Abstract
Understanding the abundance change of certain bacterial taxa is quite important for the study of soil microbiology. However, the observed differences of relative abundances by high-throughput techniques may not accurately reflect those of the actual taxon abundances. This study investigated whether soil microbial abundances coupling with microbial quantities can be more informative in describing the microbial population distribution under different locations. We analyzed relative abundances of the major species in soil microbial communities from Beijing and Tibet grasslands by using 16 S rRNA high-throughput sequencing technique, and quantified the absolute bacterial cell numbers directly or indirectly by multiple culture-independent measurements, including adenosine tri-phosphate (ATP), flow cytometry (FCM), quantitative real-time PCR (qPCR), phospholipid fatty acids (PLFA) and microbial biomass Carbon (MBC). By comparison of the relative abundance and the estimated absolute abundances (EAA) of the major components in soil microbial communities, several dominant phyla, including Actinobacteria, Bacteroidetes, Verrucomicrobia, Chloroflexi, Gemmatimonates and Planctomycetes, showed significantly different trends. These results indicated that the change in EAA might be more informative in describing the dynamics of a population in a community. Further studies of soil microbes should combine the quantification and relative abundances of the microbial communities for the comparisons among various locations.
Introduction
Microorganisms play essential roles in key biogeochemical cycles in almost all environments1. Particularly in soil, the microbial community with high taxonomic diversity and metabolic potential, can serve as a great biological indicator to reflect the major ecological processes2, 3. Therefore, to measure the abundance of some key populations is fairly important for understanding the contributions of the microbial community in soils4. However, obtaining accurate population density of key taxon is not an easy task. Often capturing individuals is an efficient mean available for macroscopic community surveys5, but rarely are all individuals in microbial community identified6, 7. With the rapid development of high-throughput molecular technologies, such as Illumina MiSeq/Hiseq sequencing, the relative abundances can be easily obtained4, 8–11. However, since relative abundance is expressed as a proportion of the total sample, the reliability of such estimates to reflect actual abundance of populations is insufficient4, 12.
Estimation of the absolute population abundances by combining the corresponding relative abundances with the absolute bacterial density, is an important prerequisite for the in-depth analysis of the structure and function of the microbial communities in natural environments12. The bacterial density is a fundamental measure in microbiology, but its assessment is often tedious13. To overcome this limitation, the rapid and reliable techniques are urgently needed to quantify the microbial populations in various environments, especially in soil with extremely high biodiversity14.
With the development of culture-independent measurements, various approaches, including direct counts of extracted bacteria, biochemical measurement and nucleic molecular approaches, have been used to quantify the microbial quantities13, 15–18. Recently, some studies have been done to compare these quantification methods. Flow cytometry (FCM) and fluorescence microscopy (EM) are two powerful direct counting methods, but the FCM is more rapid and accurate on enumerating bacterial cell numbers than visually observed EM19. Both methods use various fluorescent dyes to stain the bacterial cells, such as 4′,6-diamidino-2-phenylindole (DAPI), acridine orange (AO), propidium iodide (PI) and SYBR Green I (SYBR-I)20–22. Among of them, SYBR-I is the most commonly used fluorescent dye, because it is known to exhibit a high fluorescence quantum yield upon forming a complex with DNA molecules, and its background fluorescence is quite low23. Adenosine tri-phosphate (ATP), as the energy currency of all living cells, has been promoted as a potential indicator for viable quantities estimation for several decades24–26. The ATP measurement is fast, robust, and easy to perform, but it should be complemented with FCM or EM when it is used to estimate the bacterial cell numbers13, 15. In addition, quantitative real-time PCR (qPCR) with 16 S rRNA gene-based specific primers has been utilized as a sensitive and rapid method to quantify total bacterial community27, 28. The bacterial cell numbers measured by FCM, ATP, EM and qPCR, have been compared for diverse environments, such as drinking water29, wastewater30, activated sludge19, sediments13 and soil samples17. Almost all of these studies have proved that the FCM is more appropriate to monitor the cell numbers. Although both the FCM and ATP have been considered to be powerful measurements for the quantification of bacterial cell numbers, these two methods require a pre-suspension procedure of soil samples. Efficient detachment of bacterial cells is crucial for assessing microbial numbers31. However, there is no agreement on which procedure gives the best results with which type of substratum.
For the state of soil matrix, the soil microbial biomass, defining as the mass of the living component of soil organic matters, can be quantified by extracting a specific component of microbial biochemical substances. The classical method using chloroform fumigation to determine the microbial biomass carbon (MBC) was developed 40 years ago32, 33 but is still widely used today34, 35. Besides, the phospholipid fatty acids (PLFA) can be also served as biomarkers for bacteria in general, offering sensitive reproducible measurements for characterizing the numerically dominant portion of soil microbial community without cultivating the organisms18, 36. The PLFA analysis, based on modern biochemical techniques, shows many advantages over the conventional culturing methods, such as accuracy, quantitation, simple in operation, as well as no strict requirements for the microbial physiological status and sample conservation duration16, 37. However, the significant differences existing in those measurements, such as FCM and PLFA, make it difficult to compare the microbial quantities with the same criterion.
With the development of high-throughput nucleic molecular technologies and various quantitative measurements of microbial quantities, the relative abundances of microbial communities and absolute microbial quantities become easy to be estimated13, 38. Props et al. combined the high-throughput sequencing approach (16 S rRNA gene) with FCM to quantify the absolute taxon abundances on a cooling water system, which provided evidence that the increase in relative abundances could not necessarily relate to the increase in absolute abundances39. Stammler et al. demonstrated that combining the next-generation 16 S rRNA gene sequencing with qPCR could provide novel insights into the structure and the dynamics of intestinal microbiomes40. Smets et al. proposed a method with a simple addition of an internal standard at the DNA extraction step, to simultaneously measure the soil bacterial abundances and community composition via 16 S rRNA gene sequencing41. However, there is still a lack of systematic studies combining the absolute and relative measures to reveal the differences of microbial communities in diverse soil ecosystems. Besides, soils with distinct climates were good to identify major comparative consequences of the microbial communities42.
In the present study, distinct grassland soils from Tibet plateau and Songshan mountain in Beijing were collected which were over thousands of miles away. The relative abundances of microbial communities were analyzed by sequencing microbial 16 S rRNA gene amplicons using Illumina MiSeq technology. Diverse measurements, including ATP, FCM, qPCR, PLFA and MBC, were systematically compared and applied to quantify the total numbers of all individuals. Our results suggested that the relative abundances of species is insufficient to accurately figure out the absolute abundances of those in the studies of microbial communities, thus the quantification analysis of the bacterial absolute abundances is essential for the comprehensive analyses of the microbial community.
Results
Relative abundance of species in grassland soils from Beijing and Tibet
A total of 1,118,928 paired-end sequences were obtained from 20 samples of these two sampling sites, with 24,931-83,244 sequences per sample, and the sequencing depth proved to be close to saturation (Supplementary Fig. S1). Finally, random re-sampling was performed with 24,931 sequences per sample, resulting in 498,620 OTUs at 97% identity level.
The detrended correspondence analysis (DCA) showed that the structure of the microbial communities was significantly different in the two sampling sites, which has been confirmed by all three non-parametric dissimilarity tests (MRPP, ANOSIM, and ADONIS) (Supplementary Fig. S2, Table S2). Further comparison of the microbial taxonomic composition at the phylum level showed that 24 out of 46 phyla were shared by all samples, of which nine phyla represented over 90% of sequences in each sample (Fig. 1). Additionally, the composition of these nine dominant phyla differed significantly (P < 0.05) between Beijing and Tibet samples (Supplementary Table S2). The Proteobacteria and Actinobacteria were the most abundant bacterial phyla in both Beijing and Tibet grasslands (Fig. 1). The other seven phyla dominated in both sites, including Acidobacteria, Bacteroidetes, Verrucomicrobia, Planctomycetes, Chloroflexi, Gemmatimonadetes, and Cyanobacteria. Response ratio analyses at 95% confidence interval (CI) were performed on the relative abundances and absolute abundances of the major phyla between those two sampling sites (Fig. 2a, Supplementary Fig. S3a). For the relative abundances of the major phyla, the phylum Proteobacteria in Beijing was significantly higher than that in Tibet, while the phyla Verrucomicrobia, Planctomycetes, and Cyanobacteria were significantly lower than those in Tibet (Fig. 2a). Yet other phyla were not remarkably different.
Figure 1.
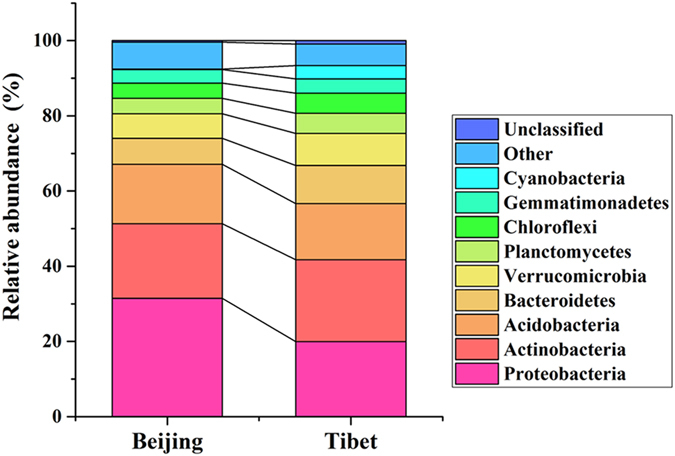
Relative abundances of nine dominant phyla in two sampling sites.
Figure 2.
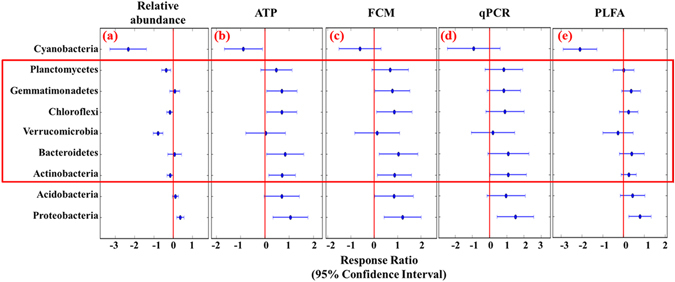
Response ratio for the differences between the relative abundances and estimated absolute abundances (EAA) of the major phyla in Beijing and Tibet grasslands. Significance was determined using the response ratio analysis at a 95% confidence level. The 95% CI of a response variable without overlapping with zero represent a significant result, otherwise, with non-significance. The response ratio more than zero means the abundances of Beijing more than Tibet samples, and vice versa.
Absolute quantification of microbial quantities by multiple measurements
The absolute microbial biomass quantified by PLFA and MBC methods were 16.4 ± 10.75 nmol and 213.02 ± 164.56 μg per gram of dry soil, respectively (Table 1). Based on an empirical conversion that 1.40 × 10–8 nmol bacterial PLFA cell−1 approximately equal to 2.0 × 10−5 μg bacterial C per cell18, the bacterial cell numbers derived from bacterial PLFA concentrations was an average of 1.17 × 109 ± 7.68 × 108 cells g−1 dry soil, as well an average of 1.10 × 1010 ± 8.20 × 109 cells g−1 dry soil was derived from the MBC concentrations. The absolute bacterial cell numbers directly determined by FCM averaged 9.47 × 108 ± 1.07 × 109 cells g−1 dry soil, whereas averages of 4.98 × 108 ± 4.53 × 108, and 3.90 × 109 ± 5.24 × 109 cells g−1 dry soil were derived from ATP and qPCR methods respectively, after some appropriate transformations (Table 1). All the tests of ATP, FCM, qPCR and PLFA quantification methods showed a good repeatability, but the MBC method showed obvious inconsistency with others (Fig. 3, Supplementary Table S3). Except MBC, the cell abundances based on FCM and qPCR showed greater variations within 10 replicates than the cell numbers obtained by ATP and PLFA assay between methods (Supplementary Fig. S4).
Table 1.
Absolute quantification of microbial biomass in the soil samples determined by different measurements.
| ATP (cells g−1 dry soil) | FCM (cells g−1 dry soil) | qPCR (copies g−1 dry soil) | PLFA (cells g−1 dry soil) | MBC (cells g−1 dry soil) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| All | 4.98 × 108 | 4.53 × 108 | 9.47 × 108 | 1.07 × 109 | 3.90 × 109 | 5.24 × 109 | 1.04 × 109 | 5.27 × 108 | 1.07 × 1010 | 8.23 × 109 |
| Beijing | 8.03 × 108 | 4.54 × 108 | 1.65 × 109 | 1.07 × 109 | 6.18 × 109 | 5.88 × 109 | 1.27 × 109 | 6.49 × 108 | 7.56 × 109 | 3.96 × 109 |
| Tibet | 1.93 × 108 | 1.42 × 108 | 2.47 × 108 | 4.20 × 108 | 1.62 × 109 | 3.43 × 109 | 8.06 × 108 | 2.09 × 108 | 1.37 × 1010 | 1.03 × 1010 |
Figure 3.
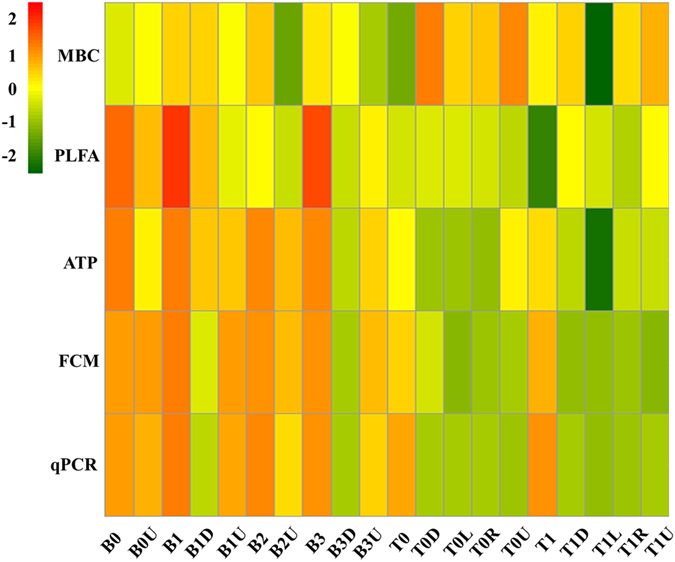
Comparison of the bacterial cell numbers derived from different measurements. Differences were visualized by heatmap. FCM: flow cytometry, ATP: adenosine tri-phosphate, qPCR: quantitative PCR, PLFA: phospholipid fatty acids, MBC: microbial biomass Carbon.
Correlation analyses, including Pearson correlation, Spearman and Kendall rank correlation showed that the measurements from ATP, FCM, qPCR, and PLFA were significantly (P < 0.05) positive correlations with each other, however, all of them showed negative correlation with MBC (Table 2, Supplementary Table S3).
Table 2.
Pearson correlations (r values, n = 20) between different measurements. (**P < 0.01).
| Pearson correlation | ATP | FCM | qPCR | PLFA | MBC |
|---|---|---|---|---|---|
| ATP | 1 | 0.871** | 0.827** | 0.799** | −0.16 |
| FCM | 1 | 0.911** | 0.720** | −0.257 | |
| qPCR | 1 | 0.599** | −0.163 | ||
| PLFA | 1 | −0.076 | |||
| MBC | 1 |
Microbial quantities in different locations
Different measurements showed that the microbial quantities were significantly different between the two sampling sites (Table 1, Fig. 4). Bacterial cell numbers based on ATP measurement, were significantly higher (P < 0.001) in Beijing sampling site (8.03 × 108 ± 4.54 × 108 g−1 dry soil, Beijing) than those in Tibet (1.93 × 108 ± 1.42 × 108 g−1 dry soil), similar with the results from FCM, qPCR and PLFA methods. However, the cell numbers derived from MBC were shown significantly lower in Beijing (7.56 × 109 ± 3.96 × 109 g−1 dry soil) than those in Tibet (1.37 × 1010 ± 1.03 × 1010 g−1 dry soil), which was not consistent with other methods. Besides, the Pearson correlation tests showed that all of these measurements, except for MBC, were significantly correlated with the environmental factors (soil moisture and TOC) (Supplementary Table S4). Thus the MBC measurement result was excluded from the following calculations.
Figure 4.
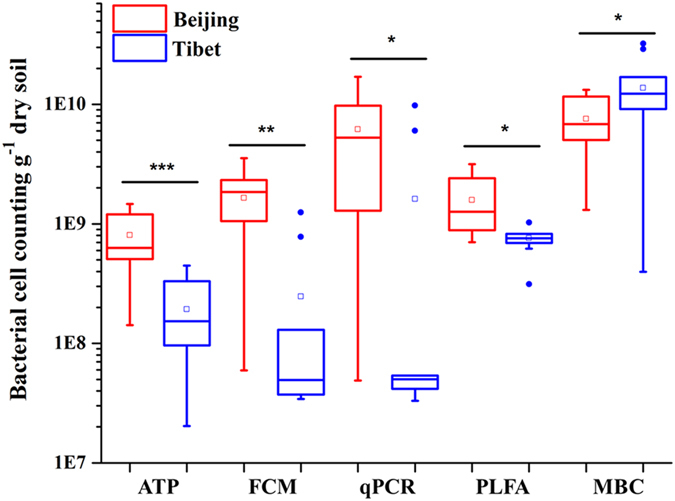
Comparison of the bacterial cell numbers between two sampling sites. (*P < 0.05; **P < 0.01; ***P < 0.001). Horizontal lines show median values, □ indicate mean values, boxes denote values comprised within the lower and upper quartile of the data, the vertical lines represent ranges, and • indicate outliers.
EAA of major phyla and classes in Beijing and Tibet sampling sites
We combined the bacterial quantities measured by the four different methods (ATP, FCM, qPCR and PLFA) with the relative abundances of those dominant phyla. Since the absolute microbial cell numbers in Beijing is significantly higher than that in Tibet, the estimated absolute abundances (EAA) of the major phyla showed distinct trends with the corresponding relative abundances, for example, by using ATP measured (Figs 1 and 5). Besides, the other three approaches, including FCM, qPCR and PLFA, showed very consistent trends with ATP measurement in different phyla (Fig. 2b–e). For Proteobacteria, both the relative abundances and EAA showed significantly higher in Beijing than those in Tibet. In addition, the Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria classes showed similar result that the relative abundances and EAA in Beijing were significantly higher than those in Tibet (Supplementary Fig. S3). However, dramatic changes were found in the EAA of some major phyla than their relative abundances. For example, the relative abundances of the phyla Actinobacteria, Bacteroidetes, Chloroflexi and Gemmatimonates, in Beijing samples were no significant difference with those in Tibet, but the EAA of those phyla were significantly higher in Beijing than those in Tibet; additionally, the phyla Verrucomicrobia and Planctomycetes, of which the relative abundances in Beijing were significantly higher than those in Tibet, showed no distinct difference in the EAA in Beijing and Tibet (Fig. 2).
Figure 5.
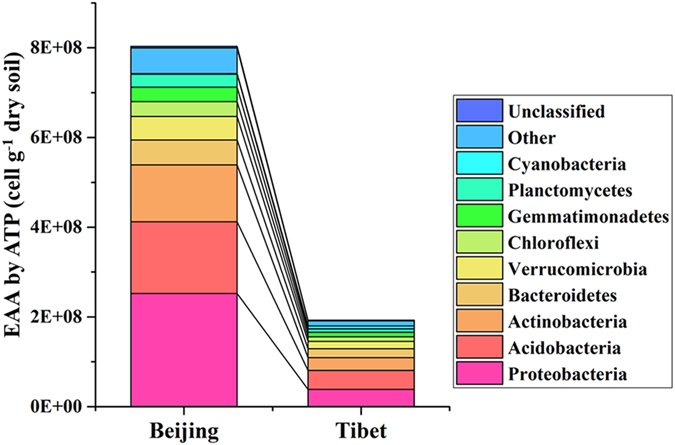
Comparison of the estimated absolute abundances (EAA) of the major phyla based on the ATP measurement in the two sampling sites. ATP: adenosine tri-phosphate.
Discussions
As same as the macro-ecology, the differences of relative abundances could be inappropriate to interpret the real difference in abundances of microbial community in natural conditions4. In this study, we measured the EAA of the compositions in soil microbial communities by combining the corresponding relative abundances from the high-throughput sequencing approach with the absolute microbial quantities by various quantitative measurements. According to our results, we propose a conceptual model for the relationship between the EAA with the relative abundances and the quantitative detection (Fig. 6). For instance, in the given community 1 and 2, when we only considered the relative abundances of different taxa based on sampling, we could only obtain the composition of microbial community relatively, but when those compositions have been integrated with the quantitative estimation, the taxa abundance changes could be depicted more close to real abundance changes.
Figure 6.
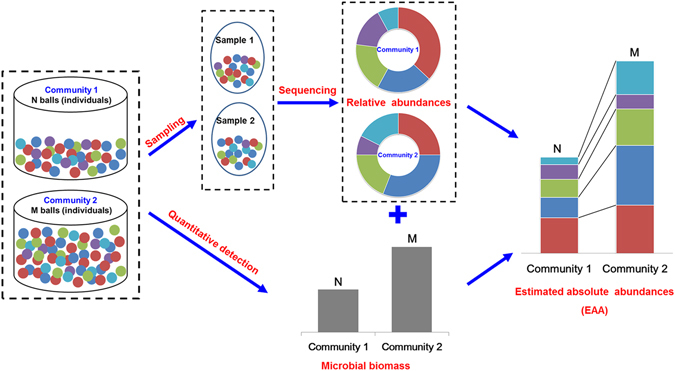
Conceptual model of the relationship between the estimated absolute abundances (EAA) with the relative abundances and the quantitative detection. The total number of balls in each community is quantified as N balls in community 1 and M balls in community 2, where N is about half of M. Meanwhile, the relative abundance of each color of ball is also calculated by regarding each community as an integral whole. The EAA could be obtained by combining the quantitative detection and the relative abundances.
In soil matrices, bacteria are closely associated with various physical components, and thus some quantitative methods, like ATP and FCM assays, should suspend the bacterial cells as a pretreatment step. In the present study, a simple but efficient method was selected, which was previously described and calibrated for natural soil samples17, 43. While, as reported by Frossard et al.13, the bacterial cells were detached and separated from soil matrix particles by treatment with an ultrasonic probe, centrifugation in tubes receiving sample suspensions and Histodenz® solution, which was also suitable for sediments and sludge samples. Although the detachment procedures were different, we both got a rough same amounts of cells for assessing the absolute bacterial quantities in natural soils. Some studies have discussed the detachment methods for the bacterial extraction in diverse environments, such as soils, sediments, activated sludge, and plant litter, however, the number of released bacterial cells could vary moderately among different detachment devices19, 30, 31, 43. Since the efficient detachment of bacterial cells is crucial for assessing bacterial cell numbers in natural environments, we have chosen the simple but quite stable way in this study and the final bacterial cell numbers obtained here are reasonable.
The absolute quantification of the microbial quantities is a crucial step for estimating absolute abundance of each taxon, where the accurate quantitative method should be evaluated key prerequisite. By comparison of bacterial cell numbers from total 20 soil samples, our results showed that the MBC would be unsuitable for this quantitative study (Table 2, Fig. 3), possibly because the soil microbial quantities measured by MBC were overestimated by fungi and protozoa2, 44. Except for the MBC method, bacterial cell numbers counted by the four quantitative methods, including ATP, FCM, qPCR and PLFA, showed a good repeatability and significant correlations (Fig. 3, Table 2). This observation showed that those four techniques focusing on the same biological component could provide conclusions with minor differences. The ATP, FCM and HPC methods have been applied to measure the microbial cells in drinking water and other aquatic environments, suggesting that the ATP analysis is a more rapid and easy tool for counting the intact microbial quantities15. Most studies comparing various quantitative measurements consider the FCM analysis as a most potential and robust single-cell enumeration technology for diverse environments, such as drinking water systems29, 45, cooling water system39, agricultural soil17, sediments13 and activated sludge19, 30. But it is possible that the FCM led to an underestimation of bacterial numbers, since in soil and sediment matrices bacteria are closely associated with physical components17. One study showed the estimation based on the ATP assay yielded significantly higher average bacterial abundances than the FCM method13. However, our study showed that the counting results from ATP assay were similar and significantly correlated (P < 0.001) with FCM (Tables 1 and 2). The reason for this difference may be due to the pretreatment step for FCM and ATP. Furthermore, the qPCR carried out with the same DNA with the amplicon sequencing analyses may be more appealing. A significant positive association was observed between FCM counts and qPCR quantification in the present study, whereas the absolute number detected by FCM was lower than qPCR method within 1–2 orders of magnitude, which was consistent with the study by Bressan et al.17. The difference between FCM and qPCR could be due to the combination of the inherent biases of all experimental steps, such as the efficiency of cell or DNA extraction, PCR inhibition due to the amplification efficiency and primer specificity, or underestimated counts of bacterial cells with FCM. In the previous study, qPCR relied upon total DNA extraction and could overestimate the bacterial cell numbers, since the exact number of copies of the 16 S rRNA gene in any given bacterial species varies28, 46. Few studies have compared PLFA measurements with the ATP, FCM and qPCR methods in the estimation of absolute microbial quantities. Our results from the biomarker PLFA for bacteria showed significantly correlated with the other three methods, and lower variation than that from qPCR. The PLFA profile is widely used to measure microbial biomass and community composition in soil and other types of environmental samples, but PLFA analysis is somewhat slow and expensive to carry out47.
According to the results of those four measurements, the soil microbial quantities appeared significantly higher in Beijing than those in Tibet. The correlation tests showed that the measurements, including ATP, FCM, qPCR and PLFA, were significantly correlated with the soil moisture and TOC (Supplementary Table S4). Soil moisture plays an important role for the terrestrial ecosystem processes, and also could determine the diversity of soil microbial communities48–50. We found that the moistures in Beijing were significantly higher than those in Tibet, which might lead to the higher bacterial cell numbers in Beijing sampling sites. On the other hand, the significantly higher of TOC, which might provide more substrates for the growth of microorganisms, also resulted in the higher the bacterial quantities in Beijing than those in Tibet51.
The most important question we tried to address is whether the changes in the absolute abundances of the components in microbial communities are inconformity with the shifts in relative abundances. Since the relative abundances of microbial community compositions was expressed as a proportion of the total sample, it had ignored the absolute abundances of species under different conditions6. To further verify the limitations of the relative abundances, we quantified the EEA of two distinct sampling sites by combining with the absolute microbial quantities, Beijing and Tibet. Several major phyla, including Actinobacteria, Bacteroidetes, Verrucomicrobia, Chloroflexi, Gemmatimonates and Planctomycetes, showed significant differences between the EAA with the relative abundances. For instance, the relative abundance of phylum Actinobacteria in Beijing is lower than that in Tibet with no significance, while the EEA of this phylum is significantly higher in Beijing and has significant correlation with moisture and TOC (Supplementary Table S5). It has been reported that the vast majority of Actinobacteria are important saprophytes capable of breaking down a wide range of plant and animal debris in the process of decomposition52. In addition, the phylum Proteobacteria, which has been recognized as a most dominant composition in soil microbial community53, shows significantly high in Beijing than Tibet, no matter relative abundances and EAA. Even though, from our result we are able to show that the quantification of the EEA for major phyla is essential and reliable, and has the potential to reflect the actual phenomenon of microbial communities in natural environments.
The conceptual model proposed in this study (Fig. 6) shows that the EEA of the different taxa change dramatically when the quantitative detection has been taken into account in the relative abundances. As described by Finnegan et al., any change in the relative abundance of a taxon could be explained in two ways12: (i) a change in its absolute abundance will obviously cause a corresponding change in its relative abundance; (ii) the shift in a taxon’s relative abundance may be driven by changes in the absolute abundance of other taxa in the assemblage, with little changes in the absolute abundance of the taxon itself. From the study of Fang et al., warming significantly altered the soil microbial community compositions, but did not significantly affect soil microbial biomass in all aggregate fractions54. For many ecological processes, the relative abundances of equivalent species on local and regional scales will not be influenced by changes in environmental conditions, while the fitness, such as soil quality, will only depend on the total microbial quantities55, 56. It has been reported that the microbial quantities could present sensitive respond to diverse environmental changes with decreasing in stress, such as heavy metal, or increasing in nutrient, including different fertilizations57, 58, while relative abundances of the microbial lineages showed distinct shifts39. Berga et al. demonstrated that with increasing salinity stress, the active bacterial community composition did not show significant changes directly, however, the bacterial abundances decreased significantly after the disturbance59. The possible reason might be that the change rates of the species in microbial communities showed distinct differences along with the microbial quantities responding to the environmental changes, resulting in that relative abundances of some species increase under stress, while others fall in nutrients. Therefore, understanding the ecological processes that shapes the absolute and relative abundances across the landscape and through time is ultimately required.
In summary, this study has demonstrated that the information reflected by the relative abundances could be insufficient to interpret the actual abundances, and the absolute bacterial quantities have been proved to be essential for the comprehensive analysis of the microbial communities in natural environment. Our study is also the first attempt to quantify the absolute abundances of the major components in the microbial communities by combining the relative abundances and bacterial quantities through diverse measurements.
Material and Methods
Site description and sample collection
Grassland soils from the Tibet plateau (29°36′21″N, 85°45′09″E) and the Songshan mountain located in Beijing (42°31′10″N, 115°49′32″E), China, were used in this study. At each site, we collected 10 parallel samples, each of which was homogeneously mixed by five surface soil cores (5–10 cm depth) within a 1 m*1 m quadrat. Soils were kept on ice until shipped to our laboratory for soil geochemical variables and microbial measurements.
The soils were all sieved by a 2 mm mesh, and the geochemical variables were measured as follows: pH was determined with a glass electrode in water-to-soil ratio of 2.5:1 (v/w). Soil moisture was measured by putting 2.0 g soil into 105 °C oven until constant weight was reached, and the percentage of weight loss after oven dry to the original weight was calculated as soil moisture content (%). The soil characteristics of the two sampling sites were summarized (Supplementary Table S1).
Whole cell extraction from soil samples
The cell extraction was performed as described by Bressan et al. 17. Briefly, soil samples (5 g) were diluted in 45 mL of NaCl 0.85% solution (filtered at 0.22 μm) and homogenized by vortexing for 5 min at full speed. This suspension was centrifuged at 130 × g (gravities) for 5 min to exclude the large soil particle. The supernatant was then filtered by cell strainer (40 μm; BD FALCON), to remove particles prior to the further ATP and FCM analyses.
Adenosine tri-phosphate (ATP) analysis
ATP was measured using the BacTiter-GloTM Microbial Cell Viability Assay (G8231, Promega Corporation, Dübendorf, CH) and a GloMax 20/20 Luminometer (Turner BioSystems, Sunnyvale, CA, USA). We applied an optimized protocol as described by Hammes et al.15, comparing with the manufacturer’s protocol for this product. Total ATP was measured as follows: 2 mL of the supernatant for each sample was transferred into a 2 mL Eppendorf tube and 50 μl of the ATP reagent was transferred to a separate sterile 1.5 mL reaction tube (Axygen). Both sample and reagent were heated for at least 1 min in a heating block at 38 °C. Then, 500 μl of the sample was transferred to the 50 μl reagent, and the mixture was incubated for a further 20 s in the heating block (38 °C). The luminescence was subsequently measured as an integral over 10 s, expressed in relative light units (RLU). The RLU values were converted to ATP concentrations using a calibration curve (R2 > 0.99), which was prepared with pure ATP standard (10 mM; Promega Corporation) diluted in ATP free, sterile water (Milli-Q) to different concentrations (10−2–104 nM). A conversion factor of 1.75 × 10−10 nmol ATP per cell was applied to calculate the bacterial cell number15. All ATP measurements were done in triplicate, and all procedures were conducted under a sterile condition.
Fluorescence staining and flow cytometry (FCM) analysis
All the soil supernatants (1 mL) with 10,000-time dilution were stained with 10 μL of SYBR® Green I (100 × in DMSO; Invitrogen S7563), and then incubated for 15 min in the dark at room temperature before usage30. The flow cytometry (FCM) analyses were carried out on the BD FACSCalibur equipped with a 488 nm argon laser. Green fluorescence was collected in the FL1 channel at 600 nm, and red fluorescence was collected in the FL3 channel at 650 nm. All samples were collected as logarithmic signals and were triggered on the green fluorescence channel (FL1). The collection of data as FL1/FL3 dot plots allowed for optimal distinction between the stained microbial cells and instrument noise or sample background. In order to count the absolute number of bacteria in soil samples, the stained soil supernatants (450–700 μL) were added into the BD TruCount Absolut Count Tubes (BD Biosciences), and detected cell numbers after mixing. The FCM results were calculated according to the manufacturer’s protocol.
Quantitative PCR (qPCR)
Total soil DNA was extracted from 0.5 g of moist sieved soil using the FastDNA® SPIN Kit for Soil (MP-Biomedicals), following the manufacturer’s protocol. DNA was finally resuspended in 50 μL DES (DNase/Pyrogen-Free Water) and measured by Qubit® 3.0 Fluorometer. Each PCR reaction mixture (20 μL) contained 10 ng of template DNA, 10 μL SuperReal PreMix Plus (SYBR Green, TIANGEN, FP205), and 1.5 μL of each primer (10 μM; Forward: 5’ -CCTACGGGAGGCAGCAG- 3′; Reverse:5′- TTACCGCGGCTGCTGGCAC- 3′). The qPCR reactions were performed in triplicate under thermal cycler conditions of 15 min at 95 °C, and 39 cycles of 10 s at 95 °C, 30 s at 55 °C and 32 s at 72 °C in a CFX ConnectTM Real-Time PCR Detection System (BioRad). All results were normalized and calculated using the ΔCt method.
Phospholipid fatty acids (PLFA) analysis
Lipids were extracted from the soil samples using the modified Bligh-Dyer method34. Briefly, 2 g of each fresh soil sample was incubated in a 2:1:0.8 solution of methanol, chloroform, and phosphate buffer. The soil extracts were filtered and the chloroform phases collected. The phospholipids were separated from glycolipids and neutral lipids through silicic acid chromatography, subsequently saponified and methylated to fatty-acid methyl esters (FAME). FAME were then separated on a capillary gas chromatograph and identified using a gas chromatography/mass spectrometry. All procedures of PLFA analysis were performed in the Institute of Soil Science, Chinese Academy of Sciences (Nanjing, China).
A total of 109 different PLFA were detected and identified. Specific PLFA (15:0, i16:0, 17:0, i15:0, a15:0, i17:0, a17:0, cy17:0, 16: 1w5, 16: lw7t, 16:lw9, 18:lw7, and cy19:0), were used as biomarkers of bacterial origin, following the study of Frostegard16. The absolute bacterial biomass of the soils was calculated by adding the absolute content of each bacterial PLFA biomarkers. Bacterial cell numbers derived from bacterial PLFA concentrations are based on a conversion factor of 1.40 × 10−8 nmol per bacterial PLFA cell18.
Microbial biomass Carbon (MBC) analysis
A rapid chloroform-fumigation extraction method was used to measure soil microbial biomass carbon (MBC) as previously described60, 61. Briefly, 15 g of each fresh soil sample was fumigated with alcohol-free CHCl3 for 24 h. Extraction for CHCl3-C was done with K2SO4 immediately after fumigation, and total microbial biomass Carbon with 10-fold dilution was detected by a MultiNC3100TOC analyzer. All procedures of MBC analysis were performed in the Institute of Applied Ecology, Chinese Academy of Sciences (Shenyang, China).
Illumina sequencing analysis of 16 S rRNA gene amplicons
Microbial genomic DNA was extracted following the protocol of the FastDNA® SPIN Kit for Soil. DNA quality and final concentrations were assessed by the ratios of 260 nm/280 nm, and 260 nm/230 nm using a NanoDrop Spectrophotometer (Nano-100, Aosheng Instrument Co Ltd.). All extracted DNA were stored at −80 °C for further amplifications. The primers 515 F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806 R (5′-GGACTACHVGGGTWTCTAAT-3′) targeting the V4 hypervariable regions of microbial 16 S rRNA genes were selected62, and tagged with paired barcode sequence (12 mer) for pooling of multiple samples in one sequencing run. All primers were synthesized by Sangon Biotech (Shanghai, China). Following amplification, the PCR product was used for agarose gel (1%) electrophoresis and purified by recovering the gel following the protocol of Gel Extraction Kit (D2500–02, OMEGA BioTek). From each sample, 150 ng of the PCR product, which was quantified by NanoDrop Spectrophotometer combining with Gel Image System (Taxon-1600), was collected and pooled with other samples. The sequencing run was conducted on MiSeq (Illumina) for 2 × 250 bp paired-end sequencing in Central South University (Changsha, China).
Raw data was processed using an in-house pipeline (http://mem.rcees.ac.cn:8080) which was built on the Galaxy platform but integrated various software tools related to sequencing processing. The operational taxonomic units (OTUs) were classified using UClust at 97% similarity level, and taxonomic annotations were assigned to each OTU’s representative sequence by the RDP 16 S Classifier63. Random resample was performed with the lowest sequences (>20,000 sequences) among samples. This resampled OTUs summary table was used for further various statistical analyses.
Estimated absolute abundance (EAA) of each major taxon
We defined the estimated absolute abundance (EAA) as the product of a corresponding relative abundance multiplying with the total absolute microbial cell numbers. The relative abundance of this taxon was obtained from the 16 S rRNA sequencing analysis, while the absolute microbial cell numbers was quantified by the measurements mentioned previously.
Statistical analysis
Non-parametric permutational multivariate analysis of variance of the Adonis function (ANONIS), analysis of similarities (ANOSIM), and multiresponse permutation procedure (MRPP) were used to evaluate the significant differences of the microbial community compositions between different sampling sites. Response ratio was performed to measure the relationship between the relative abundances and the EAA of major phyla64. The correlation between the bacterial cell numbers quantified by different measurements was conducted in SPSS22 using single-tailed Pearson correlation test, Kendall’s tau_b correlation, and Spearman correlation. Besides, the significance of difference between the microbial quantities from Beijing and Tibet was determined by a two-tailed T test.
Electronic supplementary material
Acknowledgements
This project was supported by Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (Grant XDB15010302), the National Natural Science Foundation of China (Grant No. 31540071) and CAS 100 talent program, the Open Project Program of State Key Laboratory of Applied Microbiology Southern China (SKLAM001-2015).
Author Contributions
Z.Z. and Y.D. designed and performed this study. Z.Z. conducted the experiments, analyzed the data, created the figures and wrote the manuscript. S.L., K.F., S.W. and W.C. participated in sample and data collection, contributed to the manuscript writing. Y.L. performed the phospholipid fatty acid experiments. H.L. performed the microbial biomass Carbon experiments. H.Y. performed the 16 S rRNA gene sequencing experiments. Y.Q., M.X. and Y.D. provided critical revision for substantial intellectual content of the manuscript. All authors contributed to the revision of manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05260-w
Accession codes: Raw sequences were submitted to the NCBI Sequence Read Archive (SRA) under the accession number SRP092803.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuanyuan Qu, Email: qyy@dlut.edu.cn.
Ye Deng, Email: yedeng@rcees.ac.cn.
References
- 1.Zhou JZ, et al. Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. P Natl Acad Sci USA. 2014;111:E836–E845. doi: 10.1073/pnas.1324044111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schloter M, Dilly O, Munch JC. Indicators for evaluating soil quality. Agr Ecosyst Environ. 2003;98:255–262. doi: 10.1016/S0167-8809(03)00085-9. [DOI] [Google Scholar]
- 3.Roesch LF, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. Isme J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou, J. et al. High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. Mbio6 (2015). [DOI] [PMC free article] [PubMed]
- 5.Hopkins HL, Kennedy LL. An assessment of indices of relative and absolute abundance for monitoring populations of small mammals. Wildlife Soc B. 2004;32:1289–1296. doi: 10.2193/0091-7648(2004)032[1289:AAOIOR]2.0.CO;2. [DOI] [Google Scholar]
- 6.Zhou JZ, et al. Reproducibility and quantitation of amplicon sequencing-based detection. Isme J. 2011;5:1303–1313. doi: 10.1038/ismej.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM. Counting the uncountable: Statistical approaches to estimating microbial diversity. Appl Environ Microb. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huber JA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 9.Degnan PH, Ochman H. Illumina-based analysis of microbial community diversity. Isme J. 2012;6:183–194. doi: 10.1038/ismej.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Widder S, et al. Challenges in microbial ecology: building predictive understanding of community function and dynamics. Isme J. 2016;10:2557–2568. doi: 10.1038/ismej.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou, J. Z. et al. Temperature mediates continental-scale diversity of microbes in forest soils. Nature communications7 (2016). [DOI] [PMC free article] [PubMed]
- 12.Finnegan S, Droser ML. Relative and absolute abundance of trilobites and rhynchonelliform brachiopods across the Lower/Middle Ordovician boundary, eastern Basin and Range. Paleobiology. 2005;31:480–502. doi: 10.1666/0094-8373(2005)031[0480:RAAAOT]2.0.CO;2. [DOI] [Google Scholar]
- 13.Frossard, A., Hammes, F. & Gessner, M. O. Flow Cytometric Assessment of Bacterial Abundance in Soils, Sediments and Sludge. Frontiers in microbiology7 (2016). [DOI] [PMC free article] [PubMed]
- 14.Davis C. Enumeration of probiotic strains: Review of culture-dependent and alternative techniques to quantify viable bacteria. J Microbiol Meth. 2014;103:9–17. doi: 10.1016/j.mimet.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 15.Hammes F, Goldschmidt F, Vital M, Wang YY, Egli T. Measurement and interpretation of microbial adenosine tri-phosphate (ATP) in aquatic environments. Water Res. 2010;44:3915–3923. doi: 10.1016/j.watres.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Frostegard A, Baath E, Tunlid A. Shifts in the Structure of Soil Microbial Communities in Limed Forests as Revealed by Phospholipid Fatty-Acid Analysis. Soil Biol Biochem. 1993;25:723–730. doi: 10.1016/0038-0717(93)90113-P. [DOI] [Google Scholar]
- 17.Bressan M, et al. A rapid flow cytometry method to assess bacterial abundance in agricultural soil. Appl Soil Ecol. 2015;88:60–68. doi: 10.1016/j.apsoil.2014.12.007. [DOI] [Google Scholar]
- 18.Frostegard A, Baath E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fert Soils. 1996;22:59–65. doi: 10.1007/BF00384433. [DOI] [Google Scholar]
- 19.Abzazou T, et al. Assessment of total bacterial cells in extended aeration activated sludge plants using flow cytometry as a microbial monitoring tool. Environ Sci Pollut R. 2015;22:11446–11455. doi: 10.1007/s11356-015-4372-3. [DOI] [PubMed] [Google Scholar]
- 20.Weinbauer MG, Beckmann C, Hofle MG. Utility of green fluorescent nucleic acid dyes and aluminum oxide membrane filters for rapid epifluorescence enumeration of soil and sediment bacteria. Appl Environ Microb. 1998;64:5000–5003. doi: 10.1128/aem.64.12.5000-5003.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klauth P, Wilhelm R, Klumpp E, Poschen L, Groeneweg J. Enumeration of soil bacteria with the green fluorescent nucleic acid dye Sytox green in the presence of soil particles. J Microbiol Meth. 2004;59:189–198. doi: 10.1016/j.mimet.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Porter J, Deere D, Hardman M, Edwards C, Pickup R. Go with the flow - use of flow cytometry in environmental microbiology. Fems Microbiol Ecol. 1997;24:93–101. doi: 10.1016/S0168-6496(97)00038-X. [DOI] [Google Scholar]
- 23.Morono, Y. & Kallmeyer, J. In Developments in Marine Geology Vol. Volume 7 (eds Donna K. Blackman Fumio Inagaki Ruediger Stein & Larsen Hans-Christian) 65-83 (Elsevier, 2014).
- 24.Holmhansen O. Determination of Microbial Biomass in Ocean Profiles. Limnol Oceanogr. 1969;14:740. doi: 10.4319/lo.1969.14.5.0740. [DOI] [Google Scholar]
- 25.Venkateswaran K, Hattori N, La Duc MT, Kern R. ATP as a biomarker of viable microorganisms in clean-room facilities. J Microbiol Meth. 2003;52:367–377. doi: 10.1016/S0167-7012(02)00192-6. [DOI] [PubMed] [Google Scholar]
- 26.Velten S, Hammes F, Boller M, Egli T. Rapid and direct estimation of active biomass on granular activated carbon through adenosine tri-phosphate (ATP) determination. Water Res. 2007;41:1973–1983. doi: 10.1016/j.watres.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 27.Castillo M, et al. Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet Microbiol. 2006;114:165–170. doi: 10.1016/j.vetmic.2005.11.055. [DOI] [PubMed] [Google Scholar]
- 28.Smith CJ, Osborn AM. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. Fems Microbiol Ecol. 2009;67:6–20. doi: 10.1111/j.1574-6941.2008.00629.x. [DOI] [PubMed] [Google Scholar]
- 29.Hammes F, et al. Flow-cytometric total bacterial cell counts as a descriptive microbiological parameter for drinking water treatment processes. Water Res. 2008;42:269–277. doi: 10.1016/j.watres.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 30.Foladori P, Bruni L, Tamburini S, Ziglio G. Direct quantification of bacterial biomass in influent, effluent and activated sludge of wastewater treatment plants by using flow cytometry. Water Res. 2010;44:3807–3818. doi: 10.1016/j.watres.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 31.Buesing N, Gessner MO. Comparison of detachment procedures for direct counts of bacteria associated with sediment particles, plant litter and epiphytic biofilms. Aquat Microb Ecol. 2002;27:29–36. doi: 10.3354/ame027029. [DOI] [Google Scholar]
- 32.Jenkinson DS, Powlson DS. Effects of Biocidal Treatments on Metabolism in Soil. 1. Fumigation with Chloroform. Soil Biol Biochem. 1976;8:167–177. doi: 10.1016/0038-0717(76)90001-8. [DOI] [Google Scholar]
- 33.Vance ED, Brookes PC, Jenkinson DS. An Extraction Method for Measuring Soil Microbial Biomass-C. Soil Biol Biochem. 1987;19:703–707. doi: 10.1016/0038-0717(87)90052-6. [DOI] [Google Scholar]
- 34.Brant JB, Sulzman EW, Myrold DD. Microbial community utilization of added carbon substrates in response to long-term carbon input manipulation. Soil Biol Biochem. 2006;38:2219–2232. doi: 10.1016/j.soilbio.2006.01.022. [DOI] [Google Scholar]
- 35.Cao YF, Li Y, Li CH, Huang G, Lu GH. Relationship between presence of the desert shrub Haloxylon ammodendron and microbial communities in two soils with contrasting textures. Appl Soil Ecol. 2016;103:93–100. doi: 10.1016/j.apsoil.2016.03.011. [DOI] [Google Scholar]
- 36.Kaur A, Chaudhary A, Kaur A, Choudhary R, Kaushik R. Phospholipid fatty acid - A bioindicator of environment monitoring and assessment in soil ecosystem. Curr Sci India. 2005;89:1103–1112. [Google Scholar]
- 37.Tunlid A, Hoitink HAJ, Low C, White DC. Characterization of Bacteria That Suppress Rhizoctonia Damping-Off in Bark Compost Media by Analysis of Fatty-Acid Biomarkers. Appl Environ Microb. 1989;55:1368–1374. doi: 10.1128/aem.55.6.1368-1374.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franzosa EA, et al. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat Rev Microbiol. 2015;13:360–372. doi: 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Props, R. et al. Absolute quantification of microbial taxon abundances. Isme J (2016). [DOI] [PMC free article] [PubMed]
- 40.Stammler, F. et al. Adjusting microbiome profiles for differences in microbial load by spike-in bacteria. Microbiome4 (2016). [DOI] [PMC free article] [PubMed]
- 41.Smets W, et al. A method for simultaneous measurement of soil bacterial abundances and community composition via 16S rRNA gene sequencing. Soil Biol Biochem. 2016;96:145–151. doi: 10.1016/j.soilbio.2016.02.003. [DOI] [Google Scholar]
- 42.Liang Y, et al. Long-term soil transplant simulating climate change with latitude significantly alters microbial temporal turnover. Isme J. 2015;9:2561–2572. doi: 10.1038/ismej.2015.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calbrix R, Laval K, Barray S. Analysis of the potential functional diversity of the bacterial community in soil: a reproducible procedure using sole-carbon-source utilization profiles. Eur J Soil Biol. 2005;41:11–20. doi: 10.1016/j.ejsobi.2005.02.004. [DOI] [Google Scholar]
- 44.Danovaro, R., Molari, M., Corinaldesi, C. & Dell’Anno, A. Macroecological drivers of archaea and bacteria in benthic deep-sea ecosystems. Science Advances2 (2016). [DOI] [PMC free article] [PubMed]
- 45.Prest EI, Hammes F, Kotzsch S, van Loosdrecht MCM, Vrouwenvelder JS. Monitoring microbiological changes in drinking water systems using a fast and reproducible flow cytometric method. Water Res. 2013;47:7131–7142. doi: 10.1016/j.watres.2013.07.051. [DOI] [PubMed] [Google Scholar]
- 46.Sun DL, Jiang X, Wu QLL, Zhou NY. Intragenomic Heterogeneity of 16S rRNA Genes Causes Overestimation of Prokaryotic Diversity. Appl Environ Microb. 2013;79:5962–5969. doi: 10.1128/AEM.01282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buyer JS, Sasser M. High throughput phospholipid fatty acid analysis of soils. Appl Soil Ecol. 2012;61:127–130. doi: 10.1016/j.apsoil.2012.06.005. [DOI] [Google Scholar]
- 48.Xu, W. F. et al. A meta-analysis of the response of soil moisture to experimental warming. Environ Res Lett8 (2013).
- 49.Ollivier J, et al. Bacterial community structure in soils of the Tibetan Plateau affected by discontinuous permafrost or seasonal freezing. Biol Fert Soils. 2014;50:555–559. doi: 10.1007/s00374-013-0869-4. [DOI] [Google Scholar]
- 50.Wang, X. B. et al. Scale-dependent effects of climate and geographic distance on bacterial diversity patterns across northern China’s grasslands. Fems Microbiol Ecol91 (2015). [DOI] [PubMed]
- 51.He ZL, et al. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett. 2010;13:564–575. doi: 10.1111/j.1461-0248.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- 52.Ul-Hassan, A. & Wellington, E. M. Actinobacteria. Encyclopedia of Microbiology 25-44 (2009).
- 53.Jangid K, et al. Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biol Biochem. 2008;40:2843–2853. doi: 10.1016/j.soilbio.2008.07.030. [DOI] [Google Scholar]
- 54.Fang X, et al. Warming effects on biomass and composition of microbial communities and enzyme activities within soil aggregates in subtropical forest. Biol Fert Soils. 2016;52:353–365. doi: 10.1007/s00374-015-1081-5. [DOI] [Google Scholar]
- 55.Leibold MA, McPeek MA. Coexistence of the niche and neutral perspectives in community ecology. Ecology. 2006;87:1399–1410. doi: 10.1890/0012-9658(2006)87[1399:COTNAN]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 56.Lakzayi M, Moradi H, Sabbagh E, Rigi K. Effect of vermicomposting on microbial biomass in contaminated soil. Journal of Biodiversity and Environmental Science. 2015;6:17. [Google Scholar]
- 57.Ahmad SS, Nawaz M, Wahid A, Butt A. Response of soil microbial biomass and respiration in heavy metal contaminated soil of Multan. International Journal of Biosciences (IJB) 2015;7:68–77. doi: 10.12692/ijb/7.4.68-77. [DOI] [Google Scholar]
- 58.Wang YP, et al. The influence of soil heavy metals pollution on soil microbial biomass, enzyme activity, and community composition near a copper smelter. Ecotox Environ Safe. 2007;67:75–81. doi: 10.1016/j.ecoenv.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 59.Berga, M., Szekely, A. J. & Langenheder, S. Effects of Disturbance Intensity and Frequency on Bacterial Community Composition and Function. Plos One7 (2012). [DOI] [PMC free article] [PubMed]
- 60.Brookes PC, Landman A, Pruden G, Jenkinson DS. Chloroform Fumigation and the Release of Soil-Nitrogen - a Rapid Direct Extraction Method to Measure Microbial Biomass Nitrogen in Soil. Soil Biol Biochem. 1985;17:837–842. doi: 10.1016/0038-0717(85)90144-0. [DOI] [Google Scholar]
- 61.Witt C, Gaunt JL, Galicia CC, Ottow JCG, Neue HU. A rapid chloroform-fumigation extraction method for measuring soil microbial biomass carbon and nitrogen in flooded rice soils. Biol Fert Soils. 2000;30:510–519. doi: 10.1007/s003740050030. [DOI] [Google Scholar]
- 62.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo YQ, Hui DF, Zhang DQ. Elevated CO2 stimulates net accumulations of carbon and nitrogen in land ecosystems: A meta-analysis. Ecology. 2006;87:53–63. doi: 10.1890/04-1724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


