136–2 nup82–2 plants. (E) Siliques of wild-type (WT), nup82–1, nup136–2, and nup136–2 nup82–1 plants.
Nup82 and Nup136 are required for the plant immune response
Since Nup82 and Nup136 shared amino acid sequences and were suggested to function redundantly, we next examined the transcriptome of the nup82 nup136 double mutants to clarify the molecular function of the nucleoporins. Total RNA was isolated from 14-day-old seedlings grown under normal conditions and labeled for a microarray analysis (Supplemental Table 2). An enrichment analysis of the differentially expressed genes in the WT and the mutant identified 12 Gene Ontology (GO) terms in the biologic process category (the minimum hypergeometric (mHG) model, P < 0.05; Benjamini–Hochberg false discovery rate (FDR) < 0.33), all of which were identified among the downregulated genes in the mutant (Supplemental Table 3). On the contrary, none of GO terms were enriched in upregulated genes in the mutant. The 12 GO terms were further analyzed using REViGO (reduce and visualize gene ontology)39 to cluster semantic functional redundancies across these lists of significantly overrepresented GO terms (Fig. 3A). The most prominent and significantly overrepresented biologic processes among the downregulated genes in the mutant were a defense response to other organisms (Fig. 3A and Supplemental Table 3). Fig. 3B shows a list of the top 10 defense response genes that were downregulated in the mutant compared with the WT. Among them, publically available microarray data (AtGenExpress. http://jsp.weigelworld.org/AtGenExpress/resources/) revealed that the expressions of 6 genes were upregulated significantly in response to attack by either virulent or avirulent bacteria (Fig. 3B). To verify the microarray data, we performed quantitative real-time RT-PCR (qRT-PCR) analysis of 14-day-old Arabidopsis leaf tissues under normal growth condition. In the mutants, the expression levels of the top 3 genes in the list (At2g14610, At4g12490 and At4g07820) were significantly reduced compared with WT (Fig. 3C). This reduction was in a gene dose-dependent manner, suggesting that bacterial response pathway in the mutant might be affected by deficiency of both Nup136 and Nup82.
Figure 3.
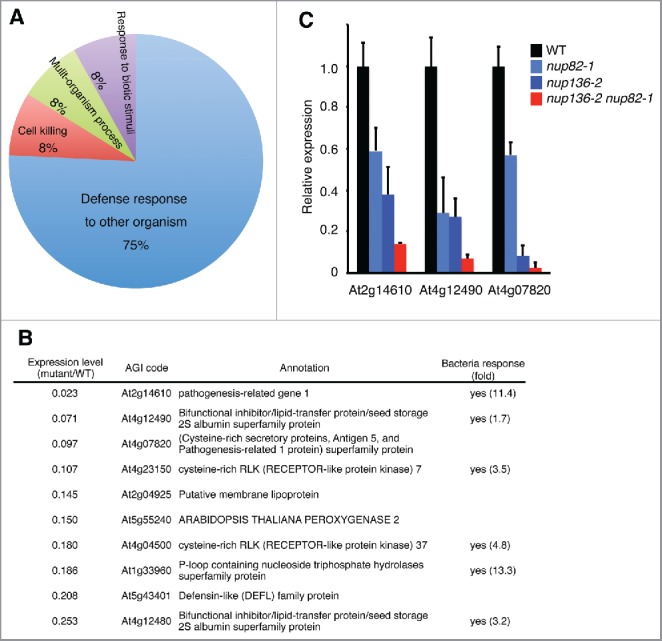
Defense-related genes are downregulated in the nup82 nup136 double mutant. (A) An enrichment analysis of Gene Ontology (GO) terms in the biologic processes category. REViGO amalgamated GO terms associated with the downregulated genes in the nup136 nup82 double mutant. (B) The top 10 genes in the group of defense response, which were downregulated in the nup82 nup136 double mutant. Bacterial response indicates whether gene expression is induced by bacterial infection, according to a public microarray analysis (AtGeneExpress). fold, fold change in gene expression between Control P.s. at 24 h vs Nonhost P.s. at 24 h. (C) Quantitative RT-PCR of defense-related genes in 14-day-old seedlings of the wild-type (WT), nup82–1, nup136–2, and nup82–1 nup136–2 plants. Data represent mean values of 3 independent experiments with standard deviations.
The reduced basal level of defense gene expression in the mutant raised the possibility that the mutant is more susceptible to pathogens than the WT. We first challenged the nup136, nup82, and nup82 nup136 mutants with a virulent bacteria, Pseudomonas syringae pv tomato DC3000 (Pto DC3000). Two days after inoculation, there was significant increase in Pto DC3000 growth in each mutant (Fig. 4A). In particular, deficiency of both nup136 and nup82 led to the strongest phenotype. To understand the immune response against Pto DC3000 in the mutants, we treated them with exogenous benzothiadiazole (BTH), which is an analog of salicylic acid (SA) to trigger global transcriptional reprogramming and resistance to a broad spectrum of biotrophic and hemibiotrophic pathogens.40 Consistent with a previous report,41 BTH treatment induced disease resistance, resulting in a substantial reduction of bacterial growth in the WT an the mutants (Fig. S4). Bacterial growth on the nup82 single mutant showed no difference compared with the WT (Fig. 4B). By contrast, the nup136 single and the nup82 nup136 double mutant had statically higher numbers of the bacteria than the WT and nup82 single mutant (p < 0.05) (Fig. 4B). We also found that the nup82 nup136 double mutant was compromised in BTH-dependent expression of immune-related genes (Fig 4C). These results suggest that the functions of Nup136 and Nup82 are partially redundant in SA-dependent resistance against virulent bacteria.
Figure 4.
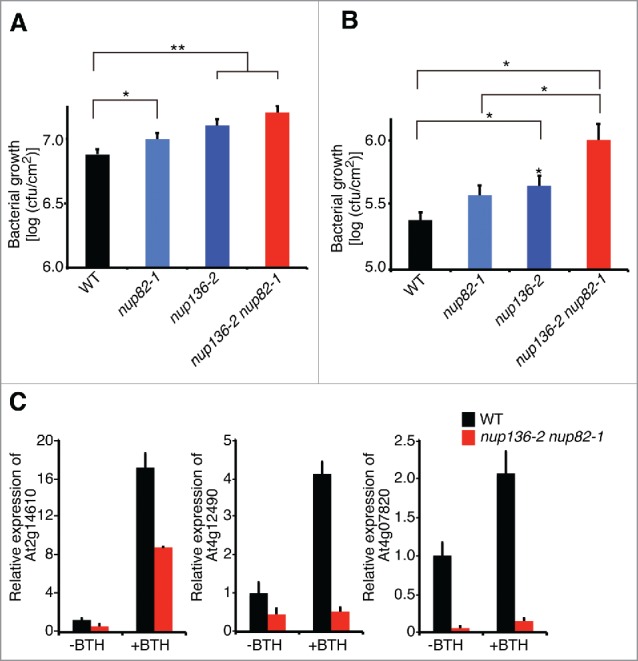
Nup136 and Nup82 are involved in benzothiadiazole (BTH)-induced resistance against Pseudomonas syringae pv tomato virulent strain DC3000 (Pto DC3000). (A and B) Bacterial growth at 2 d after inoculation with Pto DC3000 (OD600 = 0.0001) in the leaves of nup136–2, nup82–1, and nup136–2 nup82–1 mutants that were pretreated without (A) or with (B) 150 µM BTH for 1 day. *, p < 0.05; **, p < 0.01 compared with the wild-type plants. Data represent mean values of 2 independent experiments (n = 16). Error bars indicate standard errors (Student's t test, *p < 0.05, **p < 0.01). (C) Quantitative RT-PCR of defense-related genes in 14-day-old seedling of WT and nup82–1 nup136–2 with (+BTH) and without benzothiazole (-BTH) treatment. Data represent mean values of 3 independent experiments with standard deviations.
How do Nup82 and Nup136 control the expression of defense-related genes? One possibility is direct association of the NPC with the genes to regulate their activity. The nuclear basket components of the NPC in Drosophila bind up to 25% of the genome domains that are enriched in active transcription chromatin markers.42 The other possibility is NPC-mediated nucleocytoplasmic transport of specific cargos, which induce the expression of defense-related genes. Several Arabidopsis nucleoporins have been found to be involved in immune responses. Arabidopsis nup9621 and nup8822 mutants were deficient in SA accumulation, defense-related gene expression and pathogen resistance. In the nup88 mutants, the nuclear accumulations of defense-signaling components, either NPR1 or EDS1, were impaired specifically,22 suggesting that nucleocytoplasmic transport of particular cargos plays a key role in the immune response. Consistent with this, Nup160 and Seh1, both of which are components of the Nup107-Nup160 subcomplex, were also required for the nuclear accumulation of EDS1 in response to bacterial infection.23 It would be interesting to know the functional relationships between these nucleoporins and Nup82/Nup136 in plant immune responses.
Considering the generality of NPC's function in plant cells, it is particularly interesting that a specific set of genes is regulated by certain nucleoporins. Transcriptome analyses have been performed for various Arabidopsis nucleoporin mutants, including tpr,30 hos134, nup6212 and nup160.12 Interestingly, each nup mutation affected the expressions of a different set of genes, suggesting the functional diversity of nucleoporins. For example, in the hos1 mutant, multiple functional categories of genes were affected, possibly resulting from disruption of mRNA export from the nucleus.34,43 By contrast, both nup160 and nup62 mutants upregulated groups of genes involved in nucleoporins and nuclear transport factors, implying feedback regulation to compensate for the loss of the NPC activity.12 It is hypothesized that such specificity of nucleoporin-mediated gene expression plays an important role in response to multiple signaling pathways in plants.
Materials and methods
Plant materials
Wild type and mutant derived form Arabidopsis thaliana (accession Col-0) were used. The T-DNA insertion mutants, SALK_001707 (nup82–1), SALK_024526 (nup82–2), and SAIL_796_H02 (nup136–2) were obtained from the Arabidopsis Biological Resource Center at Ohio State University.
RT-PCR
Total RNA was isolated from 14-day-old seedlings using the RNeasy Plant Mini Kit (74904, Qiagen). cDNA was synthesized using Read-To-Go RT-PCR Beads (27925901, GE Healthcare) with an oligo (dT)12–18 primer. DNA fragments were amplified using specific primers (ACT2; 5′-AGAGATTCAGATGCCCAGAAGTCTTGTTCC-3′ and 5′-GAGTATGATGAGGCAGGTCCAGGAATCGTT-3′, full length of Nup82; 5′- ATGGCCACTCAAGGAGAAGCGACG-3′ and 5′-TTAACAGCAGACACCGTCACTTC-3′, primer set 1 of Nup82; 5′- ATGGCCACTCAAGGAGAAGC-3′ and 5′- CACTGGGATCATTGTCCTGTGG-3′, and primer set 2 of Nup82; 5′- GGACCCAGTAGAAGAACTCGTC-3′ and 5′- CTCATCATGCATGCTTCTGG-3′). ACT2 was amplified using 26 and Nup82 using 38 PCR cycles, respectively. qRT-PCR was performed with specific primers (At2g14610; 5′-CGAACACGTGCAATGGAGTT-3′ and 5′-CACTTTGGCACATCCGAGTC-3′, At4g12490; 5′-CCCTACGCCAGTCATTCCTC- 3′ and 5′- AGGTGATGGCTGTCCCAACT-3′, and At4g07820; 5′-AGGCGTGAGTCCCTTGATGT-3′ and 5′-TAGGGCCCACCTGAGAGAAA- 3′) and SYBR Premix Ex Taq II (RR820A, Takara).
Imaging
The Nup82 fragment was generated using specific primers (5′-AACCAATTCAGTCGACATGGCCACTCAAGGAGAAGCGAC-3′ and 5′-AAGCTGGGTCTAGATATCCACAGCAGACACCGTCACTTCCT-3′) and cloned into the pENTR1A vector (11813–011, Invitrogen). To generate the DNA constructs for GFP fusions, the cloned DNA fragment was transferred from the entry clone to the pGWB405 Gateway destination vector44 by an in vitro recombination attL × attR reaction. Transient and stable expression of Nup82–GFP in Arabidopsis was performed as described previously.35 Confocal images were obtained using laser scanning microscopes (Zeiss LSM 780 and Zeiss LSM 510 META; Carl Zeiss) equipped with a 488-nm 40-mW Ar/Kr laser, and a 100×1.45 N.A. oil immersion objective (α Plan-Fluar, 000000–1084–514, Carl Zeiss), or 63×1.2 N.A. water immersion objective (C-Apochromat, 441777–9970–000, Carl Zeiss). Image analysis was performed using either LSM image examiner software (Carl Zeiss) or Fiji software.
Interactome analysis
Immunoprecipitation was performed with µMACS Epitope Tag Protein Isolation Kits (Miltenyi Biotec), as described by previously.35 LC-MS/MS analyses were performed using the LTQ-Orbitrap XL-HTC-PAL system (Thermo Fisher Scientific, Bremen, Germany), as described previously.45 The Mascot (Matrix Science Ltd, UK) search parameters were set as follows: threshold of the ion-score cut-off, 0.05; peptide tolerance, 10 ppm; MS/MS tolerance, ± 0.8 Da; and peptide charge, 2+ or 3+. The search was also set to allow one missed cleavage by trypsin, a carboxymethylation modification of cysteine residues, and a variable oxidation modification of methionine residues.
Bimolecular fluorescence complementation (BiFC)
To generate the DNA constructs for a BiFC assay, the cloned DNA fragments were transferred from the entry clone to the Gateway destination vectors (pB4NY2 and pB4CY2) by an in vitro recombination attL × attR reaction. Transient expression in tobacco was performed as described previously.45 Histone-RFP fusion was coexpressed as a marker for visualizing nuclei in the transformed cells.
Bacterial strains and preparation of the inoculant
Pseudomonas syringae pv tomato virulent strain DC3000 (Pto DC3000) was grown overnight in King's B medium supplemented with 50 µg/ml of rifampicin. The bacteria were harvested by centrifugation, washed and diluted to the desired density with water.
Bacterial growth assay
Bacterial growth assays were performed as described previously.46 Briefly, bacterial suspensions (OD600 = 0.0001) were infiltrated into leaves of 4 to 5-week-old plants that had been treated with or without 150 µM BTH for 1 day using a needleless syringe. Colony-forming units (cfu) per cm2 of the leaf surface area were counted from leaf tissue collected 2 d after infiltration.
Microarray and GO analysis
Total RNA (200 ng) isolated from 14-day-old seedlings was labeled with Cy3 and hybridized to an Agilent 4×44k Gene Expression Array (Arabidopsis thaliana Ver. 4), according to the manufacturer's instructions. Functional categorization and GO enrichment of the differentially expressed genes in the mutant were performed using GOrilla.47 We considered a GO category significant if it had a hypergeometric test P-value <0.05. REViGO39 was used to account for the functional and semantic redundancies among the identified of GO terms, using a simple clustering algorithm.
Supplementary Material
Abbreviations
- NPC
Nup
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by Grant-in-Aid for Scientific Research to K.T. (no. 26711017 and 15K14545), a Specially Promoted Research for Grant-in Aid for Scientific Research to I.H.-N. (no. 22000014) from JSPS, and by grants from US National Science Foundation (IOS-1121425 and MCB-1518058) to F.K.
References
- [1].Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, et al.. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 2012; 149:832-46; PMID:22579286; https://doi.org/ 10.1016/j.cell.2012.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Roberts K, Northcote DH. Structure of the nuclear pore in higher plants. Nature 1970; 228:385-6; PMID:5473989; https://doi.org/ 10.1038/228385a0 [DOI] [PubMed] [Google Scholar]
- [3].Fiserova J, Goldberg MW. Relationships at the nuclear envelope: lamins and nuclear pore complexes in animals and plants. Biochem Soc Trans 2010; 38:829-31; PMID:20491671; https://doi.org/ 10.1042/BST0380829 [DOI] [PubMed] [Google Scholar]
- [4].Fiserova J, Kiseleva E, Goldberg MW. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J 2009; 59:243-55; PMID:19392704; https://doi.org/ 10.1111/j.1365-313X.2009.03865.x [DOI] [PubMed] [Google Scholar]
- [5].Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 2000; 148:635-51; PMID:10684247; https://doi.org/ 10.1083/jcb.148.4.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 2002; 158:915-27; PMID:12196509; https://doi.org/ 10.1083/jcb.200206106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al.. Determining the architectures of macromolecular assemblies. Nature 2007; 450:683-94; PMID:18046405; https://doi.org/ 10.1038/nature06404 [DOI] [PubMed] [Google Scholar]
- [8].Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al.. The molecular architecture of the nuclear pore complex. Nature 2007; 450:695-701; PMID:18046406; https://doi.org/ 10.1038/nature06405 [DOI] [PubMed] [Google Scholar]
- [9].Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure 2009; 17:1156-68; PMID:19748337; https://doi.org/ 10.1016/j.str.2009.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Grossman E, Medalia O, Zwerger M. Functional architecture of the nuclear pore complex. Ann Rev Biophys 2012; 41:557-84; PMID:22577827; https://doi.org/ 10.1146/annurev-biophys-050511-102328 [DOI] [PubMed] [Google Scholar]
- [11].Tamura K, Hara-Nishimura I. The molecular architecture of the plant nuclear pore complex. J Exp Bot 2013; 64:823-32; PMID:22987840; https://doi.org/ 10.1093/jxb/ers258 [DOI] [PubMed] [Google Scholar]
- [12].Parry G. Components of the Arabidopsis nuclear pore complex play multiple diverse roles in control of plant growth. J Exp Bot 2014; 65:6057-67; PMID:25165147; https://doi.org/ 10.1093/jxb/eru346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol 2010; 11:490-501; PMID:20571586; https://doi.org/ 10.1038/nrm2928 [DOI] [PubMed] [Google Scholar]
- [14].Raices M, D'Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol 2012; 13:687-99; PMID:23090414; https://doi.org/ 10.1038/nrm3461 [DOI] [PubMed] [Google Scholar]
- [15].Van de Vosse DW, Wan Y, Lapetina DL, Chen WM, Chiang JH, Aitchison JD, Wozniak RW. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell 2013; 152:969-83; PMID:23452847; https://doi.org/ 10.1016/j.cell.2013.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Engelhardt P, Pusa K. Nuclear pore complexes: “press-stud” elements of chromosomes in pairing and control. Nat New Biol 1972; 240:163-6; PMID:4564189; https://doi.org/ 10.1038/newbio240163a0 [DOI] [PubMed] [Google Scholar]
- [17].Liang Y, Hetzer MW. Functional interactions between nucleoporins and chromatin. Curr Opin Cell Biol 2011; 23:65-70; PMID:21030234; https://doi.org/ 10.1016/j.ceb.2010.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sarma NJ, Willis K. The new nucleoporin: regulator of transcriptional repression and beyond. Nucleus 2012; 3:508-15; PMID:23047597; https://doi.org/ 10.4161/nucl.22427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Smith S, Galinha C, Desset S, Tolmie F, Evans D, Tatout C, Graumann K. Marker gene tethering by nucleoporins affects gene expression in plants. Nucleus 2015; 6:471-8; PMID:26652762; https://doi.org/ 10.1080/19491034.2015.1126028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Parry G. Assessing the function of the plant nuclear pore complex and the search for specificity. J Exp Bot 2013; 64:833-45; PMID:23077202; https://doi.org/ 10.1093/jxb/ers289 [DOI] [PubMed] [Google Scholar]
- [21].Zhang Y, Li X. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 2005; 17:1306-16; PMID:15772285; https://doi.org/ 10.1105/tpc.104.029926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cheng YT, Germain H, Wiermer M, Bi D, Xu F, Garcia AV, Wirthmueller L, Despres C, Parker JE, Zhang Y, et al.. Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 2009; 21:2503-16; PMID:19700630; https://doi.org/ 10.1105/tpc.108.064519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wiermer M, Cheng YT, Imkampe J, Li M, Wang D, Lipka V, Li X. Putative members of the Arabidopsis Nup107-160 nuclear pore sub-complex contribute to pathogen defense. Plant J 2012; 70:796-808; PMID:22288649; https://doi.org/ 10.1111/j.1365-313X.2012.04928.x [DOI] [PubMed] [Google Scholar]
- [24].Kanamori N, Madsen LH, Radutoiu S, Frantescu M, Quistgaard EM, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, et al.. A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci U S A 2006; 103:359-64; PMID:16407163; https://doi.org/ 10.1073/pnas.0508883103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Saito K, Yoshikawa M, Yano K, Miwa H, Uchida H, Asamizu E, Sato S, Tabata S, Imaizumi-Anraku H, Umehara Y, et al.. NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 2007; 19:610-24; PMID:17307929; https://doi.org/ 10.1105/tpc.106.046938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Groth M, Takeda N, Perry J, Uchida H, Draxl S, Brachmann A, Sato S, Tabata S, Kawaguchi M, Wang TL, et al.. NENA, a Lotus japonicus homolog of Sec 13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 2010; 22:2509-26; PMID:20675572; https://doi.org/ 10.1105/tpc.109.069807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Verslues PE, Guo Y, Dong CH, Ma W, Zhu JK. Mutation of SAD2, an importin beta-domain protein in Arabidopsis, alters abscisic acid sensitivity. Plant J 2006; 47:776-87; PMID:16889648; https://doi.org/ 10.1111/j.1365-313X.2006.02833.x [DOI] [PubMed] [Google Scholar]
- [28].Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M. The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 2006; 18:1590-603; PMID:16751346; https://doi.org/ 10.1105/tpc.106.041566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Boeglin M, Fuglsang AT, Luu DT, Sentenac H, Gaillard I, Cherel I. Reduced expression of AtNUP62 nucleoporin gene affects auxin response in Arabidopsis. BMC Plant Biol 2016; 16:2; PMID:26728150; https://doi.org/ 10.1186/s12870-015-0695-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jacob Y, Mongkolsiriwatana C, Veley KM, Kim SY, Michaels SD. The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiol 2007; 144:1383-90; PMID:17535820; https://doi.org/ 10.1104/pp.107.100735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ishitani M, Xiong L, Lee H, Stevenson B, Zhu JK. HOS1, a genetic locus involved in cold-responsive gene expression in arabidopsis. Plant Cell 1998; 10:1151-61; PMID:9668134; https://doi.org/ 10.1105/tpc.10.7.1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dong CH, Hu X, Tang W, Zheng X, Kim YS, Lee BH, Zhu JK. A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol Cell Biol 2006; 26:9533-43; PMID:17030626; https://doi.org/ 10.1128/MCB.01063-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo–cytoplasmic partitioning. Genes Dev 2001; 15:912-24; https://doi.org/ 10.1101/gad.866801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Macgregor DR, Gould P, Foreman J, Griffiths J, Bird S, Page R, Stewart K, Steel G, Young J, Paszkiewicz K, et al.. HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 Is Required for circadian periodicity through the promotion of nucleo-cytoplasmic mRNA export in Arabidopsis. Plant Cell 2013; 25:4391-404; PMID:24254125; https://doi.org/ 10.1105/tpc.113.114959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 2010; 22:4084-97; PMID:21189294; https://doi.org/ 10.1105/tpc.110.079947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tamura K, Hara-Nishimura I. Involvement of the nuclear pore complex in morphology of the plant nucleus. Nucleus 2011; 2:168-72; PMID:21818409; https://doi.org/ 10.4161/nucl.2.3.16175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 2004; 6:1114-21; PMID:15502822; https://doi.org/ 10.1038/ncb1184 [DOI] [PubMed] [Google Scholar]
- [38].Tamura K, Hara-Nishimura I. Functional insights of nucleocytoplasmic transport in plants. Front Plant Sci 2014; 5:118; PMID:24765097; https://doi.org/ 10.3389/fpls.2014.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 2011; 6:e21800; https://doi.org/ 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gorlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel KH, Oostendorp M, Staub T, Ward E, Kessmann H, et al.. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 1996; 8:629-43; PMID:8624439; https://doi.org/ 10.1105/tpc.8.4.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J 1996; 10:71-82; PMID:8758979; https://doi.org/ 10.1046/j.1365-313X.1996.10010071.x [DOI] [PubMed] [Google Scholar]
- [42].Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet 2010; 6:e1000846; PMID:20174442; https://doi.org/ 10.1371/journal.pgen.1000846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].MacGregor DR, Penfield S. Exploring the pleiotropy of hos1. J Exp Bot 2015; 66:1661-71; PMID:25697795; https://doi.org/ 10.1093/jxb/erv022 [DOI] [PubMed] [Google Scholar]
- [44].Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 2007; 104:34-41; PMID:17697981; https://doi.org/ 10.1263/jbb.104.34 [DOI] [PubMed] [Google Scholar]
- [45].Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, Nishimura M, Hara-Nishimura I. Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr Biol 2013; 23:1776-81; PMID:23973298; https://doi.org/ 10.1016/j.cub.2013.07.035 [DOI] [PubMed] [Google Scholar]
- [46].Hatsugai N, Hillmer R, Yamaoka S, Hara-Nishimura I, Katagiri F. The mu Subunit of Arabidopsis Adaptor Protein-2 Is Involved in Effector-Triggered Immunity Mediated by Membrane-Localized Resistance Proteins. Mol Plant Microbe Interact 2016; 29:345-51; PMID:26828402; https://doi.org/ 10.1094/MPMI-10-15-0228-R [DOI] [PubMed] [Google Scholar]
- [47].Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 2009; 10:48; PMID:19192299; https://doi.org/ 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4:406-25; PMID:3447015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


