Abstract
Gene duplication is the primary mechanism leading to new genes and phenotypic novelty, but the proximate evolutionary processes underlying gene family origin, maintenance, and expansion are poorly understood. Although sub- and neofunctionalization provide clear long-term advantages, selection does not act with foresight, and unless a redundant gene copy provides an immediate fitness advantage, the copy will most likely be lost. Many models for the evolution of genes immediately following duplication have been proposed, but the robustness and applicability of these models is unclear because of the lack of data at the population level. We used qPCR, protein expression data, genome sequencing, and hybrid enrichment to test three competing models that differ in whether selection favoring the spread of duplicates acts primarily on expression level or sequence diversity for specific toxin-encoding loci in the eastern diamondback rattlesnake (Crotalus adamanteus). We sampled 178 individuals and identified significant inter- and intrapopulation variation in copy number, demonstrated that copy number was significantly and positively correlated with protein expression, and found little to no sequence variation across paralogs in all populations. Collectively, these results demonstrate that selection for increased expression, not sequence diversity, was the proximate evolutionary process underlying gene family origin and expansion, providing data needed to resolve the debate over which evolutionary processes govern the fates of gene copies immediately following duplication.
Keywords: copy number, gene expression, gene family, selection
GENE families are groups of genes descended from a single gene through duplication and are often similar in sequence and related in function (Nei and Rooney 2005). The rate of gene duplication (∼1 gene−1 million years−1 in eukaryotes; Lynch and Conery 2000) is similar to the nucleotide substitution rate, and of all human genes are the result of duplication (Zhang 2003). Gene families participate in nearly all aspects of organismal physiology and function (Hughes 1999), such as oxygen transport (Efstratiadis et al. 1980) and immunity (Ota and Nei 1994), but the proximate evolutionary processes underlying gene family origin, maintenance, and expansion are poorly understood (Hahn 2009; Innan and Kondrashov 2010) because most studies of gene duplication have focused on macroevolutionary patterns of ancient duplication events (i.e., interspecific comparisons across millions of years of divergence; Nguyen et al. 2006; Dumas et al. 2007; Sudmant et al. 2010) when the origination of gene families is very much a microevolutionary process (i.e., occurs at the population level; Hahn 2009).
The fate of a duplicate immediately following duplication can be influenced by selection or random effects (Nozawa et al. 2007; McCarroll et al. 2008; Nei et al. 2008), but evidence linking copy number variation in specific loci to selection or drift is sparse because how duplications affect specific phenotypes is often unclear (Hastings et al. 2009; Conrad et al. 2010). Because of their genetic tractability (Casewell et al. 2009; Wagstaff et al. 2009; Rodrigues et al. 2012; Margres et al. 2014), venoms allow us to connect genotype and phenotype for specific duplications. Venoms are complex, polygenic, ecologically important traits that collectively function in prey capture, digestion, and defense. Venoms comprise large, multigene families, and these gene familes are believed to be the result of gene duplication and positive selection (Casewell et al. 2011; Rokyta et al. 2012) by means of the birth-and-death model of protein evolution (Fry et al. 2008; Rokyta et al. 2015b). Because venoms are secretions, changes in mRNA expression levels directly alter protein amounts in the venom (Aird et al. 2013; Rokyta et al. 2015a) and, therefore, directly influence venom efficacy (Casewell et al. 2011; Margres et al. 2016a); these specific attributes allow us to directly examine the phenotypic effects of copy number variation for a trait with direct contributions to fitness.
We characterized the degree of copy number variation and identified the proximate evolutionary process underlying gene family origin and expansion for a specific toxic protein, myotoxin (i.e., crotamine), within and between 11 populations of the eastern diamondback rattlesnake (Crotalus adamanteus). Myotoxin is a small basic peptide that causes skeletal muscle spasms and paralysis following envenomation (Ponce-Soto et al. 2010; Peigneur et al. 2012) and can be one of the more highly expressed toxins in C. adamanteus (Margres et al. 2015a,b). Previous work on this species (and others; Margres et al. 2016b) demonstrated that the expression levels of highly expressed proteins evolve under purifying selection, and rapid, adaptive differentiation in expression is restricted to low-expression proteins because of reduced selective and physiological constraints relative to high-expression proteins. However, despite these evolutionary constraints on high-expression proteins, Margres et al. (2015a,b) demonstrated that myotoxin exhibits the greatest range of expression values (0–37% of total venom expression among adults) of any toxin across C. adamanteus, similar to patterns in other species (Schenberg 1959; Aird 1985; Bober et al. 1988), and this variation may be the result of adaptive copy number variation (Margres et al. 2015b). This exception to the normal pattern of toxin expression evolution (Margres et al. 2016b), in combination with the rapid accumulation of variation, implies the action of selection, potentially in response to dietary differences and/or predator–prey coevolution.
Although examples of positive dosage effects (Gonzalez et al. 2005) and adaptive sequence divergence (Zhang et al. 1998) have been described following duplication, systematic comparisons of multiple models with different proximate evolutionary processes responsible for the fixation of a duplicate across populations are largely lacking (but see Cardoso-Moreira et al. 2016). We used qPCR, protein expression data, genome sequencing, and hybrid enrichment to test the following three models outlined by Innan and Kondrashov (2010): (1) positive dosage model, (2) permanent heterozygote model, and (3) the multi-allelic diversifying selection model. Under the positive dosage model, an increase in expression is beneficial, and the gene duplicate is initially fixed by positive selection; if selection for increased expression is strong, the sequences of both copies evolve under purifying selection. Here, we expect the number of copies to be positively correlated with expression level, and paralogs should be identical (or nearly so) in sequence. The permanent heterozygote model describes a scenario where overdominance, or heterozygote advantage, leads to the fixation and maintenance of the duplicate because the duplication fixes heterosis and eliminates the segregation load (i.e., the reduced fitness of homozygotes) found in the single-locus case; here we expect two loci, each homozygous for a different allele. The multi-allelic diversifying model is similar to the permanent heterozygote model but describes a scenario where selection favors genetic variability and, therefore, >2 alleles are maintained in the preduplication phase and expected following duplication.
Materials and Methods
Sampling
We collected venom and blood samples from 116 live-caught specimens and recorded snout–vent length (SVL) and total length (TL) for 110 of these individuals. We collected muscle and/or liver samples from an additional 66 individuals, sampling 182 individuals in total across the entire range of C. adamanteus. Population designations were based on putative biogeographic barriers as well as geographic distance as outlined in previous studies (Margres et al. 2015a,b, 2016a) and were as follows: ANF, Apalachicola National Forest and the surrounding region east of the Apalachicola River and west of the Suwannee River, FL (); CAL, Caladesi Island, FL (); EGL, Eglin Air Force Base and the surrounding region east of Pensacola Bay and west of the Apalachicola River, FL (); ENP, Everglades National Park, FL (); JEK, Jekyll Island, GA (); JNS, Joseph W. Jones Ecological Research Center, GA (); LSG, Little St. George Island, FL (); MS, Camp Shelby and southern Mississippi (); ONF, Osceola National Forest and northern peninsular Florida east of the Suwannee River, FL (); PRS, Parris Island, SC (); SAP, Sapelo Island, GA (). We also collected additional samples from the following locations: St. Vincent Island, FL (), southern Alabama (), Wayne County, GA (), and Miller County, GA (). Samples were collected under the following permits: Florida Fish and Wildlife Conservation Commission (FWC) LSSC-13-00004 and LSSC-09-0399, Eglin Air Force Base 96 SFS/SFOIRP, Everglades National Park – EVER-2012-SCI-0053, Florida Department of Environmental Protection Division of Recreation and Parks – Permits #04101310 and #03211410, St. Vincent National Wildlife Refuge – Permit #41650-2012-08, Mississippi Department of Wildlife, Fisheries, and Parks Salvage Permit, and Sapelo Island NERR Research Projects collaboration. The above procedures were approved by the Florida State University Institutional Animal Care and Use Committee under protocols #0924 and #1333.
Reversed-phase high-performance liquid chromatography
Reversed-phase high-performance liquid chromatography was performed on a Beckman System Gold HPLC (Beckman, Fullerton, CA) equipped with Beckman 32 Karat Software Version 8.0 for peak quantification as described previously (Margres et al. 2015a,b). Twenty-five RP-HPLC peaks per venom sample were quantified as previously described (Margres et al. 2015a,b; Wray et al. 2015). Briefly, relative amounts of individual peaks were determined by measuring the area under each peak relative to the total area of all identified peaks (Margres et al. 2014, 2015a). The Lambert–Beer law states that this relative amount corresponds to the percentage of total peptide bonds in the sample (McNaught and Wilkinson 1997) and provides a robust estimate of the relative amount of a specific protein by weight (Gibbs et al. 2009). Myotoxin elutes in the second peak (Margres et al. 2014, 2015a, 2016b), and the percentage of this peak was used in all analyses.
Myotoxin gene copy number estimation
We followed the method of Margres et al. (2015b) and the general approach of Oguiura et al. (2009) to estimate myotoxin gene copy number by means of qPCR for all 182 C. adamanteus samples. Genomic DNA was extracted from whole blood samples drawn from the caudal vein or from muscle/liver tissue using the Omega Bio-tek E.Z.N.A Tissue DNA Kit according to the manufacturer’s protocol. DNA quality was checked on a 2% agarose gel; all samples possessed high-quality genomic DNA. Myotoxin exon 2 was amplified from 13 ng of DNA in 20 μl qPCR runs using the primers from Margres et al. (2015b) at 0.5 μM and the Invitrogen SYBR Green PCR Master Mix. qPCR was performed on the Applied Biosystems 7500 Fast Real-Time PCR System under the following thermal cycling protocol: 2 min at 50°, 10 min at 95°, followed by 40 cycles of 15 sec at 95°, 30 sec at 58°, and 30 sec at 70°. All samples were run in triplicate, and melting curve analysis was performed with a temperature gradient from 60 to 95° at default settings. Total DNA amounts per sample replicate were quantified based on a NanoDrop Spectrophotometer reading of the NotI-linearized myoEx/TA clone, which was diluted 10× serially. Relative copy number estimates were reported as the mean of the total DNA amount quantified across sample replicates in femtograms (i.e., fg mean). This quantity is not meaningful in absolute copy number amounts, but allows us to make relative comparisons across all samples. Raw data are provided in Supplemental Material, Table S1.
Processing the raw qPCR data
qPCR samples were run in triplicate across four different sets (raw data are provided in Table S1). Although the same standard was used for each set and replicates within a set were tightly correlated (i.e., when plotted against one another, all replicates within a set exhibited a best-fit line with where slope), slight variations in the standard led to a bias in our fg estimate. This bias was evident when the data across sets were better explained by fitting two separate lines (i.e., one line for each set) rather than a single line as found in all within-set comparisons. To account for this bias, we rescaled the values of particular sets using the ratio of the slopes of the two lines (smaller over larger m value) as a multiplicative factor; for the sample set where the DNA amount quantified per sample > the mean DNA amount quantified per sample, we used the multiplicative factor to obtain our new estimates of the DNA amount quantified per sample. All corrections were performed comparing replicates two and three, and following these corrections, all replicates were well correlated ( when comparing replicates one and two, when comparing replicates one and three, and when comparing replicates two and three; Figure S1 in File S1). Additionally, we removed four individuals with coefficients of variation >0.40 (KW1906 from ONF, KW1757 from MS, and MM0090 and MM0100 from JEK). The final qPCR data set contained 178 individuals. The final, corrected data are in Table S2.
Testing for copy number variation
To test for interpopulation variation in myotoxin copy number, we ran an ANOVA on the fg mean values and included the 11 populations with samples. To test for intrapopulation variation in myotoxin copy number, we used the replicate fg measures for each individual and ran individual ANOVAs on each of the 11 populations discussed above. We used a Bonferroni correction to account for multiple tests and used a significance threshold of
Estimating total copy number
We sequenced a low-coverage genome for a single, high-copy-number specimen to establish baseline single-copy-gene and myotoxin coverage levels. The sequenced animal (KW0529) was a juvenile female from Wakulla County, FL (ANF) weighing 393 g with a SVL of 79.2 cm and a TL of 84.4 cm. The venom-gland transcriptome of this individual has been extensively characterized (Rokyta et al. 2012, 2013; Margres et al. 2014, 2015a; Rokyta et al. 2015a). We extracted genomic DNA as described above and used one lane of 150 nt paired-end sequencing on a HiSeq 2000 in the Florida State University College of Medicine Translational Science Laboratory. We targeted a 200 nt insert size and generated 164,596,064 pairs of reads; the average phred quality of the forward reads was 34, and the average phred quality of the reverse reads was 28. We merged 113,736,319 pairs of reads using PEAR (Zhang et al. 2014), and the composite reads had an average length of 200 nt and an average phred quality score of 37. We failed to merge 50,641,581 read pairs, and 218,164 read pairs were discarded.
To identify a set of presumably single-copy exons, we selected a random set of 200 nontoxin transcripts from the previously described C. adamanteus venom-gland transcriptome (Rokyta et al. 2012). We merged read pairs using PEAR (Zhang et al. 2014) and aligned the merged genome reads against the coding sequences of the 200 random transcripts using the local alignment option of bowtie2 (Langmead and Salzberg 2012). Exon boundaries were identified on the basis of consistent read-trimming across aligned reads at particular sequence positions of the coding sequence. Such trimming occurs in local alignments when a genomic read spans a boundary between exons, because part of the read will match the exon sequence and part will match the missing intron. The same approach was used to confirm the exons of myotoxin, for which the genic structure had been previously described in another rattlesnake species, C. durissus, by Oguiura et al. (2009). According to Oguiura et al. (2009), the gene was 1.8 kbp and organized into three exons and two introns: exon 1 was the first 19 aa of the signal peptide, exon 2 encoded 42 aa (3 aa of the signal peptide and 39 aa of the mature protein), and exon 3 encoded the last 3 aa of the mature protein. We again aligned the merged genome reads against the coding sequence of the myotoxin transcript to identify exon boundaries in C. adamanteus. The precursor coding region was 213 nt, and the predicted signal peptide by SignalP (Bendtsen et al. 2004) was the first 66 nt; we identified the following three exons, although the exact starting and ending location of each exon was not certain due to slight ambiguities in the ending points: exon 1 was 57 nt and encoded the first 19 aa of the signal peptide, exon 2 was 126 nt and encoded 42 aa (3 aa of the signal peptide and 39 aa of the mature protein), and exon 3 was 30 nt and encoded the last 10 aa of the mature protein. All exon sequences are provided in Table S3.
We mapped merged and unmerged reads to the 109 nontoxin exons and myotoxin exon 2 using SeqMan NGen version 12.2 and a 95% minimum match percentage. Similar to other assemblers (e.g., bwa), SeqMan NGen performs a local alignment and trims unmatched regions of the aligned read. We interpreted the difference between nontoxin coverage and myotoxin exon 2 coverage as differences in copy number and extrapolated these results (following a second, independent confirmation; see Results and Discussion) to infer a total gene copy estimate for each individual. We next used the Mclust package in R (Fraley et al. 2012) and Bayesian information criterion to fit a Gaussian mixture model to characterize the frequency and density of different copy number classes in the whole data set (i.e., 178 individuals).
Determining the relationship between copy number and protein expression
To determine the relationship between copy number and protein expression, we used a linear model to compare the fg mean and proteome percentage for 106 individuals. Because of the ontogenetic shift in venom composition (Margres et al. 2015b), we separated individuals into adult () and juvenile () age classes based on SVL (Waldron et al. 2013). Myotoxin proteome percentage is usually much larger in juvenile venoms than adults from the same population because of the simpler composition of juvenile venoms and the constant-sum constraint associated with compositional traits (Aitchison 1986). Specimens 102 cm were classified as adults and individuals <102 cm were classified as juveniles (Waldron et al. 2013), consistent with previous analyses (Margres et al. 2015a,b). Because we separated our samples into age classes, we ran an ANCOVA using age class as a factor to determine if the regression lines for adults and juveniles had different or similar slopes. Because the ontogenetic shift in venom composition affects only protein expression and not genomic content, all other analyses did not separate individuals by age class.
Probe design
Hybrid enrichment, or sequence capture, involves hybridizing short probe sequences to fragmented genomic DNA with subsequent enrichment of those regions prior to sequencing. This approach allows the target loci to be separated from the nontarget regions of the genome. Because enrichment efficiency depends largely on the level of sequence divergence between the probe and target sequences, we aimed to design probes based on sequences across multiple pit viper species; probes representing the diversity of sequences, particularly toxin sequences, found in pit vipers allowed us to target more variable probe regions with higher efficiency as well as account for unidentified gene duplications (Hamilton et al. 2016), which are common in toxin gene families (Casewell et al. 2009). We designed 120 bp probes for toxin (94 long exons, 112 short exons; see below), nontoxin (), anchored (), and anonymous (short long ) loci. Because this study focused on a single toxin locus, we describe probe design only for the toxin loci below.
For the toxin probes, the venom-gland transcriptomes and low-coverage genomes (∼20–25×) for five individuals across three genera of pit vipers were utilized as a starting point: C. adamanteus, the cottonmouth (Agkistrodon piscivorus), the pygmy rattlesnake (Sistrurus miliarius), and two timber rattlesnakes (C. horridus) with divergent venom types (i.e., type A and type B venoms; Rokyta et al. 2015b). All venom-gland transcriptomes were sequenced on a HiSeq 2000 PE 100 and assembled and annotated as described by Rokyta et al. (2015b); toxin sequences were identified on the basis of homology to known snake toxins. Each genome was sequenced on a single lane on a HiSeq 2000 PE 150, and reads were merged as previously described (Rokyta et al. 2012). We sequenced 155–169 million read pairs for each genome. We first aligned the genome reads, merged and unmerged, against the corresponding toxin transcript coding sequences using bowtie2 (Langmead and Salzberg 2012). We used local alignments and a seed length of 22; any exon shorter than 22 nt was, therefore, not considered. The resulting alignments were visualized in Geneious version 6 (Kearse et al. 2012) for the identification of exon–intron boundaries on the basis of consistent trimming locations of aligned genomic reads.
Exons longer than 120 nt (i.e., longer than the length of the probes) were aligned against homologous transcripts from an additional nine pit viper species: the copperhead (A. contortrix), western diamondback rattlesnake (C. atrox), sidewinder rattlesnake (C. cerastes), rock rattlesnake (C. lepidus), speckled rattlesnake (C. mitchellii), black-tailed rattlesnake (C. molossus), Mojave rattlesnake (C. scutulatus), tiger rattlesnake (C. tigris), and massasauga rattlesnake (S. catenatus). These venom-gland transcriptomes were also sequenced on a HiSeq 2000 PE 100 and assembled as previously described (Rokyta et al. 2015b). Incorporating these toxin sequences increased the diversity of sequences used in probe design and, therefore, should result in higher capture efficiency. These alignments were trimmed at the exon boundaries.
Because the capture probes were 120 bp, exons <120 nt had to be extended into the flanking introns for probe design. For these exons, we built intron sequences by realigning the originally aligned genomic reads from the five sequenced genomes and the identified exon sequence using the SeqMan Pro assembler from the DNASTAR Lasergene software suite with a match size of 22 and a minimum match percentage of 95. Intron sequence extending from the ends of the exon sequence with at least 2× coverage was retained. For probe design for these short exons, we only had intron sequences for the five individuals for which we sequenced the genome. To incorporate the variation in the exons of the remaining nine species, the intron sequence from the most similar (i.e., minimum sequence divergence) genome-sequenced species exon was copied to the exons of the secondary species. We identified 94 long toxin exons (i.e., >120 nt) and 112 short toxin exons (i.e., <120 nt).
For each sequence in each alignment, probes were tiled uniformly at ∼4× density (new probe beginning every 30 bp), with ambiguities being resolved randomly in order to produce probes containing no ambiguities. Tiling probes across all sequences allowed the probes to represent the sequence diversity across the reference species. As this process for all locus types (i.e., not just toxin loci) generated 173,188 probes, greater than the number that can be contained in one standard Agilent SureSelect kit (57,700 probes), we next thinned the probes. First all 20-mers found in all probes were loaded into a hash table, with the 20-mer as the key and the probe identification number as the value. As the k-mers were being hashed, each probe was scored as the number of 20-mers that probe shared with any of the previously evaluated probes. Finally, all probes with a score >69 (indicating that ∼69% of probe was identical to another probe) were removed from the probe list. The thinning process, which retains more probe diversity in more diverse regions of target loci, resulted in 57,292 probes being retained.
Library preparation and sequencing
Genomic DNA was extracted from whole blood samples drawn from the caudal vein or from muscle/liver tissue of 140 C. adamanteus individuals using the Omega Bio-tek E.Z.N.A Tissue DNA Kit according to the manufacturer’s protocol. DNA quality was checked on a 2% agarose gel; all samples possessed high-quality genomic DNA. Hybrid enrichment data were collected through the Center for Anchored Phylogenomics at Florida State University (www.anchoredphylogeny.com) following the general methods of Lemmon et al. (2012) and Prum et al. (2015). Briefly, each genomic DNA sample was sonicated to a fragment size of ∼175–325 bp using a Covaris E220 Focused-ultrasonicator with Covaris microTUBES. Subsequently, library preparation and indexing were performed on a Beckman-Coulter Biomek FXp liquid-handling robot following a protocol modified from Meyer and Kircher (2010). One important modification was a size-selection step after blunt-end repair using SPRIselect beads (0.9× ratio of bead to sample volume; Beckman-Coulter). Indexed samples were then pooled at equal quantities (24 samples per pool), and enrichments were performed on each multi-sample pool using an Agilent Custom SureSelect kit (Agilent Technologies) that contained the probes designed for all locus types. After enrichment, each set of enrichment reactions was pooled in equal quantities.
Forty-four C. adamanteus samples and four additional samples from other species were sequenced on one PE150 Illumina HiSeq 2500 lane. Sequencing was performed in the Translational Science Laboratory in the College of Medicine at Florida State University. For the additional 96 C. adamanteus samples, two sequencing runs were performed. Sequencing was originally performed on one PE150 Illumina HiSeq 2500 lane as described above. To increase coverage and data quality, an additional size-selection using the Pippen HT (Sage Science) was used to target only fragments 400–600 bp in length. These fragments were then sequenced on one Illumina HiSeq 2500 PE 200 lane in the Translational Science Laboratory in the College of Medicine at Florida State University. For all downstream analyses, reads from both sequencing runs were used.
Alignments and variant calling
Reads were merged with PEAR (Zhang et al. 2014). Merged and unmerged reads for each sample were aligned separately to exons 2 and 3 of the myotoxin sequence from C. adamanteus with bowtie2 (Langmead and Salzberg 2012). Reads with >3 mismatches (gaps or sequence differences) were removed from the resulting alignments. Variant discovery analysis was performed in Geneious version 6 (Kearse et al. 2012). We required a minimum coverage of five, a minimum variant frequency of 0.05, and all other parameters were default values.
Data availability
Specimen identifications, population designations, copy number (i.e., fg) estimates, myotoxin proteome percentages, and age class information are contained in Table S2. All exon sequences are provided in Table S3. Myotoxin exon 2 and 3 bam alignments were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accessions SRR5271805–SRR5271935 and SRR5271936–SRR5272066, respectively. The raw genome sequencing reads were deposited in the NCBI SRA as follows: C. adamanteus SRX1470209, C. horridus A SRR5270836, C. horridus B SRR5270839, A. piscivorus SRR5270335, and S. miliarius SRR5270354. The raw transcriptome sequencing reads were deposited in the NCBI SRA as follows: C. adamanteus SRX127425, C. horridus A SRX188936, C. horridus B SRX683475, C. atrox SRR5270430, C. cerastes SRR5270834, C. lepidus SRR5270851, C. mitchellii SRR5270850, C. molossus SRR5270852, C. scutulatus SRR5270449, C. tigris SRR5270853, A. piscivorus SRX1032570, A. contortrix SRX1032565, S. miliarius SRX1032424, and S. catenatus SRX1030301. All reads can be found in BioProject PRJNA88989.
Results and Discussion
Copy number varies within and between populations
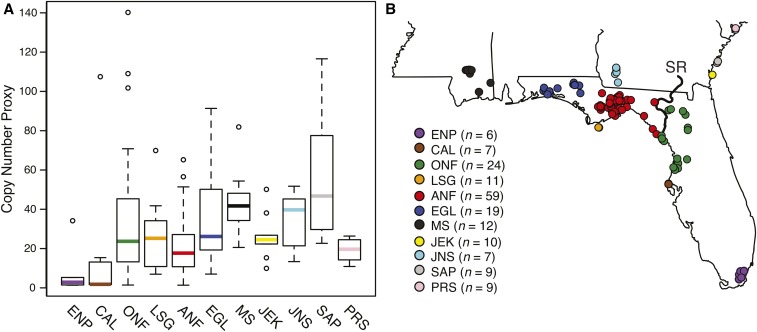
Prior to identifying the proximate evolutionary process underlying gene family origin and expansion and testing the three models outlined above, we first needed to demonstrate that duplication of the myotoxin locus had occurred. We used qPCR to estimate relative myotoxin copy number for 173 individuals across 11 populations ( for all populations) and detected significant inter- () and intrapopulation ( < P < 0.0001) variation in copy number (Figure 1); all populations exhibited significant intrapopulation variation except for PRS ( nonsignificant following a Bonferroni correction), and this extent of genetic divergence within a species at extremely small spatial and short temporal (i.e., <1 MY; Margres et al. 2015b) scales indicated the action of selection.
Figure 1.
Copy number varied significantly across and within populations. (A) We plotted the fg mean values (i.e., copy number proxy) for the 11 populations with n Populations were organized by latitude; the southernmost population (ENP) appears on the left of the plot, and the northernmost population (PRS) appears on the right of the plot. Relative to the southernmost population (ENP), all other populations were significantly different except CAL, LSG, ANF, JEK, and PRS. All populations exhibited significant intrapopulation variation except for PRS. Outliers are indicated with open circles and colors correspond to the map in (B). Whiskers extend to the most extreme data point ≤1.5× the interquartile range (i.e., the length of the box away from the box). (B) Sampling of the 11 populations analyzed for inter- and intrapopulation variation in copy number. SR, Suwannee River.
Total copy number varies by more than an order of magnitude
To estimate total copy number from our qPCR data, we sequenced a low-coverage genome for a single individual with high myotoxin expression to establish baseline single-copy-gene and myotoxin coverage levels. To identify a set of presumably single-copy exons, we selected a random set of 200 nontoxin transcripts from the previously described C. adamanteus venom-gland transcriptome (Rokyta et al. 2012). We aligned the genome reads against the coding sequences of the 200 random transcripts, and exon boundaries were identified on the basis of consistent read-trimming across aligned reads at particular sequence positions of the coding sequence. The same approach was used to confirm the exons of myotoxin, for which the genic structure had been previously described in another rattlesnake species (see Materials and Methods for details; Oguiura et al. 2009).
We chose to use coverage as a proxy for copy number, assuming that most nontoxin loci would be present as single-copy genes and, therefore, the coverage of these loci would provide a baseline of average coverage for nonduplicated genes. Because local guanine-cytosine, or GC, content is often correlated with fragment count in Illumina sequencing data and, therefore, could bias our estimates of coverage and copy number (Benjamini and Speed 2012), we limited our analysis to exons with similar GC%. Myotoxin exon 2 (the longest exon) possessed a GC% of 44%, and we limited our nontoxin data set to 109 exons with GC% For these exons, we plotted GC content vs. coverage and identified only a weak correlation ( where was the coefficient of determination and r was Pearson’s correlation coefficient; Figure S2 in File S1).
We mapped the genome reads to the 109 nontoxin exons and myotoxin exon 2; the average median coverage for single-copy-gene exons was 21.99, and the median coverage for myotoxin exon 2 was 60.79. This 2.76× difference suggested that there were 5–6 myotoxin copies in the genome animal, indicating that this individual was potentially heterozygous for copy number (e.g., three copies on one chromosome and two on the other). Because the genome animal had a fg mean of 15.94 [relative copy number estimates from the qPCR assays were reported as the mean of the total DNA amount quantified across sample replicates in femtograms (i.e., fg mean)], and our genome analysis suggested that this individual possessed 5–6 gene copies, we assumed that each gene copy corresponded to fg in our qPCR assays. According to this estimate, copy number varied extensively across individuals and ranged from 0 to 47 total copies (Table 1).
Table 1. Mixed modeling results.
| Cluster | Cluster fg mean | Cluster fg range | Total copies |
|---|---|---|---|
| 1 (n = 11) | 0.2296 | 0–0.9 | 0 |
| 2 (n = 38) | 8.1799 | 1–12 | 1–4 |
| 3 (n = 32) | 16.3949 | 13–19 | 5–6 |
| 4 (n = 34) | 22.9423 | 20–26 | 7–9 |
| 5 (n = 50) | 37.8072 | 27–59 | 10–20 |
| 6 (n = 13) | 78.2120 | 60–136 | 21+ |
To provide a second, independent confirmation of this scaling factor, we sought to identify the fg mean estimates for single-copy individuals in our qPCR data set. Gene absence was not fixed in any population, but individuals possessing zero copies were not uncommon and present in several different populations (e.g., ANF, CAL, ENP, ONF; Figure 1). Given this information, single-copy individuals should presumably be in our data set, although they may be rare. We took the three lowest fg means values above the maximum value for a confirmed zero-copy individual (i.e., 0.9; Figure S3 in File S1, Table 1) for specimens with sufficient coverage in our hybrid enrichment approach (see below). These putative single-copy individuals had a mean fg estimate of 3.1 fg (coefficient of variation of 0.16), confirming that our estimate based on the genome analysis above (2.9 fg units per gene copy) was reasonable.
We next fit a Gaussian mixture model to the data to characterize the frequency and density of different copy number classes in the whole data set (i.e., 178 individuals assuming 2.9 fg units per copy). We evaluated models for –47 and found that was the best-fit model (Figure S3 in File S1, Table 1). The relatively large variance found in cluster 6 (Figure S3 in File S1) indicated that this approach had trouble distinguishing among paralogs for high-copy-number individuals, and this is most likely the result of our correction factor; variance in a multiplicative factor will have a larger effect on larger values. The majority of our samples (i.e., %), however, were placed in clusters 2–5 (i.e., 1–20 copies; Table 1) with relatively small variances (Figure S3 in File S1), indicating a high level of confidence in the placement of most of our samples.
Copy number was positively correlated with protein expression
Under the positive dosage model, gene duplication leads to an increase in expression (Innan and Kondrashov 2010). To identify the potential functional significance of copy number variation, we first determined whether changes in copy number corresponded to changes in protein expression, or if dosage-compensation-like mechanisms that have been found in placental mammals (Lan and Pritchard 2016) ensured a specific expression level regardless of copy number (Tang and Amon 2013), eliminating the dosage model as a viable hypothesis. Toxin expression variation has direct phenotypic effects because relative amounts of venom components determine, in part, venom toxicity and activity (Margres et al. 2016a), suggesting that toxin expression levels should evolve under strong selective pressures (Margres et al. 2016b). Strong positive (Margres et al. 2015a,b) and purifying selection (Margres et al. 2016b) on toxin expression levels has been identified in these particular C. adamanteus populations, and because expression is a metabolically costly process, we expect any variation in toxin expression (due to copy number variation or any other mechanism) to be the target of selection. We used the transcriptome-proteome map previously described by Margres et al. (2014, 2015a, 2016b) to estimate myotoxin protein expression by means of reversed-phase high-performance liquid chromatography (RP-HPLC).
Because of the ontogenetic shift in venom expression (Margres et al. 2015b), we compared copy number and protein expression levels for 66 adults and 40 juveniles separately and identified a significant relationship in both adults (<0.0001) and juveniles (), where was the coefficient of determination (Figure 2A). The relatively low value for the juvenile coefficient of determination may reflect the compositional nature of venom as well as the less complex juvenile expression phenotype. Myotoxin proteome abundance can only be measured in relative amounts (Margres et al. 2014) and, therefore, is contingent on the expression of all other toxins, which also exhibit extensive variation (i.e., the up-regulation of another toxin locus decreases myotoxin abundance regardless of myotoxin expression; Margres et al. 2015a). When we account for variation at other loci by limiting our comparison to within adults from a single population where variation at other loci should be reduced, we identify a much tighter correlation between copy number and expression level ( Figure 2B). Juvenile C. adamanteus venom is also, for the most part, a less complex version of adult venoms (Margres et al. 2015b). The trendline for juveniles was much steeper than the trendline for adults because of the ontogenetic shift in venom composition (Figure 2A), but an ANCOVA analysis did not identify a significant interaction between copy number and age class as a predictor of protein expression (), although both factors were significant independently (P < 0.0001). This result indicated that the slopes for adults and juveniles were not significantly different, suggesting that the relationship between copy number and protein expression was similar across age classes. Because of these limitations in measuring absolute expression across age classes and populations, we binned the data according to the clusters identified in the mixture model above (Table 1) to visualize the relationship between copy number and expression level (Figure 2, C and D). Overall, we identified a significant positive correlation between copy number and protein expression in both adults and juveniles, indicating that duplication led to an increase in expression as predicted by the positive dosage model.
Figure 2.
Myotoxin protein expression was significantly correlated with copy number in the venom of C. adamanteus. (A) We plotted the fg mean and myotoxin proteome percentage for adult () and juvenile () age classes. We identified a significant positive correlation between copy number and protein expression in adults and juveniles, and the ANCOVA analysis indicated that the slopes for adults and juveniles were not significantly different. (B) Because venom is a compositional trait and myotoxin proteome abundance can only be measured in relative amounts, myotoxin abundance is contingent on the expression/abundance of other variable toxin loci. To account for variation at other loci, we limited the plot to the largest adult data set from a single population (ONF ) where variation at other loci should be reduced and identified a much tighter correlation between copy number and expression level. A power trendline was used to fit the data as previously described (Margres et al. 2015b). is the coefficient of determination. (C and D) We plotted myotoxin expression level (i.e., proteome abundance) for the six clusters identified in the mixed model analysis for adults (C) and juveniles (D) separately. Binning by cluster assisted in accounting for limitations in measuring protein expression for a compositional trait such as venom. No juveniles were placed in cluster 1. Outliers are indicated with open circles. Whiskers extend to the most extreme data point ≤1.5× the interquartile range.
Dosage effects, not sequence diversity, drove gene family origin and expansion
The permanent heterozygote and the multi-allelic diversifying selection models each require sequence diversity (Innan and Kondrashov 2010). Additionally, sequence variation could have impacted the effectiveness of the qPCR assay and, therefore, our estimate of copy number because divergent paralogs could have prevented the amplification of specific variants based on our single primer set. To assess the level of sequence divergence among myotoxin paralogs, we used a hybrid enrichment approach to sequence myotoxin exons 1, 2, and 3 across 140 individuals. Because we designed probes based on sequences across multiple pit viper species (see Materials and Methods), we were able to target more variable probe regions with higher efficiency as well as account for unidentified gene duplications (Hamilton et al. 2016), alleviating the potential issue of false negatives in the qPCR assay as described above; exon 1 was not included in the sequence diversity analyses because this exon codes for the signal peptide and, therefore, should not evolve under positive selection because signal peptides are cleaved and not a part of the mature protein.
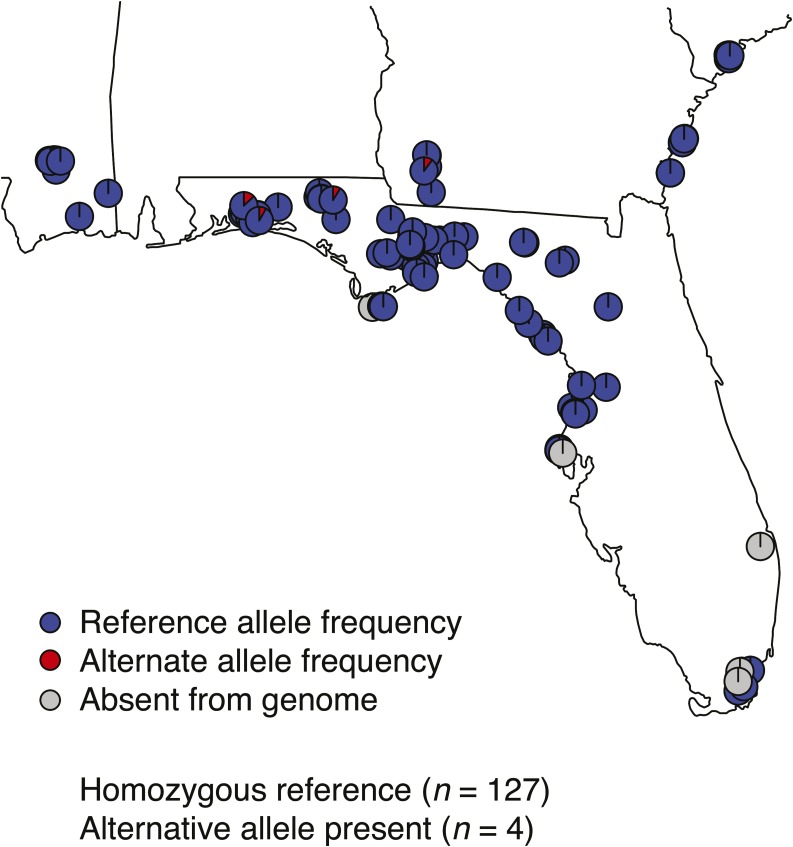
Of the 140 individuals sequenced, 131 individuals possessed mean coverage for both exons 2 and 3; no sequence was obtained for either exon across nine individuals. Eight of the nine individuals for which no sequence was obtained were included in the qPCR assays, and all eight individuals exhibited fg mean values 0.00–0.45 (Figure S3 in File S1, Table 1), consistent with a lack of the locus in the genome, thus providing an independent confirmation of the robustness of our qPCR assays. For the 131 individuals with sufficient coverage, we found no sequence variation in exon 3 and only a single SNP, a nonsynonymous substitution, in four out of 131 individuals in exon 2 (transcript position 73, CGG→TGG, R→W; Figure 3). Of the four individuals exhibiting sequence variation, three individuals were from EGL and the other individual was from JNS. The SNP was at low frequency within each of these four individuals (8.2–13.0%), indicating that the majority of paralogs were identical in sequence, even in these polymorphic individuals (Figure 3). This lack of sequence divergence eliminated the permanent heterozygote and multi-allelic diversifying selection models and suggested that this particular variant arose recently after the majority of duplicates had become widespread.
Figure 3.
Hybrid enrichment identified a single, low-frequency myotoxin SNP across the range of C. adamanteus. We sequenced myotoxin exons 2 and 3 for 140 individuals; each point represents an individual. One-hundred and thirty-one samples possessed mean coverage for both exons; no sequence was obtained for either exon for nine individuals, indicating the locus was absent from the genome (gray points). For the 131 samples with sufficient coverage, we found no sequence variation in exon 3 and only a single nonsynonymous SNP in exon 2, which was present in only four individuals. We plotted the frequency of each allele, the reference allele and the alternative, nonsynonymous variant identified in exon 2, for each individual; pie charts indicate the percentage of duplicates with a particular allele within each individual.
Conclusions
We demonstrated that selection for increased expression (i.e., the positive dosage model; Innan and Kondrashov 2010), not sequence diversity, was the proximate evolutionary process underlying myotoxin gene family origin and expansion by establishing (1) rapid divergence in copy number (Figure 1, Table 1) across populations, (2) rapid divergence in expression level (Figure 2) and, therefore, the phenotype (because toxin expression levels determine venom toxicity and activity; Margres et al. 2016a) across populations, and (3) a lack of sequence divergence (Figure 3), indicating that selection for increased expression was strong, and that the sequences of all paralogs evolved under strong purifying selection (Innan and Kondrashov 2010).
Although our results provided clear evidence of the positive dosage model, the frequency of this specific process relative to others, particularly for models requiring sequence variation (Francino 2005; Bergthorsson et al. 2007), is unclear because of the lack of data on copy number variation at the population level (Innan and Kondrashov 2010). For example, the duplication of toxin-encoding genes has been documented in cone snails (Duda and Palumbi 1999), corals (Gacesa et al. 2015), and extensively in snakes (Fry et al. 2003; Fox and Serrano 2005; Lynch 2007; Casewell et al. 2011; Margres et al. 2013; Rokyta et al. 2013) at the phylogenetic level, but the role of toxin-encoding gene duplication in population divergence was relatively unknown until the current study. To resolve the debate on the evolutionary forces governing the fates of gene copies immediately following duplication, additional studies sequencing polymorphic gene duplications across populations are needed to determine the frequency of, as well as the potential biases, affecting certain evolutionary models. Because the positive dosage model relies on well-established evidence of positive selection on expression (Shibata et al. 2012; Fraser 2013; Lamichhaney et al. 2016) whereas other models rely on balancing selection, the frequency of which is largely unclear (Fijarczyk and Babik 2015), we argue that the positive dosage model may be a more general mechanism leading to gene family origin and expansion, at least in venoms, with diversity-based mechanisms being more efficacious on the larger mutational target following duplication.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.202655/-/DC1.
Acknowledgments
The authors thank Kimberley Andrews and Joseph Colbert at the Jekyll Island Authority’s Georgia Sea Turtle Center, Jim Lee and The Nature Conservancy at Camp Shelby Military Base, Jim Mendenhall, Lora Smith and Jennifer Howze at the Joseph W. Jones Ecological Research Center at Ichuaway, and Kenneth Wray for help in acquiring venom and tissue samples. The authors thank Megan Lamb, Ethan Bourque, and Rebecca Bernard with the Florida Department of Environmental Protection and Apalachicola National Estuarine Research Reserve Bradley Smith and the St. Vincent National Wildlife Refuge Dorset Hurley and Doug Samson at the Sapelo Island National Estuarine Research Reserve Brett Williams and the Department of the Air Force, and Peter Krulder, Carl Calhoun, and Rick Coosey at Caladesi Island State Park for access to field sites. We thank Brian Washburn for his assistance with qPCR and Margaret Seavy for her assistance with RP-HPLC. We thank Michelle Kortyna and Ameer Jalal at Florida State University’s Center for Anchored Phylogenomics for assistance with hybrid enrichment data collection and analysis. This work was supported by the National Science Foundation (DEB 1145987 to A.R.L., E.M.L., and D.R.R.).
Footnotes
Communicating editor: J. A. Birchler
Literature Cited
- Aird S., 1985. A quantitative assessment of variation in venom constituents within and between three nominal rattlesnake subspecies. Toxicon 23: 1000–1004. [DOI] [PubMed] [Google Scholar]
- Aird S., Watanabe Y., Villar-Briones A., Poy M., Terada K., et al. , 2013. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis). BMC Genomics 14: 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J., 1986. The Statistical Analysis of Compositional Data. Chapman & Hall, London. [Google Scholar]
- Bendtsen J. D., Nielsen H., von Heijne G., Brunak S., 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340: 783–795. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Speed T., 2012. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 40: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U., Andersson D., Roth J., 2007. Ohno’s dilemma: evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. USA 104: 17004–17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bober M., Glenn J., Straight R., Ownby C., 1988. Detection of myotoxin-a like proteins in various snake venoms. Toxicon 26: 665–673. [DOI] [PubMed] [Google Scholar]
- Cardoso-Moreira M., Arguello J., Gottipati S., Harshman L., Grenier J., et al. , 2016. Evidence for fixation of gene duplications by positive selection in Drosophila. Genome Res. 26: 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell N. R., Harrison R. A., Wüster W., Wagstaff S. C., 2009. Comparative venom gland transcriptome surveys of the saw-scaled vipers (Viperidae: Echis) reveal substantial intra-family gene diversity and novel venom transcripts. BMC Genomics 10: 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell N. R., Wagstaff S. C., Harrison R. A., Renjifo C., Wüster W., 2011. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evol. 28: 2637–2649. [DOI] [PubMed] [Google Scholar]
- Conrad D., Pinto D., Redon R., Feuk L., Gokcumen O., et al. , 2010. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda T. F., Jr, Palumbi S. R., 1999. Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc. Natl. Acad. Sci. USA 96: 6820–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas L., Kim Y., Karimpour-Fard A., Cox M., Hopkins J., et al. , 2007. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 17: 1266–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J., Maniatis T., Lawn R., O’Connell C., et al. , 1980. The structure and evolution of the human β-globin gene family. Cell 21: 653–668. [DOI] [PubMed] [Google Scholar]
- Fijarczyk A., Babik W., 2015. Detecting balancing selection in genomes: limits and prospects. Mol. Ecol. 24: 3529–3545. [DOI] [PubMed] [Google Scholar]
- Fox J. W., Serrano S. M. T., 2005. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon 45: 969–985. [DOI] [PubMed] [Google Scholar]
- Fraley C., Raftery A., Murphy T., Scrucca L., 2012. mclust Version 4 for R: Normal mixture modeling for model-based clustering, classification, and density estimation. Technical Report 597. Department of Statistics, University of Washington, ; Seattle, WA. [Google Scholar]
- Francino M., 2005. An adaptive radiation model for the origin of new gene functions. Nat. Genet. 37: 573–577. [DOI] [PubMed] [Google Scholar]
- Fraser H., 2013. Gene expression drives local adaptation in humans. Genome Res. 23: 1089–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry B. G., Wüster W., Kini R. M., Brusic V., Khan A., et al. , 2003. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evol. 57: 110–129. [DOI] [PubMed] [Google Scholar]
- Fry B. G., Scheib H., van der Weerd L., Young B., McNaughtan J., et al. , 2008. Evolution of an arsenal. Mol. Cell. Prot. 7: 215–246. [DOI] [PubMed] [Google Scholar]
- Gacesa R., Chung R., Dunn S., Weston A., Jaimes-Becerra A., et al. , 2015. Gene duplications are extensive and contribute significantly to the toxic proteome of nematocysts isolated from Acropora digitifera (Cnidaria: Anthozoa: Scleractinia). BMC Genomics 16: 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs H. L., Sanz L., Calvete J. J., 2009. Snake population venomics: proteomics-based analyses of individual variation reveals significant gene regulation effects on venom protein expression in Sistrurus rattlesnakes. J. Mol. Evol. 68: 113–125. [DOI] [PubMed] [Google Scholar]
- Gonzalez E., Kulkarni H., Bolivar H., Mangana A., Sanchez R., et al. , 2005. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307: 1434–1440. [DOI] [PubMed] [Google Scholar]
- Hahn M., 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 100: 605–617. [DOI] [PubMed] [Google Scholar]
- Hamilton C., Lemmon A., Lemmon E., Bond J., 2016. Expanding anchored hybrid enrichment to resolve both deep and shallow relationships within the spider tree of life. BMC Evol. Biol. 16: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings P., Lupski J., Rosenberg S., Ira G., 2009. Mechanisms of change in gene copy number. Nat. Rev. Genet. 10: 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A., 1999. Adaptive Evolution of Genes and Genomes. Oxford University Press, New York. [Google Scholar]
- Innan H., Kondrashov F., 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11: 97–108. [DOI] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., et al. , 2012. Genious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S., Han F., Berglund J., Wang C., Almen M., et al. , 2016. A beak size locus in Darwin’s finches facilitated character displacement during a drought. Science 353: 470–474. [DOI] [PubMed] [Google Scholar]
- Lan X., Pritchard J., 2016. Coregulation of tandem duplicate genes slows evolution of subfunctionalization in mammals. Science 352: 1009–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon A. R., Emme S., Lemmon E. M., 2012. Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst. Biol. 61: 727–744. [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J. S., 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch V. J., 2007. Inventing an arsenal: adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes. BMC Evol. Biol. 7: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres M., Aronow K., Loyacano J., Rokyta D., 2013. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genomics 14: 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres M., McGivern J., Wray K., Seavy M., Calvin K., et al. , 2014. Linking the transcriptome and proteome to characterize the venom of the eastern diamondback rattlesnake (Crotalus adamanteus). J. Proteomics 96: 145–158. [DOI] [PubMed] [Google Scholar]
- Margres M., McGivern J., Seavy M., Wray K., Facente J., et al. , 2015a Contrasting modes and tempos of venom expression evolution in two snake species. Genetics 199: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres M., Wray K., Seavy M., McGivern J., Sanader D., et al. , 2015b Phenotypic integration in the feeding system of the eastern diamondback rattlesnake (Crotalus adamanteus). Mol. Ecol. 24: 3405–3420. [DOI] [PubMed] [Google Scholar]
- Margres M., Walls R., Suntravat M., Lucena S., Sanchez E., et al. , 2016a Functional characterizations of venom phenotypes in the eastern diamondback rattlesnake (Crotalus adamanteus) and evidence for expression-driven divergence in toxic activities among populations. Toxicon 119: 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres M., Wray K., Seavy M., McGivern J., Herrera N., et al. , 2016b Expression differentiation is constrained to low-expression proteins over ecological timescales. Genetics 202: 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll S., Kuruvilla F., Korn J., Cawley S., Nemesh J., et al. , 2008. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 40: 1166–1174. [DOI] [PubMed] [Google Scholar]
- McNaught A., Wilkinson A., 1997. Compendium of Chemical Terminology: IUPAC Recommendations, Vol. 1669, Ed. 2 Blackwell Science, Oxford. [Google Scholar]
- Meyer M., Kircher M., 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010: pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- Nei M., Rooney A. P., 2005. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39: 121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Niimura Y., Nozawa M., 2008. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat. Rev. Genet. 9: 951–963. [DOI] [PubMed] [Google Scholar]
- Nguyen D., Webber C., Ponting C., 2006. Bias of selection on human copy-number variants. PLoS Genet. 2: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M., Kawahara Y., Nei M., 2007. Genomic drift and copy number variation of sensory receptor genes in humans. Proc. Natl. Acad. Sci. USA 104: 20421–20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguiura N., Collares M. A., Furtado M. F. D., Ferrarezzi H., Suzuki H., 2009. Intraspecific variation of the crotamine and crotasin genes in Crotalus durissus rattlesnakes. Gene 446: 35–40. [DOI] [PubMed] [Google Scholar]
- Ota T., Nei M., 1994. Divergent evolution and evolution by the birth-and-death process in the immunoglobulin VH gene family. Mol. Biol. Evol. 11: 469–482. [DOI] [PubMed] [Google Scholar]
- Peigneur S., Orts D. J. B., Preito da Silva A. R., Oguiura N., Boni-Mitake M., et al. , 2012. Crotamine pharmacology revisited: novel insights based on the inhibition of channels. Mol. Pharmacol. 82: 90–96. [DOI] [PubMed] [Google Scholar]
- Ponce-Soto L., Martins-do Souza D., Marangoni S., 2010. Structural and pharmacological characterization of the crotamine isoforms III-4 (MYX4_CROCu) and III-7 (MYX7_CROCu) isolated from the Crotalus durissus cumanensis venom. Toxicon 55: 1443–1452. [DOI] [PubMed] [Google Scholar]
- Prum R., Berv J., Dornburg A., Field D., Townsend J., et al. , 2015. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526: 569–573. [DOI] [PubMed] [Google Scholar]
- Rodrigues R. S., Boldrini-França J., Fonseca F. P. P., de la Torre P., Henrique-Silva F., et al. , 2012. Combined snake venomics and venom gland transcriptome analysis of Bothropoides pauloensis. J. Proteomics 75: 2707–2720. [DOI] [PubMed] [Google Scholar]
- Rokyta D. R., Lemmon A. R., Margres M. J., Aronow K., 2012. The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus). BMC Genomics 13: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D., Wray K., Margres M., 2013. The genesis of an exceptionally deadly venom in the timber rattlesnake (Crotalus horridus) revealed through comparative venom–gland transcriptomics. BMC Genomics 14: 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D., Margres M., Calvin K., 2015a Post-transcriptional mechanisms contribute little to phenotypic variation in snake venoms. G3 5: 2375–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D., Wray K., McGivern J., Margres M., 2015b The transcriptomic and proteomic basis for the evolution of a novel venom phenotype within the Timber Rattlesnake (Crotalus horridus). Toxicon 98: 34–48. [DOI] [PubMed] [Google Scholar]
- Schenberg S., 1959. Geographical pattern of crotamine distribution in the same rattlesnake subspecies. Science 129: 1361–1363. [DOI] [PubMed] [Google Scholar]
- Shibata Y., Sheffield N. C., Fedrigo O., Babbitt C. C., Wortham M., et al. , 2012. Extensive evolutionary changes in regulatory element activity during human origins are associated with altered gene expression and positive selection. PLoS Genet. 8: e1002789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant P., Kitzman J., Antonacci F., Alkan C., Malig M., et al. , 2010. Diversity of human copy number variation and multicopy genes. Science 330: 641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Amon A., 2013. Gene copy-number alterations: a cost-benefit analysis. Cell 152: 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff S. C., Sanz L., Juárez P., Harrison R. A., Calvete J. J., 2009. Combined snake venomics and venom gland transcriptomic analysis of the ocellated carpet viper, Echis ocellatus. J. Proteomics 71: 609–623. [DOI] [PubMed] [Google Scholar]
- Waldron J. L., Welch S. M., Bennett S. H., Kalinowsky W. G., Mousseau T. A., 2013. Life history constraints contribute to the vulnerability of a declining North American rattlesnake. Biol. Conserv. 159: 530–538. [Google Scholar]
- Wray K., Margres M., Seavy M., Rokyta D., 2015. Early significant ontogenetic changes in snake venoms. Toxicon 96: 74–81. [DOI] [PubMed] [Google Scholar]
- Zhang J., 2003. Evolution by gene duplication: an update. Trends Ecol. Evol. 18: 292–298. [Google Scholar]
- Zhang J., Rosenberg H., Nei M., 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 95: 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kobert K., Flouri T., Stamatakis A., 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30: 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Specimen identifications, population designations, copy number (i.e., fg) estimates, myotoxin proteome percentages, and age class information are contained in Table S2. All exon sequences are provided in Table S3. Myotoxin exon 2 and 3 bam alignments were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accessions SRR5271805–SRR5271935 and SRR5271936–SRR5272066, respectively. The raw genome sequencing reads were deposited in the NCBI SRA as follows: C. adamanteus SRX1470209, C. horridus A SRR5270836, C. horridus B SRR5270839, A. piscivorus SRR5270335, and S. miliarius SRR5270354. The raw transcriptome sequencing reads were deposited in the NCBI SRA as follows: C. adamanteus SRX127425, C. horridus A SRX188936, C. horridus B SRX683475, C. atrox SRR5270430, C. cerastes SRR5270834, C. lepidus SRR5270851, C. mitchellii SRR5270850, C. molossus SRR5270852, C. scutulatus SRR5270449, C. tigris SRR5270853, A. piscivorus SRX1032570, A. contortrix SRX1032565, S. miliarius SRX1032424, and S. catenatus SRX1030301. All reads can be found in BioProject PRJNA88989.