ABSTRACT
Estrogen, through its receptors, regulates various aspects of spermatogenesis and male fertility. To understand the roles of estrogen receptors (ERα and ERβ) in male fertility, we have developed in vivo selective ER agonist administration models. Treatment of adult male rats with ERα or ERβ agonist for 60 d decreases fertility and litter size mainly due to increased pre- and post-implantation embryo loss. Since epigenetic mechanisms like DNA methylation play a crucial role in male fertility, we investigated the effects of the ER agonists on DNA methylation in spermatozoa. Treatment with ERβ agonist causes a significant decrease in DNA methylation both at the global level and at the H19 differentially methylated region (DMR). This could be due to decrease in DNA methyltransferases in the testis upon ERβ agonist treatment. The hypomethylation observed at the H19 DMR corroborates with aberrant expression of Igf2 and H19 imprinted genes in the resorbed embryos sired by ERβ agonist-treated males. Thus, our study demonstrates that ERβ regulates DNA methylation and methylating enzymes during adult rat spermatogenesis. Activation of estrogen signaling through ERβ could therefore cause DNA methylation defects leading to impaired male fertility. These results define a role for estrogen in epigenetic regulation of male germ line, suggesting that epigenetic insults by exposure to environmental estrogens could potentially affect male fertility.
KEYWORDS: DNA methylation, DNA methyltransferases, ERβ, estrogen, spermatozoa
Abbreviations
- DMR
differentially methylated region
- Igf2
insulin-like growth factor 2
- CTCF
CCCTC-binding factor
- Dnmt
DNA methyltransferase
- ER
estrogen receptor
- SERM
selective estrogen receptor modulator
- BPA
bisphenol-A
- PPT
4,4″,4″-(4-Propyl-[1H] pyrazole-1,3,5-triyl), DPN, 2,3-bis(4-hydroxyphenyl)-propionitrile, 5mC, 5-methylcytosine
- DTT
1,4-dithiothreitol
Introduction
DNA methylation is an epigenetic mechanism that plays an important role during spermatogenesis, the process of sperm formation.1 Besides regulating gene expression, DNA methylation is crucial for genomic imprinting. The phenomenon of genomic imprinting refers to the differential expression of a gene or chromosomal regions depending on their parental origin; such genes are called imprinted genes. They are characterized by monoallelic expression and are often clustered in evolutionarily conserved domains. Imprinted genes are regulated by distinct regions called differentially methylated regions (DMR). DNA methylation differs between DMRs in the maternal and paternal alleles, thus bringing about allelic differential expression.2
Insulin-like growth factor 2 (Igf2) and H19 are the best characterized imprinted genes, and are closely linked and reciprocally imprinted. They play a pivotal role during embryogenesis and modulate embryo and placental development.3 Their expression is regulated by a shared DMR located −2 to −4 kb relative to the H19 transcription start site in mouse4 and harbors several CCCTC-binding factor (CTCF) binding sites.5 In developing embryos, the H19 DMR on the maternal allele is unmethylated, which promotes binding of CTCF insulator protein. This creates a chromatin boundary, blocking the access of Igf2 promoter to shared enhancer elements (downstream of H19); H19 is transcribed from the maternal allele. Conversely, on the paternal allele, the H19 DMR is methylated, which prevents the binding of CTCF. This allows interaction of the enhancer with the Igf2 promoter, thus allowing transcription of Igf2 from the paternal allele.3,6-8 The H19 DMR is methylated in sperm, while it remains unmethylated in oocytes. The establishment of these imprinting patterns during gametogenesis thus determines the expression of these crucial genes during embryo development. DNA methyltransferases—mainly Dnmt3a, Dnmt3b and Dnmt3l (de novo methyltransferases) and Dnmt1 (maintenance methyltransferase)—are expressed during spermatogenesis9,10 and are involved in establishment and maintenance of imprinting patterns in male germ cells.1
Spermatogenesis is under strict endocrine regulation. Besides gonadotropins and androgen, estrogens have an important role in the regulation of various aspects of spermatogenesis. Estrogen receptors (ER) α and β are expressed in the male germ cells11,12 and regulate their development and maturation.13 Exogenous estradiol treatment to adult rats disrupts spermatogenesis and is detrimental to male fertility.14-16 To further understand the roles of the ERs during spermatogenesis, we have developed and characterized models using ER subtype-specific agonists. We have found that ERα or ERβ agonist treatment to adult male rats for 60 d causes a decrease in male fertility and litter size, mainly due to an increase in pre- and post-implantation embryo loss.17 These studies showed that activation of estrogen signaling through either of the ERs could be detrimental to male fertility. Studies have also shown that administration of selective estrogen receptor modulators (SERMs), such as tamoxifen and bisphenol-A (BPA), to male rats (at the adult and neonatal stages, respectively) causes decrease in various fertility parameters.18-20 One of the probable causes of the sub-fertility observed with these treatments has been attributed to disturbed DNA methylation patterns of the imprinted genes in male germ line.21,22 These studies implicate involvement of estrogen signaling in the establishment and maintenance of imprinting patterns in male germ cells. However, these ligands, being SERMs, can have variable agonistic or antagonistic action through either of the ERs. Thus, the mechanism by which estrogen, through its receptors, is involved in imprint acquisition is not clear. Since, we observed a decrease in male fertility upon ER agonist treatments, we sought to examine the potential effects on DNA methylation in spermatozoa after these treatments. In the present study, we have explored the effect of ER agonist treatments on global and locus-specific DNA methylation patterns in spermatozoa.
Results
Increase in post-implantation embryo loss after paternal PPT and DPN treatment
Adult male rats were treated with 4,4″,4″-(4-Propyl-[1H] pyrazole-1,3,5-triyl) (PPT), a 410-fold selective, ERα-specific agonist23 or 2,3-bis(4-hydroxyphenyl)-propionitrile (DPN), a 70-fold selective ERβ-specific agonist24 for 60 d followed by mating studies were performed. There was a significant increase in percent post-implantation embryo loss after paternal PPT (17.75 ± 2.532%) and DPN (15.44 ± 6.253%) treatments as compared with control treatment (4.695 ± 1.065%). These results are in agreement with our earlier studies.17
Decrease in global DNA methylation levels in spermatozoa on DPN treatment
We observed a significant decrease in global 5-methylcytosine (5mC) content in spermatozoa after DPN treatment as compared with controls (Fig. 1A). The methylation analysis of the LINE-1 promoter (consisting of 10 CpG sites) is depicted in Table 1. There was a modest but significant decrease in percent methylation at the first 3 analyzed CpG sites of LINE-1 promoter. Total methylation of all the 10 CpG sites studied decreased after DPN treatment, but this decrease was not statistically significant. There was no change in 5mC content or LINE-1 promoter methylation in spermatozoa after PPT treatment.
Figure 1.
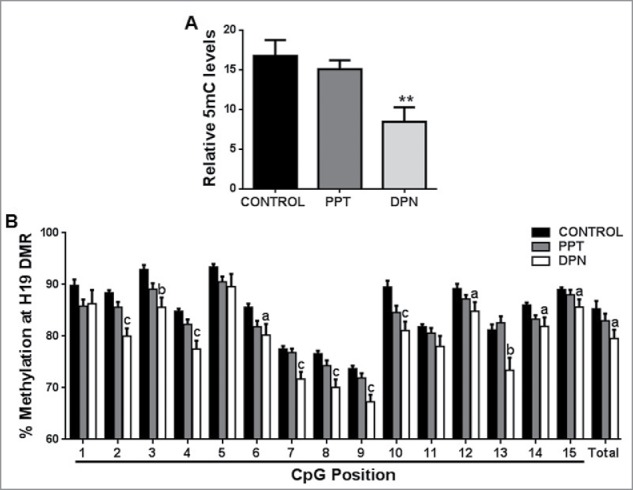
Effect of PPT and DPN treatment on DNA methylation. (A) Decrease in global 5mC content in spermatozoa after DPN treatment. All values are expressed as means ± SEM. ** represents P < 0.01. (B) Methylation mapping of 15 CpG sites and total methylation of H19 DMR by pyrosequencing after PPT and DPN treatments. ‘a’ indicates P < 0.05, ‘b’ indicates P < 0.01, and ‘c’ indicates P < 0.001. n = 8.
Table 1.
DNA methylation analysis of LINE-1 promoter by pyrosequencing after PPT and DPN treatments. Data represented as mean ± SEM n = 8. **P < 0.01; ***P < 0.001.
| CpG Position | CONTROL (% Methylation) | PPT (% Methylation) | DPN (% Methylation) |
|---|---|---|---|
| 1 | 82.1 ( ± 0.6) | 81 ( ± 0.4) | 80 ( ± 0.34)** |
| 2 | 64.1 ( ± 0.38) | 63.3 ( ± 0.3) | 61.7 ( ± 0.3)*** |
| 3 | 69.2 ( ± 0.44) | 68.9 ( ± 0.6) | 66.4 ( ± 0.31)*** |
| 4 | 84.6 ( ± 0.4) | 83.9 ( ± 0.69) | 83.7 ( ± 0.43) |
| 5 | 80.9 ( ± 0.35) | 80.9 ( ± 0.52) | 79.7 ( ± 0.48) |
| 6 | 52.1 ( ± 0.68) | 50.9 ( ± 0.51) | 52.7 ( ± 0.3) |
| 7 | 84.3 ( ± 0.34) | 84.5 ( ± 0.31) | 84.6 ( ± 0.23) |
| 8 | 75.1 ( ± 0.28) | 75 ( ± 0.26) | 74.4 ( ± 0.17) |
| 9 | 77.4 ( ± 0.31) | 77.5 ( ± 0.31) | 77.5 ( ± 0.23) |
| 10 | 82.9 ( ± 0.51) | 82.8 ( ± 0.54) | 81.9 ( ± 0.46) |
| Average | 75.27 ( ± 3.34) | 74.87 ( ± 3.43) | 74.26 ( ± 3.35) |
Hypomethylation at the H19 DMR in spermatozoa after DPN treatment
The total percent methylation at H19 DMR in spermatozoa was decreased after DPN treatment compared with controls (Fig. 1B). A significant decrease in percent methylation was observed at CpG sites 2–4, 6–10, and 12–15 after DPN treatment. Out of the 15 CpG sites studied, 12 sites showed hypomethylation after DPN treatment. Within the CTCF binding site encompassing the CpG positions 10, 11, and 12, we detected hypomethylation at CpG positions 10 and 12. There was no change in H19 DMR methylation upon PPT treatment (Fig. 1B).
Decrease in DNA methyltransferases in testis after DPN treatment
We observed a significant decrease of Dnmt1, Dnmt3a, Dnmt3b, and Dnmt3l transcripts and protein levels in the testis after DPN treatment as compared with controls. Upon PPT treatment, only Dnmt3a transcript and protein levels of were decreased (Fig. 2).
Figure 2.
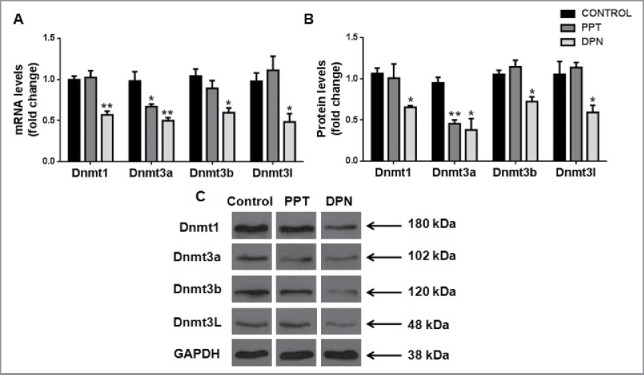
Effect of DNA methyltransferases in the testis after PPT and DPN treatments. (A) Transcript levels and (B) protein levels of Dnmts in testis after PPT and DPN treatments. (C) Representative images of protein bands of Dnmts and GAPDH. Results are expressed as means ± SEM. * indicates P < 0.05 and ** indicates P < 0.01. n = 8.
Aberrant expression of imprinted genes in resorbed embryos
Expression of Igf2 was downregulated and that of H19 was upregulated in resorbed embryos sired by DPN-treated males as compared with resorbed embryos sired by control males (Fig. 3A). Igf2 and H19 expression was not affected in resorbed embryos sired by PPT-treated males. Also, Dnmt1, Dnmt3a, Dnmt3b, and Dnmt3l transcript levels were not affected in resorbed embryos sired by PPT- or DPN-treated males (Fig. 3B).
Figure 3.
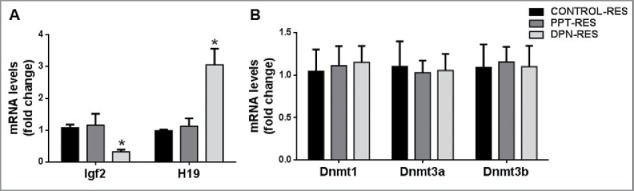
Effect of paternal PPT and DPN treatments on gene expression in resorbed embryos. (A) Expression of imprinted genes Igf2 and H19. (B) Expression of Dnmt1, Dnmt3a and Dnmt3b. ‘Control Res’ indicates resorbed embryos sired by control males; ‘PPT Res’ indicates resorbed embryos sired by PPT-treated males; ‘DPN Res’ indicates resorbed embryos sired by DPN-treated males. All the results are expressed as means ± SEM. * indicates P < 0.05. n = 8.
Discussion
Our study demonstrates that activation of estrogen signaling though ERβ leads to decrease in DNA methylation both at a global level and locally at the H19 DMR. Exposure to several endocrine disrupting compounds, such as anti-androgenic fungicide vinclozolin and estrogenic pesticide methoxychlor, in rats during gestation causes spermatogenic defects leading to a decrease in male fertility in adulthood and even in subsequent generations.25 These effects were correlated with aberrant DNA methylation patterns in the male germ line. Human studies have demonstrated that, in spermatozoa, aberrant DNA methylation patterns in imprinted genes like H19 (specifically at its CTCF binding site) are associated with oligozoospermia and azoospermia.26-28 These studies indicate that defects in DNA methylation patterns can potentially disrupt spermatogenesis and male fertility. Establishment of DNA methylation patterns at imprinted genes is initiated in the developing gonads and continues throughout adulthood in male germ cells.29,30 Therefore, disturbances in establishment and maintenance of these imprinting marks, along with defects in global DNA methylation observed upon ERβ agonist (DPN) treatment can impair male fertility and the subsequent embryogenesis. In our study, we show that ERβ is specifically involved in the process of imprint acquisition at the H19 DMR in the male germ line. Interestingly, ERβ is associated with the estrogen response element (ERE) at the H19 DMR in rat testes and colocalizes with Dnmt1 in male germ cells.31 These observations further support our findings that ERβ is involved in imprint acquisition and maintenance at the H19 DMR.
A decrease in global 5mC content in spermatozoa along with an apparent decrease in methylation levels of the first 3 CpG sites of LINE-1, which is generally used as an indicator of global methylation, were observed.32 We therefore analyzed the levels of DNA methyltransferase enzymes Dnmt1, Dnmt3a, Dnmt3b, and Dnmt3l in the testis. We found that testicular expression of all Dnmts was reduced at both transcript and protein level upon DPN treatment. Dnmt3a and Dnmt3b catalyze active de novo methylation33 and are involved in establishing imprinting marks. Although Dnmt3l lacks catalytic activity, it regulates DNA methylation through its association with Dnmt3a and Dnmt3b.10 In adult rat testes, Dnmt3a and Dnmt3b both are expressed in pre-meiotic cells (such as spermatogonia); however, only Dnmt3a is localized to meiotic spermatocytes.9 Expression of Dnmt3l is strikingly similar to that of Dnmt3a and Dnmt3b, indicating that it is required for their activity and function.10 The DNA methylation patterns established are faithfully maintained and transmitted during DNA replication by the maintenance methyltransferase Dnmt1, due to its affinity for hemimethylated CpGs.33 It is expressed in almost all male germ cells (spermatogonia, spermatocytes, and post-meiotic round spermatids) and it associates with DNA replication foci.34 Germ line specific knockout studies have shown that Dnmt3a and Dnmt3l are required for the establishment of maternal and paternal imprinting marks. Loss of either Dnmt3a or Dnmt3l in male germ cells leads to arrest in spermatogenesis and sterility. Importantly, their deletion leads to hypomethylation at the LINE-1 promoter and paternally methylated DMRs of H19, Dlk-Gtl2, and Rasgrf.10,35-37 Drawing parallels to our observations, decreased testicular levels of Dnmt3a and Dnmt3l could have led to the observed decrease in global DNA methylation and hypomethylation at the H19 DMR. Additionally, decreased levels of Dnmt1 could lead to passive demethylation in dividing germ cells, which would contribute to the decreased 5mC content. Since all Dnmts were reduced, it is possible that DNA methylation at other loci could also be affected; more detailed studies are required to explore the possible effects on other imprinted genes. Studies by Jue et al.38 have showed increased expression of Dnmt1 transcripts upon hypophysectomy in adult rats, indicating that it could be hormonally regulated. Besides this, there is little information on the regulation of Dnmts during spermatogenesis. We here show for the first time that estrogen, through ERβ, regulates Dnmts in adult rat testis.
Loss of global DNA methylation upon deletion of Dnmt3l leads to defects in formation of a compact heterochromatin state, which could cause failure of chromosomes to align in meiotic spermatocytes.37 Bourc'his et al. observed that, on a Dnmt3l knockout, non-synapsed chromosomal regions in spermatocytes trigger activation of an apoptotic checkpoint leading to apoptosis and germ cell loss.36 Intriguingly, in our earlier studies, we have observed increased spermatocyte apoptosis after ERβ agonist treatment.13 Therefore, the decrease in Dnmts and the resultant decrease in global methylation could have contributed to the increased spermatocyte apoptosis observed. There were no defects in global DNA methylation or at H19 DMR in the spermatozoa upon ERα agonist treatment. Also, the transcript and protein levels of all the Dnmts were unaffected in PPT treated testis, except for Dnmt3a, which was downregulated. This indicates that ERα may play a dispensable role in the regulation of DNA methylation during spermatogenesis. However, since we did find a decrease in Dnmt3a levels, effects on DNA methylation at other loci cannot be ruled out.
Igf2 is expressed mainly in the embryonic and placental tissues, where it acts as a mitogen regulating their overall growth and development.3 H19 encodes a noncoding RNA, which is a precursor for a microRNA postulated to downregulate specific mRNAs during embryonic development.39 Aberrant expression of these imprinted genes during embryogenesis, due to defects in methylation at the H19 DMR regulating their expression, has been associated with the resorbed embryo phenotype.40,22 We thus evaluated Igf2 and H19 gene expression in resorbed embryos sired by PPT- or DPN-treated males as compared with those sired by controls. An increased number of resorbed embryos were observed in females mated with PPT- or DPN-treated males as compared with controls, which indicates increased intensity of post-implantation loss, as observed in our earlier studies.17 In the present study, we observed decreased expression of Igf2 transcripts while expression of H19 was decreased, specifically in the resorbed embryos sired by DPN-treated males as compared with controls. This implies that hypomethylation observed at the H19 DMR in spermatozoa upon DPN treatment could have lead to aberrant expression of Igf2 and H19 genes in developing embryos. It is interesting to note that, even though the decrease in total H19 DMR methylation is about 6%, it could potentially deregulate the expression of imprinted genes. Also, we observed hypomethylation at the CpG sites encompassing the CTCF binding site in the H19 DMR in spermatozoa after DPN treatment. This could potentially affect the differential allelic expression of Igf2 and H19 in developing embryos. Expression of Igf2 and H19 genes was not altered in resorbed embryos sired by PPT-treated males, which agrees with the fact that no methylation defects were observed at the H19 DMR in spermatozoa of PPT-treated males. The possible reasons for increased post-implantation loss on PPT treatment are not clear and are currently being investigated. Therefore, the methylation defects in spermatozoa at the H19 DMR can be correlated with aberrant expression of Igf2 and H19 genes during embryo development.
Several studies have underlined the crucial role of DNA methylation and DNA methyltransferases during embryo development. During embryogenesis, loss of Dnmt1 has disastrous consequences, with Dnmt1 knockouts being lethal after gastrulation due to significant loss of global DNA methylation and 5mC content.41 Dnmt3a knockouts are partially viable but die shortly after birth, whereas deletion of Dnmt3b leads to embryonic lethality.42 However, Dnmt3l knockout embryos are viable and develop up until adulthood without any obvious morphological abnormalities.10 Decreased expression of Dnmts has been reported in resorbed embryos sired by males treated with BPA neonatally.43 We therefore analyzed Dnmt1, Dnmt3a, and Dnmt3b expression in resorbed embryos and found no changes in their levels in resorbed embryos sired by PPT- or DPN-treated males as compared with those sired by controls. Thus, the increased incidence of resorbed embryos upon DPN treatment to males was not due to inherent low levels of Dnmts during embryogenesis.
There are rising concerns regarding exposure to endocrine disrupting compounds and decreasing sperm counts and declining semen quality in men, leading to longer times taken to conceive.44 Exposure to several environmental estrogens such as DDT, methoxychlor, BPA, and natural soy isoflavones impairs the reproductive health of humans and wildlife.45 It is thus important to understand the effects that activation of estrogen signaling cause on the sperm epigenome, which is crucial for male fertility.
Conclusion
Taken together, our data demonstrate that ERβ is specifically involved in the regulation of DNA methyltransferases in the testis. Activation of estrogen signaling through ERβ therefore affects DNA methylation globally and at the imprinted H19 DMR. These epigenetic disturbances in the male germ line could be detrimental to subsequent embryo development and could cause decrease in male fertility. These observations shed light on the endocrine regulation of epigenetic pathways during spermatogenesis.
Materials and methods
Animals
Randomly bred adult Holtzman strain male (75 d old) and female (90 d old) rats were used for the study. Animals were maintained under controlled temperature (23 ± 1°C) and humidity (55 ± 5%) conditions with a 14:10 h light:darkness cycle. Animals were supplied with a diet of soy-free, in-house prepared rat pellets and water ad libitum. Prior approval for the use of animals was obtained from the Institutional Animal Ethics Committee.
In vivo agonist treatments
PPT (> 99%, Axon Medchem) and DPN (> 99%, Tocris Biosciences) were administered at the dose of 0.05 mg/kg/day to adult male rats for 60 d. This was the lowest dose at which the fertility parameters were decreased after 60 d of treatment in our earlier studies.17 Drugs were dissolved completely in vehicle, DMSO saline (75:25; Sigma) and administered subcutaneously as described earlier.17 Animals in the control group received vehicle alone. There were at least 8 males in each of the treatments and control groups.
Mating studies and sample collection
Control and treated rats were cohabited with normal cycling females, at a ratio of one male: 2 females, a week before completion of 60 d of treatment. Females showing sperm-positive vaginal smear were considered to be pregnant and assigned day 0 of gestation. Pregnant females mated with control and treated males were killed at day 17–18 of gestation and uterine horns were exposed. Resorbed embryos were counted, dissected out, and immediately snap frozen in liquid nitrogen and stored at −80°C for RNA extraction. Embryos collected were sired by different males within the same group. Post-implantation loss was calculated as described previously.17
After the mating experiment, control and treated males were killed, both the testes were excised out and immediately snap frozen in liquid nitrogen and stored at -80°C for RNA and protein extraction. For collection of caudal spermatozoa, cauda epididymides were excised in PBS [0.01 M phosphate buffer containing 0.154 M NaCl (pH 7.4)], given superficial cuts and incubated at 37°C for 30 min to allow sperms to diffuse into the buffer. The spermatozoa were passed through a 40 μm cell strainer and washed twice with hypotonic buffer (0.45% NaCl) to lyse any contaminating cells. The spermatozoa were then washed with PBS, counted and stored at -80°C until further use. An aliquot of 2 million spermatozoa was stained by propidium iodide and analyzed by flow cytometry to check for homogeneity of the sample (data not shown). After verifying the purity of the sample, the spermatozoa were used for global and locus-specific methylation analysis.
Genomic DNA extraction from spermatozoa
Genomic DNA was extracted from 10 × 106 caudal spermatozoa using the HiPurA Sperm Genomic DNA Purification Kit (HiMedia) as per manufacturer's instructions. Briefly, the spermatozoa were lysed using sperm lysis buffer containing proteinase K and 1,4-dithiothreitol (DTT) and incubated at 55°C for 2 h with intermittent vortexing. The resulting lysate was subjected to RNase A treatment and the DNA was precipitated using chilled ethanol, washed, and eluted in nuclease free water. Concentration was determined by UV spectrophotometer by calculating ratio of absorbance of DNA at 260 and 280 nm. Pure DNA samples (with 260/280 ratios of ∼1.6–1.8) were processed further for global 5mC content estimation and bisulfite modification.
Global DNA methylation
Global 5mC content of caudal spermatozoa was estimated using Imprint Methylated DNA Quantification Kit (Sigma-Aldrich) as per manufacturer's instructions. Briefly, 10 ng of genomic DNA from control and treated samples was diluted in binding buffer and immobilized on a microtiter plate (in duplicates) at 37°C for 1 h, followed by blocking. A standard curve was used, ranging from 0 to 100 ng of Methylated DNA Control provided in the kit. Diluted capture antibodies were added to each well and incubated at room temperature for 1 h, followed by washing. Diluted detection antibodies were added followed by washing and detection reaction was performed using developing solution provided and the reaction was terminated using stop solution. The absorbance was read at 450 nm on a plate reader and relative global methylation levels were calculated from the standard curve. The amount of methylated DNA present in the sample is proportional to the absorbance measured.
Methylation analysis of spermatozoa DNA by bisulfite pyrosequencing
Sperm genomic DNA was subjected to bisulphite modification using MethylCode Bisulfite Conversion Kit (Invitrogen) as per manufacturer's protocol. Briefly, 1 μg of genomic DNA was subjected sequentially to denaturation, sulphonation, deamination, and desulphonation to convert the unmethylated cytosines to uracil, while the methylated cytosines were unchanged. Modified DNA was eluted in 10 μl nuclease free water and used for PCR amplification of LINE-1 and H19 DMR. PCR was performed with primers specific for modified sequence of LINE-1 and H19 DMR (Pyromark Custom Assay; Qiagen) using Pyromark PCR Amplification Kit (Qiagen) as per manufacturer's instructions. Unmodified genomic DNA samples were used to confirm primer specificity for modified DNA during primer optimization. For both loci, pyrosequencing was done in both directions to cover all the CpG sites (10 for LINE-1 and 15 for H19 DMR) in the PCR product. PCR was done with 2 sets of primers: in set 1 the reverse primer was biotinylated and pyrosequencing was done in the forward direction; vice versa for set 2. Primer sequences and amplicon sizes are listed in Supplementary Table 1. Amplification conditions used: initial denaturation for 15 min at 95°C, followed by 45 cycles of denaturation at 95°C for 30 sec, primer annealing for 30 sec, extension at 72°C for 30 sec, and final extension at 72°C for 10 min. The PCR products were then bound to streptavidin-coated Sepharose Beads (GE Healthcare) in the presence of binding buffer (Qiagen). Bound PCR products were then denatured and the non-biotinylated strand was washed off. The bound biotinylated strand was then annealed to the sequencing primer and the reaction mixture was subjected to pyrosequencing in PyroMark Q96 ID (Qiagen). The sequencing results were analyzed using the Pyromark Q96 software.
RNA extraction and qRT-PCR
Total testicular RNA was extracted from 100 mg of frozen testis using TRI pure (Roche Diagnostics) according to the manufacturer's instructions. Similarly, total RNA was extracted from entire frozen resorbed embryos after removing the uterine covering. At least 8 samples were used per treatment group. RNA concentration was determined by the absorbance at 260 nm (Nanovue, GE). The purity and integrity of the RNA were checked by measuring the optical density at 260 and 280 nm followed by electrophoresis on a 1.5% agarose gel stained with 0.01% ethidium bromide. The RNA was treated with 30 μg/ml DNase I for 1 h at 37°C. RNA extracted (2 μg) was then reverse transcribed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's protocol. qRT-PCR was performed in a Light Cycler 96 real time PCR System (Roche) using Takyon SYBR Green Master Mix (Eurogentec). Relative expression levels of Dnmt1, Dnmt3a, Dnmt3b, and Dnmt3l (for testis and resorbed embryos), and Igf2 and H19 (for resorbed embryos alone) were determined in relation to the reference gene 18S rRNA, using gene specific primers (Sigma; Supplementary Table 2). Amplification reactions were set (20 μl) containing 1.6 μl cDNA, 10 pM of respective primers and SYBR green master mix with the thermal cycling conditions of initial denaturation of 10 min at 95°C followed by 40 cycles of 95°C for 10 sec, primer annealing temperature for 10 sec, and extension at 72°C for 10 sec. Amplification reactions were prepared in duplicate and a no template control was included. To check for specificity, melt curve analysis was performed and all products obtained yielded the predicted melting temperature. Primer PCR efficiencies (listed in Supplementary Table 3) were determined using a standard curve. Pfaffl method was used to calculate the relative gene expression.46 Real-time PCR procedures and analysis followed the MIQE guidelines47 (Supplementary Table 4).
Immunoblotting
Total protein from frozen control, PPT-, and DPN-treated testes was extracted in radioimmunoprecipitation assay lysis buffer (RIPA) (150 mM NaCl, 50 mM Tris (pH 7.5), 5 mM EDTA, 1% Nonidet P-40, 1% deoxycholic acid, 10% sodium dodecyl sulfate) containing protease inhibitors. Fifty micrograms of protein was resolved on 8% SDS-PAGE (for Dnmt1, Dnmt3a, and Dnmt3b) and 12% SDS-PAGE (for Dnmt3l and GAPDH), and transferred onto nitrocellulose membrane (GE). Blots were washed with PBS, blocked with 5% non-fat dry milk, and incubated separately with primary antibodies, namely anti-Dnmt1 (1:200; Abcam, cat. no. ab13537), anti-Dnmt3a (1:100; Abcam, cat. no. ab13888), anti-Dnmt3b (1:100; Abcam, cat. no. ab13604), anti-Dnmt3l (1:100; Santa Cruz, cat. no. sc 10241) and anti-GAPDH (1:1000; Sigma, cat. no. G9545) in 1% non-fat dry milk at 4°C overnight. Membranes were washed with PBS containing 0.1% Tween 20 and incubated with the appropriate secondary antibodies conjugated to horseradish peroxidase (Sigma). Blots were developed with ECL Plus (GE) or Femto reagent (Pierce Biotechnology). Bands were visualized and quantified by densitometry using Gene Tools Version 3.06 (Syngene) and normalized relative to housekeeping protein GAPDH.
Statistical analysis
Data were analyzed by one-way ANOVA followed by a Dunnett test to compare the means of the treatment groups with control using GraphPad Prism Software Version 6.0 (GraphPad). All data are represented as means ± SEM. The level of significance was set as P < 0.05.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors are grateful for technical help and expertise Mr. S. M. Mandavkar in animal handling and dissections. The authors acknowledge technical assistance of Mr. D. Shelar.
Funding
This study has been funded from National Institute of Research in Reproductive Health (NIRRH) core budget (RA/446/12–2016) to NHB. The authors acknowledge Indian Council of Medical Research (ICMR) for providing financial support and fellowship to Ms. K. Dumasia.
References
- 1.Trasler J. Epigenetics in spermatogenesis. Mol Cell Endocrinol 2009; 306:33-6; PMID:19481683; https://doi.org/ 10.1016/j.mce.2008.12.018 [DOI] [PubMed] [Google Scholar]
- 2.Inbar-Feigenberg M, Choufani S, Butcher D, Roifman M, Weksberg R. Basic concepts of epigenetics. Fertil Steril 2013; 99:607-15; PMID:23357459; https://doi.org/ 10.1016/j.fertnstert.2013.01.117 [DOI] [PubMed] [Google Scholar]
- 3.DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 1991; 64:849-59; PMID:1997210; https://doi.org/ 10.1016/0092-8674(91)90513-X [DOI] [PubMed] [Google Scholar]
- 4.Thorvaldsen J, Duran K, Bartolomei M. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Gene Dev 1998; 12:3693-702; PMID:9851976; https://doi.org/ 10.1101/gad.12.23.3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell A, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 2000; 405:482-85; PMID:10839546; https://doi.org/ 10.1038/35013100 [DOI] [PubMed] [Google Scholar]
- 6.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature 1991; 351:153-55; PMID:1709450; https://doi.org/ 10.1038/351153a0 [DOI] [PubMed] [Google Scholar]
- 7.Tremblay KD, Saam JR, Ingram RS, Tilghman SM. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nature 1995; 9:407-13; PMID:7795647; https://doi.org/ 10.1038/ng0495-407 [DOI] [PubMed] [Google Scholar]
- 8.Kaffer CR, Grinberg A, Pfeifer K. Regulatory mechanisms at the mouse Igf2/H19 locus. Mol Cell Biol 2001; 21:8189-96; PMID:11689707; https://doi.org/ 10.1128/MCB.21.23.8189-8196.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu HX, Qin JZ, Zhang KY, Zeng WX. Dynamic expression profile of DNA methyltransferases in rat testis development. Pol J Vet Sci 2015; 18:549-56; PMID:26618587; https://doi.org/ 10.1515/pjvs-2015-0071 [DOI] [PubMed] [Google Scholar]
- 10.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 2002; 129:1983-93; PMID:11934864 [DOI] [PubMed] [Google Scholar]
- 11.Van Pelt AM, de Rooij DG, van der Burg B, van der Saag PT, Gustafsson JA, Kuiper GG. Ontogeny of estrogen receptor-beta expression in rat testis. Endocrinology 1999; 140:478-83; PMID:9886860; https://doi.org/ 10.1210/endo.140.1.6438 [DOI] [PubMed] [Google Scholar]
- 12.Bois C, Delalande C, Nurmio M, Parvinen M, Zanatta L, Toppari J, Carreau S. Age- and cell-related gene expression of aromatase and estrogen receptors in the rat testis. J Mol Endocrinol 2010; 45:147-59; PMID:20554652; https://doi.org/ 10.1677/JME-10-0041 [DOI] [PubMed] [Google Scholar]
- 13.Dumasia K, Kumar A, Deshpande S, Sonawane S, Balasinor NH. Differential roles of estrogen receptors, ESR1 and ESR2, in adult rat spermatogenesis. Mol Cell Endocrinol 2016; 428:89-100; PMID:27004961; https://doi.org/ 10.1016/j.mce.2016.03.024 [DOI] [PubMed] [Google Scholar]
- 14.Robaire B, Duron J, Hales B. Effect of estradiol-filled polydimethylsiloxane subdermal implants in adult male rats on the reproductive system, fertility, and progeny outcome. Biol Reprod 1987; 37:327-34; PMID:3676392; https://doi.org/ 10.1095/biolreprod37.2.327 [DOI] [PubMed] [Google Scholar]
- 15.Gill-Sharma M, D'Souza S, Padwal V, Balasinor N, Aleem M, Parte P, Juneja H. Antifertility effects of estradiol in adult male rats. J Endocrinol Invest 2001; 24:598-607; PMID:11686542; https://doi.org/ 10.1007/BF03343900 [DOI] [PubMed] [Google Scholar]
- 16.D'Souza R, Gill-Sharma M, Pathak S, Kedia N, Kumar R, Balasinor N. Effect of high intratesticular estrogen on the seminiferous epithelium in adult male rats. Mol Cell Endocrinol 2005; 241:41-48; PMID:15936871; https://doi.org/ 10.1016/j.mce.2005.04.011 [DOI] [PubMed] [Google Scholar]
- 17.Dumasia K, Kumar A, Kadam L, Balasinor N. Effect of estrogen receptor-subtype-specific ligands on fertility in adult male rats. J Endocrinol 2015; 225:169-80; PMID:25869617; https://doi.org/ 10.1530/JOE-15-0045 [DOI] [PubMed] [Google Scholar]
- 18.Gill-Sharma M, Gopalkrishnan K, Balasinor N, Parte P, Jayaraman S, Juneja H. Effects of tamoxifen on the fertility of male rats. J Reprod Fertil 1993; 99:395-402; PMID:8107021; https://doi.org/ 10.1530/jrf.0.0990395 [DOI] [PubMed] [Google Scholar]
- 19.Balasinor N, Parte P, Gill-Sharma M, Juneja H. Effect of tamoxifen on sperm fertilising ability and preimplantation embryo development. Mol Cell Endocrinol 2001; 178:199-206; PMID:11403910; https://doi.org/ 10.1016/S0303-7207(01)00428-2 [DOI] [PubMed] [Google Scholar]
- 20.Salian S, Doshi T, Vanage G. Perinatal exposure of rats to Bisphenol A affects the fertility of male offspring. Life Sci 2009; 85:742-52; PMID:19837096; https://doi.org/ 10.1016/j.lfs.2009.10.004 [DOI] [PubMed] [Google Scholar]
- 21.Pathak S, Kedia-Mokashi N, Saxena M, D'Souza R, Maitra A, Parte P, Gill-Sharma M, Balasinor N. Effect of tamoxifen treatment on global and insulin-like growth factor 2-H19 locus-specific DNA methylation in rat spermatozoa and its association with embryo loss. Fertil Steril 2009; 91:2253-63; PMID:18778817; https://doi.org/ 10.1016/j.fertnstert.2008.07.1709 [DOI] [PubMed] [Google Scholar]
- 22.Doshi T, D'souza C, Vanage G. Aberrant DNA methylation at Igf2–H19 imprinting control region in spermatozoa upon neonatal exposure to bisphenol A and its association with post implantation loss. Mol Biol Rep 2013; 40:4747-57; PMID:23653003; https://doi.org/ 10.1007/s11033-013-2571-x [DOI] [PubMed] [Google Scholar]
- 23.Stauffer S, Coletta C, Tedesco R, Nishiguchi G, Carlson K, Sun J, Katzenellenbogen B, Katzenellenbogen J. Pyrazole ligands: Structure and affinity/activity relationships and estrogen receptor a-selective agonists. J Med Chem 2000; 43:4934-47; PMID:11150164; https://doi.org/ 10.1021/jm000170m [DOI] [PubMed] [Google Scholar]
- 24.Meyers M, Sun J, Carlson K, Marriner G, Katzenellenbogen B, Katzenellenbogen J. Estrogen receptor-β potency-selective ligands: structure and activity relationship studies of diarylpropionitriles and their acetylene and polar analogues. J Med Chem 2001; 44:4230-51; PMID:11708925; https://doi.org/ 10.1021/jm010254a [DOI] [PubMed] [Google Scholar]
- 25.lAnway M, Cupp A, Uzumcu M, Skinner M. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005; 308:1466-69; PMID:15933200; https://doi.org/ 10.1126/science.1108190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, Sasaki H, Yaegashi N, Arima T. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet 2007; 16:2542-51; PMID:17636251; https://doi.org/ 10.1093/hmg/ddm187 [DOI] [PubMed] [Google Scholar]
- 27.Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, Sousa M. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod 2008; 14:67-74; PMID:18178607; https://doi.org/ 10.1093/molehr/gam093 [DOI] [PubMed] [Google Scholar]
- 28.Marques J, Francisco T, Sousa S, Carvalho F, Barros A, Sousa M. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil Steril 2010; 94:585-94; PMID:19338988; https://doi.org/ 10.1016/j.fertnstert.2009.02.051 [DOI] [PubMed] [Google Scholar]
- 29.Davis T, Yang G, McCarrey J, Bartolomei M. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet 2000; 9:2885-94; PMID:11092765; https://doi.org/ 10.1093/hmg/9.19.2885 [DOI] [PubMed] [Google Scholar]
- 30.Oakes CC, Salle S, Smiraglia DJ, Robaire B, Trasler JM. Developmental acquisition of genome-wide DNA methylation occurs prior to meiosis in male germ cells. Dev Biol 2007; 307:368-79; PMID:17559830; https://doi.org/ 10.1016/j.ydbio.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 31.Pathak S, D'Souza R, Ankolkar M, Gaonkar R, Balasinor N. Potential role of estrogen in regulation of the insulin-like growth factor2-H19 locus in the rat testis. Mol Cell Endocrinol 2010; 314:110-17; PMID:19683557; https://doi.org/ 10.1016/j.mce.2009.08.005 [DOI] [PubMed] [Google Scholar]
- 32.Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 2004; 32:e38; PMID:14973332; https://doi.org/ 10.1093/nar/gnh032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bestor T. The DNA methyltransferases of mammals. Hum Mol Genet 2000; 9:2395-02; PMID:11005794; https://doi.org/ 10.1093/hmg/9.16.2395 [DOI] [PubMed] [Google Scholar]
- 34.Jue K, Bestor T, Trasler J. Regulated synthesis and localization of DNA methyltransferase during spermatogenesis. Biol Reprod 1995; 53:561-69; PMID:7578680; https://doi.org/ 10.1095/biolreprod53.3.561 [DOI] [PubMed] [Google Scholar]
- 35.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 2004; 429:900-03; PMID:15215868; https://doi.org/ 10.1038/nature02633 [DOI] [PubMed] [Google Scholar]
- 36.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 2004; 431:96-99; PMID:15318244; https://doi.org/ 10.1038/nature02886 [DOI] [PubMed] [Google Scholar]
- 37.Webster K, O'Bryan M, Fletcher S. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc Natl Acad Sci U S A 2005; 102:4068-73; PMID:15753313; https://doi.org/ 10.1073/pnas.0500702102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jue K, Benoit G, Alcivar-Warren A, Trasler J. Developmental and hormonal regulation of DNA methyltransferase in the rat testis. Biol Reprod 1995; 52:1364-71; PMID:7632844; https://doi.org/ 10.1095/biolreprod52.6.1364 [DOI] [PubMed] [Google Scholar]
- 39.Cai X, Cullen B. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007; 13:313-16; PMID:17237358; https://doi.org/ 10.1261/rna.351707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pathak S, Saxena M, D'Souza R, Balasinor NH. Disrupted imprinting status at the H19 differentially methylated region is associated with the resorbed embryo phenotype in rats. Reprod Fertil Dev 2010; 22:939-48; PMID:20591328; https://doi.org/ 10.1071/RD09154 [DOI] [PubMed] [Google Scholar]
- 41.Li E, Bestor T, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992; 69:915-26; PMID:1606615; https://doi.org/ 10.1016/0092-8674(92)90611-F [DOI] [PubMed] [Google Scholar]
- 42.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999; 99:247-57; PMID:10555141; https://doi.org/ 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- 43.Doshi T, D'Souza C, Dighe V, Vanage G. Effect of neonatal exposure on male rats to bisphenol A on the expression of DNA methylation machinery in the postimplantation embryo. J Biochem Mol Toxicol 2012; 26:337-43; PMID:22730197; https://doi.org/ 10.1002/jbt.21425 [DOI] [PubMed] [Google Scholar]
- 44.Gore A, Chappell V, Fenton S, Flaws J, Nadal A, Prins G, Toppari J, Zoeller R. EDC-2: The endocrine society's second scientific statement on endocrine-disrupting chemicals. Endocr Rev 2015; 36:E1-150; PMID:26414233; https://doi.org/ 10.1210/er.2015-1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daston G, Gooch J, Breslin W, Shuey D, Nikiforov A, Fico T, Gorsuch J. Environmental estrogens and reproductive health: A discussion of the human and environmental data. Reprod Toxicol 1997; 11:465-81; PMID:9241667; https://doi.org/ 10.1016/S0890-6238(97)00014-2 [DOI] [PubMed] [Google Scholar]
- 46.Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45; PMID:11328886; https://doi.org/ 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bustin S, Benes V, Garson J, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl M, Shipley G, et al.. The MIQE Guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009; 55:611-22; PMID:19246619; https://doi.org/ 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


