Abstract
Background
Total immunoglobulin E (IgE) is a therapeutic target in allergic diseases. DNA methylation in white blood cells (WBCs) was associated with total IgE in an epigenome-wide association study (EWAS) of Caucasians. Whether DNA methylation of eosinophils explains those findings is insufficiently understood.
Methods
We tested for association between genome-wide DNA methylation in WBCs and total IgE in two studies of Hispanic children: the Puerto Rico Genetics of Asthma and Lifestyle Study (PR-GOAL, n = 306) and the Genes-environments and Admixture in Latino Americans (GALA II, n = 573). Whole-genome methylation of DNA from WBCs was measured using the Illumina Infinium HumanMethylation450 BeadChip. Total IgE was measured using the UniCAP 100 system. In PR-GOAL, WBC types (i.e. neutrophils, eosinophils, basophils, lymphocytes, and monocytes) in peripheral blood were measured using Coulter-Counter techniques. In GALA II, WBC types were imputed. Multivariable linear regression was used for the analysis of DNA methylation and total IgE, which was first conducted separately for each cohort, and then combining results from the two cohorts in a meta-analysis.
Results
CpG sites in multiple genes, including novel findings and results previously reported in Caucasians, were significantly associated with total IgE. However, adjustment for WBC types resulted in markedly fewer significant sites. Top findings from this adjusted meta-analysis were in genes ZFPM1 (P=1.5×10−12), ACOT7 (P=2.5×10−11), and MND1 (P=1.4×10−9).
Conclusions
In an EWAS adjusted for WBC types (including eosinophils), methylation changes in genes enriched in pathways relevant to asthma and immune responses were associated with total IgE among Hispanic children.
Keywords: EWAS, total IgE, Hispanics, children
INTRODUCTION
Allergic diseases affect millions of people worldwide. In subjects with allergic diseases, secretion of interleukin (IL)-4 and IL-13 by T helper 2 (Th2) cells increases production of total immunoglobulin E (IgE) by B cells. Total IgE is a key intermediate phenotype and a therapeutic target in allergic diseases such as asthma and allergic rhinitis1.
Genome-wide association studies have identified SNPs linked with total IgE, but their joint estimated effect accounts for a very small proportion of the trait’s heritability2–5. Environmental exposures affect total IgE6,7, and thus epigenetic mechanisms such as DNA methylation may explain the “missing heritability” of total IgE.
A recent epigenome-wide association study (EWAS) using DNA from white blood cells (WBCs) reported that methylation in several genes is implicated on total IgE level in Caucasians8, with methylation of three genes (LPCAT2, IL5RA, and ZNF22) accounting for ~13% of total IgE variance. Whether those findings are explained by DNA methylation of eosinophils (which play a major role in allergic inflammation) or other WBC is insufficiently understood.
In the United States, Puerto Ricans are heavily affected with allergic diseases, while Mexican Americans have a lower but non-negligible burden from allergies9–11. We report the first EWAS of total IgE with adjustment for (percentage of) WBC types, as well as the first EWAS of total IgE in Hispanics.
METHODS
Please also see the Online Supplement.
Subject recruitment and study procedures
Puerto Rico Genetics Of Asthma and Lifestyle Study (PR-GOAL)
From March of 2009 to June of 2010, 678 children with and without asthma (defined as physician-diagnosed asthma and at least one episode of wheeze in the prior year) were recruited in San Juan (Puerto Rico), as described elsewhere12. Eligibility criteria included age 6 to 14 years and having four Puerto Rican grandparents. Of the 562 subjects who had blood samples with sufficient DNA for genome-wide genotyping, 306 subjects had additional data (including WBC count, total IgE, and a genome-wide study of DNA methylation) and were thus included in this analysis. Total serum IgE was measured using the UniCAP 100 system (Pharmacia & Upjohn, Kalamazoo, MI), and total WBC count and percentages of the five possible cell types (i.e. neutrophils, eosinophils, basophils, lymphocytes, and monocytes) in peripheral blood were measured using Coulter-Counter techniques. There were no significant differences in age, gender, asthma status or total IgE between subjects with available DNA who were (n=306) and were not (n=256) included in the current analysis.
Following bisulfite conversion of DNA from WBCs using the EZ DNA methylation kit (Zymo Research, Irvin CA), whole-genome DNA methylation levels were measured using the HumanMethylation 450K BeadChip system (Illumina, San Diego, CA). In brief, 200 ng of bisulfite-converted DNA was whole-genome amplified for 23 hours, followed by end-point fragmentation. The fragmented DNA was precipitated, denatured, and hybridized to the BeadChips for 18 hours at 48°C. The BeadChips were then washed, and the hybridized primers were extended and labeled prior to scanning the BeadChips using the Illumina iScan system (Illumina, San Diego, CA).
Principal components were calculated from genome-wide SNP data, which were genotyped using the HumanOmni2.5 BeadChip platform [Illumina Inc., San Diego, CA), as previously described12. Genome-wide gene expression in a subset of 121 whole-blood samples was measured at the University of Pittsburgh Genomics and Proteomics Core Laboratories, using the Illumina HumanHT-12 v4 Expression BeadChip, as described in our previous work13.
Written parental consent was obtained for participating children, from whom written assent was also obtained. The study was approved by the Institutional Review Boards of the University of Puerto Rico (San Juan, PR), Brigham and Women’s Hospital (Boston, MA) and the University of Pittsburgh (Pittsburgh, PA).
Genes-environments & Admixture in Latino Americans (GALA II)
The GALA II study is a multicenter case-control study of asthma in 1,858 Latino children and adolescents14. Cases and healthy controls were recruited from five centers throughout the U.S. (Chicago, Bronx, Houston, San Francisco Bay Area, and Puerto Rico). Subjects were eligible if they were 8 to 22 years of age, self-identified all four grandparents as Latino, and had <10 pack-years of smoking history. Asthma was defined as physician-diagnosed asthma and report of symptoms and medication use in the previous two years. Participants were classified into three Latino subgroups (Puerto Rican, Mexican and other Latino), according to the self-reported ancestry of their four grandparents. All participants were genotyped on the Axiom® LAT1 array (World Array 4, Affymetrix, Santa Clara, CA), as described elsewhere15. A subset of 573 participants (219 Puerto Ricans, 269 Mexicans and 85 other Latinos) with data for genome-wide DNA methylation and total IgE were included in the current analysis; there were no significant differences in age, gender, asthma status or total IgE between subjects who were (n=573) and were not (n=1,285) included in the analysis. Total serum IgE was measured using the ImmunoCAP™ 100 system (Phadia, Kalamazooo, MI)4. WBC DNA was bisulfite-converted and hybridized with Infinium HumanMethylation 450K BeadChips to measure methylation levels.
The study was approved by the Institutional Review Boards of the University of California at San Francisco and all other participating centers. All children and their parents provided written informed assent and consent, respectively.
Preprocessing and quality control for 450K DNA methylation data
We performed the same preprocessing and quality control for 450K DNA methylation data in PR-GOAL and GALA II. We read methylation data from the raw IDAT files using the R package methylumi, and calculated the β-value for each CpG as β = M/(M + U + α), where M and U represent methylated and un-methylated signal intensities at the specific site and α is an arbitrary offset (usually 100) intended to stabilize β-values where fluorescent intensities are low. Next, the methylation data were preprocessed by color balance adjustment, background correction and quantile-normalization with the lumi R package, and followed by Beta-Mixture Quantile (BMIQ) normalization with the wateRmelon package. This normalization strategy first quantile-normalized the intensities of methylation signals among all arrays, and then used BMIQ dilation to normalize the β-values within each array16–18.
In each data set, we removed poor-quality probes with detection P-value > 0.01 in at least 20% of the samples. We also removed 11,656 probes with CpG loci located on sex chromosomes and 19,344 probes that had rs* names or located at 0 distance to known SNPs, according to the Illumina product annotation of Infinium HD Methylation SNP List. This QC process left 454,552 CpGs in both data sets. We then filtered CpG sites with an overall mean β-value > 0.9 or < 0.1 (considering extreme values as noise signals and to reduce multiple tests), which left 188,368 CpGs for data analysis.
Comparison to previous findings in Caucasians (see also Online Supplement)
We compared our results in Hispanic children to those of Liang et al. in Caucasians8. The comparison sample consists of three cohorts of Caucasians (Table E1), as follows: (1) The Medical Research Council Asthma (MRCA) panel, a cohort of 355 subjects (183 male) with mean age of 28 years (range 2–61 years), of whom 175 had doctor-diagnosed asthma, (2) a subset of the Poblogaeth Asthma Prifysgol Abertawe (PAPA) study, including 149 subjects (77 male, 34 with doctor-diagnosed asthma) with mean age of 21 years (range, 18 to 30 years), selected equally from the top and bottom deciles of the total IgE distribution from the entire PAPA study of 1,614 unselected volunteers, and (3) a subset of the Saguenay-Lac-Saint-Jean (SLSJ) asthma familial collection, including 160 subjects (80 male) with mean age of 29 years (range 5–79 years), of whom 69 had asthma. MRCA used the Illumina HumanMethylation 27K chip, while PAPA and SLSJ used the Illumina HumanMethylation 450K chip. We used the results presented in the original report, more detailed information can be found in Liang et al.8. Of note, we could not directly compare effect sizes across our study and those studies (which used transformed methylation values), but the directions of effect estimates were comparable across studies.
Statistical analysis
Total IgE was transformed to a log10 scale as a natural value in medical studies for data analysis. Linear regression was used for the multivariable analysis of DNA methylation and total IgE, separately for PR-GOAL and GALA II. Both analyses were adjusted for age, sex, asthma, batch (or plate), population subgroup (in GALA II) and the first five principal components (PCs) of the genotypic data, i.e., EWAS regression model (1) is: Log10Ige ~ CpG methylation + Age + Gender + Asthma + Batch + Population + 5 PCs. PCs were calculated using common SNPs (minor allele frequency > 5%) by using EIGENSTRAT19, separately for PR-GOAL and GALA II. We next performed a meta-analysis to combine summary statistics from PR-GOAL and GALA II, using inverse variance weighting to generate the combined effect size and P-value for each CpG site. A Bonferroni-corrected P < 2×10−7 was used as the cutoff for genome-wide statistical significance.
We then carried out a similar multivariable analysis of DNA methylation and total IgE in PR-GOAL, with additional adjustment for the percentages of WBC types (only neutrophils, eosinophils, lymphocytes, and monocytes are included, since the sum of the percentages of these four cell types determines the percentage of the fifth possible type) to uncover IgE regulatory pathways that are independent of eosinophil count. Hence, our EWAS regression model (2) is: Log10Ige ~ CpG methylation + Age + Gender + Asthma + Batch + Population + 5 PCs + Percentages of 4 WBC types. Since WBC percentages were not available in GALA II, cell type percentages were inferred based on the method described by Houseman et al.20. In brief, a Lasso regression model for each of the five cell types was first trained from the observations of cell percentage and DNA methylation in PR-GOAL, and then the most predictive 200 CpG sites (see Table E2) for the five cell types were selected as reference CpGs for the application of a multiple imputation method21 to impute the cell percentages, which were adjusted for in the multivariable analysis of GALA II. Of note, we validated the performance of the Houseman imputation-based method using data from PR-GOAL. In PR-GOAL, the imputed and true cell percentages were significantly and highly correlated (see Figure E1). Thus, methodologically, this imputation should work for GALA II samples, whose imputed WBC types are likely valid and without significant bias.
Next, we conducted a meta-analysis of total IgE in PR-GOAL and GALA II, after additional adjustment for WBC types. Genes containing the top 1000 CpG sites associated with total IgE in this meta-analysis were examined in a KEGG pathway enrichment analysis, using Fisher’s exact test, the multiple testing p-values were adjusted with FDR. Moreover, we performed an expression Quantitative Trait Methylation (eQTM) analysis for the top 20 findings from the meta-analysis adjusted for WBC types (using PR-GOAL data), to test for association between DNA methylation and cis-gene (< 1M bases) expression in whole blood, adjusting for age, sex, asthma status (and WBC types), i.e. Gene expression ~ CpG methylation + Age + Gender + Asthma + 5 PCs +(Percentages of 4 WBC types).
RESULTS
The characteristics of participants in the Puerto Rico Genetics Of Asthma and Lifestyle Study (PR-GOAL) and the Genes-environments & Admixture in Latino Americans Study (GALA II) are shown in Table I. Compared to participants in GALA II, those in PR-GOAL were younger and had a higher total IgE. DNA methylation levels were similar across studies (Figure E2).
Table I.
| Study | PR-GOAL (n=306) | GALA II (n=573) |
|---|---|---|
|
| ||
| Age (years) | 9 (6–15) | 13 (8–22) |
|
| ||
| Gender (female) | 131 (42.8%) | 287 (50.1%) |
|
| ||
| Asthma | 173 (56.5%) | 311 (54.3%) |
|
| ||
| Total IgE (IU/mL) | 694 (2–10178) | 371 (1–8545) |
|
| ||
| Average global genetic ancestry | ||
| European | 63% | 53% |
| African | 25% | 17% |
| Native American | 12% | 30% |
Data shown as mean (range) for continuous variables and number (percentage) for categorical variables
PR-GOAL: Puerto Rican Genetics of Asthma and Lifestyle Study
GALA II: Genes-environments & Admixture in Latino Americans Study
We conducted a multivariable linear regression analysis of DNA methylation and total IgE, first separately for each cohort, and then combining results from the two cohorts in a meta-analysis. The meta-analysis identified 1,326 CpGs associated with total IgE at P < 2×10−7, and 2,992 CpGs at FDR < 0.01 (See Manhattan and QQ plots in Figure E3). Table E3 lists the top 200 CpG sites (by P-value) in this meta-analysis, as well as their results of separate analysis for each cohort. The direction of association between CpG sites and total IgE was consistent across studies. PR-GOAL had 640 significant CpGs with P < 2×10−7, while GALA II had 404 significant CpGs, and their correlation of overall effect size or -log10(P-value) were also significant (Figure E4). In a sensitivity analysis, we performed a mega-analysis by combining the datasets from PR-GOAL and GALA II into a single dataset, using a similar approach to that for the meta-analysis, with additional adjustment for study site. This mega-analysis yielded similar results to those from the meta-analysis (Table E3).
Next, we compared the results of our meta-analysis unadjusted for WBC types with those from an EWAS of total IgE in Caucasians8. In spite of differences in ethnicity and age, most of the top significant CpG sites in our EWAS had the same direction of association as in Caucasians (Table E3). Of the top 200 CpG sites in our meta-analysis, 189 were also associated with total IgE (at P < 0.01) in either the PAPA or SLSJ study; and all 9 CpG sites that overlapped with those in the MRCA study (which had fewer CpG sites from the Illumina 27K Chip) were significantly associated with total IgE in that study. Moreover, we confirmed the top findings from the previous EWAS in Caucasians8 in our study (Table E4).
We then repeated the multivariable meta-analysis of our study cohorts after adjustment for WBC types. Fig 1 shows Manhattan and QQ plots of results in this adjusted meta-analysis, the number of significant CpG sites is markedly reduced comparing to the unadjusted results (Figure E3).
Fig 1.
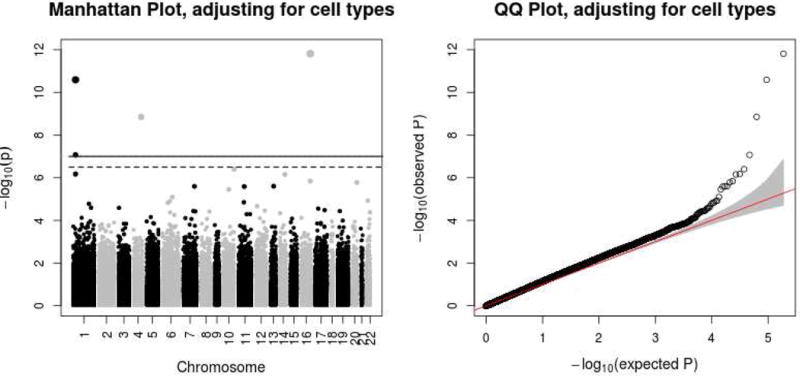
Manhattan and QQ plots for the results of meta-analyses adjusting for WBC types. The Manhattan plot shows the chromosomal locations of –log10(P-value) for association at each CpG site; the solid line illustrates a threshold of P-value=2×10−7 and the dashed line illustrates a threshold of FDR=0.01. The QQ plot shows observed versus expected –log10(P-value) for association at all loci. The gray area illustrates the 95% confidence band.
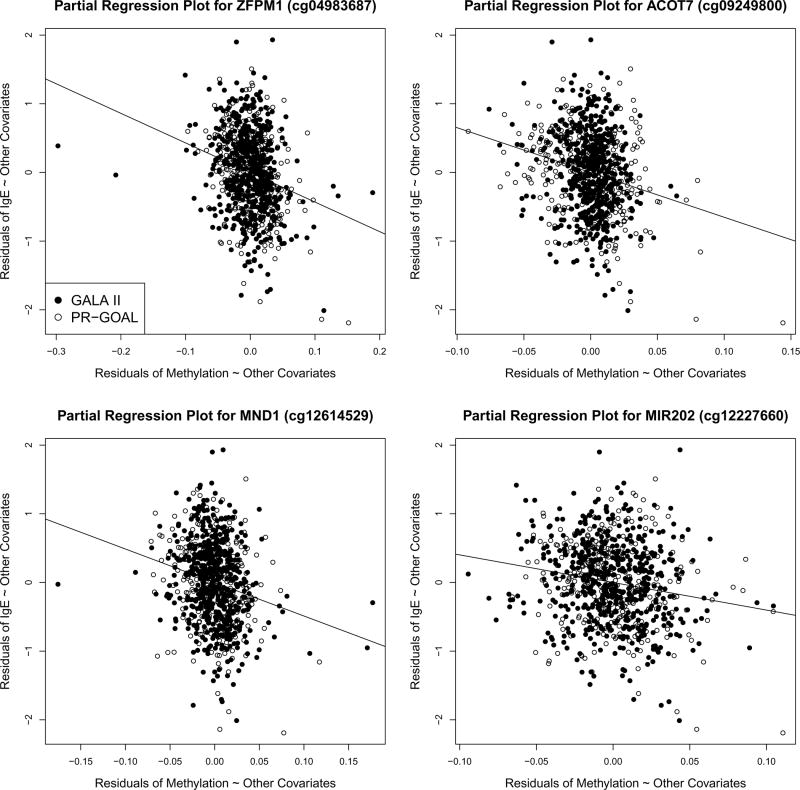
Table II lists the top twenty CpG sites, and four (cg09249800, cg04983687, cg12614529 and cg21220721) of them were significantly associated with total IgE at P < 2×10−7, and the top two CpGs overlap with those from the meta-analysis without adjusting for WBC types. Partial regression plots in Fig 2 show the associations between CpG methylation values and total IgE, conditional on all other covariates, for the top four genes. Table E5 lists the top 200 CpG sites as well as the results of this cell-adjusted analysis for each cohort, and similar results were obtained when a mega-analysis was conducted instead of a meta-analysis.
Table II.
Top twenty CpG sites associated with total IgE in the meta-analysis adjusting for WBC types
| ProbeID | Gene | CHR | Position | PR-GOAL | GALA II | Meta-analysis | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Effect* | P-value | Effect | P-value | Effect | P-value | FDR | ||||
| cg04983687 | ZFPM1 | chr16 | 88558223 | −5.27 | 4.69×10−7 | −3.82 | 3.61×10−7 | −4.31 | 1.53×10−12 | 2.88×10−7 |
| cg09249800 | ACOT7 | chr1 | 6341287 | −6.81 | 5.32×10−8 | −6.09 | 1.06×10−4 | −6.53 | 2.55×10−11 | 2.41×10−6 |
| cg12614529 | MND1 | chr4 | 154269418 | −5.25 | 7.38×10−5 | −4.67 | 4.39×10−6 | −4.88 | 1.40×10−9 | 8.76×10−5 |
| cg21220721 | ACOT7 | chr1 | 6341230 | −5.31 | 1.26×10−7 | −1.84 | 1.92×10−3 | −2.74 | 8.45×10−8 | 3.98×10−3 |
| cg12227660 | MIR202 | chr10 | 135061670 | −4.26 | 1.21×10−3 | −3.89 | 9.19×10−5 | −4.02 | 3.96×10−7 | 1.49×10−2 |
| cg11699125 | ACOT7 | chr1 | 6341327 | −5.77 | 5.34×10−6 | −3.19 | 1.43×10−2 | −4.52 | 6.68×10−7 | 1.88×10−2 |
| cg01000631 | EVL | chr14 | 100610667 | −4.65 | 1.68×10−4 | −3.88 | 1.09×10−3 | −4.25 | 6.97×10−7 | 1.88×10−2 |
| cg08940169 | ZFPM1 | chr16 | 88540241 | −5.49 | 8.37×10−5 | −4.13 | 4.14×10−3 | −4.83 | 1.43×10−6 | 3.38×10−2 |
| cg01458054 | HELZ2 | chr20 | 62200603 | −3.34 | 2.75×10−3 | −2.99 | 1.78×10−4 | −3.11 | 1.66×10−6 | 3.48×10−2 |
| cg15017119 | SPRY2 | chr13 | 82585793 | −4.84 | 7.04×10−5 | −2.96 | 5.45×10−3 | −3.78 | 2.49×10−6 | 4.06×10−2 |
| cg00405825 | HIPK2 | chr7 | 139474807 | 4.10 | 2.84×10−3 | 3.35 | 2.47×10−4 | 3.58 | 2.54×10−6 | 4.06×10−2 |
| cg25087851 | PTGDR2 | chr11 | 60623918 | −4.43 | 2.41×10−3 | −4.30 | 3.29×10−4 | −4.35 | 2.58×10−6 | 4.06×10−2 |
| cg12103951 | WAPL | chr10 | 88162314 | 0.77 | 7.00×10−2 | 1.28 | 1.18×10−5 | 1.12 | 3.55×10−6 | 5.15×10−2 |
| cg12427941 | TJAP1 | chr6 | 43457177 | 5.40 | 7.88×10−3 | 4.56 | 3.11×10−4 | 4.79 | 7.99×10−6 | 1.08×10−1 |
| cg06623197 | MTMR3 | chr22 | 30400763 | 2.99 | 5.23×10−3 | 1.64 | 3.59×10−4 | 1.86 | 1.18×10−5 | 1.48×10−1 |
| cg12296550 | HLA-DQB2 | chr6 | 32728862 | 0.54 | 5.89×10−2 | 0.91 | 4.77×10−5 | 0.77 | 1.26×10−5 | 1.48×10−1 |
| cg15700636 | PRG2 | chr11 | 57156050 | −5.59 | 9.80×10−4 | −4.41 | 4.07×10−3 | −4.94 | 1.41×10−5 | 1.57×10−1 |
| cg13074835 | NFKBIL1 | chr6 | 31515196 | −2.05 | 1.22×10−1 | −3.32 | 3.72×10−5 | −2.98 | 1.51×10−5 | 1.58×10−1 |
| cg15492834 | C1orf226 | chr1 | 162351057 | −5.26 | 1.55×10−3 | −3.11 | 1.79×10−3 | −3.68 | 1.66×10−5 | 1.64×10−1 |
| cg20481287 | ETV6 | chr12 | 12224457 | −2.10 | 3.73×10−2 | −2.58 | 1.95×10−4 | −2.43 | 2.14×10−5 | 2.02×10−1 |
Effect is the regression parameter for methylation data (beta value) at each CpG site
Fig 2.
Partial regression plots of CpG methylation and total IgE for the top four genes in the meta-analysis adjusting for WBC types. The x and y axis respectively stand for the residuals of regressing methylation and IgE against all other covariates, which are adjusted in the multivariable regressions. The solid and hollow circles indicate GALA II and PR-GOAL samples.
We then performed a KEGG pathway enrichment analysis on the genes closest to the top 1000 CpG sites from the meta-analysis adjusted for WBC types. Of the seven pathways enriched for these genes at FDR ≤ 0.01 (Table III), the first one (“hsa05310 Asthma”) is highly related to total IgE. Some significant pathways are relevant to immune responses, such as “hsa04612 Antigen processing and presentation”, “hsa05320 Autoimmune thyroid disease” and “hsa04672 Intestinal immune network for IgA production”.
Table III.
KEGG pathways enriched by genes closest to the top CpG sites in the meta-analysis adjusting for WBC types
| Pathway | Odds Ratio |
P-value | FDR | Included.Genes |
|---|---|---|---|---|
| hsa05310 Asthma | 7.41 | 1.72×10−05 | 0.002 | PRG2,HLA-DOA,HLA-DQA1,HLA-DQB1,IL4,HLA-DRB5,IL13,HLA-DQA2,HLA-DPA1 |
| hsa05330 Allograft rejection | 6.06 | 6.46×10−05 | 0.004 | HLA-DOA,HLA-DQA1,HLA-G,HLA-DQB1,IL4,HLA-DRB5,HLA-DQA2,GZMB,HLA-DPA1 |
| hsa04612 Antigen processing and presentation | 4.15 | 2.02×10−04 | 0.009 | HLA-DOA,PSME3,HSPA2,HLA-DQA1,HLA-G,HLA-DQB1,CTSB,TAP2,HLA-DRB5,HLA-DQA2,HLA-DPA1 |
| hsa05320 Autoimmune thyroid disease | 4.88 | 2.66×10−04 | 0.009 | HLA-DOA,HLA-DQA1,HLA-G,HLA-DQB1,IL4,HLA DRB5,HLA-DQA2,GZMB,HLA-DPA1 |
| hsa05332 Graft versus-host disease | 5.22 | 3.89×10−04 | 0.01 | HLA-DOA,HLA-DQA1,HLA-G,HLA-DQB1,HLA-DRB5,HLA-DQA2,GZMB,HLA-DPA1 |
| hsa04750 Inflammatory mediator regulation of TRP channels | 3.29 | 4.25×10−04 | 0.01 | ADCY4,PRKCE,PIK3CG,PLA2G4C,IL1RAP,ADCY8,TRPM 8,ADCY5,PRKCA,PPP1CA,PRKCB,PIK3CB,ADCY3 |
| hsa05323 Rheumatoid arthritis | 3.43 | 5.06×10−04 | 0.01 | ATP6V0A2,HLA-DOA,HLA-DQA1,TNFSF11,CSF2,HLA-DQB1,ATP6V0E2,HLA-DRB5,TGFB3,HLA-DQA2,ANGPT1,HLA-DPA1 |
Next, we performed eQTM analyses for the twenty CpG sites in Table II and their cis-genes, using the DNA methylation and gene expression data from PR-GOAL. From the analysis unadjusted for WBC types, cg25087851 methylation was significantly associated with expression of the gene for the prostaglandin D2 receptor 2 (PTGDR2, P = 5.16×10−17, see Table E7)). cg25087851 is 473 nucleoside bases upstream of PTGDR2, which encodes a G-protein-coupled receptor that is preferentially expressed in CD4+ effector Th2 cells. This receptor mediates pro-inflammatory chemotaxis of eosinophils, basophils, and Th2 lymphocytes generated during allergic inflammation22, and a PTGDR2 antagonist reduces eosinophilic airway inflammation (“experimental asthma”) in murine models23. DNA methylation of another CpG site (cg12227660) was significantly associated with expression of the gene polyamine oxidase (PAOX, P = 2.04×10−13); cg12227660 is 131,071 bases upstream of PAOX. PAOX particles can be used as a drug delivery vehicle for the treatment of acute inflammatory diseases such as asthma and acute lung injury24,25. After adjusting for cell types, the observed associations between methylation of CpG sites and gene expression became non-statistically significant, following a pattern similar to that for the EWAS results (Table E7).
GALA II participants belong to three Latino subgroups, captured by PCA of genotypic data, while the effect of population composition on DNA methylation is negligible, according to PCA of methylation data (Figure E5). Since PR-GOAL participants are all Puerto Rican, we conducted a secondary EWAS of total IgE after selecting only Puerto Ricans from GALA II (n = 219). Figure E6 shows the Manhattan and QQ plots for all CpG sites, and the results of top 200 sites are listed in Table E8 for the EWAS without cell type adjustment and Table E9 for the EWAS with cell type adjustment. Findings in this analysis in Puerto Ricans were similar to those in all Hispanics, the correlations of resultant effect size or −log10(Pval) were significant (Figure E7); and the 4 significant CpG sites in the meta-analyses with adjustment for WBC types were the same as those in all Hispanics.
DISCUSSION
Our meta-analysis without adjustment for WBC types identified 1,326 CpG sites that achieved a genome-wide significant association with total IgE at P < 2×10−7. There are some novel findings on top of the results (Table E3), including methylation in the genes for microRNA 202 (MIR202), lipin 1 (LPIN1), and prostaglandin D2 receptor 2 (PTGDR2). MIR202 is differentially expressed in cultured airway epithelial cells of monkeys in response to ozone exposure and lipopolysaccharide (LPS) treatment26, and is decreased in bronchoalveolar lavage (BAL) of patients with sarcoidosis27. LPIN1, which has been implicated in lipid metabolism and rhabdomyolisis, encodes a magnesium-ion-dependent phosphatidic acid phosphohydrolase enzyme. Expression of LPIN1 is low in subjects with atopic eczema, who often have other atopic diseases such as allergic asthma and allergic rhino-conjunctivitis28.
After additional adjustment of our meta-analysis for WBC types, there was a marked reduction in the number of CpG sites associated with total IgE. Since only eosinophils were consistently and significantly associated with total IgE in our study (Table IV), our overall findings suggest that DNA methylation in eosinophils explain most results for DNA methylation and total IgE in the analyses unadjusted for WBC types, thus emphasizing the importance of adjusting for cell types in future studies.
Table IV.
Association between WBC type percentages and total IgE
| IgE ~ Cell Percentage | PR-GOAL | GALA II | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Control | Case | All | Control | Case | All | |||||||
|
| ||||||||||||
| Effect | P-value | Effect | P-value | Effect | P-value | Effect | P-value | Effect | P-value | Effect | P-value | |
| Neutrophil | −0.007 | 0.216 | −0.009 | 0.091 | −0.008 | 0.050 | −0.007 | 0.061 | −0.004 | 0.319 | −0.008 | 0.007 |
| Lymphocyte | −0.011 | 0.076 | −0.001 | 0.885 | −0.008 | 0.057 | 0.001 | 0.792 | −0.009 | 0.055 | −0.002 | 0.573 |
| Monocyte | 0.045 | 0.128 | −0.023 | 0.419 | 0.007 | 0.756 | 0.021 | 0.237 | −0.012 | 0.566 | −0.008 | 0.575 |
| Eosinophil | 0.116 | 2.1×10−15 | 0.040 | 6.0×10−05 | 0.067 | 2.1×10−16 | 0.112 | 5.1×10−11 | 0.090 | 4.2×10−15 | 0.108 | 1.0×10−29 |
| Basophil | 0.491 | 0.047 | −0.058 | 0.756 | 0.194 | 0.200 | 0.449 | 0.010 | 0.115 | 0.519 | 0.258 | 0.050 |
Our four genome-wide significant findings in the meta-analysis adjusted for WBC types were not among the top 62 top loci associated with total IgE (at FDR <0.005, with consistent direction of association across cohorts) in the meta-analysis of three cohorts included in the prior EWAS of total IgE in Caucasians8. These four CpG sites were located in the genes for zinc-finger protein multi-type 1 (ZFPM1), acyl-CoA thioesterase 7 (ACOT7), and meiotic nuclear division 1 (MND1). ZFPM1 suppresses expression of interleukin 4 (IL-4) and induces expression of IFN-γ in CD4+ T cells, thus reducing Th2 cell differentiation29. Moreover, WBC DNA methylation in both ZFPM1 and ACOT7 has been associated with asthma30, and WBC DNA methylation of ZFPM1 was consistently associated with atopy (defined as at least one positive IgE to allergens) or high total IgE (≥200 kU/L) across two Caucasian cohorts included in a prior study31. In that study, DNA methylation of acyl-CoA thioesterase 7 (ACOT7) was significantly associated with atopy or high total IgE in the discovery cohort but not in a replication cohort. Thus, herein we first report consistent evidence of a genome-wide significant association between ACOT7 methylation and total IgE.
Of the top findings in our analysis unadjusted for cell types, CpG sites in ZFPM1, ACOT7, MIR202, EVL, PTGDR2 and PGR2 remained among the top twenty results in the analysis adjusted for WBC types. While not genome-wide significant, the WBC-adjusted analysis revealed some new genes among our top twenty findings, including MND1, sprouty RTK signaling antagonist 2 (SPRY2), major histocompatibility complex, class II, DQ beta 2 (HLA-DQB2) and ETS variant 6 (ETV6) genes. SPRY2 differentially regulates lymphoid and myeloid cell functions relevant to allergic asthma32. HLA-DQB2 is biologically relevant to immune responses and has previously associated with asthma33. ETV6-ABL gene rearrangement can result in chronic myeloid leukemia associated with eosinophilia34.
We recognize several study limitations. First, WBC types were not measured in GALA II. Although we applied validated methods to perform imputation of WBC percentages, the imputation uncertainty may still have reduced our statistical power. It is unlikely to have biased our results. Second, WBC types such as lymphocytes include multiple cell types (e.g., Th1, Th2, Th17) and we lack data to identify whether methylation of a specific cell type accounts for our findings in the meta-analysis adjusted for cell types. Third, we had data for gene expression in whole-blood (and not WBCs) in only a subset of participants in PR-GOAL, likely reducing our statistical power for the eQTM analysis of correlation between DNA methylation and gene expression. Finally, we lack functional analyses, but our top CpG sites are located within biologically plausible genes, as demonstrated by combined evidence from prior publications, and the results from our enrichment and eQTM analyses.
In summary, we demonstrate novel WBC DNA methylation changes associated with total IgE in an EWAS of Hispanic children, while replicating prior findings in Caucasians8. Our findings also emphasize the importance of adjusting for cell types (particularly eosinophils) in future studies of WBC DNA methylation and total IgE or related phenotypes (e.g., asthma).
Supplementary Material
Clinical Implications.
Among Hispanic children, methylation changes in the genes ZFPM1, ACOT7, and MND1 are associated with total IgE. ACOT7 and ZFPM1 have been implicated in the pathogenesis of asthma or atopy.
Capsule Summary.
In an EWAS adjusted for WBC types, few DNA methylation changes were significantly associated with total IgE among Hispanic children. Future EWAS of total IgE should account for DNA methylation of WBC types, particularly eosinophils.
Acknowledgments
We thank the participants and staff of the PR-GOAL and GALA II studies for their contributions to this work. The PR-GOAL Study was supported by grants HL079966 and HL117191 from the U.S. National Institutes of Health (NIH), and by The Heinz Endowments. The GALA II study was supported by grants HL088133, HL004464, HL117004, ES015794, ES24844, TRDRP 24RT 0025, MD006902, and GM007546 from the U.S. NIH. Dr. Burchard was also supported by the RWJF Amos Medical Faculty Development Award, the American Asthma Foundation and the Sandler Foundation. Dr. Forno’s contribution was supported by grant HL125666 from the U.S. NIH. Dr. Laprise is the chairholder of the Canada Research Chair in the Environment and genetics of respiratory disorders and allergy. Dr. Pino-Yanez was supported by the grant AC15/00015 by Instituto de Salud Carlos III within the ERACoSysMed 1st Joint Transnational Call (SysPharmPedia 99) from the European Union, under the Horizon 2020.
Abbreviations
- EWAS
epigenome-wide association study
- IgE
immunoglobulin E
- WBC
white blood cell
- PR-GOAL
Puerto Rico Genetics of Asthma and Lifestyle Study
- GALA II
Genes-environments and Admixture in Latino Americans study
- eQTM
expression Quantitative Trait Methylation
- PC
principle component
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: Study concept and design: E.G.B., and J.C.C.; data collection, and material and technical support: N.B., E.A-P., C.E., C.L., G.A.D., J.M.H., M.M., W.O.C., and G.C.; statistical analysis: W.C., T.W., M.P-Y., A.B., D.H., E.F., Q.Y., D.W. and L.L.; drafting of the manuscript for intellectual content: W.C., T.W., E.G.B. and J.C.C. All authors approved the final version of the manuscript prior to submission.
References
- 1.Stokes JR, Casale TB. The use of anti-IgE therapy beyond allergic asthma. J Allergy Clin Immunol Pract. 2015 Mar-Apr;3(2):162–166. doi: 10.1016/j.jaip.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010 Sep 23;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Granada M, Wilk JB, Tuzova M, et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J Allergy Clin Immunol. 2012 Mar;129(3):840–845 e821. doi: 10.1016/j.jaci.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pino-Yanes M, Gignoux CR, Galanter JM, et al. Genome-wide association study and admixture mapping reveal new loci associated with total IgE levels in Latinos. J Allergy Clin Immunol. 2015 Jun;135(6):1502–1510. doi: 10.1016/j.jaci.2014.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yatagai Y, Sakamoto T, Masuko H, et al. Genome-wide association study for levels of total serum IgE identifies HLA-C in a Japanese population. PLoS One. 2013;8(12):e80941. doi: 10.1371/journal.pone.0080941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nickel R, Illi S, Lau S, et al. Variability of total serum immunoglobulin E levels from birth to the age of 10 years. A prospective evaluation in a large birth cohort (German Multicenter Allergy Study) Clin Exp Allergy. 2005 May;35(5):619–623. doi: 10.1111/j.1365-2222.2005.02237.x. [DOI] [PubMed] [Google Scholar]
- 7.Feleszko W, Ruszczynski M, Jaworska J, Strzelak A, Zalewski BM, Kulus M. Environmental tobacco smoke exposure and risk of allergic sensitisation in children: a systematic review and meta-analysis. Arch Dis Child. 2014 Nov;99(11):985–992. doi: 10.1136/archdischild-2013-305444. [DOI] [PubMed] [Google Scholar]
- 8.Liang L, Willis-Owen SA, Laprise C, et al. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature. 2015 Apr 30;520(7549):670–674. doi: 10.1038/nature14125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosser FJ, Forno E, Cooper PJ, Celedon JC. Asthma in Hispanics. An 8-year update. Am J Respir Crit Care Med. 2014 Jun 1;189(11):1316–1327. doi: 10.1164/rccm.201401-0186PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs TS, Forno E, Brehm JM, et al. Underdiagnosis of allergic rhinitis in underserved children. J Allergy Clin Immunol. 2014 Sep;134(3):737–739 e736. doi: 10.1016/j.jaci.2014.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celedon JC, Sredl D, Weiss ST, Pisarski M, Wakefield D, Cloutier M. Ethnicity and skin test reactivity to aeroallergens among asthmatic children in Connecticut. Chest. 2004;125(1):85–92. doi: 10.1378/chest.125.1.85. [DOI] [PubMed] [Google Scholar]
- 12.Brehm JM, Acosta-Perez E, Klei L, et al. African ancestry and lung function in Puerto Rican children. J Allergy Clin Immunol. 2012 Jun;129(6):1484–1490 e1486. doi: 10.1016/j.jaci.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, Brehm JM, Lin J, et al. Expression quantitative trait loci (eQTL) mapping in Puerto Rican children. PLoS One. 2015;10(3):e0122464. doi: 10.1371/journal.pone.0122464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishimura KK, Galanter JM, Roth LA, et al. Early-life air pollution and asthma risk in minority children. The GALA II and SAGE II studies. Am J Respir Crit Care Med. 2013 Aug 1;188(3):309–318. doi: 10.1164/rccm.201302-0264OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pino-Yanes M, Thakur N, Gignoux CR, et al. Genetic ancestry influences asthma susceptibility and lung function among Latinos. J Allergy Clin Immunol. 2014 Sep 13; doi: 10.1016/j.jaci.2014.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang T, Guan W, Lin J, et al. A systematic study of normalization methods for Infinium 450K methylation data using whole-genome bisulfite sequencing data. Epigenetics. 2015;10(7):662–669. doi: 10.1080/15592294.2015.1057384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013 Jan 15;29(2):189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marabita F, Almgren M, Lindholm ME, et al. An evaluation of analysis pipelines for DNA methylation profiling using the Illumina HumanMethylation450 BeadChip platform. Epigenetics. 2013 Mar;8(3):333–346. doi: 10.4161/epi.24008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006 Aug;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 20.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C, Demerath EW, Pankow JS, et al. Imputation of Missing Covariate Values in Epigenome-Wide Analysis of DNA Methylation Data. Epigenetics. 2016 Feb 18;:0. doi: 10.1080/15592294.2016.1145328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuoka T, Hirata M, Tanaka H, et al. Prostaglandin D2 as a mediator of allergic asthma. Science. 2000;287(5460):2013–2017. doi: 10.1126/science.287.5460.2013. [DOI] [PubMed] [Google Scholar]
- 23.Uller L, Mathiesen JM, Alenmyr L, et al. Antagonism of the prostaglandin D2 receptor CRTH2 attenuates asthma pathology in mouse eosinophilic airway inflammation. Respiratory research. 2007;8:16. doi: 10.1186/1465-9921-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H, Kim Y, Guk K, et al. Fully biodegradable and cationic poly(amino oxalate) particles for the treatment of acetaminophen-induced acute liver failure. Int J Pharm. 2012 Sep 15;434(1–2):243–250. doi: 10.1016/j.ijpharm.2012.05.067. [DOI] [PubMed] [Google Scholar]
- 25.Seong K, Seo H, Ahn W, et al. Enhanced cytosolic drug delivery using fully biodegradable poly(amino oxalate) particles. J Control Release. 2011 Jun 10;152(2):257–263. doi: 10.1016/j.jconrel.2011.02.025. [DOI] [PubMed] [Google Scholar]
- 26.Clay CC, Maniar-Hew K, Gerriets JE, et al. Early life ozone exposure results in dysregulated innate immune function and altered microRNA expression in airway epithelium. PLoS One. 2014;9(3):e90401. doi: 10.1371/journal.pone.0090401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dyskova T, Fillerova R, Novosad T, et al. Correlation Network Analysis Reveals Relationships between MicroRNAs, Transcription Factor T-bet, and Deregulated Cytokine/Chemokine-Receptor Network in Pulmonary Sarcoidosis. Mediators Inflamm. 2015;2015:121378. doi: 10.1155/2015/121378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saaf AM, Tengvall-Linder M, Chang HY, et al. Global expression profiling in atopic eczema reveals reciprocal expression of inflammatory and lipid genes. PLoS One. 2008;3(12):e4017. doi: 10.1371/journal.pone.0004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitamura N, Mori A, Tatsumi H, Nemoto S, Hiroi T, Kaminuma O. Zinc finger protein, multitype 1, suppresses human Th2 development via downregulation of IL-4. Int Arch Allergy Immunol. 2011;155(Suppl 1):53–56. doi: 10.1159/000327292. [DOI] [PubMed] [Google Scholar]
- 30.Yang IV, Pedersen BS, Liu A, et al. DNA methylation and childhood asthma in the inner city. J Allergy Clin Immunol. 2015 Jul;136(1):69–80. doi: 10.1016/j.jaci.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Everson TM, Lyons G, Zhang H, et al. DNA methylation loci associated with atopy and high serum IgE: a genome-wide application of recursive Random Forest feature selection. Genome Med. 2015;7(1):89. doi: 10.1186/s13073-015-0213-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorentla BK,RA. The Adapter Protein Sprouty 2 (Spry 2) Differentially Regulates Lymphoid and Myeloid Cell Function and Is Important for Allergic Asthma. J Allergy Clin Immunol. 2015;135(2):AB283. [Google Scholar]
- 33.Yang IV, Tomfohr J, Singh J, et al. The clinical and environmental determinants of airway transcriptional profiles in allergic asthma. Am J Respir Crit Care Med. 2012 Mar 15;185(6):620–627. doi: 10.1164/rccm.201108-1503OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon D, Simon HU. Eosinophilic disorders. J Allergy Clin Immunol. 2007 Jun;119(6):1291–1300. doi: 10.1016/j.jaci.2007.02.010. quiz 1301–1292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.