Abstract
The increasing characterization of childhood acute lymphoblastic leukemia (ALL) has led to the identification of multiple molecular targets, but have yet to translate into more effective targeted therapies, particularly for high-risk, relapsed T-cell ALL. Searching for master regulators controlling multiple signaling pathways in T-ALL, we investigated the multi-functional protein redox factor-1 (Ref-1/APE1), which acts as a signaling “node” by exerting redox regulatory control of transcription factors important in leukemia. Leukemia patients’ transcriptome databases showed increased expression in T-ALL of Ref-1 and other genes of the Ref-1/SET interactome. Validation studies demonstrated that Ref-1 is expressed in high-risk leukemia T-cells, including in patient biopsies. Ref-1 redox function is active in leukemia T-cells, regulating the Ref-1 target NF-κB, and inhibited by the redox-selective Ref-1 inhibitor E3330. Ref-1 expression is not regulated by Notch signaling, but is upregulated by glucocorticoid treatment. E3330 disrupted Ref-1 redox activity in functional studies and resulted in marked inhibition of leukemia cell viability, including T-ALL lines representing different genotypes and risk groups. Potent leukemia cell inhibition was seen in primary cells from ALL patients, relapsed and glucocorticoid-resistant T-ALL cells, and cells from a murine model of Notch-induced leukemia. Ref-1 redox inhibition triggered leukemia cell apoptosis and down-regulation of survival genes regulated by Ref-1 targets. For the first time, this work identifies Ref-1 as a novel molecular effector in T-ALL and demonstrates that Ref-1 redox inhibition results in potent inhibition of leukemia T-cells, including relapsed T-ALL. These data also support E3330 as a specific Ref-1 small molecule inhibitor for leukemia.
Keywords: T-cell acute lymphoblastic leukemia, Ref-1/APE1, redox signaling, molecular targeting, relapse leukemia
INTRODUCTION
Clinical studies using selective inhibitors of signaling molecules support the notion that, in most cancers, effective, curative therapeutic strategies require the concurrent disruption of multiple molecular targets or pathways (1, 2). Accordingly, the focus of targeted therapies is shifting to the identification of combinations of signaling inhibitors that optimally promote tumor cytotoxicity and overcome drug resistance on defined cancers, while preventing the emergence of tumor variants that may lead to cancer recurrence. In this context, the clinical development of drugs that disrupt master molecular regulators controlling the activity of multiple non-recurrent signaling pathways or transcription programs has great potential.
Ref-1 (redox factor-1; AP endonuclease 1, APE1; APEX1) is a multifunctional protein acting as a unique nuclear reduction-oxidation (redox) factor and a DNA repair endonuclease (3). In contrast to general reduction-oxidation systems, Ref-1 directly reduces cysteine residues in the DNA binding and activation domains of target transcription factors (TFs) (4), thus controlling their binding to target promoter sequences and their downstream transcriptional programs (3, 5–7). The redox targets of Ref-1 include NF-κB, AP-1, STAT3, p53, HIF-1 and other TFs (6, 8–10) that have been shown to play critical roles in tumorigenesis, cancer cell survival, proliferation, drug resistance and other functions. In solid tumors, increased expression of Ref-1 has been associated with drug-resistance and poor prognosis (6). Blockade of Ref-1 by E3330 (also called APX3330), a small molecule inhibitor that specifically recognizes the redox function of Ref-1, (11, 12) results in significant reduction or abrogation of the transcriptional activity of NF-κB, AP-1, and HIF-1 (5, 7), and inhibition of tumor cell functions (5, 8, 10). This compound has IND approval and will enter Phase 1 clinical trials this year.
Despite successes in the treatment of pediatric T-cell acute lymphoblastic leukemia (ALL), leukemia relapse, refractory disease and induction failure continue to pose significant therapeutic challenges (13–16). In adult T-ALL, current therapies are much less effective, with high relapse rates and poor outcomes. Marked progress in the molecular characterization of T-cell ALL raised the prospect of a new generation of more efficient, selective therapies, particularly for high-risk and relapsed patients. This included the targeting of Notch signaling, which seems to function as an oncogenic driver in the majority of T-ALL (17–19). However, clinical trials with Notch pathway inhibitors showed limited efficacy and were hampered by significant toxicities, and studies showed that presence of Notch mutations is not associated with poor prognosis and that those patients may be in fact more responsive to conventional therapies (20). Other signaling effectors or pathways were identified as potential therapeutic targets for pediatric T-ALL, including PI3K/Akt, mTOR/TORC1, NF-κB, Gfi-1, CXCL12/CXCR4 and others (21–25). Inhibitory compounds targeting these pathways are in preclinical development or in clinical studies, but the use of signaling antagonists has yet to lead to transformative clinical successes in ALL patients. Therefore, the need remains for more effective, less toxic targeted therapies for high-risk, refractory and relapsed T-cell ALL.
In search for master molecular regulators of transcriptional programs in T-cell ALL, we investigated the potential role of Ref-1, as it exerts redox control of multiple transcription factors, including the leukemia-associated NF-κB, AP-1 and p53. We observed that Ref-1 is expressed in T-ALL specimens and cell lines, and that its redox function is active in leukemia T-cells. Blockade of Ref-1 by the redox-specific inhibitor E3330 resulted in potent inhibition of viability of leukemia T-cells, including primary cells, relapsed and chemotherapy-resistant cells, and cells from a mouse model of T-ALL. Ref-1 redox inhibition promoted leukemia cell apoptosis, which was associated with down-regulation of pro-survival genes. Although some studies have addressed the potential role of general redox systems (thioredoxin) in ALL (26), they have not led to clinical interventions and lack the target-selectivity of the Ref-1 redox function. Overall, our results demonstrate a role for Ref-1 in the regulation of multiple transcriptional programs in T-cell ALL, and suggest that disruption of Ref-1 redox function represents an efficient strategy to target leukemia T-cells, including high-risk, relapsed leukemias.
MATERIALS AND METHODS
Leukemia T-cells and Specimens
No clinical studies were performed in this work. Leukemia specimens were collected according with the protocol approved by the Indiana University Institutional Review Board. All leukemia specimens were unidentified, and no patient information was collected or used in this study. TAIL7, a human IL-7-dependent leukemia T-cell line that mimics the functional and molecular properties of primary T-ALL cells, was established in our laboratory in 2004 (27). TAIL7 cells were maintained in RPMI-1640 supplemented with 10% FBS (RPMI-10) and IL-7 (10ng/ml; R&D Systems, Minneapolis, MN), and viable cells were separated on Ficoll every 7 days and recultured in fresh media. The immortalized T-ALL cell lines Jurkat E6-1, MOLT-4, CCRF-CEM, Loucy, TALL-104 (IL-2-dependent) were obtained from ATCC, and SUP-T1 cells were obtained from a collaborator. All cell lines were obtained in 2014 and are validated for authenticity on a bi-annual basis. The Jurkat/Bcl2 and Jurkat/Neo are sublines derived from transfection of Jurkat cells respectively with a psFFV-neo expressing vector containing human BCL2 or empty vector, and were obtained from ATCC in 2014. TAIL7-ICN subline was generated by stable transduction of TAIL7 cells with constitutively-active Notch1 (ICN) construct, leading to persistence activation of Notch signaling and significant induction or upregulation of the expression of Notch target genes (Batista A, Cardoso AA, unpublished data) in 2014. TAIL7-DexaR is a subline resistance to high-dose Dexamethasone (up to 2μM) and was generated by exposure of TAIL7 cells to increasing doses of Dexamethasone in 2015. Primary T-ALL cells were obtained from diagnostic specimens of pediatric patients with high leukemia involvement (>90%) in 2015. After gradient centrifugation, cells were washed in RPMI-10.
Animal model of Notch-induced T-ALL and xenograft model of human T-ALL
Animal models of leukemia (Notch-induced T-ALL; xenograft model of human T-ALL) were performed using protocols approved by the Indiana University School of Medicine IACUC. For the Notch-induced leukemia model, hematopoietic progenitor Lin- cells were purified from donor C57BL/6 mice (CD45.2+), and transduced with MSCV-ICN/GFP (ICN) viral particles (28). Equal numbers of transduced Lin-GFP+ICN+ cells (20,000/mice) were injected I.V. into lethally irradiated 8-wk old recipient BoyJ (CD45.1+) admixed with a radio-protective dose of BM cells (CD45.1+). This model has 100% penetrance, with leukemia progression correlating with increased WBC counts, circulating blasts and splenomegaly. Mice were bled weekly for WBC counts and quantification of leukemia cells, and were sacrificed at stage of terminal disease, at which they exhibit high content of blasts in PB, BM and spleen, with most leukemia cells being GFP+ CD4+ CD8+ (DP) T-cells. Cells were isolated from harvested femur bones and spleens, and processed for biochemical and functional studies.
For the xenograft human T-ALL model, TAIL7 cells (1×106) were transplanted i.v. into NOD/SCID or NSG mice (7–9wk old) (27, 29). Mice were bled weekly for presence of human blasts in the PB, by flow cytometry. Animals exhibiting >2% circulating human leukemia blasts were randomly allocated into experimental groups, and initiated treatment with Vincristine (i.p., 0.5mg/Kg, every 4 days for 3 weeks) or control vehicle. Mice were sacrificed at stage of terminal disease (very high leukemia cell content in BM), and leukemia cells were isolated from harvested femurs, and processed for functional studies.
Bioinformatics Analyses
Publicly available databases of transcriptome studies of pediatric ALL patients’ specimens were assessed and analyzed using Oncomine® 3.0 (30). Relative expression of Ref-1/APEX1 or genes of the Ref-1 interactome was compared in T-ALL vs. BM from healthy donors, or in T-ALL vs. B-ALL. The Ref-1 interactome was defined based on the Human Protein Reference Database (HPRD, release 9; Institute of Bioinformatics, Johns Hopkins University) (31).
Immunoblotting
Cell lysates were prepared in RIPA lysis buffer system (Santa Cruz Biotechnology, Dallas, TX), as described (21, 22). All experiments with TAIL7 cells were performed using IL-7 (10ng/ml). For studies of Ref-1 regulation by glucocorticoids, TAIL7 cells were incubated with Dexamethasone for the timepoints indicated. Equal amounts of protein (20–50mg/sample) were resolved by SDS-PAGE, transferred onto nitrocellulose membranes, and immunoblotted with antibodies for Ref-1 (Novus Biologicals, Littleton, CO), or for Actin (Thermo Fisher Scientific, Waltham, MA) as loading control. Immunodetection was performed by incubation with HRP-conjugated anti-mouse IgG antibodies (EMD Millipore, Billerica, MA), followed by chemiluminescence developing using WesternBright Quantum Western blotting detection kit (Advansta, Menlo Park, CA). Determination of relative protein intensity was performed using Quantity One software (Bio-Rad, Hercules, CA).
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue samples from pediatric patients with T-ALL at the time of original diagnosis were used for immunohistochemistry. Immunoperoxidase staining was performed by an automated immunostainer (DAKO, Carpinteria, CA, USA) using a standard streptavidin–biotin–peroxidase complex technique and the Ref-1 Ab (1:200; Novus Biologicals). The primary antibody was followed by HRP-conjugated goat-anti-mouse Ab, with an irrelevant IgG2 antibody (Southern Biotech) used as isotype control. Images were acquired on an Olympus microscope using an Olympus DP71 camera.
Ref-1 Intracellular Staining and Flow Cytometry
Analysis of Ref-1 expression in leukemia T-cells was performed also using intracellular staining and flow cytometry. Briefly, leukemia cells were fixed in Cytofix/Cytoperm solution (BD Biosciences) for 30min, and then washed in Perm/Wash buffer (BD Biosciences) with 3% BSA. Cells were incubated with anti-Ref-1 antibody (Novus Biologicals) at 1:125 dilution for 2hrs, washed with Perm/Wash buffer, followed by incubation with PE-conjugated or FITC-conjugated secondary antibodies (Southern Biotech, Birmingham, Alabama) at 1:500 dilution for 1.5hrs. Staining with fluorochrome-conjugated secondary antibody alone was used as negative control. Cells were acquired in a FACSCalibur flow cytometer (BD, Franklin Lakes, NJ), and data analyzed using FlowJo software (FlowJo LLC; Ashland, OR).
Ref-1 Redox Inhibitor E3330
E3330, also called APX3330, is a small molecule that inhibits the redox function of Ref-1 by selectively recognizing the redox-active conformation of Ref-1 and blocking interaction with the downstream transcription factors. E3330 was synthesized by the University of Michigan Vahlteich Medicinal Chemistry Core Facility (Dr. Hollis Showalter), as previously described (7, 32). Analogs of E3330 (APX2007, APX2009 and APX2032) were synthesized by Cascade Custom Chemistry (Eugene, OR) as described (33). The synthesis steps include a common intermediate, iodolawsone (2-iodo-3-hydroxy-1,4 naphthoquinone which is reacted with methacrylic acid or 2-propylacrylic acid, along with oxalyl chloride and the corresponding amine, and with sodium methoxide in methanol to yield APX2007 [(2E)-2-[(3-methoxy-1,4-dioxo-1,4-dihydronaphthalen-2-yl)methylidene]-N,N-dimethylpentanamide], APX2009 [(2E)-2-[(3-methoxy-1,4-dioxo-1,4-dihydronaphthalen-2-yl)methylidene]-N,N-diethylpentanamide], and APX2032 [(2E)-2-(3-methoxy-1,4-dioxo-1,4-dihydronaphthalen-2-yl)-N,N,2-trimethylprop-2-enamide] (33).
Electrophoretic Mobility Shift Assay (EMSA)
Redox-EMSA were performed as previously described (34), with some modifications. Briefly, nuclear extracts were prepared from leukemia TAIL7 cells cultured in IL-7, as described (21, 22, 27). For the experiments assessing regulation of NF-kB DNA-binding by Ref-1, purified Ref-1 protein was reduced with 2mM DTT for 10min and diluted to a final amount of 4mg with 0.4mM DTT in PBS. Reduced Ref-1 was added to 15mg nuclear extract from leukemia cells. The final concentration of DTT in redox reactions was 0.02mM. A HEX-labeled double-stranded oligonucleotide DNA containing repeat consensus sequences for NF-κB was used: 5′ - AGT TGA GGG GAC TTT CCC AGG C – 3′. For the competition experiment, increasing amount of unlabeled wild-type and mutant (5′-AGT TGA GGC GAC TTT CCC AGG C-3′) probe was added to the EMSA reaction with Hex-labeled probe. For EMSA with Ref-1 redox inhibition, E3330 was preincubated with purified, reduced Ref-1 in EMSA reaction buffer for 30min, followed by addition of 3mg of TAIL7 cells’ nuclear extracts.
Gene Expression by Quantitative PCR
Quantitative PCR (qPCR) was used to quantify levels of defined mRNA transcripts in leukemia cells. RNA was extracted using the Qiagen RNeasy Mini kit (Qiagen; Valencia, CA), according to manufacturer’s instructions. cDNA was prepared using reserve transcriptase (Applied Biosystems, ABI; Foster City, CA), as described.(35) qPCR was performed using Taqman Gene Expression Assays for Ref-1, Survivin/BIRC5, and Bcl-xL expression (ABI), or SYBR® Green-based assay using the Brilliant II SYBR Green Kit (Agilent Technologies; Santa Clara, CA) for the other transcripts analyzed, in a 7900HT Fast Real-Time PCR System (ABI). Quantification was performed using the comparative Ct method, with GAPDH or 18S rRNA being used as reference, endogenous controls. In each experiment, assays were performed in duplicate or triplicate for each sample.
Viability Studies with E3330
Leukemia cell viability was determined using the CellTiter-Glo® ATP assay (Promega; Madison, WI), according to the manufacturer’s instructions. For experiments with E3330, leukemia cells were cultured with RPMI supplemented with 5% FBS (RPMI-5). TAIL7 cells were cultured with IL-7 (10ng/ml), and TALL-104 cells with IL-2 (20ng/mL; R&D Systems). Primary T-ALL cells were cultured with IL-7 plus IL-9 (10ng/ml; R&D Systems), and leukemia cells from the ICN-induced T-ALL model were cultured with IL-7 (10ng/mL), SCF (10ng/mL; R&D Systems) and FLT3L (10ng/mL; R&D Systems). All cultures were performed in triplicates, for the timepoints indicated. Plates were analyzed in a SpectraMax Gemini EM microplate reader (Molecular Devices; Sunnyvale, CA).
Apoptosis Assay
For apoptosis studies, leukemia cells were cultured with 25 or 40μM E3330, in RPMI-5. At the timepoints indicated, cells were resuspended in binding buffer, stained with FITC-conjugated Annexin V (1μl/ml; BD-Biosciences) and Propidium Iodide (PI; 5μg/ml, 15min), and analyzed by flow cytometry.
Statistics
Differences between experimental study conditions were evaluated using the statistical tests indicated in the respective Figure Legends. Differences of means were considered statistically significant for p<0.05.
RESULTS
The Ref-1 multi-function protein is expressed in leukemia T-cells
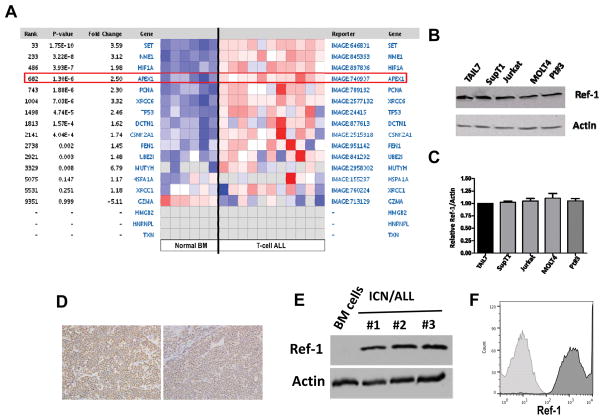
A data mining search for regulators that may control critical leukemia-associated transcriptional programs unveiled as a potential candidate Ref-1/APE1/APEX1, a multi-functional protein known to regulate the transcriptional activity of NF-κB, AP-1, p53, STAT3, HIF-1, and other transcription factors (3, 5, 6, 10). Bioinformatics analyses of transcriptome datasets using Oncomine® showed that Ref-1/APEX1 transcripts are significantly elevated in the T-ALL patients in comparison to BM from normal donors (Figure 1A; p=1.30E-6; Andersson leukemia database) (36). Other molecules of the Ref-1/APEX1 interactome (Supplementary Figure 1A) were found also to be increased in T-ALL specimens, including SET, CSNK2A1, NME1, TP53, and HDAC1 (Figure 1A; p values from 4E-4 to 1E-10). This expression pattern was confirmed in factor-dependent, patient-derived leukemia TAIL7 cells using qPCR. Relative increased expression of Ref-1 transcripts in T-cell ALL specimens was observed in other leukemia datasets (Oncomine®; Supplementary Figure 1B). Analyses of pediatric ALL specimens resistant to frontline chemotherapy drugs showed expression of Ref-1 transcripts, although with no statistically significant differences in comparison to drug-sensitive cases (Supplementary Figure 1C; ref. (37)). Next, we performed immunoblotting using leukemia T-cells representing distinct molecular genotypes, and including high-risk and relapsed T-cell ALL (Supplementary Table 1). Ref-1 protein was detected in all T-ALL cell lines tested, as well as in primary T-ALL specimens (Figure 1B, 1C). Using a new flow cytometry assay to detect intracellular Ref-1, we confirmed marked expression of Ref-1 protein in all T-ALL lines tested (Supplementary Figure 2A). Importantly, immunohistochemistry analyses of biopsies from patients diagnosed with T-ALL confirmed expression of Ref-1 in leukemia cells (Figure 1D; two representative cases shown). We investigated the expression of Ref-1 in leukemia cells from a mouse model of T-ALL. In this model, transplantation of donor hematopoietic stem/progenitor cells (HSPC) transduced with constitutively active, oncogenic Notch1 leads to the development of a malignancy in recipient mice that recapitulates human T-ALL (28, 38, 39). Immunoblotting using BM cells from mice with terminal leukemia (more than 95% leukemia cell content) showed high expression of Ref-1 protein (Figure 1E), which was confirmed in circulating leukemia cells (identified as expressing GFP, positivity for donor CD45.2 and CD4+ CD8+ phenotype) using intracellular staining and flow cytometry (Figure 1F). These results are in congruence with those found in databases demonstrating a low level of Ref-1 in normal BM cells, and higher in leukemic cells (Figure 1A).
Figure 1. Ref-1 expression in leukemia T-cells.
A) Heatmap displaying relative expression levels of transcripts for Ref-1/APEX1 and Ref-1-interacting proteins in patients with T-cell ALL and BM from normal donors; data obtained the Andresson Leukemia database, and analysis performed in Oncomine 3.0, using the Ref-1 interactome as defined by the Human Protein Reference Database (HPRD). B) Immunoblotting for Ref-1 in leukemia T-cell lines and primary cells from a T-ALL patient (Pt#3). Actin was used as loading control. C) Relative expression of Ref-1 protein in T-ALL cells by immunoblotting, determined by densitometry, and normalized to Actin expression levels. D) Immunohistochemistry using Ref-1 Ab in biopsies from two patients diagnosed with T-ALL. E) Immunoblotting for Ref-1 in leukemia cells harvested from the BM of mice exhibiting terminal Notch-induced T-cell ALL and control normal mice BM cells, Actin was used as loading control. F) Intracellular staining with Ref-1 Ab of leukemia cells from mice carrying Notch-induced T-ALL; at least 10,000 events were acquired by flow cytometry and analyzed using FlowJo, with leukemia cells gated based on GFP positivity. Dark gray histogram indicates Ref-1 staining and light gray histogram indicates isotope IgG negative control.
Ref-1 redox function and regulation in leukemia T-cells
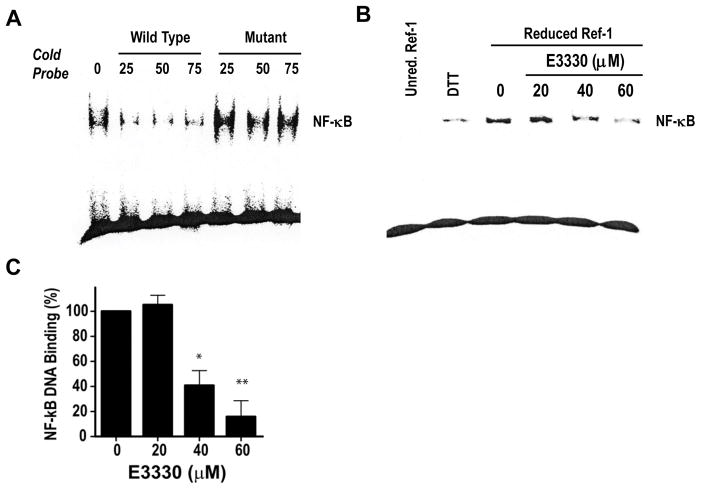
To investigate the activity of Ref-1 redox in leukemia T-cells, we performed redox-EMSA assays in TAIL7 cells, assessing the DNA binding of the transcription factor NF-κB, a known redox target of Ref-1 and an important transcriptional effector in T-cell ALL (23, 24, 40). First, we performed a competitive EMSA experiment using excess cold probe (WT) to block NF-κB DNA binding or a NF-κB binding-defective probe (mutant), as control. Excess WT competitor probe resulted in marked inhibition of the NF-κB binding to DNA in the presence of reduced Ref-1 protein (over 90%), which was not observed using the mutated, binding-defective probe (Figure 2A). Second, we performed EMSA assays for NF-κB binding in the presence of the Ref-1 inhibitor E3330, which has been shown to inhibit selectively the redox activity of Ref-1, without affecting its DNA repair function (7). As shown in Figure 2B, incubation of reduced Ref-1 with increasing amounts of E3330 resulted in a dose-dependent inhibition of binding to NF-κB consensus sequence, in nuclear extracts from leukemia TAIL7 cells. Quantification showed that E3330 inhibits NF-κB binding with an IC50 of 37.9 ± 2.6μM (Figure 2C). Overall, these studies demonstrate that the Ref-1 redox function is preserved in leukemia T-cells, controlling the binding of NF-κB to DNA target sequences.
Figure 2. Ref-1 redox controls NF-κB DNA binding in leukemia T-cells.
A) Competitive EMSA for NF-κB using nuclear extracts from TAIL7 cells in the presence of WT or NF-κB-binding-defective cold probe (25-, 50- or 75-fold). B) NF-κB redox-EMSA in extracts from TAIL7 cells treated with increasing doses of E3330. DTT final dose was 0.02mM. C). Densitometry quantification of effect of E3330 on NF-κB DNA binding in TAIL7 cells; data shown as mean ± SEM from two independent experiments. *p<0.05, **p<0.01, using T-test.
Subsequently, we performed experiments to analyze the potential regulation of Ref-1 protein expression in leukemia T-cells. The majority of T-ALL patients exhibit genomic alterations activating the Notch pathway, and multiple studies showed that Notch signaling an important role in the biology of primary T-ALL cells (18, 19). To investigate whether increased Notch signaling impacts on Ref-1 expression, we performed immunoblotting in a TAIL7 cell subline transduced to overexpress an activated form of Notch1 (TAIL7-ICN; exhibits increased Notch activation and expression of Notch target genes; Batista A, Cardoso AA, unpublished data) in comparison to parental TAIL7 cells. We observed that increased Notch signaling does not affect the levels of Ref-1 protein in leukemia T-cells (Figure 3A), and no changes were observed in Ref-1 transcript levels. In addition, analysis of a public dataset assessing transcriptome changes in T-ALL cell lines following blockade of Notch signaling via gamma-secretase inhibition (compound E), showed no effects on Ref-1/APEX1 mRNA relative expression (Supplementary Figure 3A; data collected from Palomero et al. using Oncomine; ref. (41)).
Figure 3. Ref-1 expression in leukemia cells is not influenced by Notch signaling, but is upregulated by Dexamethasone.
A) Immunoblotting of Ref-1 expression in leukemia TAIL7 cells and the TAIL7-ICN subline, which overexpresses ICN and exhibit increased expression of Notch target genes; three independent replicates for each condition. Actin was used as loading control. B) Immunoblotting of Ref-1 expression in TAIL7 cells treated with Dexamethasone (200nM) for the periods indicated; TAIL7-DexaR is a subline that is resistant to high-dose Dexamethasone, three independent replicates for each condition, Actin was used as loading control. C) Relative expression of Ref-1 protein TAIL7 cells treated with Dexamethasone (200nM) for the periods indicated, by immunoblotting; determined by densitometry, and normalized to Actin expression levels. Data shown as mean ± SEM. *p<0.05, ** p<0.01 Mann-Whitney test. D) Intracellular staining with Ref-1 Ab in TAIL7 cells treated with increasing doses of Dexamethasone (125, 250, 500nM); at least 8,000 events were acquired by flow cytometry and analyzed using FlowJo. Ref-1 histograms are depicted in pink (125nM), orange (250nM) and red (500nM), and light gray histogram indicates isotype negative control.
Refractory, relapsed leukemia remains a significant problem in T-ALL, and resistance to glucocorticoids is a predictor of poor prognosis in pediatric ALL, including T-ALL (42). Therefore, we investigated whether glucocorticoid exposure affected Ref-1 expression. Dexamethasone treatment of TAIL7 cells resulted in upregulation of Ref-1 protein, which was observed as early as 6 hrs, remaining elevated at 7 days post-treatment (Figure 3B–C). Flow cytometry analysis of intracellular Ref-1 following stimulation with Dexamethasone confirmed upregulation of Ref-1 protein expression in factor-dependent TAIL7 cells, in a dose-dependent manner (Figure 3D). qPCR analysis showed that Dexamethasone treatment does affect the levels of Ref-1 transcripts in leukemia cells, but only at the 48 hour time point, and the level of elevated expression, while statistically significant is only a 0.25 fold increase and not of major biological relevance (Supplementary Figure 3B), suggesting that the effect of glucocorticoids on Ref-1 expression involves more of a protein increase or stabilization of Ref-1 rather than a transcriptional regulation mechanism. Overall, these studies suggest that Ref-1 expression in leukemia T-cells is not regulated by the Notch signaling, but protein levels can be modulated by exposure to glucocorticoids.
Selective blockade of Ref-1 redox potently inhibit leukemia T-cells, including drug-resistant, relapsed T-ALL cells
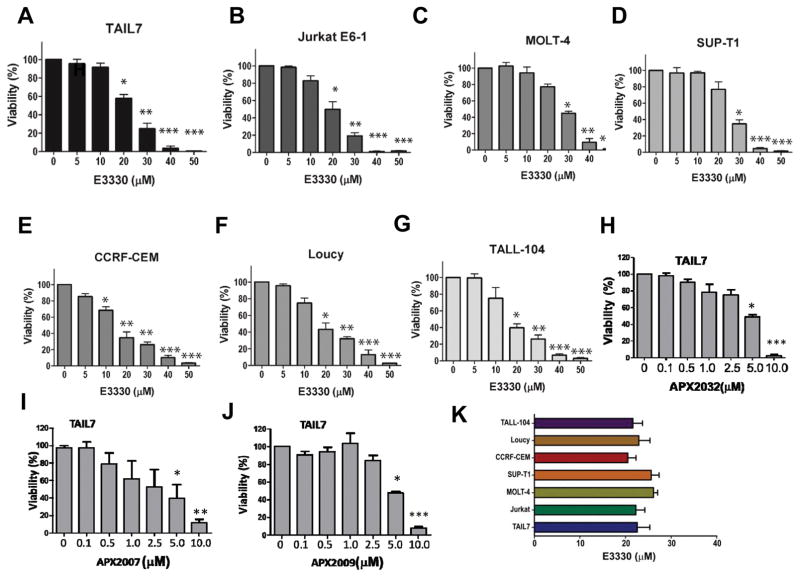
Functional studies were performed to determine the impact of Ref-1 blockade in leukemia T-cells, by using E3330, a highly selective compound that recognizes a redox-active Ref-1 protein (11, 12, 43). First, we evaluated the effect of E3330 on leukemia cell viability, using the T-ALL cell lines described above (Supplementary Table 1). Treatment with E3330 resulted in potent, dose dependent inhibition of the survival of TAIL7 cells (Figure 4A), with an IC50 of around 22.5 μM (Figure 4K). Importantly, as shown in Figure 4B–G, E3330 showed comparable inhibitory effects in leukemia cell lines representing distinct genotypes, including high-risk ETP leukemia (as LOUCY), or derived from patients in leukemia relapse. Although some variability was seen between experiments, the IC50s calculated for the E3330 activity in the different T-ALL lines were within a limited dose range (from 20.5 to 26.1μM; Figure 4K), which correspond to clinically achievable concentrations (5). We also evaluated the effect of second generation Ref-1 inhibitors APX2007, APX2009 and APX2032 on leukemia cell viability on TAIL7 cell (Figure 4H–J). The IC50s ranged from 2.5 to 5.0 μM for the three compounds and is consistent with our evaluations of these agents, particularly APX2009, demonstrating 5–10 increased efficacy (33).
Figure 4. Blockade of Ref-1 redox by E3330 or second generation Ref-1 inhibitors markedly inhibits human T-ALL cell lines.
A) ATP-based viability assay using cytokine-dependent TAIL7 cells, in the presence of increasing doses of the Ref-1 redox inhibitor E3330 (5 to 50μM); TAIL7 cells were cultured in the presence of IL-7 (10ng/mL), and analyses performed at 96hrs. Data shown as mean ± SEM, from 9 independent experiments. *p<0.05, **p<0.01, ***p<0.001, using T-test. B-G) ATP viability assessing effect of E3330 on various human leukemia T-ALL lines, representing different genotypes and risk groups. Data shown as mean ± SEM, from respectively 5 (Sup-T1), 4 (Jurkat, MOLT-4, CCRF-CEM, Loucy) or 2 (TALL-104) independent experiments. H-J) ATP viability assessing effect of second generation Ref-1 inhibitors on TAIL7 cell line. Data shown as mean ± SEM, from four independent experiments *p<0.05, **p<0.01, ***p<0.001, using T-test. K) IC50 graph of E3330 in cell lines tested.
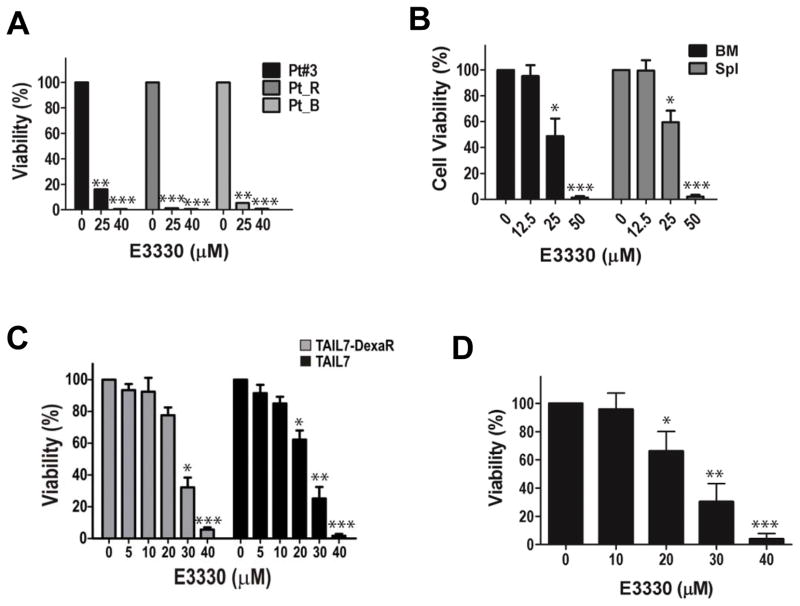
We next evaluated the activity of Ref-1 redox blockade by E3330 in primary cells from leukemia patients, using the ATP assay. As seen in Figure 5A, E3330 markedly inhibited leukemia T-cell viability in all cases tested, including a specimen from a T-ALL relapse patient (Pt_B). We then investigated the efficacy of E3330 in leukemia cells obtained from mice exhibiting terminal ICN-induced T-cell leukemia, as described above (38, 39). Cells were isolated from the BM and spleen of mice with terminal leukemia, which is characterized by massive splenomegaly, hepatomegaly, and high leukemia cell content in these organs. As shown in Figure 5B, murine leukemia T-cells were sensitive to E3330 treatment, with significant inhibition of cell survival even in the presence of stimulatory cytokines (IL-7, SCF, FLT3L). Since Ref-1 has been associated with drug resistance (6) and we observed that glucocorticoid exposure upregulates Ref-1 expression, we investigated the activity of Ref-1 redox blockade by E3330 in leukemia TAIL7 cells selected for resistance to high-dose Dexamethasone (TAIL7-DexaR), which express increased levels of Ref-1 protein (Figure 3B; Supplementary Figure 3C). Inhibition of Ref-1 redox resulted in marked inhibition of TAIL7-DexaR cells (Figure 5C), albeit with small reduction in efficacy in comparison to parental TAIL7 cells (which are moderately resistant to Dexamethasone). Additionally, we investigated the potency of E3330 in inhibiting leukemia T-cells from a xenograft model of leukemia relapse post-chemotherapy (Figure 5D). In this model, mice transplanted with TAIL7 cells were treated with Vincristine at evidence of circulating leukemia, which resulted in increased overall survival (Supplementary Data Figure 3D; p<0.001); however, following disease remission, leukemia recurrence is observed in all animals, which developed terminal leukemia around d57 to d61, thus recapitulating human T-cell ALL relapse. As shown in Supplementary Data Figure 3D leukemia T-cells harvested from mice in leukemia relapse post-Vincristine therapy retained their sensitivity to Ref-1 redox blockade, as shown by their marked inhibition by E3330 treatment(Figure 5D). These studies demonstrate that leukemia TAIL7 cells resistant to frontline drugs in in vivo therapeutic studies retain their sensitivity to Ref-1 redox inhibition, and support the use of animal models of chemotherapy for the pre-clinical evaluation of more potent, selective inhibitors of Ref-1 redox activity.
Figure 5. E3330 is a potent inhibitor of leukemia T-cells, including refractory, relapsed T-ALL.
A) Effect of E3330 on primary leukemia cells from T-ALL patients, using the ATP assay. Cells were cultured in the presence of IL-7 (10ng/mL) and IL-9 (10ng/mL), and analyses performed at 96hrs. *p<0.05, **p<0.01, ***p<0.001, using T-test. B) Effect of E3330 on BM or spleen (Spl) leukemia cells from mice carrying Notch-induced T-ALL, using the ATP assay. Cells were cultured with IL-7, SCF, and FLT3L, and analyses performed at 96h. Data shown as mean ± SD, n=6; *p<0.05, ***p<0.001 for BM and Spl, using 1-way ANOVA. C) ATP viability assay assessing effect of E3330 on TAIL7 cells resistant to high-dose Dexamethasone (TAIL7-DexaR) in comparison to parental TAIL7 cells; cells were cultured with IL-7, and analyzed at 96 hrs. Data shown as mean ± SEM, from three independent experiments. *p<0.05, **p<0.01, ***p<0.001, using T-test D) Cell viability assessing the effect of E3330 on leukemia cells from xenograft mice exhibiting relapse T-ALL, following remission induced by Vincristine therapy. Cells were cultured with IL-7, and analyzed at 96hrs using the ATP assay. Data shown as mean ± SD, n=4; *p<0.05, **p<0.01, ***p<0.001, using 1-way ANOVA.
Blockade of Ref-1 redox function promotes leukemia cell apoptosis
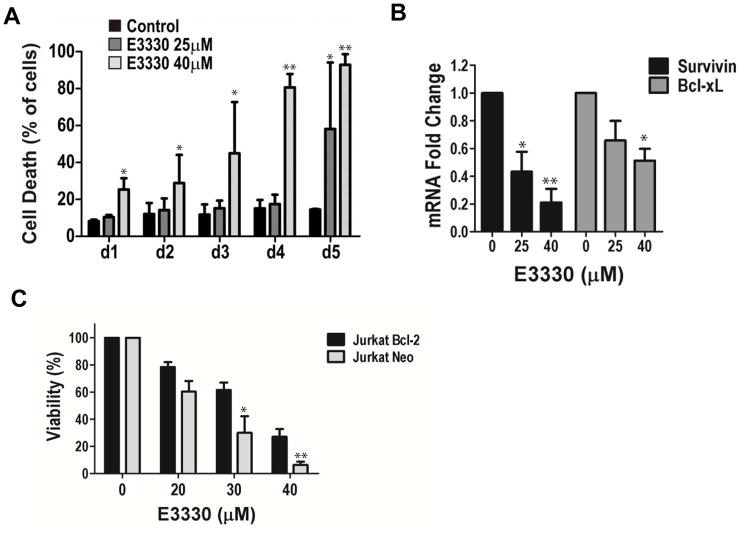
To define the mechanism by which Ref-1 blockade exerts its inhibitory effects on leukemia T-cells, we investigated the impact of E3330 treatment on leukemia cell viability. Using TAIL7 cells as a model, cell death was assessed using the Annexin V/PI apoptosis assay. As shown in Figure 6A, E3330 treatment result in marked increase in leukemia cell death, in a dose dependent manner, with massive apoptosis at d4 and d5 for 40μM dose. Next, we evaluated the effect of E3330 treatment on the expression of the pro-survival genes Bcl-xL and Survivin/BIRC5, which have been implicated in ALL biology and are known transcriptional targets of Ref-1-regulated TFs. We observed that treatment with E3330 results in significant inhibition in the expression of Survivin and Bcl-xL (Figure 6B; p<0.0001 and p=0.0001, respectively). Finally, since Bcl-2 functions as a survival effector in ALL cells (29, 44) and its transcription is also regulated by Ref-1-sensitive TFs as NF-κB (45), we evaluated whether Bcl-2 overexpression impacted on the inhibitory effects of E3330 in leukemia T-cells. Here, Jurkat/Bcl-2 cells transfected with a psFFV-neo expressing vector containing human Bcl-2 were compared to control Jurkat/Neo cells containing the empty vector; these two sublines expressed comparable levels of Ref-1 protein. As shown in Figure 6C, Jurkat/Bcl2 cells are partially rescued from the inhibitory effects of E3330 in comparison to the control Jurkat/Neo cells (p<0.01). Taken together, these studies show that blockade of Ref-1 redox triggers apoptosis of leukemia T-cells, which is associated with downregulation the transcription of the pro-survival genes Bcl-xL and Survivin, and that overexpression of Bcl-2 partially protects leukemia T-cells from the inhibitory effects of E3330.
Figure 6. Ref-1 redox signaling inhibition by E3330 promotes apoptosis of leukemia T-cells, inhibiting expression of anti-apoptotic genes.
A) Cell death as defined by the Annexin V/PI assay in leukemia TAIL7 cells treated with E3330 at the doses indicated. Data is shown as mean ± SD. *p<0.05, **p<0.01, using T-test. B) qPCR for mRNA expression of Survivin/BIRC5, Bcl-xL in TAIL7 cells treated with E3330, at the doses indicated. GAPDH was used as endogenous control, and the assays performed using TaqMan probes. Data shown as mean ± SEM, n=4; *p<0.05, **p<0.01, using 1-way ANOVA. C) ATP viability assay with E3330 in Jurkat cells overexpressing Bcl-2 (Jurkat/Bcl2) in comparison to control, vector-expressing cells (Jurkat/Neo); analyses at 96hrs. Data shown as mean ± SEM, from 4 independent experiments; *p<0.05, **p<0.01, for Jurkat/Bcl2 vs. Jurkat/Neo, using 2-way ANOVA.
DISCUSSION
The pursuit for potential multi-pathway regulators in pediatric T-cell ALL led us to the identification of Ref-1 redox as a novel molecular effector in this malignancy, including for high-risk and refractory, relapsed disease. Here we demonstrate that leukemia T-cells express functional Ref-1 protein, which regulates through its redox activity the binding of TFs to promoters (e.g. NF-κB), and the transcription of critical target genes (as Survivin and Bcl-xL). Blockade of Ref-1 signaling activity by E3330 results in potent inhibition of proliferation and survival of T-ALL cells, by promoting leukemia cell apoptosis and downregulation of pro-survival genes. To the best of our knowledge, this represents the first demonstration that Ref-1 redox plays a significant functional role in T-cell ALL, supporting Ref-1 as a potential new target for this malignancy.
Ref-1 functions as a redox signaling factor as well as having endonuclease activity repairing DNA lesions induced by oxidative and alkylating agents in the DNA base excision repair (BER) pathway. In its redox signaling function, Ref-1 exerts a selective transcriptional control on a restricted number of TFs containing cysteine residues in their DNA-binding domains, which are reduced in a specific manner, requiring the Cys-65 residue of human Ref-1, along with Cys-93 and Cys-99 (4, 11, 12). Thus, selective blockade of Ref-1 redox function represents a strategy to target critical leukemia-associated TFs and their downstream transcriptional targets, including pro-survival, mitogenic and cell cycle progression effectors. In particular, Ref-1 exerts redox control of: i) NF-κB, which is activated by Notch signaling in T-ALL, and regulates the transcription of anti-apoptotic genes (23, 24); ii) p53, which has been associated with the pro-apoptotic activity of signaling antagonists effectively inhibiting T-ALL cells (22, 25), and; iii) AP-1, which is constitutively activated in leukemia cells from patients with HTLV-I induced adult T-cell leukemia (46). Also, we have determined that Ref-1 redox-regulated TF STAT3 is essential for T-cell leukemogenesis, and that disruption of STAT3 potently inhibits T-ALL cell survival (Ding, Cardoso; manuscript in preparation). Therefore, blockade of Ref-1 redox in T-ALL cells seems to impact on multiple pathways playing important roles in leukemia cell biology, permitting more robust anti-leukemia responses and potentially reducing the risk of selection of resistant, non-responsive tumor variants.
General reduction-oxidation systems that help maintain intracellular homeostasis by scavenging reactive oxygen species (ROS) (47, 48) may affect transcriptional programs by globally reducing TFs, in a non-selective manner. Changes in redox biology are often observed in cancer, and studies showed that oncogenic events can influence redox homeostasis, altering the expression/activity of anti-oxidant pathways (49, 50). Thus, therapeutic approaches have been proposed to disrupt the oxidative state of cancer cells and to increase ROS levels towards promoting cell death. Several frontline therapeutic agents that produce ROS are used in the clinic, including chemotherapy agents employed in ALL treatment (as Vincristine, anthracyclines, cytarabine) and targeted molecular antagonists (as HDAC inhibitors, proteasome inhibitors). However, the cells’ oxidative state is dictated by a delicate balance between pro- and anti-oxidant pathways, and the effect of ROS-producing agents on cancer cells is influenced by anti-oxidant enzyme systems (as HMOX1, GSH/glutathione, thioredoxin/Trx, peroxiredoxin, SOD/superoxide dismutase), which can affect therapeutic efficacy and drug resistance (49). Multiple studies showed that leukemia cells exhibit altered redox biology, with alterations in the expression and activity of ROS and anti-oxidant effectors, and these systems have been implicated in leukemogenesis. In ALL, it has been reported that the levels of GSH, Trx and peroxiredoxin are increased, whereas SOD is reduced (26, 51–53). In particular, thioredoxin is increased in T-ALL specimens, namely in cases with high WBC counts, and it has been suggested that selective inhibition of Trx may represent an effective strategy to overcome drug resistance and to sensitize leukemia T-cells to conventional chemotherapy drugs (26). Interestingly, it has been shown that Trx can reduce and thereby activate Ref-1 (54), which could increase the transcriptional activity of Ref-1-controlled TFs. Targeting of general oxidative systems such as Trx poses challenges of target selectivity, and no strategies specifically disrupting general redox systems have translated yet into more efficient therapies for ALL. Our approach is dramatically different, targeting specific protein-protein redox signaling and effectively blocking TF function.
E3330 is a small molecule that was initially described as an inhibitor of NF-κB activation (55), and then correctly identified as a specific inhibitor of Ref-1 redox function that blocked NF-κB activation (43). E3330 recognizes a redox active conformation of Ref-1 and inhibiting its redox activity (11). Although multiple signaling pathways are modulated by Ref-1 redox function, no unacceptable toxicity has been observed in in vivo preclinical studies using E3330 (Ref. (5), and FDA IND submission), as well as in human clinical trials for a non-cancer indication performed previously by Eisai Co.
Little is known on the stimuli and molecular mechanisms regulating the expression of Ref-1, or regulating its redox activity. Ref-1 regulates the expression of cytokines, such as IL-6, IL-8, IL-12 and other inflammatory mediators that are downstream targets of TFs under the redox control of Ref-1 (56, 57). However, it is not known whether cytokines can influence the expression of Ref-1 protein in tumor cells. We have found that Ref-1 levels are not affected by IL-7 stimulation in leukemia cells, or by IL-6 treatment of pancreatic cancer cells (10, 58). As shown in this study, Ref-1 expression in leukemia cells does not seem to be directly regulated by Notch signaling, and it remains to be determined which mechanism(s) mediate the increased levels of Ref-1 transcripts seen in T-ALL specimens. Increased expression of Ref-1 has been associated with drug resistance (6), and it has been reported that patients with high Ref-1 protein levels show increased resistance to cisplatin treatment and other chemotherapeutic agents (59). Here, we show that Ref-1 protein levels are upregulated by glucocorticoid treatment. It has been reported that Ref-1 protein can be regulated by MDM2 and there is evidence of a molecular link between glucocorticoid receptor signaling and MDM2 activity (60); future studies are necessary to determine whether the effect of glucocorticoids seen in T-ALL cells involves MDM2-mediated regulation of Ref-1 ubiquitination and degradation. Finally, it has been reported that the redox activity of Ref-1 is augmented by its phosphorylation by the casein kinase CK2, enhancing the DNA binding of AP-1 to target genes (61). This is noteworthy as we have shown that CK2 activity is increased in T-ALL, and selective blockade of CK2 results in inhibition of leukemia cell survival (62). Although these effects have been related to CK2 activity in the regulation of the PTEN/PI3K axis, it would be interesting to assess in future studies whether the inhibitory effects of CK2 blockade in T-ALL are mediated through inhibition of Ref-1 redox function.
In summary, we demonstrate that the redox factor Ref-1 plays an important regulatory role in the biology of T-cell ALL. This represents the first approach targeting T-ALL cells by the selective blockade of a key regulator of redox-controlled transcription programs, affecting multiple signaling pathways. This work supports Ref-1 redox function as a novel molecular target and “node” in leukemia T-cells, and the potential translation into clinical trials for refractory, relapsed T-ALL using high-affinity Ref-1 redox inhibitors. Initial steps in this direction are being taken with the beginning of a Phase 1 trial using E3330 (APX3330) in human cancer patients.
Supplementary Material
Acknowledgments
Financial support for this work was provided by the National Cancer Institute [CA122298 (M.L. Fishel), CA138798 (M.L. Fishel and M.R. Kelley), CA205166 (M.R. Kelley), and the National Institutes of Health, [R21NS091667 (M.R. Kelley)]. Additional financial support was provided by Ralph W. and Grace M. Showalter Research Trust Fund (M.L. Fishel), the Earl and Betty Herr Professor in Pediatric Oncology Research, Hyundai Hope on Wheels, Jeff Gordon Children’s Foundation and the Riley Children’s Foundation (M.R. Kelley).
Footnotes
Disclosure of Potential Conflict of Interest: Mark R. Kelley has licensed E3330 (APX3330) through Indiana University Research and Technology Corporation to Apexian Pharmaceuticals. Angelo Cardeso was employed by Apexian Pharmaceuticals at one point in the past. Apexian Pharmaceuticals had neither control nor oversight of the studies or the study design, results or interpretation, or presentation of the data in this manuscript. They did not have to approve the manuscript in any way prior to its submission.
References
- 1.Faivre S, Djelloul S, Raymond E. New paradigms in anticancer therapy: targeting multiple signaling pathways with kinase inhibitors. Semin Oncol. 2006;33:407–20. doi: 10.1053/j.seminoncol.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10:130–7. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelley MR, Georgiadis MM, Fishel ML. APE1/Ref-1 role in redox signaling: translational applications of targeting the redox function of the DNA repair/redox protein APE1/Ref-1. Curr Mol Pharmacol. 2012;5:36–53. doi: 10.2174/1874467211205010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhakat KK, Mantha AK, Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxidants & redox signaling. 2009;11:621–38. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fishel ML, Jiang Y, Rajeshkumar NV, Scandura G, Sinn AL, He Y, et al. Impact of APE1/Ref-1 redox inhibition on pancreatic tumor growth. Mol Cancer Ther. 2011;10:1698–708. doi: 10.1158/1535-7163.MCT-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The Many Functions of APE1/Ref-1: Not Only a DNA Repair Enzyme. Antioxidants & redox signaling. 2009;11:601–20. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo M, Delaplane S, Jiang A, Reed A, He Y, Fishel M, et al. Role of the multifunctional DNA repair and redox signaling protein Ape1/Ref-1 in cancer and endothelial cells: small-molecule inhibition of the redox function of Ape1. Antioxidants & redox signaling. 2008;10:1853–67. doi: 10.1089/ars.2008.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med. 2007;28:375–95. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Luo M, He H, Kelley MR, Georgiadis MM. Redox regulation of DNA repair: implications for human health and cancer therapeutic development. Antioxid Redox Signal. 2010;12:1247–69. doi: 10.1089/ars.2009.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardoso AA, Jiang Y, Luo M, Reed AM, Shahda S, He Y, et al. APE1/Ref-1 regulates STAT3 transcriptional activity and APE1/Ref-1-STAT3 dual-targeting effectively inhibits pancreatic cancer cell survival. PloS one. 2012;7:e47462. doi: 10.1371/journal.pone.0047462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su D, Delaplane S, Luo M, Rempel DL, Vu B, Kelley MR, et al. Interactions of Apurinic/Apyrimidinic Endonuclease with a Redox Inhibitor: Evidence for an Alternate Conformation of the Enzyme. Biochemistry. 2011;50:82–92. doi: 10.1021/bi101248s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo M, Zhang J, He H, Su D, Chen Q, Gross ML, et al. Characterization of the redox activity and disulfide bond formation in apurinic/apyrimidinic endonuclease. Biochemistry. 2012;51:695–705. doi: 10.1021/bi201034z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conter V, Arico M, Basso G, Biondi A, Barisone E, Messina C, et al. Long-term results of the Italian Association of Pediatric Hematology and Oncology (AIEOP) Studies 82, 87, 88, 91 and 95 for childhood acute lymphoblastic leukemia. Leukemia. 2010;24:255–64. doi: 10.1038/leu.2009.250. [DOI] [PubMed] [Google Scholar]
- 14.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011;29:551–65. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pui CH, Pei D, Sandlund JT, Ribeiro RC, Rubnitz JE, Raimondi SC, et al. Long-term results of St Jude Total Therapy Studies 11, 12, 13A, 13B, and 14 for childhood acute lymphoblastic leukemia. Leukemia. 2010;24:371–82. doi: 10.1038/leu.2009.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunger SP, Lu X, Devidas M, Camitta BM, Gaynon PS, Winick NJ, et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children’s oncology group. J Clin Oncol. 2012;30:1663–9. doi: 10.1200/JCO.2011.37.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 18.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6:347–59. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 20.Ferrando A. NOTCH mutations as prognostic markers in T-ALL. Leukemia. 2010;24:2003–4. doi: 10.1038/leu.2010.237. [DOI] [PubMed] [Google Scholar]
- 21.Barata JT, Silva A, Brandao JG, Nadler LM, Cardoso AA, Boussiotis VA. Activation of PI3K is indispensable for interleukin 7-mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells. J Exp Med. 2004;200:659–69. doi: 10.1084/jem.20040789. (AAC and VAB contributed equally for the supervision of this work) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batista A, Barata JT, Raderschall E, Sallan SE, Carlesso N, Nadler LM, et al. Targeting of active mTOR inhibits primary leukemia T cells and synergizes with cytotoxic drugs and signaling inhibitors. Exp Hematol. 2011;39:457–72. e3. doi: 10.1016/j.exphem.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Espinosa L, Cathelin S, D’Altri T, Trimarchi T, Statnikov A, Guiu J, et al. The Notch/Hes1 pathway sustains NF-kappaB activation through CYLD repression in T cell leukemia. Cancer cell. 2010;18:268–81. doi: 10.1016/j.ccr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vilimas T, Mascarenhas J, Palomero T, Mandal M, Buonamici S, Meng F, et al. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat Med. 2007;13:70–7. doi: 10.1038/nm1524. [DOI] [PubMed] [Google Scholar]
- 25.Khandanpour C, Phelan JD, Vassen L, Schutte J, Chen R, Horman SR, et al. Growth factor independence 1 antagonizes a p53-induced DNA damage response pathway in lymphoblastic leukemia. Cancer Cell. 2013;23:200–14. doi: 10.1016/j.ccr.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao L, Diccianni MB, Tanaka T, Gribi R, Yu AL, Pullen JD, et al. Thioredoxin expression in primary T-cell acute lymphoblastic leukemia and its therapeutic implication. Cancer Res. 2001;61:7333–8. [PubMed] [Google Scholar]
- 27.Barata JT, Boussiotis VA, Yunes JA, Ferrando AA, Moreau LA, Veiga JP, et al. IL-7-dependent human leukemia T-cell line as a valuable tool for drug discovery in T-ALL. Blood. 2004;103:1891–900. doi: 10.1182/blood-2002-12-3861. [DOI] [PubMed] [Google Scholar]
- 28.Stier S, Cheng T, Dombkowski D, Carlesso N, Scadden DT. Notch1 activation increases hematopoietic stem cell self-renewal in vivo and favors lymphoid over myeloid lineage outcome. Blood. 2002;99:2369–78. doi: 10.1182/blood.v99.7.2369. [DOI] [PubMed] [Google Scholar]
- 29.Veiga JP, Costa LF, Sallan SE, Nadler LM, Cardoso AA. Leukemia-stimulated bone marrow endothelium promotes leukemia cell survival. Exp Hematol. 2006;34:610–21. doi: 10.1016/j.exphem.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nyland RL, Luo M, Kelley MR, Borch RF. Design and Synthesis of Novel Quinone Inhibitors Targeted to the Redox Function of Apurinic/Apyrimidinic Endonuclease 1/Redox Enhancing Factor-1 (Ape1/Ref-1) Journal of medicinal chemistry. 2010;53:1200–10. doi: 10.1021/jm9014857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley MR, Wikel JH, Guo C, Pollok KE, Bailey BJ, Wireman R, et al. Identification and Characterization of new chemical entities targeting Apurinic/Apyrimidinic Endonuclease 1 for the prevention of chemotherapy-induced peripheral neuropathy (CIPN) The Journal of pharmacology and experimental therapeutics. 2016;359:300–9. doi: 10.1124/jpet.116.235283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Georgiadis MM, Luo M, Gaur RK, Delaplane S, Li X, Kelley MR. Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat Res. 2008;643:54–63. doi: 10.1016/j.mrfmmm.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maia S, Pelletier M, Ding J, Hsu YM, Sallan SE, Rao SP, et al. Aberrant expression of functional BAFF-system receptors by malignant B-cell precursors impacts leukemia cell survival. PLoS One. 2011;6:e20787. doi: 10.1371/journal.pone.0020787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersson A, Ritz C, Lindgren D, Eden P, Lassen C, Heldrup J, et al. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia. 2007;21:1198–203. doi: 10.1038/sj.leu.2404688. [DOI] [PubMed] [Google Scholar]
- 37.Holleman A, Cheok MH, den Boer ML, Yang W, Veerman AJ, Kazemier KM, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351:533–42. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 38.Allman D, Karnell FG, Punt JA, Bakkour S, Xu L, Myung P, et al. Separation of Notch1 promoted lineage commitment and expansion/transformation in developing T cells. J Exp Med. 2001;194:99–106. doi: 10.1084/jem.194.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–91. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staal FJ, Langerak AW. Signaling pathways involved in the development of T-cell acute lymphoblastic leukemia. Haematologica. 2008;93:493–7. doi: 10.3324/haematol.12917. [DOI] [PubMed] [Google Scholar]
- 41.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–10. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liem NL, Papa RA, Milross CG, Schmid MA, Tajbakhsh M, Choi S, et al. Characterization of childhood acute lymphoblastic leukemia xenograft models for the preclinical evaluation of new therapies. Blood. 2004;103:3905–14. doi: 10.1182/blood-2003-08-2911. [DOI] [PubMed] [Google Scholar]
- 43.Shimizu N, Sugimoto K, Tang J, Nishi T, Sato I, Hiramoto M, et al. High-performance affinity beads for identifying drug receptors. Nat Biotechnol. 2000;18:877–81. doi: 10.1038/78496. [DOI] [PubMed] [Google Scholar]
- 44.Coustan-Smith E, Kitanaka A, Pui CH, McNinch L, Evans WE, Raimondi SC, et al. Clinical relevance of BCL-2 overexpression in childhood acute lymphoblastic leukemia. Blood. 1996;87:1140–6. [PubMed] [Google Scholar]
- 45.Feuillard J, Schuhmacher M, Kohanna S, Asso-Bonnet M, Ledeur F, Joubert-Caron R, et al. Inducible loss of NF-kappaB activity is associated with apoptosis and Bcl-2 down-regulation in Epstein-Barr virus-transformed B lymphocytes. Blood. 2000;95:2068–75. [PubMed] [Google Scholar]
- 46.Mori N, Fujii M, Iwai K, Ikeda S, Yamasaki Y, Hata T, et al. Constitutive activation of transcription factor AP-1 in primary adult T-cell leukemia cells. Blood. 2000;95:3915–21. [PubMed] [Google Scholar]
- 47.Holmgren A. Thioredoxin structure and mechanism: conformational changes on oxidation of the active-site sulfhydryls to a disulfide. Structure. 1995;3:239–43. doi: 10.1016/s0969-2126(01)00153-8. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura H, Nakamura K, Yodoi J. Redox regulation of cellular activation. Annu Rev Immunol. 1997;15:351–69. doi: 10.1146/annurev.immunol.15.1.351. [DOI] [PubMed] [Google Scholar]
- 49.Irwin ME, Rivera-Del Valle N, Chandra J. Redox control of leukemia: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2013;18:1349–83. doi: 10.1089/ars.2011.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat Rev Cancer. 2014;14:709–21. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kearns PR, Pieters R, Rottier MM, Pearson AD, Hall AG. Raised blast glutathione levels are associated with an increased risk of relapse in childhood acute lymphocytic leukemia. Blood. 2001;97:393–8. doi: 10.1182/blood.v97.2.393. [DOI] [PubMed] [Google Scholar]
- 52.Nishiura T, Suzuki K, Kawaguchi T, Nakao H, Kawamura N, Taniguchi M, et al. Elevated serum manganese superoxide dismutase in acute leukemias. Cancer Lett. 1992;62:211–5. doi: 10.1016/0304-3835(92)90098-g. [DOI] [PubMed] [Google Scholar]
- 53.Zhang P, Liu B, Kang SW, Seo MS, Rhee SG, Obeid LM. Thioredoxin peroxidase is a novel inhibitor of apoptosis with a mechanism distinct from that of Bcl-2. J Biol Chem. 1997;272:30615–8. doi: 10.1074/jbc.272.49.30615. [DOI] [PubMed] [Google Scholar]
- 54.Hirota K, Matsui M, Iwata S, Nishiyama A, Mori K, Yodoi J. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc Natl Acad Sci U S A. 1997;94:3633–8. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goto M, Yamada K, Katayama K, Tanaka I. Inhibitory effect of E3330, a novel quinone derivative able to suppress tumor necrosis factor-alpha generation, on activation of nuclear factor-kappa B. Mol Pharmacol. 1996;49:860–73. [PubMed] [Google Scholar]
- 56.Jedinak A, Dudhgaonkar S, Kelley MR, Sliva D. Apurinic/Apyrimidinic endonuclease 1 regulates inflammatory response in macrophages. Anticancer Res. 2011;31:379–85. [PMC free article] [PubMed] [Google Scholar]
- 57.Cesaratto L, Codarin E, Vascotto C, Leonardi A, Kelley MR, Tiribelli C, et al. Specific inhibition of the redox activity of ape1/ref-1 by e3330 blocks tnf-alpha-induced activation of IL-8 production in liver cancer cell lines. PLoS One. 2013;8:e70909. doi: 10.1371/journal.pone.0070909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Logsdon DPGM, Luo M, Shahda S, Jiang Y, Tong Y, Yu Z, Zyromski N, Schipani E, Carta F, Supuran CT, Korc M, Ivan M, Kelley MR, Fishel ML. Regulation of HIF1a under Hypoxia by APE1/Ref-1 Impacts CA9 Expression: Dual-Targeting in Patient-Derived 3D Pancreatic Cancer Models. Molecular cancer therapeutics. 2016;15:2722–32. doi: 10.1158/1535-7163.MCT-16-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang D, Xiang DB, Yang XQ, Chen LS, Li MX, Zhong ZY, et al. APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells. Lung Cancer. 2009;66:298–304. doi: 10.1016/j.lungcan.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 60.Busso CS, Iwakuma T, Izumi T. Ubiquitination of mammalian AP endonuclease (APE1) regulated by the p53-MDM2 signaling pathway. Oncogene. 2009;28:1616–25. doi: 10.1038/onc.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fritz G, Kaina B. Phosphorylation of the DNA repair protein APE/REF-1 by CKII affects redox regulation of AP-1. Oncogene. 1999;18:1033–40. doi: 10.1038/sj.onc.1202394. [DOI] [PubMed] [Google Scholar]
- 62.Silva A, Yunes JA, Cardoso BA, Martins LR, Jotta PY, Abecasis M, et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J Clin Invest. 2008;118:3762–74. doi: 10.1172/JCI34616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.