Abstract
The mammalian Y chromosome is considered a symbol of maleness, as it encodes a gene driving male sex determination, Sry, as well as a battery of other genes important for male reproduction. We previously demonstrated in the mouse that successful assisted reproduction can be achieved when the Y gene contribution is limited to only two genes, Sry and spermatogonial proliferation factor Eif2s3y. Here, we replaced Sry by transgenic activation of its downstream target Sox9, and Eif2s3y, by transgenic overexpression of its X chromosome–encoded homolog Eif2s3x. The resulting males with no Y chromosome genes produced haploid male gametes and sired offspring after assisted reproduction. Our findings support the existence of functional redundancy between the Y chromosome genes and their homologs encoded on other chromosomes.
Many sexual characteristics are influenced by sex chromosome constitution, with mammalian females typically carrying XX and males XY. We recently reported that in the mouse, only two Y-chromosome genes—testis-determinant Sry and spermatogonial proliferation factor Eif2s3y—are needed for successful assisted reproduction (1). Here, we asked if these two genes could be replaced by transgenic activation of their homologs encoded on other chromosomes.
For Sry replacement, we chose Sox9 (Sry-related high-mobility–group box gene 9), a direct target of SRY (2). Prior work showed that transgenic overexpression of Sox9 driven by the Wt1 promoter results in female-to-male sex reversal in XX mice (3). We placed the Wt1-Sox9 transgene in the context of a single X chromosome carrying the Eif2s3y transgene (fig. S1A) (4) and found that it generated males (XEOSox9). In these males, the Y-chromosome gene contribution is limited to Eif2s3y (table S1).
XOSry males, which carry an autosomally encoded Sry transgene, develop testes containing spermatogonia that are unable to proliferate, which results in seminiferous tubules appearing empty when compared with those from males with an intact Y chromosome (XY) (Fig. 1, A and B). This defect can be overcome by transgenic Eif2s3y addition to the X chromosome (XEOSry) (table S1 and fig. S4A) (1, 5). To replace Eif2s3y, we transgenically overexpressed its X chromosome–encoded homolog, Eif2s3x (fig. S2). We then placed the Eif2s3x transgene in the context of XOSry (fig. S1B and supplementary text). The resulting XOSry,Eif2s3x males (carrying autosomally encoded Sry and Eif2s3x transgenes) had the Y-chromosome contribution limited to Sry (table S1).
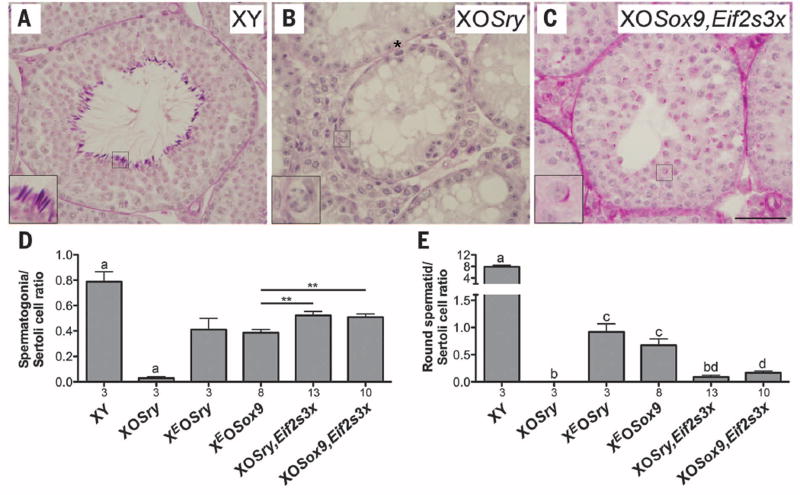
Fig. 1. Testis histology analysis.
Tubules of periodic acid–Schiff–hematoxylin–stained sections of testis from XY (A), XOSry (B), and XOSox9,Eif2s3x (C) males. XY have normal spermatogenesis with expected germ cell types present, including step 16 spermatids [inset (A)]. XOSry have spermatogonial proliferation arrest, resulting in tubules lacking germ cells except for occasional normal [inset (B)] and abnormal (*) spermatogonia. XOSox9,Eif2s3x have meiotic and postmeiotic arrests that occasionally allow formation of round spermatids [insets (C)], arresting at step 7. Scale bar, 50 µm; insets, ×3 magnification. (D and E) Quantitative analysis of spermatogenesis progression. Bars are averages ± SEM, with n under the x axis. Statistical significance (t test): (D) bars marked witha are different from all others (P < 0.05), **P < 0.01; (E) bars with different letters are significantly different (P < 0.05).
XEOSox9 and XOSry,Eif2s3x males had small testes (fig. S3), but spermatogenesis was initiated and progressed through meiosis and arrested at the round spermatid stage (fig. S4, B and C). Spermatogonia/Sertoli ratios in XEOSox9 and XOSry,Eif2s3x and spermatid/Sertoli ratio in XEOSox9 were comparable to XEOSry but lower than those in XY (Fig. 1, D and E). Round spermatids in XOSry,Eif2s3x were dramatically depleted; XEOSry and XY had 10 and 88 times as many, respectively (Fig. 1E and table S2). The spermatids from both XEOSox9 and XOSry,Eif2s3x males were functional in assisted fertilization, and live offspring were obtained after embryo transfer (Table 1).
Table 1. The results of ROSI with spermatids from males with a single or no Y-chromosome genes.
Column heads 2 to 4: Males with round spermatids identifiable in live testicular cell suspension out of males examined. Males yielding progeny out of the number of males that yielded embryos used for embryo transfer and induced pregnancy. Live offspring as a percentage of embryos transferred.
| Male genotype | Y gene contribution | Males with round spermatids (%) (no.) |
Males yielding progeny | Live offspring (%) (no.) |
|---|---|---|---|---|
| XEOSox9 | Eif2s3y | 80 (8/10) | 7/8 | 15.7 (18/115) |
| XOSry,Eif2s3x | Sry | 55 (17/31) | 16/17 | 22.5 (57/253) |
| XOSox9,Eif2s3x | None | 27 (13/48)a | 9/10 | 20.7 (46/222) |
| XY control | Intact Y | 100 (6/6) | 6/6 | 25.0 (22/88) |
Statistical significance (Fisher’s exact test, P < 0.05):
Different from all others.
We next tested whether spermatogenesis can take place in males with a complete absence of Y-chromosome genes. We used the same transgenes that were successful in single–Y gene substitutions (Wt1-Sox9 and Eif2s3x Tg1) to generate mice transgenic for Sox9 and Eif2s3x in the XO context (XOSox9,Eif2s3x) (fig. S1C and table S1) The majority (35 out of 48) of XOSox9,Eif2s3x males had testicular defects and essentially no germ cells (fig. S5 and supplementary text). In the remaining males, spermatogonial proliferation arrest was overcome (Fig. 1C and fig. S5), and spermatogenesis progression was comparable to that of XOSry,Eif2s3x (Fig. 1, D and E, and table S2). Using assisted reproduction [round spermatid injection (ROSI)], we injected oocytes with spermatids from 13 males and obtained zygotes with two well-developed pronuclei and normal two-cell embryos (fig. S6, A to C, and movie S1). Embryos from 11 males were used for transfer. Ten resulted in pregnancy, and nine yielded offspring (Table 1). Among the males that yielded progeny, there were F1, F2, and F3 generation XOSox9,Eif2s3x ROSI males (fig. S6D). ROSI offspring from males with one or no Y-chromosome genes were all normal and healthy (figs. S7 to S9 and supplementary text).
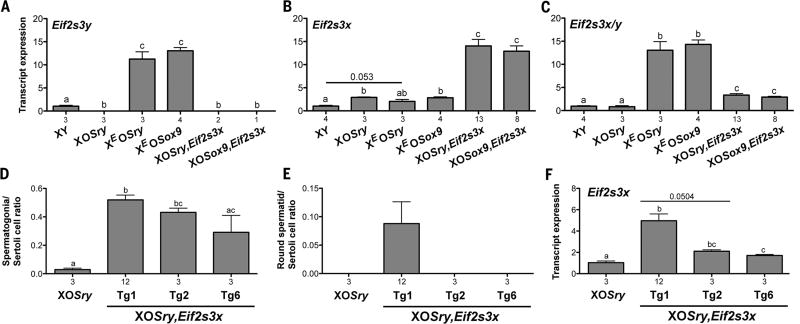
The quantification of Eif2s3x/y transcripts in males transgenic for Eif2s3y or Eif2s3x revealed a correlation between spermatogenesis progression and Eif2s3x/y expression level (Fig. 2, figs. S10 to S12, and supplementary text). All transgenic males had their respective transgene transcript levels elevated when compared with XY (Fig. 2, A and B). Compared with Eif2s3x transgenic males, Eif2s3y transgenic males showed higher Eif2s3x/y transcript levels (Fig. 2C) and increased incidence of round spermatids (Fig. 1E). When spermatogenesis and Eif2s3x expression were examined in XOSry,Eif2s3x males with varying numbers of Eif2s3x transgene copies, one (Tg2 and Tg6) and four (Tg1), no differences in spermatogonia/Sertoli cell ratio were observed, but round spermatids were found only in Tg1 males, in which Eif2s3x transcript levels are 2.4 to 2.9 times those in Tg2 and Tg6 males (Fig. 2, D to F, and fig. S4, D to G).
Fig. 2. Relation between Eif2s3x/y expression and spermatogenesis progression.
(A to C) Transcript levels of endogenous and transgenic spermatogonial proliferation factors quantified by real-time polymerase chain reaction with Actb as a loading control and XY serving as reference control. (D to F) Analysis of spermatogenesis progression (D and E) and Eif2s3x expression (F) in XOSry,Eif2s3x males with 4 (Tg1) and 1 (Tg2 and Tg6) Eif2s3x transgene copies. Means ± SEM, with n under the x axis; bars with different letters are statistically different (t test, P < 0.05).
We have shown that a male mouse without any Y-chromosome genes but with transgenically activated Sox9 and Eif2s3x can generate haploid gametes and father offspring with the help of assisted fertilization. Sox9 is not unique in being able to take over the Sry function in sex determination. Manipulation of expression of other genes can lead to sex-fate change [reviewed in (6–8)]. A surrogate sex-determination mechanism can also be activated without human input, as shown by two rodent species that lost the Y chromosome and Sry (9, 10).
Eif2s3y and Eif2s3x represent a typical, formerly autosomal, single-copy, X-Y homologous gene pair and were hypothesized to have interchangeable function (1, 5). Our data support this hypothesis: A single additional copy of Eif2s3x can functionally replace Eif2s3y in spermatogenesis initiation. For progression through meiosis, however, at least four Eif2s3x transgene copies are necessary, and the number of global Eif2s3x/y transcripts must reach a certain threshold. Our data also suggest that Eif2s3x/y may play roles in gonad formation. We observed severe abnormalities of mature testes, indicative of impaired gonadal development, in XOSox9,Eif2s3x but not XEOSox9 males. Because the global Eif2s3x/y expression is lower in the former, this suggests that a critical level of Eif2s3x/y may be required for efficient testis differentiation.
Our data support a model where Eif2s3y and Eif2s3x are functionally interchangeable in spermatogenesis, but each homolog has evolved a distinct expression level. Eif2s3y transcript amounts are ~5 to 7 times those in premeiotic and meiotic cells (11), which explains why the addition of one Eif2s3x transgene copy could not replace the function of endogenous Eif2s3y in driving spermatogenesis through meiosis. Our finding that a single Eif2s3x transgene copy was sufficient to substitute for Eif2s3y in overcoming spermatogonial proliferation arrest suggests that the strong Eif2s3y expression in spermatogonia is required for the subsequent meiotic stages but not for mitotic proliferation. Our observations contradict the accepted dogma of X-Y gene pairs evolving by decay on the Y chromosome and compensation on the X chromosome (12), because here, it is a beneficial overexpression of the Y gene Eif2s3y and not its X homolog that appears to have evolved to meet the needs of spermatogenesis. This might be the result of a selective advantage during oogenesis for reduced Eif2s3x levels or a selection for male germ cell beneficial effects on the Y chromosome.
It is generally believed that widely expressed genes on the human Y chromosome with X homologs that escape X inactivation are dosage-sensitive (13). Dosage sensitivity explains why genes are conserved on the Y chromosome: A certain combined X-Y dose is critical at certain stages in certain tissues, and the X-gene dose cannot simply be increased globally to compensate for the loss of the Y gene because this would have detrimental effects in females. To lose the Y gene safely, the X gene, or the genome, or the developmental systems must adapt. Possible strategies might include incremental increases in X-gene dosage, relaxing constraints on dose-sensing, or retrogenes (14). The mouse Eif2s3x/y gene pair is not sensitive to overexpression; substantial elevation of Eif2s3y (5) or Eif2s3x in the XY context (this study) has no obvious somatic effects, nor does it affect spermatogenesis and fertility. These findings suggest that dosage sensitivity may appear mainly in association with underexpression and/or may vary between different X-Y gene pairs.
Altogether, our analyses of the Eif2s3x/y gene pair support their importance for spermatogenesis. It will now be imperative to explore the mechanisms whereby Eif2s3x/y factors exert their functions during testicular development and spermatogenesis. Our work also paves the way and prompts future evaluations of other ancestral X-Y gene pairs to clarify the dosage requirements for spermatogenesis and beyond. Finally, our demonstration that offspring can be obtained from males with no Y-chromosome genes shows that for assisted reproduction in the mouse, the Y chromosome is no longer necessary. However, there is extensive evidence from both phenotype characterization (15–17) and genomic analyses (13, 14, 18) unequivocally supporting the importance of Y-chromosome genes for normal, unassisted fertilization. So, although our data demonstrate that it is possible to bypass the requirement for the Y chromosome in male assisted reproduction, the Y clearly remains the genetic determinant of full natural masculinity.
Supplementary Material
Acknowledgments
The work was supported by NIH HD072380 and Hawaii Community Foundation 14ADVC-64546 grants to M.A.W. and INSERM core funding to M.J.M.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/351/6272/514/suppl/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S12
Tables S1 to S4
References (19–50)
Movie S1
REFERENCES AND NOTES
- 1.Yamauchi Y, Riel JM, Stoytcheva Z, Ward MA. Science. 2014;343:69–72. doi: 10.1126/science.1242544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sekido R, Lovell-Badge R. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- 3.Vidal VP, Chaboissier MC, de Rooij DG, Schedl A. Nat. Genet. 2001;28:216–217. doi: 10.1038/90046. [DOI] [PubMed] [Google Scholar]
- 4.Materials and methods and further details are available as supporting material on Science Online.
- 5.Mazeyrat S, et al. Nat. Genet. 2001;29:49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- 6.Eggers S, Ohnesorg T, Sinclair A. Nat. Rev. Endocrinol. 2014;10:673–683. doi: 10.1038/nrendo.2014.163. [DOI] [PubMed] [Google Scholar]
- 7.Quinn A, Koopman P. Semin. Reprod. Med. 2012;30:351–363. doi: 10.1055/s-0032-1324718. [DOI] [PubMed] [Google Scholar]
- 8.Warr N, Greenfield A. Dev. Biol. 2012;1:559–577. doi: 10.1002/wdev.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Just W, et al. Nat. Genet. 1995;11:117–118. doi: 10.1038/ng1095-117. [DOI] [PubMed] [Google Scholar]
- 10.Soullier S, Hanni C, Catzeflis F, Berta P, Laudet V. Mamm. Genome. 1998;9:590–592. doi: 10.1007/s003359900823. [DOI] [PubMed] [Google Scholar]
- 11.Gan H, et al. Nat. Commun. 2013;4:1995. doi: 10.1038/ncomms2995. [DOI] [PubMed] [Google Scholar]
- 12.Jegalian K, Page DC. Nature. 1998;394:776–780. doi: 10.1038/29522. [DOI] [PubMed] [Google Scholar]
- 13.Bellott DW, et al. Nature. 2014;508:494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes JF, Skaletsky H, Koutseva N, Pyntikova T, Page DC. Genome Biol. 2015;16:104. doi: 10.1186/s13059-015-0667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cocquet J, et al. PLOS Biol. 2009;7:e1000244. doi: 10.1371/journal.pbio.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riel JM, et al. J. Cell Sci. 2013;126:803–813. doi: 10.1242/jcs.114488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamauchi Y, et al. Biol. Reprod. 2009;81:353–361. doi: 10.1095/biolreprod.109.076307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soh YQ, et al. Cell. 2014;159:800–813. doi: 10.1016/j.cell.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.