Abstract
Copper is an essential element for proper organismal development and is involved in a range of processes, including oxidative phosphorylation, neuropeptide biogenesis, and connective tissue maturation. The copper transporter (Ctr) family of integral membrane proteins is ubiquitously found in eukaryotes and mediates the high-affinity transport of Cu+ across both the plasma membrane and endomembranes. Although mammalian Ctr1 functions as a Cu+ transporter for Cu acquisition and is essential for embryonic development, a homologous protein, Ctr2, has been proposed to function as a low-affinity Cu transporter, a lysosomal Cu exporter, or a regulator of Ctr1 activity, but its functional and evolutionary relationship to Ctr1 is unclear. Here we report a biochemical, genetic, and phylogenetic comparison of metazoan Ctr1 and Ctr2, suggesting that Ctr2 arose over 550 million years ago as a result of a gene duplication event followed by loss of Cu+ transport activity. Using a random mutagenesis and growth selection approach, we identified amino acid substitutions in human and mouse Ctr2 proteins that support copper-dependent growth in yeast and enhance copper accumulation in Ctr1−/− mouse embryonic fibroblasts. These mutations revert Ctr2 to a more ancestral Ctr1-like state while maintaining endogenous functions, such as stimulating Ctr1 cleavage. We suggest key structural aspects of metazoan Ctr1 and Ctr2 that discriminate between their biological roles, providing mechanistic insights into the evolutionary, biochemical, and functional relationships between these two related proteins.
Keywords: copper, copper transport, evolution, metal homeostasis, yeast, Ctr1, Ctr2
Introduction
Copper is a critical metal for all forms of eukaryotic life because of its ability to shuttle electrons and cycle between oxidation states, Cu+ and Cu2+, under biologically relevant conditions. As such, this redox property has been harnessed during the evolution of copper-containing enzymes to participate in many diverse functions, such as respiration, superoxide disproportionation, pigmentation, connective tissue maturation, and neuropeptide biogenesis (1–3). Because of the critical importance of copper, life has evolved methods to ensure appropriate acquisition, distribution, and storage. Defects in copper metabolism are linked with pathologies that include neutropenia, cardiomyopathy, Menkes syndrome, Wilson's disease, and peripheral neuropathy (4–7).
The baker's yeast Saccharomyces cerevisiae has yielded numerous insights into the biology of copper acquisition. For example, S. cerevisiae was instrumental in the discovery of copper transporter 1 (Ctr1),3 a homotrimeric plasma membrane-spanning protein expressed in eukaryotes and responsible for the uptake of extracellular copper (8, 9). Ctr1 contains a large extracellular amino-terminal metal-binding domain rich in methionine residues that facilitates high-affinity Cu+ transport through the transmembrane pore (10). Lining the transmembrane pore is an M-X3-M motif present on transmembrane domain two that is proposed to function as a Cu+ selectivity filter (11, 12). Mutation of these residues to leucine results in a protein incapable of Cu+ transport. The third transmembrane domain contains a glycine zipper motif that is required for multimerization of Ctr1 monomers into functional Cu+-transporting homotrimers (13, 14). Because of the high selectivity of Cu+, Ctr1 requires a cell-surface reductase to reduce extracellular Cu2+ to Cu+ for transport (15, 16). Loss of Cu2+ reductase activity results in diminished Ctr1-mediated Cu+ transport. S. cerevisiae also expresses a redundant high-affinity Cu+ transporter, Ctr3 (17). Although Ctr3 also possesses a critical MXXXM motif and glycine zipper, the amino-terminal metal-binding domain is significantly smaller than that of Ctr1. Also, although the amino-terminal domain of Ctr1 contains several methionine-rich regions, the Ctr3 amino-terminal domain lacks these and instead relies on cysteine residues for Cu+ transport. Other fungi also possess redundant high-affinity Cu+ transporters, such as Ctr4 and Ctr5 from Schizosaccharomyces pombe and Ctr1 and Ctr4 from the distantly related Cryptococcus neoformans (18, 19). When in the cytosol, copper is inserted into copper-dependent enzymes for which it serves as a co-factor, shuttled to the secretory compartment or mitochondria, stored inside the vacuole, or bound to the Ace1 metalloregulatory transcription factor, where it activates expression of the Cu+-binding and detoxifying metallothioneins. In S. cerevisiae, vacuolar Cu+ is mobilized to the cytosol by Ctr2, an integral membrane protein structurally similar to Ctr1, in concert with a vacuole-localized metalloreductase (20, 21).
Multiple organisms, including the fission yeast S. pombe, the alga Chlamydomonas reinhardtii, the woodlouse Oniscus asellus, and the American lobster Homarus americanus, store copper in a vacuole-like compartment (22–26). Indeed, lysosomal/endosomal copper stores have been identified in hepatocytes of a mouse model for Wilson's disease, a genetic disorder characterized by increased hepatic copper levels (27). Presumably this copper is trapped in storage vesicles because of the inability to export copper into the bile. Although there is widespread evidence for vesicular copper stores in metazoans, it is currently unclear how endosomal copper stores are mobilized to the cytoplasm. A protein related to the mammalian Ctr1 high-affinity Cu+ transporter, Ctr2, was originally identified based on its sequence homology to Ctr1 (28). Like Ctr1, the Ctr2 protein possesses three membrane-spanning domains, assembles into a homomultimer, and possesses an MXXXM motif in trans-membrane domain two that is absolutely required for Cu+ transport by all bona fide Ctr1-like Cu+ transporter proteins. Moreover, Ctr1 and Ctr2 associate in a complex in vivo, show similar tissue selectivity for high expression levels, and the Ctr1 and Ctr2 genes are genetically linked in the mouse and human genome (29). However, Ctr1 and Ctr2 have distinct functions in copper metabolism based on a number of features. Although initially proposed to function directly as a low-affinity copper importer at the plasma membrane or as a lysosomal copper exporter, work in our laboratory suggests that Ctr2 does not function as a direct copper transporter but rather functions as a regulator of Ctr1 copper transport activity (30–32). First, although Ctr1 localizes to both the plasma membrane and intracellular vesicles, endogenous Ctr2 is confined to an endosomal compartment (33–37). Second, in contrast to loss of Ctr1, which causes a reduction in copper accumulation, loss of Ctr2 in mouse tissues and in isolated mouse embryonic fibroblasts (MEFs) results in increased cell-associated copper. Third, Ctr2 expression modulates cellular copper accumulation even when both methionine residues of the essential MXXXM motif, are mutated to LXXXL (29). Fourth, unlike all other Ctr1-like proteins analyzed from fungi, plants, reptiles, and mammals, expression of the mouse or human Ctr2 protein fails to complement the copper deficiency phenotype of S. cerevisiae cells lacking a functional copper transporter (38, 39).
Consistent with the loss of Ctr2 resulting in increased cellular and tissue copper, Ctr2 stimulates the cleavage of the Ctr1 copper-binding ecto-domain, in a mechanism involving an initial, rate-limiting ectodomain cleavage by the Cathepsin L endosomal protease (40). Cleavage of the Ctr1 ecto domain results in a truncated form of Ctr1 (tCtr1) that supports reduced cellular Cu accumulation as compared with full-length Ctr1. Therefore, loss of Ctr2 maintains full-length Ctr1, thereby driving increased Cu uptake at the plasma membrane. In MEFs and some cell types the increased cell-associated Cu is preferentially localized to endosomal compartments, suggesting a role for Ctr2 in the mobilization of endosomal Cu stores. The observation that tCtr1 is capable of mediating the release of vesicular copper stores, whereas the full-length Ctr1 is not, suggests that Ctr2-mediated generation of tCtr1 functions to regulate both copper uptake at the plasma membrane and endosomal copper mobilization by modulating the cleavage of the Ctr1 ecto-domain (41, 42). Given the requirement for both Ctr2 and Cathepsin L for Ctr1 ecto-domain cleavage and the copper hyperaccumulation phenotypes because of loss of either component, these observations support an indirect role for Ctr2 in regulating Ctr1-mediated copper transport at the plasma membrane and from endosomal compartments. Together, these observations raise the question of the origin of metazoan Ctr2.
Here we report a biochemical and molecular phylogenetic analysis of eukaryotic Ctr1 and Ctr2 proteins suggesting that metazoan Ctr2 arose ∼550 million years ago through a gene duplication event involving the Ctr1 genomic locus. The resulting Ctr2 has since experienced a more rapid exploration of mutagenic space, a hallmark of neo-functionalization, losing the ability to transport copper but gaining the ability to regulate Ctr1 cleavage. By performing an unbiased mutagenic screen followed by biochemical analyses, we isolated single point mutants of mammalian Ctr2 that restore copper transport activity without loss of its function in Ctr1 ecto-domain cleavage. Interestingly, these point mutations change Ctr2 residues that are highly conserved among metazoan Ctr2 into the corresponding highly conserved Ctr1 residues, thus reversing the direction of evolution. These studies define key residues involved in copper transport and shed light on the evolutionary biochemical history of metazoan copper homeostasis.
Results
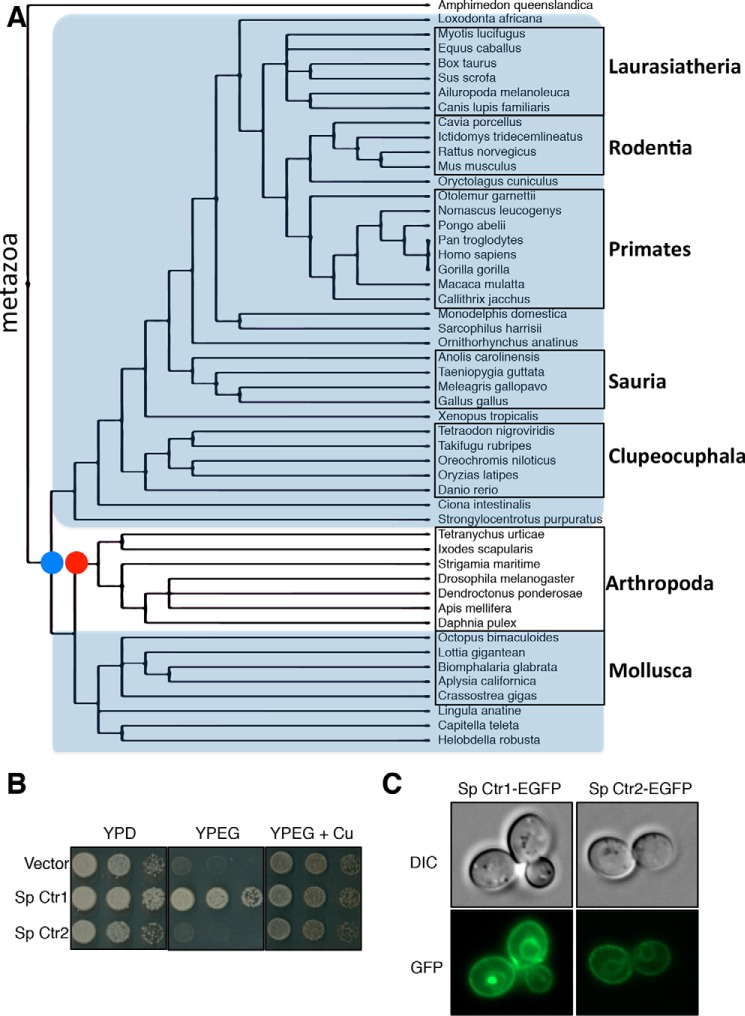
In an effort to understand the origins of mammalian Ctr2, 125 Ctr protein sequences from a wide variety of eukaryotic organisms were analyzed via a molecular phylogenetic approach. Rather than solely constructing a sequence alignment and performing maximum likelihood estimation, we sought to utilize well established taxonomy data to inform tree building (43). To this end, we built a Ctr phylogenetic tree guided by a species tree utilizing the program TreeBeST, coupled with curated taxonomic data from NCBI (Fig. 1A) (44). Both mismatch and Jones-Taylor-Thornton distance models yielded two clusters of proteins corresponding to either Ctr1 or Ctr2. All species analyzed contained at least one copy of Ctr1. Analysis of species harboring a Ctr2-encoding gene suggests that, at the beginning of the metazoan lineage, here represented by the sea sponge Amphimedon queenslandica, Ctr2 was not present. At the time of the protostome/deuterostome divergence, ∼550 million years ago, Ctr2 appeared and was maintained in subsequent species, with the notable exception of the phylum Arthropoda (45). This is in agreement with previous studies that identified and analyzed the Drosophila melanogaster Ctr proteins Ctr1A, Ctr1B, and Ctr1C, indicating that all three have Cu+ transport activity and suggest that they are more closely related to Ctr1 rather than Ctr2 (46–48).
Figure 1.
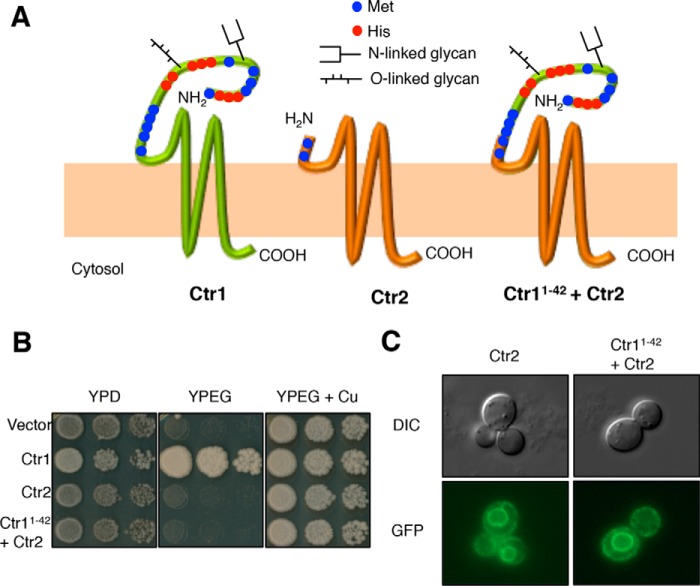
Ctr2 appearance through evolution as a uniquely metazoan gene. A, Ctr protein sequences from the indicated species were identified and retrieved from the NCBI Protein database by iterative BLAST searches. Sequences were aligned with MUSCLE and subjected to TreeBeST analysis to reveal which species possesses predicted Ctr1 and/or Ctr2. The species tree was created from the NCBI Taxonomy database. A blue dot represents the predicted appearance of Ctr2, whereas the red dot represents the predicted loss of Ctr2. Species classification is shown on the right. B, S. cerevisiae ctr1Δctr3Δ cells were transformed with the indicated plasmids and plated on the indicated medium. Sp, Strongylocentrotus purpuratus. C, the same yeast strains as in B were inspected by fluorescence microscopy and photographed. DIC, differential interference contrast.
A key functional difference between mammalian Ctr1 and Ctr2 is that Ctr1 acts as a functional Cu+ transporter when expressed in S. cerevisiae cells lacking endogenous high-affinity Cu+ transporters, whereas Ctr2 does not show Cu+ transport in this sensitive yeast assay (29). To test the Ctr1 versus Ctr2 protein assignments made by the Ctr phylogeny tree generated (Fig. 1A), the ability of Ctr1 and Ctr2 proteins to transport Cu was interrogated by their ability to support the growth of ctr1Δctr3Δ S. cerevisiae cells on non-fermentable medium (YPEG), which requires a functional, copper-loaded cytochrome c oxidase (49). The assigned Ctr1 and Ctr2 proteins from the sea urchin Strongylocentrotus purpuratus were used to test this prediction. S. purpuratus cDNAs encoding the predicted Ctr1 and Ctr2 proteins were subcloned into yeast expression vectors containing a carboxyl-terminal GFP fusion and tested for the ability to support YPEG growth in S. cerevisiae ctr1Δctr3Δ cells. In agreement with the phylogenic prediction model, sea urchin Ctr1 restored growth on YPEG, whereas expression of Ctr2 did not support copper-dependent growth (Fig. 1B). Both Ctr1 and Ctr2 were expressed and localized to the plasma membrane and the vacuole (Fig. 1C). These results suggest that the gene referred to as mammalian Ctr2 is uniquely metazoan and was not present in lower organisms but appears around the time of the protostome/deuterostome divergence.
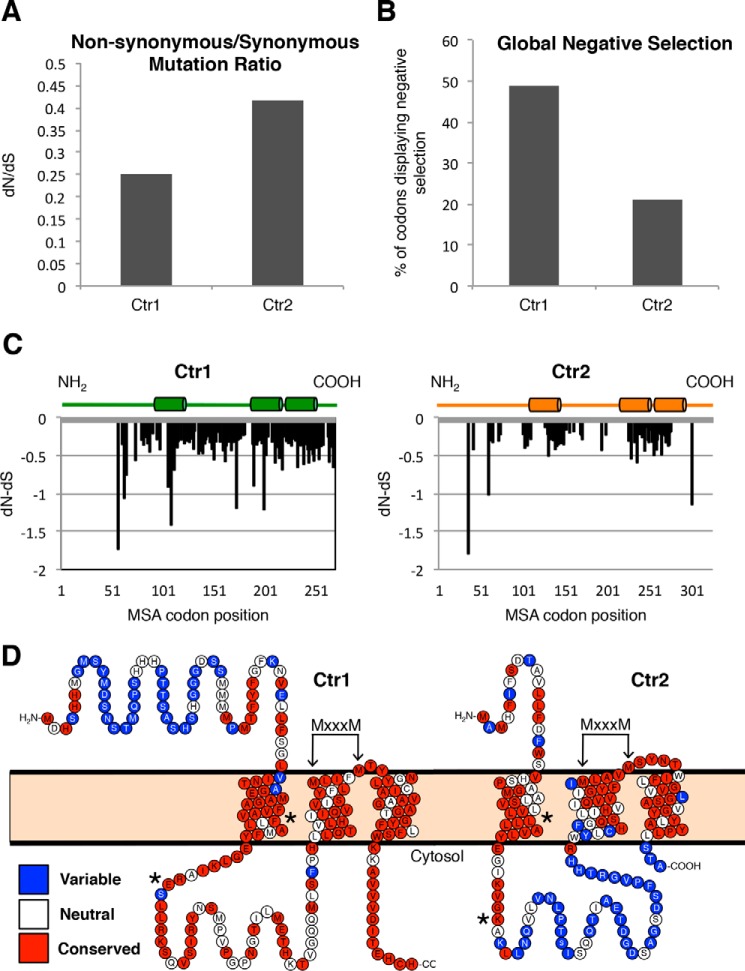
A number of mechanisms have been suggested to drive the appearance of new genes, including horizontal gene transfer, chimeric gene fusion between portions of existing genes either through recombination as DNA or via an RNA intermediate, as well as the de novo origination of genes from a previously nonfunctional genomic sequence (50, 51). Because of the sequence and overall structural similarities between the mammalian Ctr1 and Ctr2 proteins, we hypothesized that Ctr2 may have arisen from gene duplication followed by neo-functionalization (52). A hallmark of neo-functionalization is that the daughter gene undergoes more rapid mutation compared with the parental gene because of the pressure on the parental copy to maintain function, whereas the daughter copy is free to explore evolutionary space. To determine whether Ctr2 has experienced a greater mutational force over time, we compiled Ctr1 and Ctr2 sequences from 35 metazoan species and compared the ratio of non-synonymous mutations (nucleic acid changes that alter the protein sequence) with synonymous mutations (those that do not alter the protein sequence), referred to as dN/dS (Fig. 2A). Indeed, Ctr2 shows a higher global dN/dS ratio than Ctr1, as would be expected from a duplicated and neo-functionalized gene (53, 54). When dN and dS are examined on an individual codon basis, Ctr1 has a much higher percentage of codons displaying dN<dS (negative selection) than Ctr2 (Fig. 2B).
Figure 2.
Ctr2 demonstrates relaxed purifying selection. A, predicted Ctr family genes from 51 metazoan species were aligned, and single-likelihood ancestor counting analysis was performed under standard parameters using the Datamonkey server. Global dN/dS values are reported. B, the number of individual codons from the Ctr1 or Ctr2 DNA alignment that display negative selection (p ≤ 0.05) were divided by the number of total codons in the Ctr1 or Ctr2 alignment, respectively. C, normalized dN/dS values for each codon displaying selection (p ≤ 0.05) for Ctr1 and Ctr2 alignments are plotted. Cylinders represent transmembrane domains. D, protein sequences from metazoan Ctr1 and Ctr2 were aligned and analyzed for conservation with the ConSurf server using a Bayesian evolution method. Shown are the human Ctr1 and Ctr2 proteins with amino acids that represent variable (blue), neutral (white), or conserved (red) positions. Arrows point to the highly conserved MXXXM motif necessary for copper transport. Asterisks denote residues Phe-77 and Glu-91 in human Ctr1 as well as Leu-34 and Lys-47 in human Ctr2.
To identify whether there are regions of Ctr1 and Ctr2 most subject to negative selection, codons which are significantly selected for are displayed with the protein topology overlaid (Fig. 2C). This analysis shows that, with the exception of the 5′-most region, encoding the amino-terminal region of the protein, Ctr1 is subject to similar negative selection across the entirety of the gene. However, although Ctr2 was negatively selected for preferentially in regions encoding the three transmembrane domains, regions predicted to encode soluble portions of the protein, such as the amino and carboxyl termini and the loop between transmembrane domains one and two, are not subjected to negative mutational pressure. To visualize the amino acids most subject to mutation, the human Ctr1 and Ctr2 sequences were analyzed via the ConSurf server (Fig. 2D) (55). Residue conservation agrees with dN/dS analysis in that the ecto-domain of Ctr1 is highly variable over time, but the remainder of the protein is highly conserved among Ctr1 proteins. Along with the demonstration that the lack of a functional Ctr1 is embryonic lethal (56, 57), whereas mice lacking Ctr2 do not show an overt growth phenotype, this analysis suggests that Ctr2 was more free to explore mutagenic space than Ctr1. The suggestion that mutation of Ctr1 is more detrimental to organismal fitness than mutation of Ctr2 supports a model in which Ctr2 arose via Ctr1 duplication and neo-functionalization.
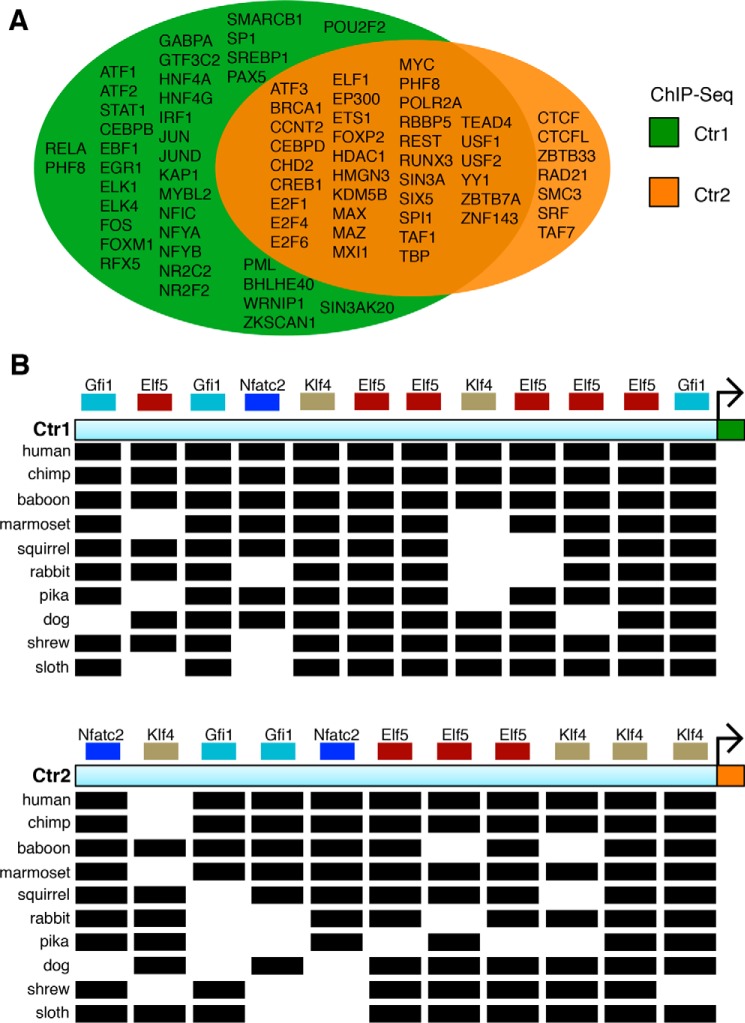
Gene duplication can occur from the duplication of a segment of DNA as well as the expression and retrotransposition of RNA inserted into chromosomal DNA (50). In the latter case, the newly duplicated gene would be expected to be under the control of an entirely different promoter compared with the parental gene. To probe the nature of the gene duplication event that gave rise to Ctr2, the genomic architecture for human Ctr1 and Ctr2 was compared. Analysis of ChIP-seq data from the human ENCODE (Encyclopedia of DNA Elements) project shows a significant overlap in the repertoire of proteins bound within a 1000-base pair region upstream of the transcription start site for Ctr1 and Ctr2 (Fig. 3A), suggesting that a similar set of transcription factors controls the expression of the Ctr1 and Ctr2 genes (58). To investigate this in more detail, 500 base pairs upstream of the transcription start sites for Ctr1 and Ctr2 were analyzed from 10 separate species with the ConTra promoter alignment tool (Fig. 3B) (59). Four transcription factors broadly involved in blood cell development, epithelial formation, immune response, and stem cell maintenance (Gfi1, Elf5, Nfatc2, and Klf4, respectively) were found to be highly conserved in the 10 species analyzed in both Ctr1 and Ctr2, suggesting similar transcriptional regulation. Consistent with this observation, we previously demonstrated that Ctr1 and Ctr2 mRNA levels are similar across a number of human and mouse tissues (28, 29). Both Ctr1 and Ctr2 transcript levels are high in the liver, which functions as the primary Cu storage organ, but relatively low in skeletal muscle, presumably because of the differentiated nature of this cell type.
Figure 3.
The Ctr1 and Ctr2 genes contain similar promoter elements. A, DNA regions 1 kb upstream of the transcription start site, located at the 5′ region of the 5′ UTR, for Ctr1 and Ctr2 were identified via the UCSC Genome Browser, and ChIP-seq data from the ENCODE Consortium were analyzed. Proteins that bound to both promoters are displayed in the center of the Venn diagram, whereas those found to occupy only one of the two promoters are shown in the non-overlapping portion of the diagram. B, Ctr1 and Ctr2 promoters from the indicated 10 species were analyzed with the ConTra v2 server under standard parameters for predicted transcription factor binding sites. Transcription factors are displayed as colored blocks placed above their binding location. A black rectangle indicates the predicted presence of a transcription factor-binding site in the indicated species.
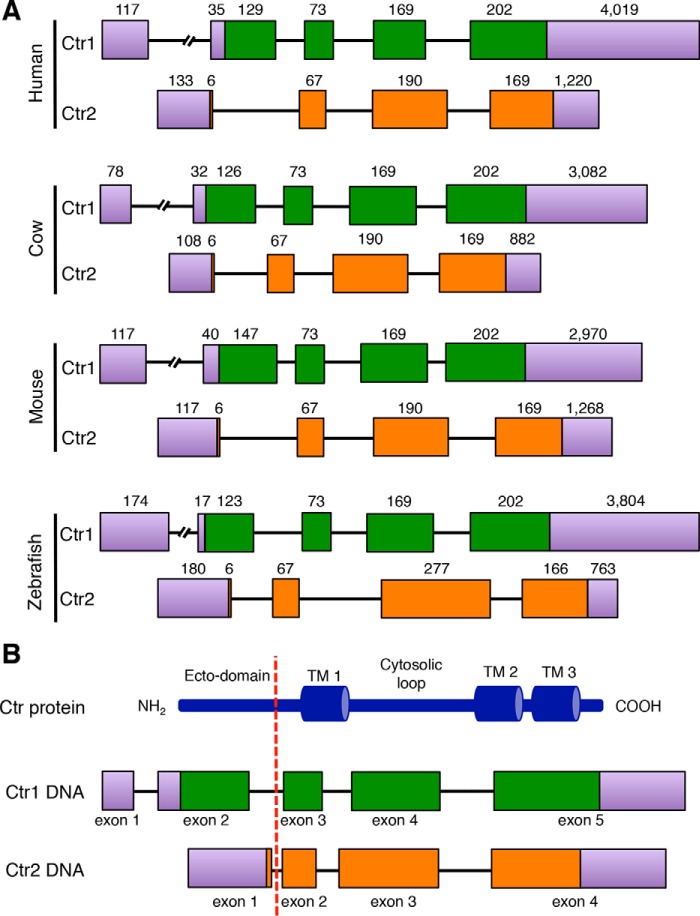
If Ctr2 arose from a duplication of the Ctr1 genomic locus rather than through the retrotransposition of an RNA intermediate, then the two genes would be expected to have a similar arrangement of introns and exons. To address this, we examined the genomic architecture of Ctr1 and Ctr2 from the fully sequenced human, cow, mouse, and zebrafish genomes (Fig. 4A). The overall intron/exon architecture shows four coding exons for both transcripts, with the Ctr1 gene containing a non-coding exon at the 5′ end of the transcript (Fig. 4B). Although the final three exons are of similar size throughout all species examined, there exists a notable difference between Ctr1 and Ctr2 in the first coding exon. For Ctr1, this exon codes for a large metal-binding domain rich in His, Met, and Cys residues. In all Ctr2 genes examined, this first exon encodes only two amino acids, one of which is the initiator methionine. It appears that, although the 5′ (promoter) and 3′ regions of the Ctr1 gene were retained in Ctr2, the first exon was largely deleted, generating a Ctr2 coding region that would lack an amino-terminal metal binding ecto-domain, perhaps explaining the lack of copper transport activity observed for Ctr2.
Figure 4.
Ctr2 genomic loci are similar to Ctr1 but lack an exon coding for the copper-binding ecto-domain. A, genomic data for Ctr1 and Ctr2 from the listed species were downloaded from Ensembl and aligned to compare intron/exon structure. Exons are represented by green (Ctr1) and orange (Ctr2) rectangles, whereas introns are displayed as black lines. Purple rectangles represent UTRs. B, depiction of the general architecture of Ctr genes with regions encoding the metal binding ecto-domain, the intracellular loop, three transmembrane domains, and the cytosolic tail. Ctr1 and Ctr2 share a similar intron/exon structure, with the notable exception that Ctr2 lacks the exon coding for a large ecto-domain. The dashed red line indicates the region of difference.
To test this possibility, a Ctr1-Ctr2 chimera was constructed in which the first 42 amino acids from human Ctr1, comprising the copper-binding ecto-domain, were fused to Ctr2 at the amino terminus (Fig. 5A). It has been suggested that the Ctr1 ecto-domain functions to concentrate copper ions at the mouth of the transmembrane pore and thus increase the local concentration of copper available to be transported (60–65). We hypothesized that this function will allow Ctr2 to rescue copper transport-dependent growth; however, addition of the Ctr1 ecto-domain to Ctr2 was not able to confer growth on non-fermentable medium (Fig. 5B) despite the observation that both Ctr2 and Ctr1 (1–42) + Ctr2 were expressed as GFP fusion proteins localized to the plasma membrane and vacuole (Fig. 5C). This suggests that deletion of the Ctr1 first coding exon, resulting in the absence of a large ecto-domain, is not solely responsible for the lack of copper transport function for Ctr2.
Figure 5.
A metal-binding ectodomain is not sufficient to induce Ctr2-mediated rescue of respiration-dependent growth. A, model showing monomeric full-length human Ctr1 and Ctr2 along with a chimeric protein consisting of the first 42 residues of Ctr1 fused to Ctr2. Metal-binding Met and His residues are indicated in blue and red, respectively, and glycans are indicated with branches. B, the indicated proteins were expressed in S. cerevisiae ctr1Δctr3Δ cells and plated onto the indicated growth medium. C, the same cells as in B were evaluated by fluorescence microscopy and photographed. DIC, differential interference contrast.
To explore sequences that account for copper transport differences between Ctr1 and Ctr2, we hypothesized that it should be possible to “reverse” these mutations to create a copper transport-competent Ctr2. An unbiased mutagenic screen was developed in which yeast-Escherichia coli shuttle plasmids encoding mouse Ctr2 were propagated in DNA repair-deficient E. coli (Fig. 6A). Sequencing validated that each Ctr2 gene harbored an average of one to two point mutations. The mutated plasmid pool was transformed into ctr1Δctr3Δ yeast cells and selected on YPEG medium in which only cells harboring copper transport activity grow. Of 26,000 colony-forming units plated on YPEG, 34 independent transformants grew. Plasmids rescued from these transformants were sequenced and represent six unique Ctr2 mutants, with each plasmid containing only one mutation per gene (Fig. 6B). Of the six unique mutations identified, two showed significantly more robust growth than the others and were examined in more detail.
Figure 6.
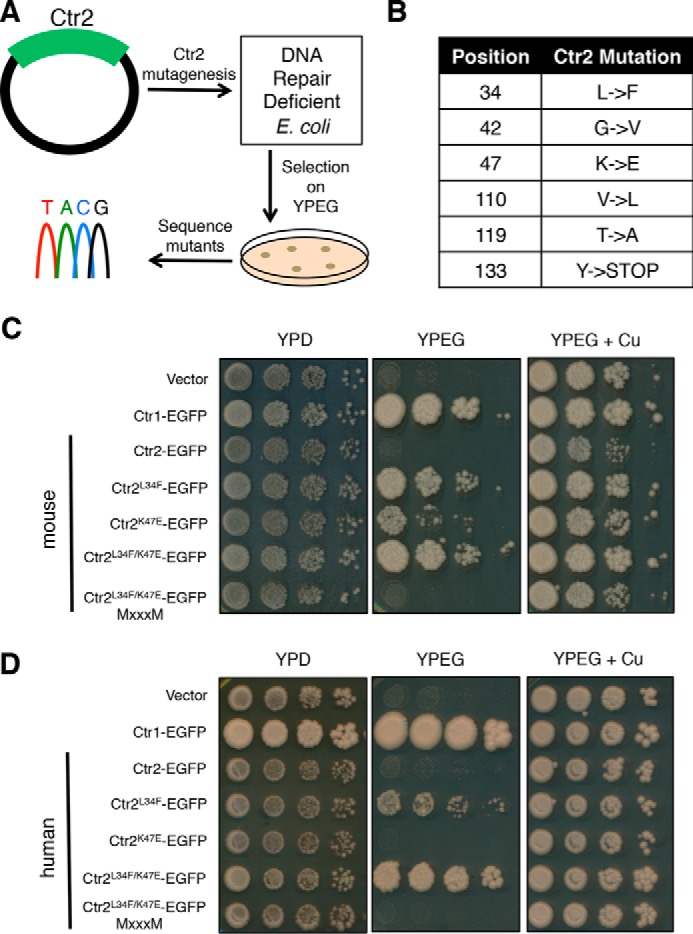
Random mutagenesis identifies Ctr2 mutants capable of rescuing respiration-dependent growth in yeast. A, scheme depicting the strategy for isolating Ctr2 mutants capable of copper-dependent growth. Ctr2 expression vectors were transformed into XL-1 E. coli and propagated for 24 h before plasmid harvest. The plasmid pool was transformed into S. cerevisiae ctr1Δctr3Δ cells and plated on YPEG medium. Colonies were isolated, and plasmids were sequenced. B, table displaying the six unique Ctr2 mutations capable of conferring growth on YPEG. C, cells containing the indicated mouse genes were spotted onto the indicated medium. MXXXM indicates mutation of transmembrane domain two methionine residues critical for Cu+ transport activity to leucine. D, mutations that were identified in the mouse Ctr2-coding region were mutated in the human Ctr2 gene and assayed for growth in a manner similar as in C.
These two variants of interest harbored Ctr2 changes at codons L34F and K47E, respectively. To verify that these mutations are solely responsible for the observed growth on YPEG, these same changes were separately introduced into the mouse Ctr2 coding region and fused with a carboxyl-terminal EGFP tag, and recipient ctr1Δ ctr3Δ cells were plated on YPEG (Fig. 6C). Although both individual mutations are able to rescue growth of this strain lacking endogenous high-affinity Cu+ transporters, K47E does not complement as well as L34F. When combined, the double mutant K47E/L34F supports enhanced growth relative to either single mutations and is dependent on the MXXXM motif in transmembrane domain 2 that is absolutely required for Cu+ transport by all known Ctr1 proteins (66, 67). As the screen was performed using mouse Ctr2, the corresponding mutations were made in a separate yeast vector expressing human Ctr2 fused to a carboxyl-terminal EGFP tag to test this observation in another mammalian Ctr2. Although expression of the human Ctr2 K47E allele was unable to rescue growth, Ctr2 L34F demonstrated copper transport-dependent growth (Fig. 6D). Moreover, ctr1Δ ctr3Δ cells expressing human Ctr2 harboring both mutations shows enhanced growth beyond either mutation alone, suggesting that the K47E mutation is necessary for the conversion of Ctr2 into a high-affinity Cu+ transport-competent molecule. As with the mouse Ctr2 variant proteins, the MXXXM motif is required for copper-dependent growth in this context.
Inspection of the K47E and L34F amino acid substitutions revealed that both of the mutations were in residues that are highly conserved in Ctr2 (Fig. 2D). This suggests that there has been a selection for these residues and that evolutionary pressure exists to maintain Ctr2 in a state that is not capable of supporting copper transport. Surprisingly, these mutations convert highly conserved Ctr2 residues into the identical residue found at the same position in Ctr1 (Fig. 7A). Moreover, these residues in Ctr1 are highly conserved, suggesting that they have been selected over time to maintain Ctr1 Cu+ transport activity. Thus, these mutations “revert” Ctr2 into a more Ctr1-like state that rescues growth on medium requiring a functional copper transporter to provide copper to cytochrome c oxidase.
Figure 7.
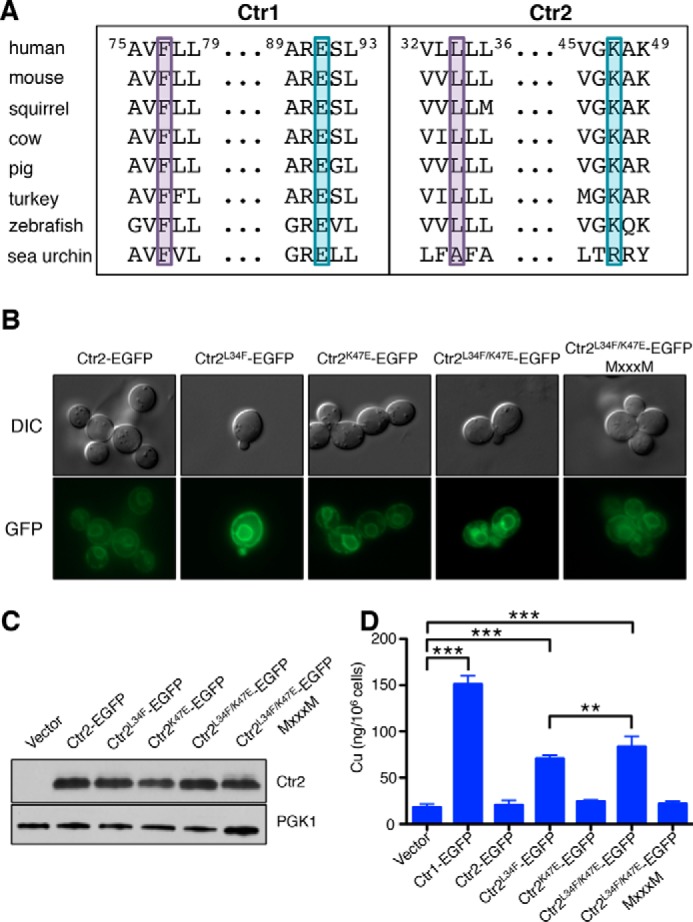
Ctr2 mutants are biochemically similar to Ctr1 and increase total cellular copper accumulation. A, Ctr1 and Ctr2 from the indicated species were aligned with MUSCLE. The two Ctr2 residues identified from the mutagenic screen, L34F and K47E, and the corresponding residues in Ctr1 are highlighted in purple and cyan, respectively. B, S. cerevisiae ctr1Δctr3Δ cells transformed with the indicated plasmids were inspected by fluorescence microscopy and photographed. DIC, differential interference contrast. C, the same cells as in B were harvested and immunoblotted with anti-GFP and PGK1 antibodies. D, the same cells as in B were analyzed by ICP-MS to determine whole-cell copper levels. Data are presented as mean ± S.E. from five biological replicates. **, p ≤ 0.01; ***, p ≤ 0.001.
To stimulate copper-dependent yeast cell growth, the Ctr2 mutants could either transport extracellular copper into the cytosol or mobilize vacuolar copper stores. Fluorescence microscopy of cells expressing the EGFP-tagged human Ctr2 and mutant derivatives revealed that wild-type and mutant Ctr2 fusion proteins localize to both the vacuole and the plasma membrane, a pattern typical for overexpressed membrane proteins (Fig. 7B). Immunoblotting for the Ctr2-EGFP fusions showed that the wild-type and mutant proteins are expressed at similar levels (Fig. 7C). Together, these data indicate that the mutations in Ctr2 do not grossly alter the stability or localization of the proteins expressed in yeast relative to wild-type Ctr2. To determine whether expression of the Ctr2 variants alters copper levels in yeast ctr1Δ ctr3Δ cells, ICP-MS analysis was used to measure total cell-associated copper levels (Fig. 7D). Human Ctr1 was expressed as a positive control and dramatically increased cellular Cu levels compared with cells harboring the empty vector. Expression of human Ctr2 L34F, but not K47E, led to increased cell-associated copper levels, which were further increased in cells expressing the Ctr2 L34F/K47E allele, in agreement with the enhanced copper-dependent growth observed with the Ctr2 double mutant. These observations suggest that the Ctr2 mutants selected for copper-dependent growth in yeast also support the accumulation of copper in yeast cells in a manner dependent on the conserved MXXXM motif, which is essential for Cu+ transport by Ctr1.
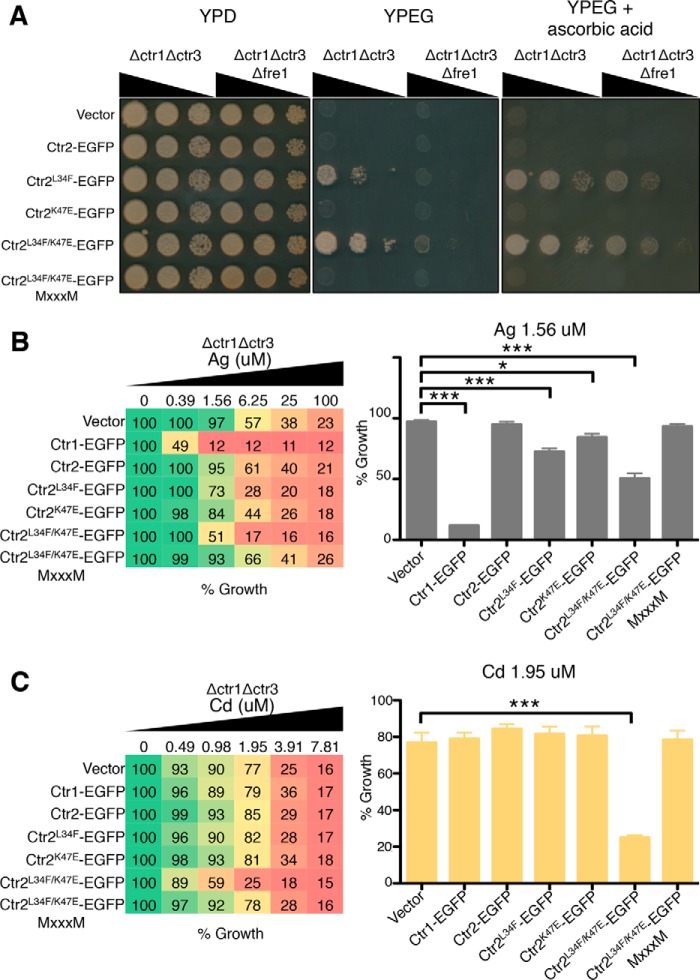
A hallmark of the Ctr1 copper transporters is their exquisite specificity for transporting Cu+ rather than Cu2+. To investigate whether the copper transport-competent Ctr2 mutants display specificity for Cu+, the human Ctr2 mutants were expressed in a ctr1Δctr3Δfre1Δ yeast strain that lacks both the endogenous high-affinity Cu+ transporters and a major cell surface Cu2+ metalloreductase, Fre1, that is required for Ctr1 and Ctr3 function (Fig. 8A) (15, 16). The Ctr2 mutants that support copper-dependent growth in ctr1Δ ctr3Δ cells did not support growth when expressed in an isogenic strain also lacking the Cu2+ metalloreductase Fre1. When the medium was supplemented with the Cu2+-reducing agent ascorbic acid, Ctr2 L34F and Ctr2 L34F/K47E rescued copper-dependent growth in both the ctr1Δ ctr3Δ and ctr1Δ ctr3Δ fre1Δ strains. This suggests that the Ctr2 mutants, like Ctr1, preferentially transport Cu+ over Cu2+.
Figure 8.
Ctr2 transport activating mutations impart selectivity for Cu+ and increase cadmium sensitivity. A, S. cerevisiae ctr1Δctr3Δ and ctr1Δctr3Δfre1Δ cells were transformed with the indicated expression plasmids and plated on the indicated medium, with wedges indicating decreasing cell density plated. B, S. cerevisiae ctr1Δctr3Δ cells were transformed with the indicated plasmids, transferred to medium containing the indicated concentration of AgNO3, and grown for 24 h with agitation at 30 °C. Data are shown as percent growth of strain grown in medium without treatment, with the histogram displaying data for growth in 1.56 μm AgNO3. C, S. cerevisiae ctr1Δctr3Δ cells were transformed with the listed plasmids and transferred to medium containing the indicated amount of CdCl2 and grown for 24 h with agitation at 30 °C. Data are shown as percent growth of strain grown in medium without treatment, with the histogram displaying data for growth in 1.95 μm CdCl2. *, p ≤ 0.05; ***, p ≤ 0.001.
Ag+ is isoelectric with Cu+, with both metals possessing thiophilic binding properties, and when administered to biological systems, Ag+ can serve as a toxic mimetic for Cu+-dependent processes (49). Ctr1 has been shown previously to mediate Ag+ transport and sensitizes cells to Ag+ toxicity (68). Therefore, to further evaluate the copper ion preferred by Ctr2 mutant proteins, yeast cells expressing the human Ctr2 Cu+ transport-competent mutants were evaluated for Ag+ resistance. Indeed, yeast ctr1Δ ctr3Δ cells expressing Ctr2 L34F or Ctr2 L34F/K47E were significantly sensitized to a range of Ag+ concentrations, although not to the same degree as cells expressing Ctr1 (Fig. 8B). This is in accordance with the observation that these cells also accumulate less copper than Ctr1-expressing cells. Interestingly, although the human Ctr2 K47E mutant does not rescue growth on YPEG or increase cellular copper, it does sensitize cells to Ag+ toxicity compared with vector, whereas WT Ctr2 does not sensitize cells compared with vector alone (Fig. 8B). This could be due to the extreme toxicity of low levels of Ag+ even when transport is minimal. To explore the metal specificity for transport by the Ctr2 mutants, cells were exposed to several transition metals over a range of concentrations and analyzed for growth. Zinc, nickel, lithium, cobalt, manganese, lead, and mercury showed no difference in toxicity as a function of wild-type or mutant Ctr2 expression (data not shown). However, the Ctr2 L34F/K47E variant, but not the Ctr2 L34F mutant, showed increased sensitivity to cadmium for growth (Fig. 8C). The increased cadmium toxicity of yeast cells expressing a Cu+ transport-competent variant of Ctr2 is not a function of increased cellular copper, as expression of Ctr1 did not increase cadmium toxicity (Fig. 8C). Moreover, the cadmium toxicity associated with Ctr2 L34F/K47E expression depends on the MXXXM motif necessary for Cu+ transport by both this Ctr2 variant and by Ctr1.
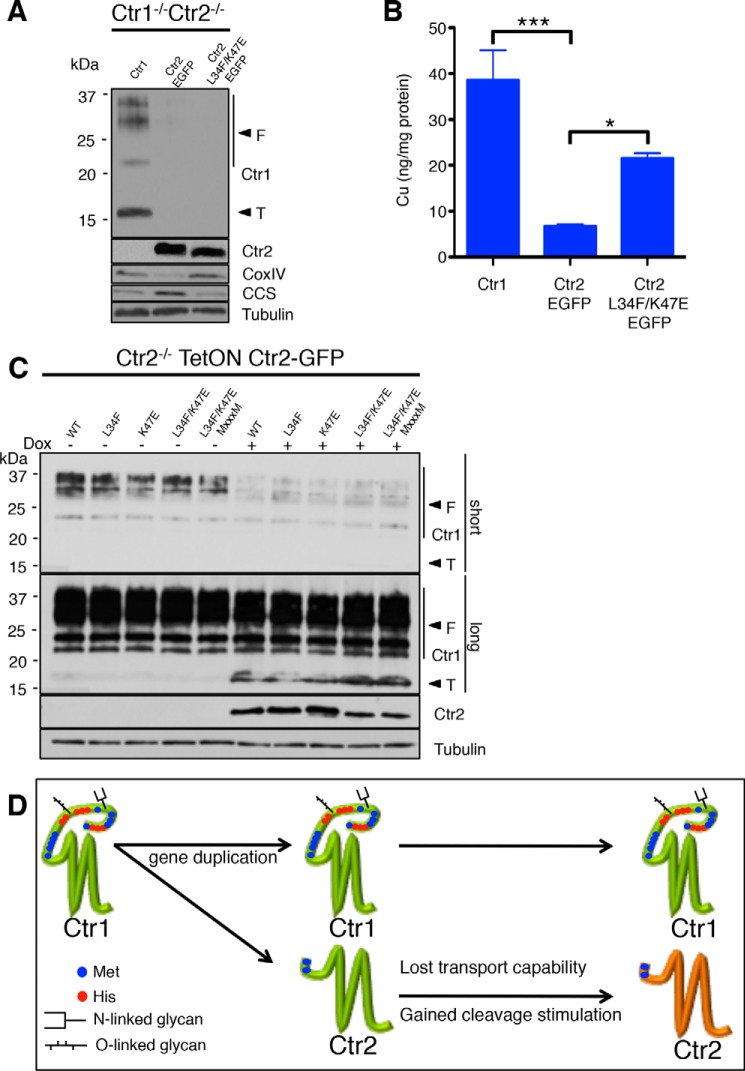
To test the copper transport function of the Ctr2 mutants that enhance yeast copper accumulation and copper-dependent growth in mammalian cells, MEFs lacking both the Ctr1 and Ctr2 genes were stably transfected with a plasmid expressing human Ctr1, wild-type human Ctr2, or the human Ctr2 L34F/K47E mutant fused to EGFP. Expression of Ctr1 and Ctr2 L34F/K47E led to increased CoxIV levels and decreased levels of CCS compared with WT Ctr2 (Fig. 9A). Furthermore, Ctr1−/− Ctr2−/− MEFs expressing Ctr1 or the Ctr2 L34F/K47E variant accumulate significantly more copper compared with cells expressing WT Ctr2 (Fig. 9B). Previous reports suggested that Ctr2 resides primarily on intracellular vesicles but also partially at the plasma membrane (29, 32). We hypothesize that the increase in Cu+ observed here is a result of extracellular Cu+ imported from the population of Ctr2 localized to the plasma membrane. Although it cannot be ruled out that the L34F/K47E mutations alter Ctr2 trafficking so that a larger percentage is present at the plasma membrane, the previous S. cerevisiae experiments suggest that the L34F/K47E mutations impart a gain of function to import extracellular Cu+ into the cell. Taken together, these results demonstrate that the Ctr2 variant that enhances copper accumulation and copper-dependent growth in yeast cells also supports enhanced copper accumulation in mammalian cells.
Figure 9.
Ctr2 transport-activating mutations increase bioavailable copper in mammalian cells while supporting Ctr1 ecto-domain cleavage. A, Ctr1−/−/Ctr2−/− MEFs containing the indicated gene were treated with 100 ng/ml doxycycline for 24 h, harvested, and immunoblotted with anti-Ctr1 (T, truncated; F, full-length), anti-GFP, anti-CoxIV, anti-CCS, and anti-tubulin antibodies. B, the same samples as in A were prepared for ICP-MS analysis of whole-cell copper levels. Data are presented as mean ± S.E. from four biological replicates. *, p ≤ 0.05; ***, p ≤ 0.001. C, Ctr2−/− MEFs containing the indicated gene were treated with 100 ng/ml doxycycline (Dox, +) or PBS (−) for 24 h, harvested, and immunoblotted. D, model for the appearance of Ctr2 during evolution. The single Ctr1 gene underwent a gene duplication to give rise to Ctr2, lacking the coding information for a copper-binding ecto-domain. The new Ctr2 gene then underwent at least two mutational events that led to a loss in the ability to transport Cu+ and a gain in the ability to stimulate the Cathepsin L-mediated cleavage of the Ctr1 ecto-domain.
Mammalian Ctr2 physically interacts with Ctr1 and stimulates the processing of the Ctr1 copper-binding ecto-domain in a manner involving cathepsin L protease as the rate-limiting step (29, 40). Although the Ctr2 L34F/K47E mutant is sufficient to support copper transport by Ctr2 in mammalian cells, the ability of this Ctr2 variant to stimulate cleavage of the Ctr1 ecto-domain was assessed. Mouse embryonic fibroblasts lacking Ctr2 but expressing endogenous Ctr1 were stably transfected with a doxycycline-inducible cassette expressing either wild-type human Ctr2 or the mutant variants (Fig. 9C). As reported previously, cells lacking Ctr2 show very low levels of tCtr1. However, when either the wild type or the mutant variants of Ctr2 were expressed by doxycycline administration, truncated Ctr1 (tCtr1) levels increased at the expense of full-length Ctr1. These data demonstrate that the amino acid changes in Ctr2 that are required for Cu+ transport are not required for Ctr2-mediated cleavage of the Ctr1 copper-binding ecto-domain.
Discussion
The ability to acquire, store, and mobilize copper ions is critical to maintaining normal growth and development under conditions of both copper sufficiency and limitation. Although several fungal species express dedicated vacuolar Cu+ efflux transporters, such as Ctr2 in S. cerevisiae and Ctr6 in S. pombe, the mechanisms by which copper is mobilized from endosomal pools is not well understood in other organisms (20, 69). Previous reports suggest that mammalian Ctr2 functions to export endosomal copper stores (30). Here we suggest that Ctr2 arose through a gene duplication event followed by neo-functionalization.
Comparative analyses coupled with mutagenesis and functional studies support a model in which the metazoan lineage originally contained a single gene, Ctr1, that functioned to transport copper into cells (Fig. 9D). Near the protostome-deuterostome split during evolution, the Ctr1 genomic locus may have largely duplicated to form a nascent Ctr2 locus. Indeed, in both the mouse and human genome, the Ctr1 and Ctr2 genes are closely linked, although this is not the case for all metazoans (29). The analysis of negative selection pressure suggests that the new Ctr2 gene explored significantly more mutagenic space than the original Ctr1 gene. Given the essentiality of copper for normal growth and for Ctr1 in embryonic development in mammals, this could largely be due to the pressure for Ctr1 to preserve residues required for Cu+ transport (56, 57). As a consequence, Ctr2 may have lost the ability to transport Cu+ while gaining the ability to stimulate cleavage of the Ctr1 copper-binding ecto-domain in concert with the cathepsin L protease. Although the changes in copper transport activity between full-length Ctr1 and tCtr1 have not yet been validated in a purified system, data suggest that Ctr2- and cathepsin L-dependent cleavage of the Ctr1 ecto-domain has two consequences. First, tCtr1 at the plasma membrane has decreased Cu+ import activity compared with Ctr1. Second, expression of tCtr1 supports the mobilization of endosomal copper pools to a greater extent than Ctr1 (29).
Although consistent with another report suggesting that Ctr2 plays a role in lysosomal/endosomal copper mobilization, the data shown here and published studies suggest that, rather than serving as a direct copper exporter, Ctr2 plays an indirect role in this process through stimulating the cleavage of Ctr1. Previous studies have demonstrated that Ctr1 ecto-domain cleavage requires the action of Ctr2, which forms a complex in vivo with Ctr1 and partially co-localizes on recycling endosomes (29, 40). This suggests that Ctr2 either hetero-oligomerizes with Ctr1, perhaps through their conserved glycine zipper domains, or that the two proteins interact as homotrimers of Ctr1 and Ctr2. Although Ctr2 facilitates Ctr1 ecto-domain cleavage independent of the MXXXM motif in the second transmembrane domain, it may have retained this motif for other functions, such as for sensing endosomal copper levels, because this region of Ctr2 is predicted to be positioned on the luminal side of the endosome.
An intriguing observation from our inspection of Ctr2 proteins in nature is that the Arthropoda lineage lacks a Ctr2 homologue. Indeed, previous studies of the three D. melanogaster Ctr proteins (Ctr1A, Ctr1B, and Ctr1C) demonstrated that they are all functional homologues of Ctr1 that have either systemic or tissue-specific functions (46–48). It is uncertain how Drosophila and other arthropods store and mobilize copper. Perhaps an explanation can be found in a report describing the role of polyphosphate-rich granules in mediating copper storage in the midgut of the velvet bean caterpillar Anticarsia gemmatalis (70). Indeed, phosphate co-localizes within the copper storage organelles in C. reinhardtii, the acidocalcisomes, and these same organelles have been shown to exist in the insect lineage (24, 71).
When considering the mobilization of intracellular copper pools, it is also informative to understand how these pools are generated. P-type Cu+-transporting ATPases both load copper into secretory vesicles and pump excess copper across the plasma membrane into the extracellular environment (72). Higher metazoans possess two distinct P-type ATPase-encoding genes, ATP7A and ATP7B, that, when mutated, give rise to Menkes syndrome or Wilson's disease, respectively (4). Our analysis, coupled with a previous report, suggests that ATP7A and ATP7B appear to have undergone a gene duplication at approximately the same time as Ctr2 (73). However, lower metazoans such as Drosophila and Caenorhabditis elegans, prior to the split of Chordata, possess a single copy of this copper pump, ATP7. Interestingly, the ATP7B and Ctr2 proteins are most highly expressed in the liver, the primary organ for copper storage. Prior to the appearance of these two genes, no species is known to possess a liver. Perhaps the evolution of the liver, which, among many other functions, serves in copper storage, necessitated the evolution of a unique set of copper-transporting proteins. Also of note is that ATP7A and ATP7B, although traditionally thought to reside at the Golgi, partially localize to intracellular vesicles (74, 75). As it is currently unknown how copper enters the vesicular storage pools, we speculate that these copper pumps may be responsible for loading the copper storage compartments. Also of interest is metallothionein, an intracellular metal binding and detoxification protein that has been implicated in copper storage (76, 77). Humans and other higher eukaryotes contain expanded repeats of metallothionein genes (78). Perhaps duplication of the copper importer, exporter, and storage genes contributed to increased organismal plasticity.
It is important to note that, although the mammalian Ctr2 and the fungal Ctr2/Ctr6 proteins both function to mobilize copper from intracellular stores, they may accomplish this goal in mechanistically different ways. Ctr2 from S. cerevisiae and Ctr6 from S. pombe directly transport copper from the vacuole lumen to the cytosol, whereas our data suggest that mammalian Ctr2 induces the proteolytic cleavage of full-length Ctr1 to generate a transporter capable of vesicular copper transport (20, 40, 69). In this sense, Ctr2 is an example of separate organisms independently evolving a similar function, vesicular copper mobilization, through two separate methods. This type of convergent evolution is now widely documented, with examples ranging from independent evolution of myoglobin net surface charge in deep-diving animals, hemoglobin function of high-altitude hummingbirds, transcriptional profiles of electric organs in fish and eels, and genes linked to auditory and visual processes involved in echolocation in dolphins and bats (79–84). The notion that biology has found multiple ways to develop a homeostatic storage mechanism underscores the importance of copper homeostasis in physiological processes.
Experimental procedures
Yeast strains and plasmids
All S. cerevisiae strains have been described previously. Cells were routinely grown in selective medium with agitation at 30 °C. All yeast plasmids were created by cloning into the p413GPD backbone at SpeI and XhoI restriction sites. The plasmid containing mouse Ctr2 used for random mutagenesis has been described previously. For the creation of plasmids containing GFP-tagged variants, codon-optimized gBlocks (Integrated DNA Technologies) were synthesized for human Ctr1-EGFP, human Ctr2-EGFP, mouse Ctr2-EGFP, sea urchin Ctr1-EGFP, and sea urchin Ctr2-EGFP. Point mutations were created using the QuikChange II site-directed mutagenesis kit (Agilent Technologies).
Phylogenetic analysis
Ctr protein sequences from the indicated species were identified and retrieved from the NCBI Protein database by iterative BLAST (Basic Local Alignment Search Tool) searches. A multiple sequence alignment (MSA) was created using MUSCLE (Multiple Sequence Comparison by Log-Expectation). A species tree was created and downloaded from the NCBI Taxonomy database. The MSA and species tree were then subjected to TreeBeST analysis under standard parameters.
Ctr2 random mutagenesis
A mouse Ctr2 expression plasmid was transformed into XL1-Red mutagenic E. coli (Stratagene), a strain deficient in three of the primary DNA repair pathways that is typically used to introduce relatively conservative single-nucleotide mutations in genes. Mutagenesis was performed according to a standard protocol, and plasmid DNA was extracted from a pooled culture of cells. Mutated plasmid DNA was transformed into ctr1Δctr3Δ yeast cells, and cells were grown on selective medium. Colonies were then replica-plated on YPEG, and colonies showing good growth were purified on fresh YPEG before plasmid rescue and sequencing to determine the nature of the Ctr2 mutation.
Growth inhibition assay
S. cerevisiae ctr1Δctr3Δ cells were transformed with the indicated plasmids and grown to exponential phase in selective medium with agitation at 30 °C. Cells were then transferred to a 96-well plate to a final concentration of metal as indicated. Cells were grown with agitation at 30 °C for 24 h before A600 was measured.
Functional complementation spot assays
ctr1Δctr3Δ or ctr1Δctr3Δfre1Δ cells were transformed with the indicated plasmids and grown to exponential phase in selective medium with agitation at 30 °C. 10-fold serial dilutions were spotted on YPD, ethanol (2%), and glycerol (3%) (YPEG) medium, YPEG containing 50 μm copper, and YPEG containing 10 mm ascorbic acid and 1.5% agar and incubated for 5–7 days at 30 °C.
Fluorescence microscopy
The ctr1Δctr3Δ strain was transformed with the indicated plasmid, grown in selective medium to exponential phase at 30 °C, washed, and resuspended in PBS before being pipetted onto a microscope slide containing an agar pad. The slide was then covered with a no. 2 coverslip and sealed with Vaseline before being imaged on a Zeiss Axio Imager.
Rates of non-synonymous and synonymous changes
MSAs were created from either Ctr1 or Ctr2 amino acid sequences via alignment with MUSCLE. Amino acid MSAs were then upload to the PAL2NAL web server along with the DNA sequences of either Ctr1 or Ctr2, respectively, to create a codon alignment (85). This codon alignment was then uploaded to the Datamonkey web server for subsequent analysis (86). The non-synonymous/synonymous mutation ratio (dN/dS) was calculated using single-likelihood ancestor counting analysis under standard parameters. Global negative selection was calculated by summing the number of positions in which dN < dS (p = 0.1) and dividing by the total number of MSA positions. Normalized dN-dS values were obtained as described previously. Amino acid alignments for both Ctr1 and Ctr2 were uploaded to the ConSurf server for conservation analysis, with the human sequence used as the query sequence and all other parameters as default (55). Sequence scores 1–3 were indicated as variable, 4–6 as neutral, and 7–9 as conserved. The sequence was then visualized using Protter to infer amino acid positions.
Genomic DNA analysis
ChIP-seq data from the ENCODE Consortium visualized on the UCSC genome browser was manually inspected for the presence of bound factors. DNA regions between the transcriptional start site and 1000 base pairs upstream were investigated. For transcription factor binding site predictions, the ConTra web server was used (59). Briefly, human Ctr1 or Ctr2 sequences were selected and 500 bases upstream of the transcriptional start site was queried under standard parameters for the presence of predicted transcription factor binding sites. Transcripts from indicated species were downloaded from Ensembl genome browser 87 and examined for location of 5′ UTR, introns, exons, and 3′UTR.
Metal analysis
Mammalian and yeast total cell copper concentrations were measured by ICP-MS (Agilent Model 7500cs, Santa Clara, CA). Briefly, log-phase yeast cells were grown in SC-His media (MP Biomedicals) normalized to cell number, washed, and harvested into acid-washed 1.5-ml micro-centrifuge tubes. Yeast pellets were dissolved 1:10 weight/volume with trace analysis grade nitric acid (Sigma). Mammalian cells were treated with 100 ng/ml doxycycline for 24 h and harvested at ∼80% confluency. Cell pellets were lysed and samples normalized to protein concentration before lysates were digested by addition of trace metal analysis grade nitric acid (Sigma) at a 1:10 volume/volume ratio. All samples were then heated at 85–95 °C for ∼1 h before analysis.
Protein extraction and immunoblotting
For the preparation of protein extracts, mammalian or yeast cell pellets were resuspended in ice-cold radioimmune precipitation assay buffer (Cell Signaling Technology) supplemented with proteinase inhibitors (Halt Protease Inhibitor Mixture, Thermo Scientific). Homogenates were vortexed for ∼10 s and centrifuged at 20,000 × g at 4 °C for 10 min, and supernatants were collected. Protein concentrations were measured with the BCA protein assay kit (Thermo Scientific). SDS-PAGE and immunoblotting were carried out following standard protocols. The antibodies used were anti-GFP (Sigma-Aldrich), anti-phosphoglycerate kinase (Invitrogen) anti-cytochrome c oxidase (CoxIV; Mitosciences, Eugene, OR), anti-copper chaperone for SOD1 (CCS, Santa Cruz Biotechnology), and anti-β-tubulin (Cell Signaling Technology, Danvers, MA). The anti-Ctr1 antibody has been described previously. HRP-conjugated anti-mouse or anti-rabbit IgG (GE Healthcare) was used as secondary antibody for immunoblotting.
Author contributions
B. L. L., L. K. W., J. L., and D. J. T. designed the experiments. B. L. L., L. K. W., and J. L. conducted the experiments, and B. L. L., L. K. W., J. L., and D. J. T. analyzed the results. B. L. L. and D. J. T. prepared the manuscript, which was reviewed and edited by L. K. W. and J. L.
Acknowledgments
We thank Drs. A. Smith and S. Garcia-Santamarina for valuable comments on this manuscript.
This work was supported by National Institutes of Health Grants DK074192 (to D. J. T.), DK079209 (to J. L.), and P30GM103335 (to the Nebraska Redox Biology Center). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Ctr
- copper transporter
- MEF
- mouse embryonic fibroblast
- tCtr
- truncated copper transporter
- dN/dS
- non-synonymous/synonymous mutation ratio
- ChIP-seq
- ChIP sequencing
- EGFP
- enhanced GFP
- CCS
- copper chaperone for SOD1
- MSA
- multiple sequence alignment
- YPEG
- YPD, ethanol, and glycerol
- YPD
- yeast peptone dextrose
- CoxIV
- cytochrome c oxidase
- ICP
- inductively coupled plasma.
References
- 1. Kim B. E., Nevitt T., and Thiele D. J. (2008) Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 4, 176–185 [DOI] [PubMed] [Google Scholar]
- 2. Nevitt T., Ohrvik H., and Thiele D. J. (2012) Charting the travels of copper in eukaryotes from yeast to mammals. Biochim. Biophys. Acta 1823, 1580–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Madsen E., and Gitlin J. D. (2007) Copper and iron disorders of the brain. Annu. Rev. Neurosci. 30, 317–337 [DOI] [PubMed] [Google Scholar]
- 4. de Bie P., Muller P., Wijmenga C., and Klomp L. W. (2007) Molecular pathogenesis of Wilson and Menkes disease: correlation of mutations with molecular defects and disease phenotypes. J. Med. Genet. 44, 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Medeiros D. M., Davidson J., and Jenkins J. E. (1993) A unified perspective on copper deficiency and cardiomyopathy. Proc. Soc. Exp. Biol. Med. 203, 262–273 [DOI] [PubMed] [Google Scholar]
- 6. Lukasewycz O. A., and Prohaska J. R. (1990) The immune response in copper deficiency. Ann. N.Y. Acad. Sci. 587, 147–159 [DOI] [PubMed] [Google Scholar]
- 7. Klevay L. M. (2000) Cardiovascular disease from copper deficiency: a history. J. Nutr. 130, 489S–492S [DOI] [PubMed] [Google Scholar]
- 8. Dancis A., Yuan D. S., Haile D., Askwith C., Eide D., Moehle C., Kaplan J., and Klausner R. D. (1994) Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell 76, 393–402 [DOI] [PubMed] [Google Scholar]
- 9. Rubino J. T., and Franz K. J. (2012) Coordination chemistry of copper proteins: how nature handles a toxic cargo for essential function. J. Inorg. Biochem. 107, 129–143 [DOI] [PubMed] [Google Scholar]
- 10. Puig S., Lee J., Lau M., and Thiele D. J. (2002) Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J. Biol. Chem. 277, 26021–26030 [DOI] [PubMed] [Google Scholar]
- 11. De Feo C. J., Aller S. G., Siluvai G. S., Blackburn N. J., and Unger V. M. (2009) Three-dimensional structure of the human copper transporter hCTR1. Proc. Natl. Acad. Sci. U.S.A. 106, 4237–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aller S. G., and Unger V. M. (2006) Projection structure of the human copper transporter CTR1 at 6-A resolution reveals a compact trimer with a novel channel-like architecture. Proc. Natl. Acad. Sci. U.S.A. 103, 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aller S. G., Eng E. T., De Feo C. J., and Unger V. M. (2004) Eukaryotic CTR copper uptake transporters require two faces of the third transmembrane domain for helix packing, oligomerization, and function. J. Biol. Chem. 279, 53435–53441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Feo C. J., Mootien S., and Unger V. M. (2010) Tryptophan scanning analysis of the membrane domain of CTR-copper transporters. J. Membr. Biol. 234, 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hassett R., and Kosman D. J. (1995) Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. J. Biol. Chem. 270, 128–134 [DOI] [PubMed] [Google Scholar]
- 16. Georgatsou E., Mavrogiannis L. A., Fragiadakis G. S., and Alexandraki D. (1997) The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 272, 13786–13792 [DOI] [PubMed] [Google Scholar]
- 17. Pena M. M., Puig S., and Thiele D. J. (2000) Characterization of the Saccharomyces cerevisiae high affinity copper transporter Ctr3. J. Biol. Chem. 275, 33244–33251 [DOI] [PubMed] [Google Scholar]
- 18. Zhou H., and Thiele D. J. (2001) Identification of a novel high affinity copper transport complex in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 276, 20529–20535 [DOI] [PubMed] [Google Scholar]
- 19. Ding C., Yin J., Tovar E. M., Fitzpatrick D. A., Higgins D. G., and Thiele D. J. (2011) The copper regulon of the human fungal pathogen Cryptococcus neoformans H99. Mol. Microbiol. 81, 1560–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rees E. M., Lee J., and Thiele D. J. (2004) Mobilization of intracellular copper stores by the ctr2 vacuolar copper transporter. J. Biol. Chem. 279, 54221–54229 [DOI] [PubMed] [Google Scholar]
- 21. Rees E. M., and Thiele D. J. (2007) Identification of a vacuole-associated metalloreductase and its role in Ctr2-mediated intracellular copper mobilization. J. Biol. Chem. 282, 21629–21638 [DOI] [PubMed] [Google Scholar]
- 22. Hopkin S. P., and Martin M. H. (1982) The distribution of zinc, cadmium, lead and copper within the hepatopancreas of a woodlouse. Tissue Cell 14, 703–715 [DOI] [PubMed] [Google Scholar]
- 23. Chavez-Crooker P., Garrido N., and Ahearn G. A. (2001) Copper transport by lobster hepatopancreatic epithelial cells separated by centrifugal elutriation: measurements with the fluorescent dye Phen Green. J. Exp. Biol. 204, 1433–1444 [DOI] [PubMed] [Google Scholar]
- 24. Hong-Hermesdorf A., Miethke M., Gallaher S. D., Kropat J., Dodani S. C., Chan J., Barupala D., Domaille D. W., Shirasaki D. I., Loo J. A., Weber P. K., Pett-Ridge J., Stemmler T. L., Chang C. J., and Merchant S. S. (2014) Subcellular metal imaging identifies dynamic sites of Cu accumulation in Chlamydomonas. Nat. Chem. Biol. 10, 1034–1042; Correction (2015) Nat. Chem. Biol.11, 235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beaudoin J., Ekici S., Daldal F., Ait-Mohand S., Guérin B., and Labbé S. (2013) Copper transport and regulation in Schizosaccharomyces pombe. Biochem. Soc. Trans. 41, 1679–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blaby-Haas C. E., and Merchant S. S. (2014) Lysosome-related organelles as mediators of metal homeostasis. J. Biol. Chem. 289, 28129–28136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ralle M., Huster D., Vogt S., Schirrmeister W., Burkhead J. L., Capps T. R., Gray L., Lai B., Maryon E., and Lutsenko S. (2010) Wilson disease at a single cell level: intracellular copper trafficking activates compartment-specific responses in hepatocytes. J. Biol. Chem. 285, 30875–30883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou B., and Gitschier J. (1997) hCTR1: a human gene for copper uptake identified by complementation in yeast. Proc. Natl. Acad. Sci. U.S.A. 94, 7481–7486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Öhrvik H., Nose Y., Wood L. K., Kim B. E., Gleber S. C., Ralle M., and Thiele D. J. (2013) Ctr2 regulates biogenesis of a cleaved form of mammalian Ctr1 metal transporter lacking the copper- and cisplatin-binding ecto-domain. Proc. Natl. Acad. Sci. U.S.A. 110, E4279–E4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van den Berghe P. V., Folmer D. E., Malingré H. E., van Beurden E., Klomp A. E., van de Sluis B., Merkx M., Berger R., and Klomp L. W. (2007) Human copper transporter 2 is localized in late endosomes and lysosomes and facilitates cellular copper uptake. Biochem. J. 407, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blair B. G., Larson C. A., Adams P. L., Abada P. B., Pesce C. E., Safaei R., and Howell S. B. (2011) Copper transporter 2 regulates endocytosis and controls tumor growth and sensitivity to cisplatin in vivo. Mol. Pharmacol. 79, 157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bertinato J., Swist E., Plouffe L. J., Brooks S. P., and L'abbé M. R. (2008) Ctr2 is partially localized to the plasma membrane and stimulates copper uptake in COS-7 cells. Biochem. J. 409, 731–740 [DOI] [PubMed] [Google Scholar]
- 33. Guo Y., Smith K., Lee J., Thiele D. J., and Petris M. J. (2004) Identification of methionine-rich clusters that regulate copper-stimulated endocytosis of the human Ctr1 copper transporter. J. Biol. Chem. 279, 17428–17433 [DOI] [PubMed] [Google Scholar]
- 34. Petris M. J., Smith K., Lee J., and Thiele D. J. (2003) Copper-stimulated endocytosis and degradation of the human copper transporter, hCtr1. J. Biol. Chem. 278, 9639–9646 [DOI] [PubMed] [Google Scholar]
- 35. Klomp A. E., Tops B. B., Van Denberg I. E., Berger R., and Klomp L. W. (2002) Biochemical characterization and subcellular localization of human copper transporter 1 (hCTR1). Biochem. J. 364, 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Molloy S. A., and Kaplan J. H. (2009) Copper-dependent recycling of hCTR1, the human high affinity copper transporter. J. Biol. Chem. 284, 29704–29713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maryon E. B., Zhang J., Jellison J. W., and Kaplan J. H. (2009) Human copper transporter 1 lacking O-linked glycosylation is proteolytically cleaved in a Rab9-positive endosomal compartment. J. Biol. Chem. 284, 28104–28114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sancenón V., Puig S., Mira H., Thiele D. J., and Peñarrubia L. (2003) Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol. Biol. 51, 577–587 [DOI] [PubMed] [Google Scholar]
- 39. Riggio M., Lee J., Scudiero R., Parisi E., Thiele D. J., and Filosa S. (2002) High affinity copper transport protein in the lizard Podarcis sicula: molecular cloning, functional characterization and expression in somatic tissues, follicular oocytes and eggs. Biochim. Biophys. Acta 1576, 127–135 [DOI] [PubMed] [Google Scholar]
- 40. Öhrvik H., Logeman B., Turk B., Reinheckel T., and Thiele D. J. (2016) Cathepsin protease controls copper and cisplatin accumulation via cleavage of the Ctr1 metal-binding ectodomain. J. Biol. Chem. 291, 13905–13916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Öhrvik H., and Thiele D. J. (2015) The role of Ctr1 and Ctr2 in mammalian copper homeostasis and platinum-based chemotherapy. J. Trace Elem. Med. Biol. 31, 178–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ohrvik H., and Thiele D. J. (2014) How copper traverses cellular membranes through the mammalian copper transporter 1, Ctr1. Ann. N.Y. Acad. Sci. 1314, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang Z., and Rannala B. (2012) Molecular phylogenetics: principles and practice. Nat. Rev. Genet. 13, 303–314 [DOI] [PubMed] [Google Scholar]
- 44. Vilella A. J., Severin J., Ureta-Vidal A., Heng L., Durbin R., and Birney E. (2009) EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19, 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fedonkin M. A., and Waggoner B. M. (1997) The late precambrian fossil Kimberella is a mollusc-like bilaterian organism. Nature 388, 868–871 [Google Scholar]
- 46. Turski M. L., and Thiele D. J. (2007) Drosophila Ctr1A functions as a copper transporter essential for development. J. Biol. Chem. 282, 24017–24026 [DOI] [PubMed] [Google Scholar]
- 47. Steiger D., Fetchko M., Vardanyan A., Atanesyan L., Steiner K., Turski M. L., Thiele D. J., Georgiev O., and Schaffner W. (2010) The Drosophila copper transporter Ctr1C functions in male fertility. J. Biol. Chem. 285, 17089–17097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou H., Cadigan K. M., and Thiele D. J. (2003) A copper-regulated transporter required for copper acquisition, pigmentation, and specific stages of development in Drosophila melanogaster. J. Biol. Chem. 278, 48210–48218 [DOI] [PubMed] [Google Scholar]
- 49. Cobine P. A., Pierrel F., and Winge D. R. (2006) Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim. Biophys. Acta 1763, 759–772 [DOI] [PubMed] [Google Scholar]
- 50. Kaessmann H. (2010) Origins, evolution, and phenotypic impact of new genes. Genome Res. 20, 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., and Postlethwait J. (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Assis R., and Bachtrog D. (2013) Neofunctionalization of young duplicate genes in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 110, 17409–17414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brust A., Sunagar K., Undheim E. A., Vetter I., Yang D. C., Casewell N. R., Jackson T. N., Koludarov I., Alewood P. F., Hodgson W. C., Lewis R. J., King G. F., Antunes A., Hendrikx I., and Fry B. G. (2013) Differential evolution and neofunctionalization of snake venom metalloprotease domains. Mol. Cell. Proteomics 12, 651–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deng C., Cheng C. H., Ye H., He X., and Chen L. (2010) Evolution of an antifreeze protein by neofunctionalization under escape from adaptive conflict. Proc. Natl. Acad. Sci. U.S.A. 107, 21593–21598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., and Ben-Tal N. (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee J., Prohaska J. R., and Thiele D. J. (2001) Essential role for mammalian copper transporter Ctr1 in copper homeostasis and embryonic development. Proc. Natl. Acad. Sci. U.S.A. 98, 6842–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kuo Y. M., Zhou B., Cosco D., and Gitschier J. (2001) The copper transporter CTR1 provides an essential function in mammalian embryonic development. Proc. Natl. Acad. Sci. U.S.A. 98, 6836–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hooghe B., Hulpiau P., van Roy F., and De Bleser P. (2008) ConTra: a promoter alignment analysis tool for identification of transcription factor binding sites across species. Nucleic Acids Res. 36, W128–W132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schwab S., Shearer J., Conklin S. E., Alies B., and Haas K. L. (2016) Sequence proximity between Cu(II) and Cu(I) binding sites of human copper transporter 1 model peptides defines reactivity with ascorbate and O2. J. Inorg. Biochem. 158, 70–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pushie M. J., Shaw K., Franz K. J., Shearer J., and Haas K. L. (2015) Model peptide studies reveal a mixed histidine-methionine Cu(I) binding site at the N-terminus of human copper transporter 1. Inorg. Chem. 54, 8544–8551 [DOI] [PubMed] [Google Scholar]
- 62. Rubino J. T., Riggs-Gelasco P., and Franz K. J. (2010) Methionine motifs of copper transport proteins provide general and flexible thioether-only binding sites for Cu(I) and Ag(I). J. Biol. Inorg. Chem. 15, 1033–1049 [DOI] [PubMed] [Google Scholar]
- 63. Haas K. L., Putterman A. B., White D. R., Thiele D. J., and Franz K. J. (2011) Model peptides provide new insights into the role of histidine residues as potential ligands in human cellular copper acquisition via Ctr1. J. Am. Chem. Soc. 133, 4427–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rubino J. T., Chenkin M. P., Keller M., Riggs-Gelasco P., and Franz K. J. (2011) A comparison of methionine, histidine and cysteine in copper(I)-binding peptides reveals differences relevant to copper uptake by organisms in diverse environments. Metallomics 3, 61–73 [PubMed] [Google Scholar]
- 65. Du X., Li H., Wang X., Liu Q., Ni J., and Sun H. (2013) Kinetics and thermodynamics of metal binding to the N-terminus of a human copper transporter, hCTR1. Chem. Commmun. 49, 9134–9136 [DOI] [PubMed] [Google Scholar]
- 66. Maryon E. B., Molloy S. A., Ivy K., Yu H., and Kaplan J. H. (2013) Rate and regulation of copper transport by human copper transporter 1 (hCTR1). J. Biol. Chem. 288, 18035–18046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Eisses J. F., and Kaplan J. H. (2005) The mechanism of copper uptake mediated by human CTR1: a mutational analysis. J. Biol. Chem. 280, 37159–37168 [DOI] [PubMed] [Google Scholar]
- 68. Bertinato J., Cheung L., Hoque R., and Plouffe L. J. (2010) Ctr1 transports silver into mammalian cells. J. Trace Elem. Med. Biol. 24, 178–184 [DOI] [PubMed] [Google Scholar]
- 69. Bellemare D. R., Shaner L., Morano K. A., Beaudoin J., Langlois R., and Labbe S. (2002) Ctr6, a vacuolar membrane copper transporter in Schizosaccharomyces pombe. J. Biol. Chem. 277, 46676–46686 [DOI] [PubMed] [Google Scholar]
- 70. Gomes F. M., Carvalho D. B., Peron A. C., Saito K., Miranda K., and Machado E. A. (2012) Inorganic polyphosphates are stored in spherites within the midgut of Anticarsia gemmatalis and play a role in copper detoxification. J. Insect Physiol. 58, 211–219 [DOI] [PubMed] [Google Scholar]
- 71. Ramos I., Gomes F., Koeller C. M., Saito K., Heise N., Masuda H., Docampo R., de Souza W., Machado E. A., and Miranda K. (2011) Acidocalcisomes as calcium- and polyphosphate-storage compartments during embryogenesis of the insect Rhodnius prolixus Stahl. PLoS ONE 6, e27276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lutsenko S., Gupta A., Burkhead J. L., and Zuzel V. (2008) Cellular multitasking: the dual role of human Cu-ATPases in cofactor delivery and intracellular copper balance. Arch. Biochem. Biophys. 476, 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gupta A., and Lutsenko S. (2012) Evolution of copper transporting ATPases in eukaryotic organisms. Curr. Genomics 13, 124–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pascale M. C., Franceschelli S., Moltedo O., Belleudi F., Torrisi M. R., Bucci C., La Fontaine S., Mercer J. F., and Leone A. (2003) Endosomal trafficking of the Menkes copper ATPase ATP7A is mediated by vesicles containing the Rab7 and Rab5 GTPase proteins. Exp. Cell Res. 291, 377–385 [DOI] [PubMed] [Google Scholar]
- 75. Polishchuk E. V., Concilli M., Iacobacci S., Chesi G., Pastore N., Piccolo P., Paladino S., Baldantoni D., van Ijzendoorn S. C., Chan J., Chang C. J., Amoresano A., Pane F., Pucci P., Tarallo A., et al. (2014) Wilson disease protein ATP7B utilizes lysosomal exocytosis to maintain copper homeostasis. Dev. Cell 29, 686–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Luza S. C., and Speisky H. C. (1996) Liver copper storage and transport during development: implications for cytotoxicity. Am. J. Clin. Nutr. 63, 812S–820S [DOI] [PubMed] [Google Scholar]
- 77. Tapia L., González-Agüero M., Cisternas M. F., Suazo M., Cambiazo V., Uauy R., and González M. (2004) Metallothionein is crucial for safe intracellular copper storage and cell survival at normal and supra-physiological exposure levels. Biochem. J. 378, 617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. West A. K., Stallings R., Hildebrand C. E., Chiu R., Karin M., and Richards R. I. (1990) Human metallothionein genes: structure of the functional locus at 16q13. Genomics 8, 513–518 [DOI] [PubMed] [Google Scholar]
- 79. Mirceta S., Signore A. V., Burns J. M., Cossins A. R., Campbell K. L., and Berenbrink M. (2013) Evolution of mammalian diving capacity traced by myoglobin net surface charge. Science 340, 1234192. [DOI] [PubMed] [Google Scholar]
- 80. Gallant J. R., Traeger L. L., Volkening J. D., Moffett H., Chen P. H., Novina C. D., Phillips G. N. Jr., Anand R., Wells G. B., Pinch M., Güth R., Unguez G. A., Albert J. S., Zakon H. H., Samanta M. P., and Sussman M. R. (2014) Nonhuman genetics: genomic basis for the convergent evolution of electric organs. Science 344, 1522–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Parker J., Tsagkogeorga G., Cotton J. A., Liu Y., Provero P., Stupka E., and Rossiter S. J. (2013) Genome-wide signatures of convergent evolution in echolocating mammals. Nature 502, 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Foote A. D., Liu Y., Thomas G. W., Vina T., Alföldi J., Deng J., Dugan S., van Elk C. E., Hunter M. E., Joshi V., Khan Z., Kovar C., Lee S. L., Lindblad-Toh K., Mancia A., et al. (2015) Convergent evolution of the genomes of marine mammals. Nat. Genet. 47, 272–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Projecto-Garcia J., Natarajan C., Moriyama H., Weber R. E., Fago A., Cheviron Z. A., Dudley R., McGuire J. A., Witt C. C., and Storz J. F. (2013) Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc. Natl. Acad. Sci. U.S.A. 110, 20669–20674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Castoe T. A., de Koning A. P., Kim H. M., Gu W., Noonan B. P., Naylor G., Jiang Z. J., Parkinson C. L., and Pollock D. D. (2009) Evidence for an ancient adaptive episode of convergent molecular evolution. Proc. Natl. Acad. Sci. U.S.A. 106, 8986–8991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Suyama M., Torrents D., and Bork P. (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pond S. L., and Frost S. D. (2005) Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533 [DOI] [PubMed] [Google Scholar]