Abstract
The aim of the present study was to analyze the interplay between gastrointestinal tract (GIT) microbiota, host genetics, and complex traits in pigs using extended quantitative-genetic methods. The study design consisted of 207 pigs that were housed and slaughtered under standardized conditions, and phenotyped for daily gain, feed intake, and feed conversion rate. The pigs were genotyped with a standard 60 K SNP chip. The GIT microbiota composition was analyzed by 16S rRNA gene amplicon sequencing technology. Eight from 49 investigated bacteria genera showed a significant narrow sense host heritability, ranging from 0.32 to 0.57. Microbial mixed linear models were applied to estimate the microbiota variance for each complex trait. The fraction of phenotypic variance explained by the microbial variance was 0.28, 0.21, and 0.16 for daily gain, feed conversion, and feed intake, respectively. The SNP data and the microbiota composition were used to predict the complex traits using genomic best linear unbiased prediction (G-BLUP) and microbial best linear unbiased prediction (M-BLUP) methods, respectively. The prediction accuracies of G-BLUP were 0.35, 0.23, and 0.20 for daily gain, feed conversion, and feed intake, respectively. The corresponding prediction accuracies of M-BLUP were 0.41, 0.33, and 0.33. Thus, in addition to SNP data, microbiota abundances are an informative source of complex trait predictions. Since the pig is a well-suited animal for modeling the human digestive tract, M-BLUP, in addition to G-BLUP, might be beneficial for predicting human predispositions to some diseases, and, consequently, for preventative and personalized medicine.
Keywords: GenPred, shared data resource, complex traits, genomic selection, gut microbial composition, microbial prediction, pig
HOST–MICROBIOTA interactions have received considerable attention in human studies in recent years, and it has been shown repeatedly that host genetics, as well as the environment, affect gut microbiota composition (Spor et al. 2011). In order to unravel host genetic effects, the use of a model organism that is kept in a stable environment with minimum environmental variations is needed (Zhao et al. 2013). Kostic et al. (2013) described different model organisms, such as mice, zebrafish, the fruit fly Drosophila, and the bobtail squid, as important sources of information on host–microbiota homeostasis. The pig can be used as a model for human-related research, as the human and porcine physiology, metabolism, and gastrointestinal tract (GIT) microbiota are similar (Heinritz et al. 2013). It has already been used as an animal model for microbiota-associated diseases such as Helicobacter pylori infections, necrotizing enterocolitis disease, obesity, and diabetes, and to formulate dietary strategies for overcoming obesity and other metabolic syndromes (Heinritz et al. 2013). Moreover, pigs are not only suitable model organisms for human-related research but also some of the most important livestock species used for meat production worldwide. Breeding is frequently carried out using genome-wide SNP data for the prediction of selection candidate breeding values (Knol et al. 2016)—a technique that is recognized as a form of genomic selection (Meuwissen et al. 2001). The underlying assumption is that each SNP affects complex traits of interest only marginally, but modeling all SNPs jointly in a prediction equation is attributed with remarkably high levels of prediction accuracy. Genomic prediction is also utilized in plant breeding (Jannink et al. 2010), and has been proposed as a tool for predicting complex genetic predispositions in humans (de los Campos et al. 2010). Inspired by the success of genomic prediction methods, Ross et al. (2013) developed a metagenomic prediction approach. These authors used human and cattle datasets with massive parallel sequencing data to form metagenomic relationship matrices, which in turn were used to predict complex traits with considerable accuracy. This study clearly highlights the potential to consider alternative high-dimensional host-related explanatory variables beyond SNP markers for prediction purposes, and, more generally, opportunities to apply quantitative-genetic methods in holistic analyses of data on microbiota, host genetics, and complex traits.
Currently, there is a lack of research in pigs regarding the genetic influences on gut microbial community, and the effect of this community on complex host traits. Besides studies on the influence of nutrition and medication on the microbiota, one study has approached the question of how the early-life pig gut microbiota impacts host phenotypes (Mach et al. 2015).
In this study, standard quantitative-genetic methods were extended and applied to analyze the interrelationship between pig GIT microbiota compositions, complex traits, and pig genomes. The specific aims were (i) to characterize GIT microbiota for pigs of a mature age, (ii) to analyze the effects of host genetics on GIT microbial composition, (iii) to investigate the role of GIT microbial composition on key host complex traits, and (iv) to evaluate genomic as well as microbial predictions of complex host traits.
Materials and Methods
Sample collection
The animal experiments were performed in accordance with German Animal Welfare legislation. All procedures regarding animal handling and treatment were approved by the University of Hohenheim Committee of Animal Welfare under authorization number S411/14TZ. The pigs belonged to the Piétrain breed. This is an important sire line breed (Stratz et al. 2014) for which genomic selection is practiced (Wellmann et al. 2013). Housing, slaughtering, and the recording of phenotypes of the pigs was performed under standardized conditions at one experimental farm. Performance testing started with a weight of 30 kg and ended with a weight of 105 kg. Animal feed intake (FI) and daily gain (DG) values were recorded during performance testing. The feed conversion (FC) value was calculated as a ratio of the consumed feed and weight gain occurring during performance testing. See Supplemental Material, Table S1 for descriptive statistics of these traits. By reaching 105 kg, the pigs were slaughtered with an average slaughter age of 188 (±14) days, and an average performance testing duration of 100 (±11) days. In total, colon and blood samples from 207 Piétrain sows were collected on 14 slaughter days. Blood samples were taken directly during the slaughtering and stored on ice. After opening of the abdomen, colon samples were collected from the mid-colon and also stored on ice. For long-term storage, blood samples were kept at −20° and colon samples at −80°.
Illumina amplicon sequencing
Colon digesta samples were thawed on ice and homogenized, and 250 mg of each sample was used to extract DNA using the FastDNA SPIN Kit for Soil (MP Biomedicals, Solon, OH) according to the manufacturer’s instructions. PCR targeting of the V1-2 region of the 16S rRNA gene was carried out as described in Camarinha-Silva et al. (2014). Based on a previous work of our group (Burbach et al. 2016), this 16S region and DNA extraction method was giving the best coverage of the microbial community. Amplicons were purified and normalized using a SequalPrep Normalization Kit (Invitrogen, Carlsbad, CA), and were pooled and sequenced with 250 bp paired-end sequencing chemistry applied on an Illumina MiSeq platform.
Bioinformatic processing of sequences was done according to Camarinha-Silva et al. (2014) with some modifications. Raw reads were assembled (Cole et al. 2014) and subsequently aligned using MOTHUR (gotoh algorithm with the SILVA reference database) prior to preclustering of the sequences with two mismatches (diffs = 2). Low abundance operational taxonomic units (OTUs), if present in <5 samples in relative abundances <0.01%, were removed. Finally, 40,379 ± 1149 sequences were obtained per sample, comprising a total of 2714 OTUs that were taxonomically assigned using the naïve Bayesian RDP classifier (Wang et al. 2007). The OTUs were then evaluated against the RDP database using Seqmatch function, which belongs to the RDP database. Note that no differences were observed in the microbiota regarding the day of DNA extraction, which was performed on 5 consecutive days by the same person.
Genotyping
In total, 207 German Piétrain sows were genotyped using an Illumina PorcineSNP60 BeadChip (Ramos et al. 2009). Genotypes from individuals were filtered with respect to call rates (removal of SNPs with a call rate of <95%), minor allele frequencies (exclusion of SNP with a minor allele frequency of <5%), significant deviations from Hardy-Weinberg equilibrium (P < 0.0001), and SNPs on the Y-chromosome were removed. The call rate across all animals was ≥0.994. After quality control measures were performed, 51,970 SNPs remained for further analysis.
Statistical analysis
Microbial community:
A multivariate dataset comprising the relative abundance of each phylotype across each sample was analyzed using v.6.1.6, PRIMER-E (Plymouth Marine Laboratory, UK; Clarke and Warwick 2001). The Bray-Curtis coefficient (Bray and Curtis 1957) was used to create a sample-similarity matrix, and microbial community structures were explored via nonmetric multidimensional scaling (MDS) (Clarke and Warwick 2001). Statistical comparisons between a priori defined groups (e.g., weight, age, DG, and FC) were drawn using analysis of similarity (ANOSIM) based on 999 permutations, and were considered significantly different at a P-value of < 0.05. Species responsible for observed differences were identified based on similarity percentages (SIMPER) (Clarke and Warwick 2001). Tax4fun (Aßhauer et al. 2015) was used to predict the functional capabilities of microbial communities detected in colons based on the 16S rRNA sequencing data.
Genetic parameters of bacterial genera:
Genetic parameters were estimated for bacterial genera rather than for OTUs, because the latter would result in numerous statistical analysis, and, thus, in a strong multiple testing problem. An analysis of variance was performed by fitting a univariate genomic mixed linear model to test for significant effects. The age and weight measured at the test station and the slaughter weight were included as fixed covariables in the model, and the slaughter day (SD) was considered as a random effect in the model. The observation vector included the relative abundance of one genus. Only genera with abundance values exceeding 0.1% were considered. By backward elimination, nonsignificant effects (significance level for the elimination of α = 0.05) were removed from the model. An univariate analysis was performed to estimate the heritability of each genus. Statistical analyses were performed using the ASReml package available through R (Butler et al. 2009). The mixed linear model is written as follows:
| (1) |
where is the observation vector, which includes the abundance of one genus, is the vector of fixed effects (described above), is the vector with random slaughter day effects with variance is a vector with random animal genetic effects, and are the corresponding design matrices, and is the residual term with residual variance Note that a random pen effect as well as a random maternal effect were not significant, and thus were not included in this model. The distribution of the random animal effect is , with being the genomic relationship matrix, and being the additive genetic variance. The G matrix was estimated using SNP genotypes following VanRaden (2008) as
where is the gene content matrix with entries 0, 1, or 2 for each SNP and each animal, and matrix contains the frequency of each SNP Narrow-sense heritability was estimated as with The p-values of the heritability estimates were calculated by conducting a likelihood-ratio test on random animal effects. The null-hypothesis, that the variance of the random effect is 0, was rejected if twice the difference in the log-likelihoods of the full model, and the reduced model without the random effect was larger than the 0.95-quantile of a –distribution with 1 d.f. We chose a significance level of P-value = 0.05. A total of 49 genera was analyzed, which resulted in a multiple testing problem. In order to judge how many false positives were among the significant results, we applied the false discovery rate (FDR) technique. We calculated for each test an FDR q-value using the software QVALUE (Storey and Tibshirani 2003). The FDR q-value of the significant genera with the lowest test statistic (P-value ≈ 0.05) provided an estimate of the proportion of false positives among the significant outcomes. Note that model (1) is a mixed linear model, which assumes normality of the data. The relative abundance of the genera were, in general, not normally distributed, and sometimes peaked at zero. However, due to the small data set we did not apply generalized linear mixed models.
Genetic and microbial parameters of host traits:
First, explanatory variables for host traits FC, DG and FI were estimated via backward elimination, using ages and weights measured upon test station arrival and slaughter weights as fixed covariables, and SD and pen as random effects. Nonsignificant effects of α = 0.05 were excluded. To estimate genetic variance components and narrow sense heritabilities of the host traits, the following model was applied
| (2) |
where is a vector of observations (FC, DG, or FI), is a vector of fixed effects (i.e., for FC and FI weights measured upon test station arrival, and for DG slaughter weights), and SD and pen are vectors of random slaughter day, and pen effects, respectively, with variance components and are corresponding design matrices, and denotes the residual term. The distribution of the random animal effect is the same as described in model (1). The heritability was estimated in the same manner as shown in model (1) for the bacteria genera. The P-value of the additive genetic variance was calculated by performing a likelihood ratio test of the animals’ random effects, as described for the random effect in model (1).
The microbial variance component was estimated by applying the following univariate microbial mixed linear model (fitted in ASReml R):
| (3) |
where the model parameters are as described in model (2) except vector which contains the random effect of the animal microbiota for each individual with where is the microbial variance. The microbial relationship matrix was calculated as follows: we have with matrix (dimension n × N where is the number of animals and is the number of OTUs), constructed from matrix (dimension n x N). The elements are the relative abundance of OTU in animal (plus 0.01). Following this, the elements in are
Thus, the off-diagonals in are calculated as and the diagonals as
Note, that it might happen that the diagonals are >1. This is the case if
i.e.,
which means that the abundances of the OTUs in animal deviate strongly from the average values, e.g., if OTUs are missing in the animal that are present in most other animals, and if OTUs are present that are rare in other animals. Note also that the microbial relationships are affected by some errors remaining in the OTU data. More research is needed regarding the effect of data screening on the precision of the microbial relationship estimation.
The fraction of the phenotypic variance explained by the microbial variance was calculated as where is the phenotypic variance. This fraction was termed microbiability by Difford et al. (2016). The P-value of the microbial variance was calculated by performing a likelihood ratio test of the animals’ random microbiota effects, as described for the random animal effect in model (1).
Genomic and microbial prediction:
To predict host traits FC, DG, and FI using genomic and microbiota data, G-BLUP and M-BLUP models were applied, respectively. For the G-BLUP predictions (VanRaden 2008), model (2) was applied with previously estimated variance components. In a similar vein, for the M-BLUP predictions, model (3) was applied. For both types of predictions, a repeated cross validation was performed with 10,000 iterations, where 80% of the individuals were sampled randomly without replacement to train the prediction model (reference population). The pig trait phenotypes (FC, DG, and FI) were predicted from the remaining 20% of the pigs based on the results of the reference population analysis (validation population). The accuracy of a prediction was defined as the correlation between predicted and observed trait phenotypes in the validation population. The mean correlation was calculated as the mean of 10,000 correlation estimates. Confidence intervals of correlations were estimated as 2.5 and 97.5% quantiles of the 10,000 ordered correlation estimates.
Effects of single OTUs on complex host traits:
To identify the drivers of prediction accuracy levels, the marginal effects of OTUs on phenotypic traits (i.e., single OTU effects not captured by the remaining OTUs) were estimated from the solutions of the M-BLUP model. To do this, an adapted version of the back solving method proposed by Strandén and Garrick (2009) was used, which is described in the supplementary information (File S1).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are fully represented within the tables and figures. Sequences are available at the European Nucleotide Archive (ENA) under accession number PRJEB18070 (http://www.ebi.ac.uk/ena/data/view/PRJEB18070). File S2 and File S3 contain genotypes and phenotypes for each individual, respectively. File S4 contains relative abundances at OTU level.
Results
Microbial community characterization and heritability of gut microbiota compositions
The main phyla that account for higher abundances were found to be Firmicutes (54%), Bacteroidetes (42%), Proteobacteria (2%), and Spirochaetes (1%). The most abundant families found in colons were Ruminococcaceae (24%), Prevotellaceae (21%), Porphyromonadaceae (8%), Clostridiaceae 1 (6%), Rikenellaceae (6%), and Lachnospiraceae (5%), with all other families found to be present at average levels of <5%. The microbial community harboring the different pigs showed a similarity of 35% between animals. Overall, the predicted KEGG pathways of higher abundance were found to be related to pathways associated with carbohydrate metabolism, such as starch and sucrose pyruvate, fructose and mannose metabolism; amino acid metabolism; environmental information processing such as membrane transport and signal transduction; genetic information processing such as replication and repair; and translation (Figure S1).
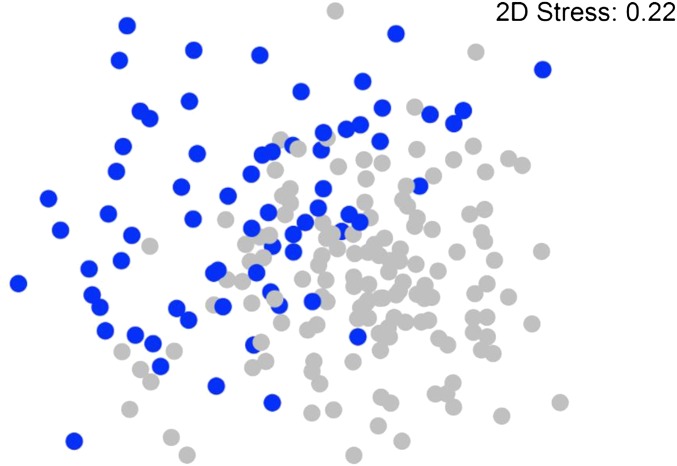
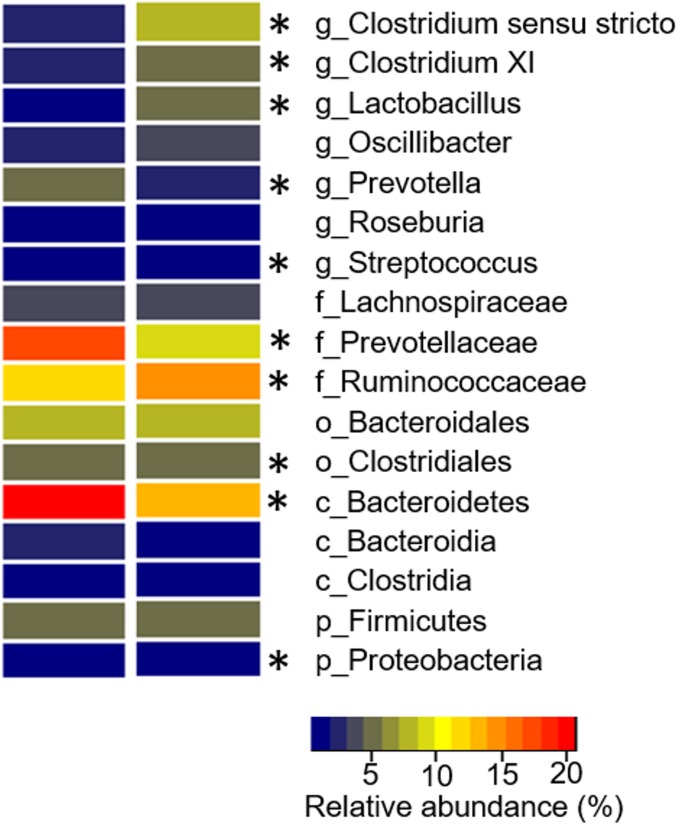
In Figure 1, a nonmetric multidimensional scaling plot is provided to illustrate similarities in the global bacterial community structure of pig colon digesta. The stress value associated with this plot measures the difficulty involved in compressing the samples relationship into two dimensions. It was 0.22, indicating some stress, but we consider this as acceptable, because larger data sets (as in our case) general result in larger stress values. A significant difference was observed between two groups of animals in this plot: one colonized at higher levels with Firmicutes (F), and another colonized with Bacteroidetes (B) (R = 0.339, P-value = 0.001). The groups showed an average dissimilarity value of 68%, where the average similarity level between all samples in group B was found to be 33%, and that for group F was 38%. Microorganisms contributing to this separation belonged to the genera Clostridium sensu stricto (B = 20.7%, F = 12.8%), Lactobacillus (B = 1.3%, F = 5.2%), Prevotella (B = 5.3%, F = 2.7%), and Clostridium XI (B = 1.9%, F = 5.8%) (Figure 2). Additional groups of microorganisms presenting significant differences between both groups are shown in Figure 2. The reasons for the separation into these two groups could not be identified and might also be due to different conditions at birth and weaning period, which were unknown to us. Despite the different colon colonization patterns found, no effect of the Firmicutes/Bacteroidetes ratio was observed for DG, FC, or FI (Figure S2). Eight genera generated significant heritability estimates (P-value < 0.05), which are shown in Table 1. The FDR of these significant results was <0.11. Heritability estimates of all 49 bacterial genera are shown in Table S2.
Figure 1.
Nonmetric multidimensional scaling (nMDS) plot illustrating similarities in the global bacterial community structure of pig colon digesta. The pigs are colonized with higher abundances of Firmicutes ( ) and Bacteroidetes (
) and Bacteroidetes ( ). While a two-dimensional (2D) stress value of 0.2 denotes some stress on the plot, this is considered acceptable since 207 samples are ordinated together.
). While a two-dimensional (2D) stress value of 0.2 denotes some stress on the plot, this is considered acceptable since 207 samples are ordinated together.
Figure 2.
Relative abundance of the most abundant genera of the Bacteroidetes (first heatmap row) and Firmicutes groups (second heatmap row). Groups with an asterisk (*) are significantly different (P < 0.05).
Table 1. Estimated heritability and P-value for the relative abundances of bacterial genera.
| Bacteria | SE | P-valuea | |
|---|---|---|---|
| Alloprevotella | 0.34 | 0.16 | 0.01 |
| Blautia | 0.33 | 0.14 | <0.01 |
| Catenibacterium | 0.39 | 0.16 | 0.01 |
| Lactobacillus | 0.34 | 0.16 | 0.02 |
| Uncultured Spirochaetales | 0.52 | 0.15 | <0.01 |
| Uncultured Spirochaetes | 0.32 | 0.17 | 0.01 |
| Uncultured Succinivibrionaceae | 0.57 | 0.14 | <0.01 |
| Uncultured Veillonellaceae | 0.33 | 0.15 | 0.01 |
All p-values showed a FDR < 0.12.
Heritability and microbiability of host traits and prediction results
The heritability and microbiability of the DG, FC, and FI traits are shown in Table 2. The heritability estimates are likely slightly underestimated, due to the use of SNP chip data instead of pedigree data. However, they are within a typical range for complex pig traits measured under standardized conditions. The heritability and microbiability estimates were significant with the exception of the heritabilities of FC and FI. For these traits, the microbiability estimates were higher than the heritability estimates. The microbial relationship matrix M, and genomic relationship matrix G, underlying these calculations are shown as heatmaps in Figure S3 and Figure S4, respectively. The mean of the diagonal values of G was 0.99, and the values ranged from 0.85 to 1.19. The mean of the diagonal values of M was 0.995, and the values ranged from 0.66 to 1.97.
Table 2. Estimated microbiability and heritability with SE and P-values for DG, FC, and feed intake.
| Trait | SE | P-value | SE | P-value | ||
|---|---|---|---|---|---|---|
| DG | 0.28 | 0.13 | 0.01 | 0.42 | 0.14 | <0.01 |
| FC | 0.21 | 0.14 | 0.01 | 0.19 | 0.13 | 0.08 |
| FI | 0.16 | 0.10 | 0.03 | 0.11 | 0.11 | 0.22 |
as defined by Difford et al. (2016).
The results of the genomic and microbial predictions are shown in Table 3. The microbial prediction generated an accuracy of 0.41 for DG, and 0.33 for FC and FI. These prediction accuracies were higher than those obtained from genomic predictions. In addition, confidence intervals showed that the accuracies were significantly >0 (as was twice the case for genomic predictions).
Table 3. Accuracy of microbial and genomic predictions of DG, FC, and FI, with C.I.
| Trait | Microbial Prediction | Genomic Prediction | ||
|---|---|---|---|---|
| 97.5% CI | 97.5% CI | |||
| DG | 0.41 | 0.18:0.62 | 0.35 | 0.08:0.58 |
| FC | 0.33 | 0.07:0.54 | 0.23 | −0.04:0.48 |
| FI | 0.33 | 0.15:0.51 | 0.20 | −0.08:0.46 |
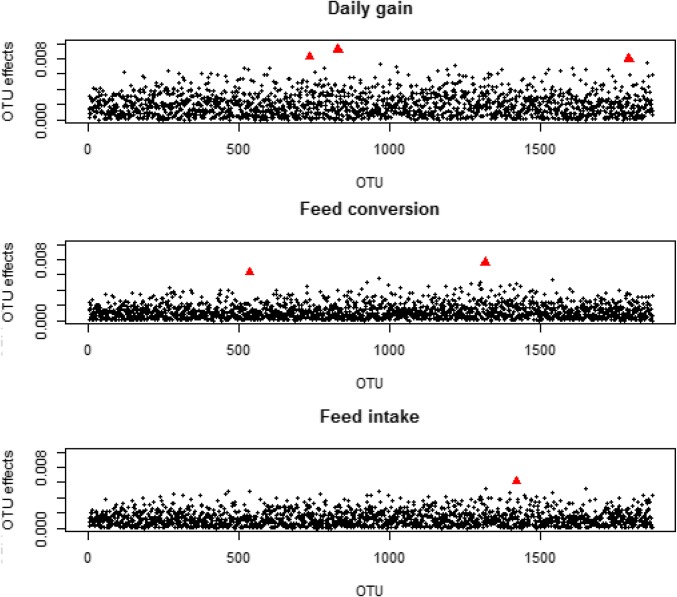
A plot of marginal OTU effects is shown in Figure 3. Some outlier effects were detected (Table 4), but none of the marginal OTU effects showed substantial effects. For DG, the outliers were assigned to uncultured Veillonellaceae, uncultured Prevotellaceae and uncultured Proteobacteria. For FC two OTUs with outlier effects were detected, which were assigned to uncultured Bacteroidales and uncultured Clostridiales. One outlier of OTU effects was found for FI, and was assigned to uncultured Clostridiales.
Figure 3.
OTU marginal effects (absolute values) of each OTU for each trait. Outliers are marked in red.
Table 4. Outliers of marginal OTU effect estimates () with designations and average abundances for DG, FC, and FI.
| OTU | Trait | Designation | Average Abundance | |
|---|---|---|---|---|
| 1284 | DG | Unc. Veillonellaceae | 0.017 | −0.009 |
| 1459 | DG | Unc. Prevotellaceae | <0.001 | −0.008 |
| 4300 | DG | Unc. Proteobacteria | 0.002 | −0.008 |
| 901 | FC | Unc. Bacteroidales | 0.029 | −0.006 |
| 2525 | FC | Unc. Clostridiales | 0.008 | 0.008 |
| 2812 | FI | Unc. Clostridiales | 0.007 | 0.006 |
Discussion
In this study, we analyzed the microbial composition of pig colon samples. The general pattern of microbiota composition is in agreement with reports on colon (Looft et al. 2014; Kim and Isaacson 2015). No interrelationships were found between the Firmicutes-Bacteroidetes ratio and DG, FC, or FI, as previously shown by Mach et al. (2015). In former studies conducted in mice and humans, this ratio was considered as an important marker for obesity (Ley et al. 2005, 2006; Turnbaugh et al. 2006). However, other studies showed contradictory results, or even no evidence of a possible effect in human obesity (Duncan et al. 2008; Schwiertz et al. 2010; Jumpertz et al. 2011).
The host genetic variance on the GIT microbiota composition was substantial for some bacterial genera as denoted by the heritability estimates (Table 1). This result is in agreement with earlier findings of Estellè et al. (2014) and O’Connor et al. (2014), who reported similar heritabilities for Blautia and Lactobacillus in a French Large White pig population, and in a segregated mouse population, respectively. The underlying mechanism of this host genetic determination remains largely unknown thus far. In general, host genetics can influence microbiota compositions through differences in immunoglobulin and antibacterial molecules secreted into gut lumen (Wen et al. 2008; Vijay-Kumar et al. 2010; Shulzhenko et al. 2011), owing to differences in mucosal gut structures (Sommer et al. 2014; Wlodarska et al. 2014) and bile acid metabolism (Ryan et al. 2014). Genome-wide association studies (GWAS) may help identify host genes affecting microbiota compositions, and, thus, derive and substantiate novel hypotheses on the genetic mechanism underlying the heritability of microbiota compositions. However, this involves the use of large datasets, and was therefore not possible in this study.
Microbiability as first defined by Difford et al. (2016) allows for a holistic view of the influence of microbiota on host traits. For all three investigated traits, microbiability levels were found to be significant, and, for FC and FI, microbiability estimates were higher than the heritability estimates (Table 3). This points to a strong effect of GIT microbiota compositions on these traits. As microbiota are partly under the control of host genes, from an animal breeder’s perspective they can be viewed as host traits. This highlights the possibility of breeding for optimized microbiota to indirectly improve complex host traits. Indeed, from the heritabilities shown in Table 1, selection responses can be expected for at least eight (from a total of 49) bacterial genera. This targeted breeding strategy might be especially beneficial for important and so-called hard-to-measure traits, for which precise data collection is restricted to few individuals. Examples include the utilization of certain nutrients in monogastric animals (Beck et al. 2016), and greenhouse gas emissions in ruminants (Hayes et al. 2013; Roehe et al. 2016).
The microbial prediction method M-BLUP is closely related to the well-known G-BLUP model, which is widely used in animal breeding. The key difference is that relationships between individuals are modeled based on relative microbiota abundances at the OTU level, for which calculations are straightforward. The M-BLUP predictions outperformed the G-BLUP predictions in terms of prediction accuracy levels (Table 3). This further underscored the importance of microbiota compositions for trait variability. Thus, it seems that, in addition to SNP data, microbiota abundances are an informative source of complex trait prediction data for pigs, which again may be of special interest for hard-to-measure traits. To identify drivers of prediction accuracy levels, we estimate the marginal effects of a single OTU. Since only few outliers were detected (Table 4), it can be tentatively concluded that many of the OTUs explain a small fraction of the trait variability and that such traits, like DG, FC, and FI, are not only polygenic in nature (Wellmann et al. 2013), but also highly polymicrobial determined in this pig population. However, this must be investigated further because this putative polymicrobial trait determination might, at least in part, also be attributable to M-BLUP model assumptions.
One limit of the microbial prediction model pertains to the fact that the GIT microbial composition for pigs is itself not constant, but changes from birth to adulthood (Kim et al. 2011; Pajarillo et al. 2014; Mach et al. 2015), and is, of course, affected by environmental conditions. In the current study, these effects were minimized by housing pigs under standardized conditions, and by collecting microbiota data on pigs of the same age, which may have additionally contributed to the high level of microbial prediction accuracy achieved. However, it remains to be determined whether GIT microbial compositions at a juvenile stage can be used as predictors of complex host traits measured at a mature age. Further, it is likely that microbial compositions collected from different locations of the GIT, or from feces, will show various complex trait predictive capacities.
The two microbiota pig breeding strategies described above (i.e., breeding for an optimized microbiome and applying microbial prediction) define the microbiota composition differently. For the former, it is treated as a quantitative host trait, whereas, for the latter, it is used as an explanatory variable for prediction purposes. Detailed investigations show that not all microbiota genera are heritable host traits, and not all microbiota OTUs are equally important for predictions (Figure 3). Hence, a comparatively detailed analysis of these two components is desirable, but larger datasets must be used. In addition to GWAS of microbiota compositions and complex host traits, this interplay may be analyzed through structural equation models as introduced to the field of livestock genetics by Gianola and Sorensen (2004). In a quantitative genetic setting, structural equation models allow for the separation of direct and indirect genetic effects shaping genetic relationships among traits. Direct genetic effects result from linkage disequilibrium between genes affecting traits or from pleiotropic effects. However, when a causal relationship between two traits exists, genes directly affecting only one trait may also affect the second trait indirectly via the causal relationship between the traits. Methods for identifying causal structures (Valente et al. 2010) would help simultaneously identify which microbiota bacteria present host genetic variance and the impact of these bacteria on complex host trait variations.
This study was conducted using pig samples. Since the pig is an animal well-suited for modeling the human digestive tract, our results may have implications for predicting human predisposition to disease, and consequently for preventative and personalized medicine. For genetically determined traits such as type-2 diabetes, G-BLUP has already been proven to be useful (de los Campos et al. 2013). This study extends the scope of predictions toward using microbiota data, which might be of special interest for traits where it is known that the microbiome plays an important role, e.g., metabolic diseases and obesity (Karlsson et al. 2013; Le Chatelier et al. 2013). Applications of M-BLUP with appropriate microbiota data may help quantify the risks of suffering from such diseases, and may further the development of personalized preventative strategies.
Methods that combine highly dimensional and correlated predictors (G-BLUP and M-BLUP) with cumulative prediction power will have to be developed in future studies. For this purpose, the present data set is too small.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.200782/-/DC1.
Acknowledgments
We thank Dr. Jörg Heinkel and the animal care staff at the experimental farm of the Landesanstalt für Schweinezucht in Boxberg, Germany, for assisting with animal sampling procedures. The authors declare no conflicts of interest.
Footnotes
Communicating editor: M. P. L. Calus
Literature Cited
- Aßhauer K. P., Wemheuer B., Daniel R., Meinicke P., 2015. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31: 2882–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck P., Piepho H. P., Rodehutscord M., Bennewitz J., 2016. Inferring relationships between Phosphorus utilization, feed per gain, and bodyweight gain in an F2 cross of Japanese quail using recursive models. Poult. Sci. 95: 764–773. [DOI] [PubMed] [Google Scholar]
- Bray J. R., Curtis J. T., 1957. An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 27: 325–349. [Google Scholar]
- Burbach K., Seifert J., Pieper D. H., Camarinha-Silva A., 2016. Evaluation of DNA extraction kits and phylogenetic diversity of the porcine gastrointestinal tract based on Illumina sequencing of two hypervariable regions. MicrobiologyOpen 5: 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D., Cullis B. R., Gilmour A. R., Gogel B. J., 2009. ASREML-R, Reference Manual Version 3. Queensland Department of Primary Industries and Fisheries, Brisbane. [Google Scholar]
- Camarinha-Silva A., Jáuregui R., Chaves-Moreno D., Oxley A. P., Schaumburg F., et al. , 2014. Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ. Microbiol. 16: 2939–2952. [DOI] [PubMed] [Google Scholar]
- Clarke K. R., Warwick R. M., 2001. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation, Ed. 1 E Ltd, Plymouth, UK. [Google Scholar]
- Cole J. R., Wang Q., Fish J. A., Chai B., McGarrell D. M., et al. , 2014. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42: D633–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Gianola D., Allison D. B., 2010. Predicting genetic predisposition in humans: the promise of whole-genome markers. Nat. Rev. Genet. 11: 880–886. [DOI] [PubMed] [Google Scholar]
- de los Campos G., Vazquez A. L., Fernando R., Klimentidis Y. C., Sorensen D., 2013. Prediction of complex human traits using the genomic best linear unbiased predictor. PLoS Genet. 9: e1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difford G. F., Lassen J., Løvendahl P., 2016. Genes and microbes, the next step in dairy cattle breeding. In Proceedings, EAAP—67th Annual Meeting, Belfast. [Google Scholar]
- Duncan S. H., Lobley G. E., Holtrop G., Ince J., Johnstone A. M., et al. , 2008. Human colonic microbiota associated with diet, obesity and weight loss. Int. J. Obes. 32: 1720–1724. [DOI] [PubMed] [Google Scholar]
- Estellè J., Mach N., Ramayo-Caldas Y., Levenz F., Lemonnier G., et al. , 2014. The influence of host’s genetics on the gut microbiota composition in pigs and its links with immunity traits. In Proceedings, 10th World Congress of Genetics Applied to Livestock Production, Vancouver. [Google Scholar]
- Gianola D., Sorensen D., 2004. Quantitative genetic models for describing simultaneous and recursive relationships between phenotypes. Genetics 167: 1407–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Lewin H. A., Goddard M. E., 2013. The future of livestock breeding: genomic selection for efficiency, reduced emissions intensity, and adaptation. Trends Genet. 29: 206–214. [DOI] [PubMed] [Google Scholar]
- Heinritz S. N., Mosenthin R., Weiss E., 2013. Use of pigs as a potential model for research into dietary modulation of the human gut microbiota. Nutr. Res. Rev. 26: 191–209. [DOI] [PubMed] [Google Scholar]
- Jannink J. L., Lorenz A. J., Iwata H., 2010. Genomic selection in plant breeding: from theory to practice. Brief. Funct. Genomics 9: 166–177. [DOI] [PubMed] [Google Scholar]
- Jumpertz R., Le D. S., Turnbaugh P. J., Trinidad C., Bogardus C., et al. , 2011. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 94: 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson F. H., Tremaroli V., Nookaew I., Bergstrom G., Behre C. J., et al. , 2013. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498: 99–103. [DOI] [PubMed] [Google Scholar]
- Kim H. B., Isaacson R. E., 2015. The pig gut microbial diversity: Understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet. Microbiol. 177: 242–251. [DOI] [PubMed] [Google Scholar]
- Kim H. B., Borewicz K., White B. A., Singer R. S., Sreevatsan S., et al. , 2011. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet. Microbiol. 153: 124–133. [DOI] [PubMed] [Google Scholar]
- Knol E. F., Nielsen B., Knap P. W., 2016. Genomic selection in commercial pig breeding. Anim. Front. 6: 15. [Google Scholar]
- Kostic A. D., Howitt M. R., Garrett W. S., 2013. Exploring host – microbiota interactions in animal models and humans. Genes Dev. 27: 701–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., et al. , 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500: 541–546. [DOI] [PubMed] [Google Scholar]
- Ley R. E., Backhed F., Turnbaugh P., Lozupone C. A., Knight R. D., et al. , 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102: 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Tunrbaugh P. J., Klein S., Gordon J. I., 2006. Microbial Ecology: Human gut microbes associated with obesity. Nat 444: 1022–1023. [DOI] [PubMed] [Google Scholar]
- Looft T., Allen H. K., Cantarel B. L., Levine U. Y., Bayles D. O., et al. , 2014. Bacteria, phages and pigs: the effects of in-feed antibiotics on the microbiome at different gut locations. ISME J. 8: 1566–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach N., Berri M., Estellé J., Levenez F., Lemonnier G., et al. , 2015. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 7: 554–569. [DOI] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor A., Quizon P. M., Albright J. E., Lin F. T., Bennett B. J., 2014. Responsiveness of cardiometabolic-related microbiota to diet is influenced by host genetics. Mamm. Genome 25: 583–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajarillo E. A. B., Chae J. P., Balolong M. P., Kim H. B., Seo K. S., et al. , 2014. Pyrosequencing-based analysis of fecal microbial communities in three purebred pig lines. J. Microbiol. 52: 646–651. [DOI] [PubMed] [Google Scholar]
- Ramos A. M., Crooijmans R. P. M. A., Affara N. A., Amaral A. J., Archibald A. L., et al. , 2009. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One 4: e6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehe R., Dewhurst R. J., Duthie C. A., Rooke J. A., McKain N., et al. , 2016. Bovine host genetic variation influences rumen microbial methane production with best selection criterion for low methane emitting and efficiently feed converting hosts based on metagenomic gene abundance. PLoS Genet. 12: e1005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross E. M., Moate P. J., Marett L. C., Cocks B. G., Hayes B. J., 2013. Metagenomic predictions: from microbiome to complex health and environmental phenotypes in humans and cattle. PLoS One 8: e73056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. K., Tremaroli V., Clemmensen C., Kovatcheva-Datchary P., Myronovych A., et al. , 2014. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature 509: 183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwiertz D., Taras K., Schäfer K., Beijer S., Bos N. A., et al. , 2010. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 18: 190–195. [DOI] [PubMed] [Google Scholar]
- Shulzhenko N., Morgun A., Hsiao W., Battle M., Yao M., et al. , 2011. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity vs. metabolism in the gut. Nat. Med. 17: 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F., Adam N., Johansson M. E. V., Xia L., Hansson G. C., et al. , 2014. Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PLoS One 9: e85254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A., Koren O., Ley R., 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9: 279–290. [DOI] [PubMed] [Google Scholar]
- Storey J. D., Tibshirani R., 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandén I., Garrick D. J., 2009. Technical note: derivation of equivalent computing algorithms for genomic predictions and reliabilities of animal merit. J. Dairy Sci. 92: 2971–2975. [DOI] [PubMed] [Google Scholar]
- Stratz P., Wimmers K., Meuwissen T. H. E., Bennewitz J., 2014. Investigations on the pattern of linkage disequilibrium and selection signatures in the genomes of German Piétrain pigs. J. Anim. Breed. Genet. 131: 473–482. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., et al. , 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- Valente B. D., Rosa G. J. M., De Los Campos G., Gianola D., Silva M. A., 2010. Searching for recursive causal structures in multivariate quantitative genetics mixed models. Genetics 185: 633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M., Aitken J. D., Carvalho F. A., Cullender T. C., Mwangi S., et al. , 2010. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science 344: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R., 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmann R., Preuß S., Tholen E., Heinkel J., Wimmers K., et al. , 2013. Genomic selection using low density marker panels with application to a sire line in pigs. Genet. Sel. Evol. 45: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Ley R. E., Volchkov P. Y., Stranges P. B., Avanesyan L., et al. , 2008. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455: 1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M., Thaiss C. A., Nowarski R., Henao-Mejia J., Zhang J. P., et al. , 2014. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 156: 1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Wang G., Siegel P., He C., Wang H., et al. , 2013. Quantitative genetic background of the host influences gut microbiomes in chickens. Sci. Rep. 3: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are fully represented within the tables and figures. Sequences are available at the European Nucleotide Archive (ENA) under accession number PRJEB18070 (http://www.ebi.ac.uk/ena/data/view/PRJEB18070). File S2 and File S3 contain genotypes and phenotypes for each individual, respectively. File S4 contains relative abundances at OTU level.